Abstract
Viral proteins reprogram their host cells by hijacking regulatory components of protein networks. Here we describe a novel property of the Epstein–Barr virus (EBV) nuclear antigen-1 (EBNA1) that may underlie the capacity of the virus to promote a global remodeling of chromatin architecture and cellular transcription. We found that the expression of EBNA1 in transfected human and mouse cells is associated with decreased prevalence of heterochromatin foci, enhanced accessibility of cellular DNA to micrococcal nuclease digestion and decreased average length of nucleosome repeats, suggesting de-protection of the nucleosome linker regions. This is a direct effect of EBNA1 because targeting the viral protein to heterochromatin promotes large-scale chromatin decondensation with slow kinetics and independent of the recruitment of adenosine triphosphate–dependent chromatin remodelers. The remodeling function is mediated by a bipartite Gly-Arg rich domain of EBNA1 that resembles the AT-hook of High Mobility Group A (HMGA) architectural transcription factors. Similar to HMGAs, EBNA1 is highly mobile in interphase nuclei and promotes the mobility of linker histone H1, which counteracts chromatin condensation and alters the transcription of numerous cellular genes. Thus, by regulating chromatin compaction, EBNA1 may reset cellular transcription during infection and prime the infected cells for malignant transformation.
INTRODUCTION
Pathogenic viruses and intracellular bacteria have evolved elaborate strategies for manipulating the host cell environment, often resorting to the production of multifunctional proteins that hijack or mimic the activity of cellular regulators. A common property of DNA tumor viruses is the establishment of non-productive infections characterized by the expression of a restricted repertoire of latency-associated viral genes. Remodeling of the infected cells by the products of these genes is an enabling feature of viral oncogenesis but, in spite of intensive research, their mechanisms of action are still poorly understood. Here we have addressed this issue in the context of cells expressing the Epstein–Barr virus (EBV) encoded nuclear antigen-1 (EBNA1).
EBV is a human gamma herpesvirus implicated in the pathogenesis of lymphoid and epithelial cell malignancies, including Burkitt’s lymphoma (BL), Hodgkin’s disease, nasopharyngeal carcinoma and post-transplant lymphoproliferative disease that arises in immunosuppressed patients (1). In healthy human carriers, the virus establishes a life-long latent infection in B-lymphocytes, where it persists as a multicopy episome that periodically reactivates to produce progeny virus (1). During latency, the EBV genome expresses a limited repertoire of proteins, non-coding RNAs and microRNAs that are required for viral genome maintenance and host-cell survival (2). EBNA1 is the only viral protein commonly expressed in dedicated latency programs that allow the persistence of EBV in B-cells during activation and differentiation and in a variety of other cells types.
The contributions of EBNA1 to virus infection and malignant transformation are only partially understood. Binding of the C-terminal domain of EBNA1 to the viral origin of latent replication, OriP, is essential for plasmid DNA replication and episome maintenance (3), while binding to viral promoters regulates transcription (4). The N-terminal domain of EBNA1 tethers the EBV episome to cellular DNA during mitosis (5–8), which is required for persistence of the episome in proliferating cells. However, only a fraction of the expressed protein is required for this activity. Thus the sites, structures and accessory proteins through which the majority of EBNA1 interacts with cellular DNA, and the purpose of such interaction, remain largely unknown. EBNA1 expression is associated with changes in the expression of cellular genes (9–11), and its binding to cellular promoters has been documented (9,12), but only in few cases the regulation of cellular promoters was validated in reporter assays. This, together with the identification of a large number of candidate DNA binding sites across the human genome, both close and far apart from transcription start sites (13), suggests that the mechanism by which EBNA1 affects transcription may be different compared with conventional transcription factors.
A defining feature of transcription factors is the capacity to recognize specific sequences in DNA and promote the local assembly of protein complexes that control gene expression. Recent genome-wide localization analyses indicate that only a small percentage of the consensus binding sites of known transcription factors is occupied at any given time (14,15), which is likely explained by the wrapping of DNA into nucleosomes and high-order chromatin structures that restrict the access of large macromolecular complexes (16). Thus, gene expression is often dependent on the capacity of transcription factors to cooperate with activators that promote chromatin decondensation through the recruitment of adenosine triphosphate (ATP)-dependent remodeling complexes and post-translational modification of histone tails (17,18). This weakens the interaction of the negatively charged DNA with positively charged histones, and allows nucleosome sliding, histone dissociation and the incorporation of histone variants (17). In addition, both local and widespread effects on the accessibility of chromatin are induced by ‘architectural factors’, such as members of the three families of High Mobility Group (HMG) non-histone proteins: HMGA, HMGB and HMGN. These accessory proteins bind in a sequence-independent manner to specific structures in DNA and cooperatively displace linker histones, which leads to a local opening of chromatin and initiation of the gene activation process (19).
Here we provide evidence for the capacity of EBNA1 to alter chromatin architecture by mimicking the activity of HMG proteins. EBNA1 binds to cellular DNA through a bipartite Gly-Arg repeat domain that resembles the chromatin-targeting module of HMGAs. We show that this domain mediates a highly dynamic interaction of EBNA1 with cellular chromatin and is sufficient to promote chromatin decompaction with a slow kinetics and independent of the recruitment of ATP-dependent remodeling complexes. These features correlate with increased mobility of histone H1 and with a broad rearrangement of the cellular transcription profile.
MATERIALS AND METHODS
Cell lines and transfection
The B lymphoma lines BJAB and sublines expressing stable or inducible EBNA (BJAB-EBNA1, BJAB-tTA, BJAB-tTA-EBNA1) (20) were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM glutamine, 10 μg/ml Ciprofloxacin (Sigma-Aldrich, St. Louis, MO, USA) (complete medium). BJAB-tTA and BJAB-tTA-EBNA1 cells were maintained in 1 µg/ml puromycin and 400 µg/ml hygromycin. The human osteosarcoma line U2OS, the murine fibroblast NIH3T3 and the NIH2/4 cell line harboring a 256 × LacO array (21) were grown in Iscove’s Modified Dulbecco’s Medium (Sigma-Aldrich) complete medium. The Chinese Hamster Ovary carcinoma line A03-1 that harbors multi-copies of the 256 × LacO array (22) was grown in DMEM/F12 (Life Technologies, Grand Island, NY, USA) complete medium. All cells were kept at 37°C in a 5% CO2 incubator. For transfection, semi-confluent monolayers were grown on Ø22 mm or Ø42 mm cover slides (VWR International, Radnor, PA, USA) and then transfected with 0.5 µg of the indicated plasmid using the JetPEI transfection kit (Polyplus Transfection, Illkirch, France), according to the instructions of the manufacturer.
Antibodies
Rabbit K67.1 anti EBNA1 (1:1000) was kindly provided by Jaap Middeldorp (Vrije Universiteit, Amsterdam). Rabbit anti-histone H3-3meK9, and mouse anti-histone H3-2meK9 and H3-acK9 (1:100) were from Abcam (Cambridge, MA, USA).
Plasmids
The pFLAG-EBNA1 and green fluorescent protein (GFP)-EBNA1 expression vectors (8) were kindly provided by Elliott Kieff (Harvard Medical School, Boston, MA, USA). To generate mCherry-EBNA1, the mCherry coding sequence was amplified by polymerase chain reaction (PCR) from pCherry-ATG5 (Addgene, Cambridge, MA, USA) using the primer pair ‘fw-AAAAAGCTTATGGTGAGCAAGGGCGAGGAG, rev-AAAAAGCTTCTTGTACAGCTCGTCCATGCCGCC’ and cloned in the HindIII site of pFLAG-EBNA1. GFP-HMGA1a was generated by inserting the GFP coding sequence amplified from pEGFP-N1 (Clontech Laboratories Inc., Mountain View, CA, USA) with the primer pair ‘fw-AAAAAGCTTATGGTGAGCAAGGGCGAGGAGC, rev-AAAAAGCTTCTTGTACAGCTCGTCCATGCCGAG’ in the HindIII site of p3 GFP-nuclear localization signal (GFP-NPS) × FLAG-HMGA1a (23). The mCherry-LacR (21), GFP-NLS, GFP-HP1β (24), GFP-H4 (25), YFP-BRG1 (26), GFP-CHD4 (27) and GFP-ALC1 (28) were described previously. The LacR-NLS gene (gift from Tom Misteli, National Cancer Institute, NIH, Bethesda, MD, USA) was amplified using the primer pair ‘fw-GATCCTGTACAAGATGGTGAAACCAGTAACGTTATAC, rev-TGAGGTACCAGATCTAACCTTCCTCTTCTTCTTAG’ and inserted as a BsrGI/BglII fragment in mCherry-C1 yielding mCherry-LacR-NLS-C1. The VP16 acidic activation domain from mCherry-VP16-N1 (gift from Roel van Driel, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, The Netherlands) was inserted as a BsrGI/MfeI fragment into mCherry-LacR-NLS-C1 yielding mCherry-LacR-NLS-VP16. The H1.0 cDNA was amplified by PCR using primer pair ‘fw-AGATCTATGACCGAGAATTCCACGTC, rev-GGATCCTCACTTCTTCTTGCCGGCCC’ and inserted into pEGFP-C1 as a BglII/BamHI fragment yielding EGFP-H1. SNF2H-GFP was kindly provided by Haico van Attikum (Leiden University Medical Center, Leiden, The Netherlands). To generate fusion proteins, the mCherry-LacR cassette was amplified from the mCherry-LacR plasmid using the primer pair ‘fw-AAAAAGCTTATGGTGAGCAAGGGCGAGGAG, rev-AAAAAGCTTAACCTTCCTCTTCTTCTTAGG’ and cloned in the HindIII site of the pCMV-3 × FLAG-EBNA1, pCMV-3 × FLAG-EBNA1ΔGA, 3 × FLAG-EBNA1-DBD and p3 × FLAG-HMGA1a vectors. The pCMV-3 × FLAG-GR1/G2-GFP plasmid was generated by inserting the GFP coding sequence amplified using the primer pair ‘fw-AAACTCGAGATGGTGAGCAAGGGCGAGGAGC, rev-TTTGCGGCCGCTTACTTGTACAGCTCGTCCATGCCGAG’ in the XhoI/NotI sites pCMV-3 × FLAG-GR1/2-NLS-VK. To generate the mCherry-LacR-GR1/2 (EBNA1 amino acid 40–86 and 324–379), -EBNA1ΔN/GA (EBNA1 amino acid 40–641Δ89–324) and -EBNA1-GR2/DBD (EBNA1 324–641) plasmids, the coding sequences were amplified from pCMV-3 × FLAG-EBNA1ΔGA with the primer pairs ‘fw-AAACTCGAGCTGGACGAGGACGGGGAAGAGGACGA, rev-TTTGGATCCTCATTCTCCACGTCCACGACCTCT’; ‘fw-AAACTCGAGCTCGACGAGGACGGGGAAGAGGACGA, rev-TTTGGATCCTCACTCCTGCCCTTCCTCACC’ and ‘fw-AAACTCGAGCTGGAGGTGGAGGCCGGGGTCGAGGA, rev-TTTGGATCCTCACTCCTGCCCTTCCTCACC’, respectively, and cloned in the XhoI/BamHI restriction sites of mCherry-LacR-C1. To generate pCMV-3 × FLAG-GFP-EBNA1-DBD, the GFP coding sequence was excised from p3 × FLAG-GFP-HMGA1a by digestion with HindIII, and the fragment was ligated into HindIII digested p3 × FLAG-EBNA1-ΔGA/GR. Plasmids expressing YFP-GCN5, -CAF, -P300, -Brd2 and -Brd4 (29) were kindly provided by Ilona Rafalska-Metcalf, The Wistar Institute, Philadelphia, PA, USA. To generate the pEGFP-USP7 plasmid, the USP7 coding sequence was PCR amplified from the pT7-USP7 plasmid (30) (kindly provided by Roger D. Everett, University of Glasgow Centre for Virus Research, Glasgow, UK) using the primer pair ‘fw-GTGAAGCTTATGAACCACCAGCAGCAGCAGCAG, rev-GGGACCGGTGGGTTATGGATTTTAATGGCCTTTTC’ and cloned in the HindIII/AgeI restriction sites of pEGFP-N1.
Immunofluorescence
Cells grown on glass cover slides were rinsed three times with phosphate buffer saline (PBS) fixed with 4% formaldehyde in PBS for 15 min and permeabilized with 0.1% Triton X-100 and 4% bovine serum albumin in PBS for 30 min. After incubation for 1 h with the indicated primary antibody, the cells were incubated for 1 h with the appropriate fluorescein isothiocyanate (FITC) or tetramethyl rhodamine isothiocyanate (TRTC) secondary antibody (DAKO, Glostrup, Denmark) and washed three times with PBS before mounting with VECTASHIELD medium containing 4′,6-Diamidino-2-Phenylindole Dihydrochloride (DAPI) (Vector Laboratories Inc. Burlingome, CA, USA). Digital images were captured with a Zeiss LSM510 META Confocal microscope and analyzed with the ImageJ 1.42q software (Wayne Rasband, National Institutes of Health, USA).
Micrococcal nuclease digestion
Cells (2 × 107) were harvested by centrifugation at 1500g for 5 min. The pellets were resuspended in 2 ml 0.5% Tween-40 in Tris buffer saline and incubated on ice for 30 min with mixing by vortexing every 5 min. After disruption of the cell membrane with a glass homogenizer, the nuclei were pelleted and resuspended in 1 ml of digestion buffer (0.32 M sucrose, 50 mM Tris–HCl pH 7.4, 4 mM MgCl2, 1 mM CaCl2 and 1 mM Phenylmethylsulfonyl fluoride (PMSF)). The amount of DNA was measured by spectrophotometer, and the nuclei were resuspended at a final DNA concentration of 0.5 mg/ml. Micrococcal nuclease (MNase) digestion was carried out for the indicated time at 37°C in the presence of 100 gel Units of enzyme (≈10 Kunitz Unit) in 100 µl. The reaction was stopped by adding 0.5 M ethylenediaminetetraacetic acid to the final concentration of 5 mM and placed on ice. The samples were clarified by centrifugation at 11 600 g for 5 min. The DNA released in the supernatant was purified with a QIAquick® kit (QIAGEN Inc., Valencia, CA, USA) and fractionated in a 1.4% agarose gel. Nucleosome size was calculated using as reference the DNA marker.
Nucleosome array conformation analysis
The A03-1 or NIH2/4 cell lines were seeded on Ø22 mm cover slides placed in six-well plate and, when 60% confluent, transfected with the indicated plasmid. After 48 h, the slides were fixed, permeabilized and mounted with VECTASHIELD medium containing DAPI. Digital images were captured with a Zeiss LSM510 META Confocal microscope and analyzed with the ImageJ software. The relative size of the array is expressed as percentage of the nuclear area. For kinetic analysis, A03-1 cells were seeded in six-well plates and cultured for 24 h in the presence of 5 mM of isopropyl-beta-d-thiogalactopyranoside (IPTG, Sigma-Aldrich) before transfection. After 24 h, the IPTG was removed by wash in PBS and the cells were cultured in complete medium for the indicated times before fixation. The size of the array was assessed in 25 cells for each time point and is expressed as percentage of the size measured at 48 h. ATP depletion was performed at the indicated time by incubating the cells for 30 min in ATP-depletion medium (137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 60 mM deoxyglucose, 30 mM NaAz, 20 mM hydroxyethyl piperazineethanesulfonic acid (HEPES) and 10% FBS) before fixation. The percentage inhibition was calculated as ratio of the dot size in the presence or absence of ATP.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) experiments were performed with a Zeiss LSM 510 META confocal microscope equipped with a CO2 chamber. For protein mobility analysis 30 pre-bleach images were acquired using Plan-Apochromat 63×/1.4 Oil Dic objective zoom 8, followed by a bleach pulse of five iterations in a strip region of 1.74 × 1.4 µm. A total of 600 imagines of the bleached area were acquired with a scan speed of 38.4 ms/cycle and a delay of 5 ms. For imaging, the power of a 25 mW argon laser (488 nm line) was set to 0.1%, and for bleaching, the laser power was set to 100%. FRAP recovery curves were generated by normalizing the fluorescence intensity in the bleached area to the initial fluorescence in the same area. For competitive FRAP analysis, five pre-bleach images were acquired using a Plan-Apochromat 63×/1.4 Oil Dic objective zoom 5, followed by a bleach pulse of 10 iterations in a spot region of 2 µm diameter. A total of 100 images were acquired with an acquisition speed of 1.97 s/cycle and no delay between acquisition cycles. Curves were generated by subtracting the background fluorescence of the slide (Region of Interest (ROI) 3), by correcting for the acquisition photobleaching measured in a non-bleached area of the nucleus (ROI 2) and by normalizing the fluorescence intensity in the bleached area (ROI 1) to the initial fluorescence of the same area. In a typical experiment, 10 cells were analyzed and each experiment was repeated at least three times. Fitting of the curves was done with the Prism5 software (GraphPad Software Inc, La Jolla, CA, USA), using non-linear regression and One-phase association equations.
Microarray gene expression analysis
Comparative gene expression profiles from BJAB and BJAB-EBNA1 transfected cells (10), three EBV negative (Ramos, ST486 and BJAB) and three EBV carrying BL cell lines expressing latency I (KEM, MUTU and ODH) retrieved from a larger study (31), cells over-expressing HMGN5 (32) and HMGA2 (33), and cells from H1 knockdown mice (34) were derived from the NCBI GEO database (GSE22964, GSE2359, GSE27546, GSE23599 and GSE3714, respectively). The data was quantile normalized and differentially expressed genes with a fold change ≥1.5 were identified using the RankProd approach (35). The SAM (significance analysis of microarrays) application (36) was used to select statistically significant differentially expressed genes with a 1.5-fold change cutoff. The Search Tool for Retrieval of Interacting Genes/Proteins (STRING) database (37) was used to build a functional interaction network of the 268 genes whose expression was concordantly regulated in EBNA1 transfectants and Latency I cells. Interactions with the highest confidence score (>0.9) were used to construct a network that included 66 protein having one or more interacting partners. The likelihood of association is based on the occurrence of one or more of the following parameters: experimental evidence, co-expressed genes, text mining evidence, protein homology, annotation in curated databases. Genes involved in transforming growth factor beta (TGFβ) and JAK/STAT signaling were identified using the KEGG (38) and Panther (39) databases.
Twist promoter reporter assays
The murine Twist promoter luciferase reporter plasmid (−1745/+209) (40) was provided by L. R. Howe (Cornell University, NY, USA). The plasmid was co-transfected in HepG2 together with 100 ng of the empty pcDNA3, 200 ng of pcDNA3-HA-hHMGA2 or pcDNA3-Flag-EBNA1 and 100 ng of the reporter plasmid pCMV-βGal for normalization, using the calcium phosphate co-precipitation method. Transfected cells were lysed in buffer containing 5 mM Tris-phosphate buffer pH 7.8, 2 mM dithiothreitol, 2 mM CDTA (trans-1,2-diaminocyclohexane-N,N,N′,N′ tetra-acetic acid), 5% glycerol and 1% Triton X-100. β-Galactosidase activity was measured by mixing the cell lysate with 100 mM sodium phosphate pH 7.3, 1 mM MgCl2, 50 mM β-mercaptoethanol and 0.67 mg/ml of ONPG (2-nitrophenyl β-d-galactopyranoside, Sigma-Aldrich) and the absorbance was monitored at 420 nm. Luciferase activity was measured using the enhanced luciferase assay kit from BD PharMingen, Inc., (Franklin Lakes, NJ, USA). Normalized luciferase activity data are shown as mean ±SD of triplicates determinations from four independent experiments.
Statistical analysis
Statistical analysis was performed with Prism5 software and the R software for microarray data. Significant probabilities are indicated as P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***).
RESULTS
EBNA1 does not accumulate on heterochromatin
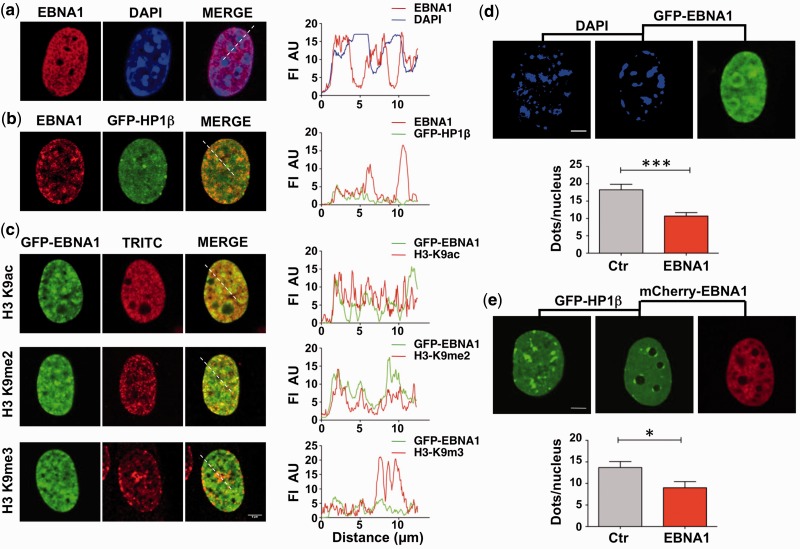
To address the mechanism by which EBNA1 may regulate the transcription of cellular genes, we first examined whether the viral protein localizes to specific chromatin domains in interphase nuclei by immunofluorescence analysis of transfected NIH3T3 mouse fibroblasts and human U2OS cells. In accordance with the diffuse staining patterns of endogenous EBNA1 in EBV infected cells, and in broad agreement with the identification of a large number of DNA binding sites by ChIP-Seq analysis (13), we found that the transfected protein does not associate with a distinct nuclear sub-compartment in mouse fibroblasts. However, EBNA1 appeared to be excluded from heterochromatic regions that are easily identified as bright foci in the nucleus of mouse cells stained with DAPI (Figure 1a). This observation was confirmed in human U2OS cells where EBNA1 failed to co-localize with the co-transfected heterochromatin-binding protein HP1β (Figure 1b). Furthermore, double immunofluorescence analysis in U2OS cells expressing GFP-tagged EBNA1 revealed that the protein is present in chromatin regions containing the euchromatin marker H3K9ac (Figure 1c, upper panel) and partially colocalizes with markers of facultative heterochromatin, such as H3K9me2 (Figure 1c, middle panel). However, we did not find accumulation in regions containing histone modifications associated with condensed heterochromatin, such as H3K9me3 (Figure 1c, lower panel). To investigate this phenomenon further, the number of heterochromatic foci was quantified in control and EBNA1-transfected cells. A significant decrease in the number of heterochromatic foci was detected by DAPI staining in NIH3T3 cells (Figure 1d) and by HP1β accumulation in U2OS cells (Figure 1e), supporting the possibility that EBNA1 may have a wide-range effect on the organization of chromatin.
Figure 1.
Localization of EBNA1 in interphase nuclei. (a) EBNA1 is visualized in interphase nuclei of transfected NIH3T3 cells by immunofluorescence staining with the rabbit K67.1 antibody (red), and DNA is visualized by DAPI staining (blue). The merged image and localization profile indicate that EBNA1 does not accumulate on heterochromatin foci that appear as bright dots in DAPI stained nuclei. FI AU = fluorescence intensity arbitrary unit. (b) Localization of EBNA1 in U2OS cells co-transfected with EBNA1 (red) and GFP-HP1β (green). EBNA1 does not accumulate in heterochromatic regions marked by the accumulation of GFP-HP1β. (c) Immunofluorescence staining of U2OS cells transfected with GFP-EBNA1 and localization profiles illustrating the partial colocalization of EBNA1 with markers of transcriptionally active euchromatin (H3-K9ac) and facultative heterochromatin (H3-K9me2) but exclusion from heterochromatin identified by accumulation of H3-K9me3. Scale bar 5 µm. (d) EBNA1 promotes the decrease of heterochromatic foci in rodent cells. Constitutive heterochromatin is visualized by DAPI staining in NIH3T3 cells 48 h after transfection with either the pcDNA3 empty vector (left panel) or GFP-EBNA1 (middle and right panels). Scale bar 5 µm. The average number of heterochromatic foci in 25 cells is shown in the lower panel. Ctr = 18.28 ± 1.59; EBNA1 = 10.7 ± 1.00; P = 0.0002. (e) EBNA1 promotes the decrease of heterochromatic foci in human cells. Constitutive heterochromatin is visualized in U2OS cells transfected with GFP-HP1β. The number of HP1β positive foci in 20 cells is shown in the lower panel. Ctr = 13.7 ± 1.39: EBNA1 = 9.0 ± 1.43; *P = 0.03.
EBNA1 alters the global organization of chromatin
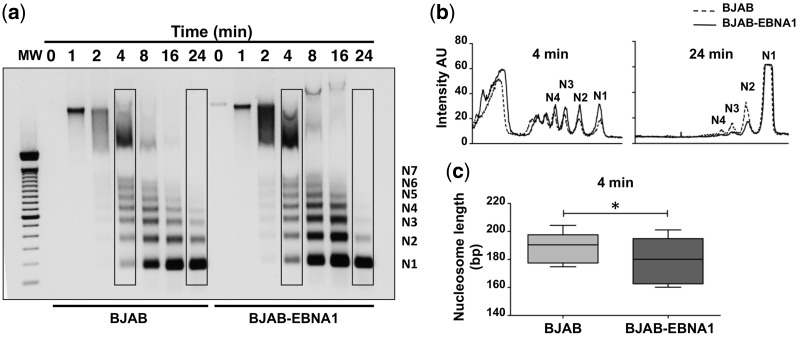
To address the effect of EBNA1 on chromatin architecture, nuclei isolated from the EBV-negative B-lymphoma line BJAB and its EBNA1-transfected subline BJAB-EBNA1 were subjected to MNase digestion, an assay commonly used to assess the compactness of chromatin (41). The rate of conversion of the chromatin fiber into progressively smaller nucleosome fragments detected in ethidium bromide–stained gels (Figure 2a, N1-N7) is an indication of the accessibility of linker DNA to the enzyme. As illustrated by the representative experiment shown in Figure 2b, the digestion of nuclei exposed to 10 U of MNase proceeded faster in EBNA1-positive cells. A more pronounced smear of high molecular weight fragments was reproducibly observed in BJAB-EBNA1 already after 2 min, after 4 min the relative intensity of the N1, N2, N3 and N4 bands was stronger and digestion was virtually complete down to the N1 size after 24 min. This process was clearly delayed in the EBNA1-negative BJAB. It is noteworthy that, although relatively small, the effect of EBNA1 is highly significant because it reflects the sum of events occurring across the entire genome. Analysis of the average nucleosome size after 4 min of digestion revealed the production of shorter fragments in BJAB-EBNA1 cells (Figure 2c and Supplementary Figure S1), further supporting the conclusion that EBNA1 promotes a global decompaction of cellular chromatin and increased accessibility of linker DNA to MNase.
Figure 2.
EBNA1 alters the global organization of chromatin. (a) Representative MNase digestion assay illustrating the faster digestion of DNA in BJAB-EBNA1 compared with BJAB, which indicates increased accessibility of the linker DNA region and global chromatin decompaction. (b) Nucleosome peaks profile after digestion for 4 and 24 min. A substantially increased intensity of the N1, N2, N3 and N4 fragments can be appreciated in BJAB-EBNA1 after 4 min and digestion is virtually complete after 24 min. One representative experiment out of three is shown. (c) EBNA1 expression is associated with decreased average nucleosome size. The size of N2, N3, N4 and N5 fragments was calculated in three independent experiments (see also Supplementary Figure S1) relative to the migration of the molecular weight marker. The average number of bases was calculated by dividing the length in bases by the number of nucleosomes in each fragment. BJAB = 189.1 ± 3.73; BJAB-EBNA1 = 180 ± 5.75; *P = 0.016.
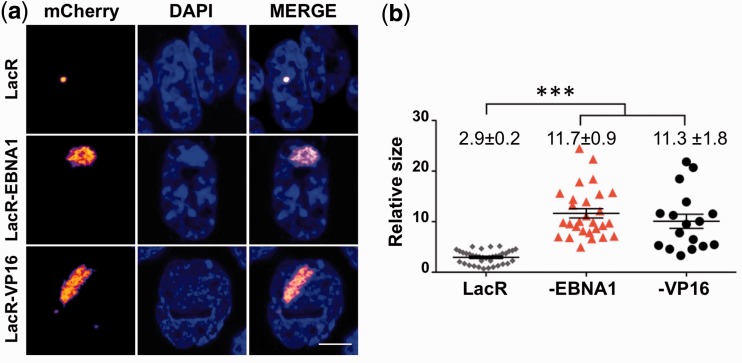
To directly probe the capacity of EBNA1 to affect chromatin organization in living cells, we took advantage of the A03-1 reporter cell line that carries multiple copies of a 256-repeats array of the Lac operon (LacO) integrated in a ≈80 Mb heterochromatin region that appears as an intensely fluorescent dot in cells expressing a chimeric Lac repressor (LacR) fused to GFP or its derivatives. Targeting of chromatin remodelers to the array by fusion to LacR is accompanied by large-scale chromatin decondensation that can be quantified as increased size of the fluorescent area relative to the size of the nucleus (42). The expression of the mCherry-LacR-EBNA1 fusion protein had a striking effect on the organization of the heterochromatic region, indicated by a highly significant 4-fold increase in the size of the fluorescent area (Figure 3a and b). An even stronger effect was observed in the NIH2/4 reporter cell line that carries a shorter LacO array (Supplementary Figure S2a and S2b). The degree of array decondensation was comparable with that induced by a chimeric mCherry-LacR fused to the herpes simplex virus transactivator VP16, a prototype inducer of large-scale chromatin unfolding in mammalian cells (22).
Figure 3.
EBNA1 induces decondensation of the LacO chromatin array. (a) A03-1 cells were transfected with mCherry-LacR, or mCherry-LacR fused to either EBNA1 or the herpes simplex-1 trans-activator VP16. The condensed array appears as a compact dot in cells expressing mCherry-LacR. Tethering of either EBNA1 or VP16 to the array induced the formation of extended structures of irregular shape. Scale bar 5 µm. (b) Quantification of the array size expressed as the percentage of nuclear area occupied by the array. EBNA1 and VP16 induced a comparable 4-fold increase in the size of the array. LacR = 2.9 ± 0.2; LacR-EBNA1 = 11.7 ± 0.9; LacR-VP16 = 11.3 ± 1.8, ***P < 0.0001.
EBNA1 promotes large-scale chromatin decondensation without recruitment of ATP-dependent remodelers
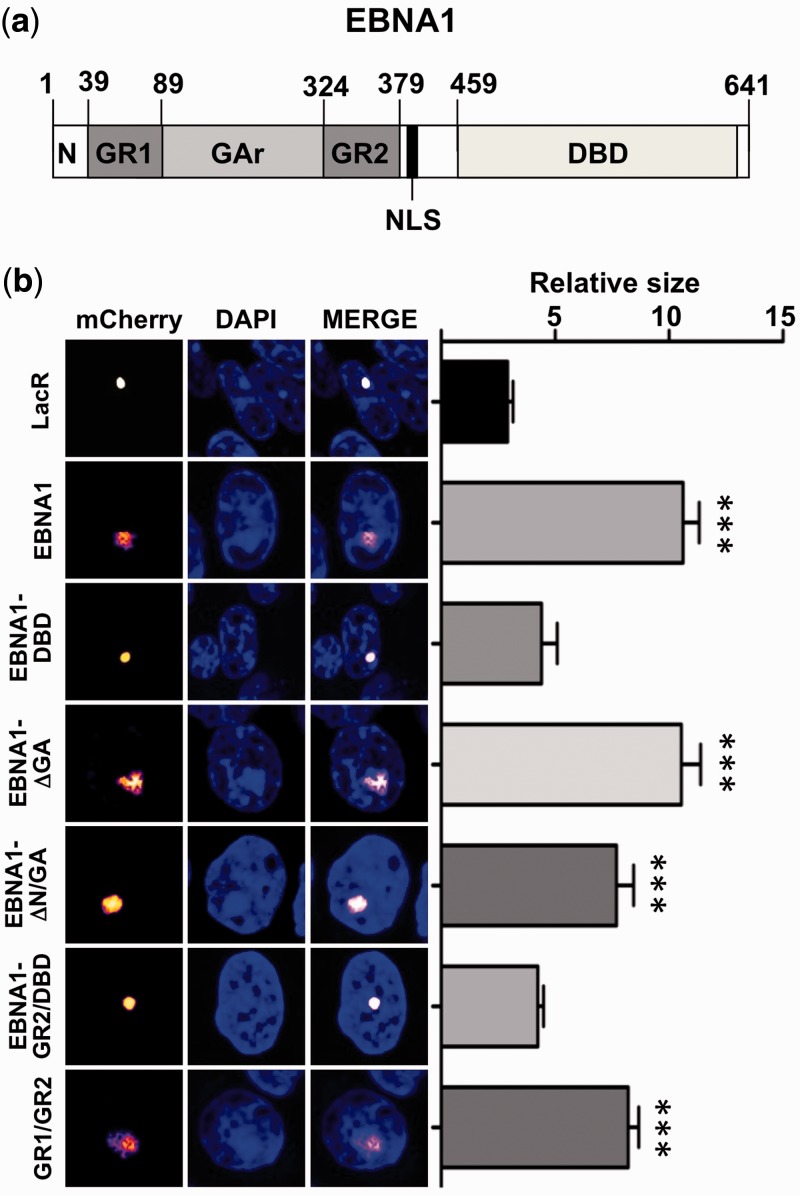
To explore the mechanisms by which EBNA1 induces chromatin decompaction, we first sought to map the protein domain responsible for the effect. A schematic illustration of the relevant domains of EBNA1 is shown in Figure 4a. Expression of mCherry-LacR fused to the C-terminal viral-DNA binding domain (EBNA1-DBD) did not promote chromatin decondensation (Figure 4b). This excludes the involvement of numerous cellular proteins that bind to this domain, including the deubiquitinating enzyme USP7 that promotes remodeling of the viral episome through deubiquitination of histone H2B (43). Notably, USP7 was efficiently recruited to the condensed array in cells expressing mCherry-LacR-EBNA1-DBD (Supplementary Figure S3), which validates the recruitment assay and confirms the failure of USP7 to cooperate in the decondensation of cellular chromatin. Deletion of the internal Gly-Ala repeat (EBNA1-ΔGA) had no effect, and decondensation was also unaffected by further deletion of the 39–amino acid long N-terminal domain (EBNA1-ΔN/GA). The N-terminal domain precedes the first of two Gly-Arg rich regions (indicated as GR1 and GR2, also known as linking regions LR1 and LR2) that flank the GA repeat and are required for the interaction of EBNA1 with cellular chromatin in interphase nuclei and condensed chromosomes (5–8). The GR1 and GR2 domains contain multiple copies of the Arg-Gly-Arg peptide and resemble the AT-hook of HMGA proteins (Supplementary Figure S4). In line with the possibility that interaction with DNA via the bipartite GR1/GR2 may be required for the capacity of EBNA1 to induce chromatin remodeling, further deletion of the GR1 (EBNA1-GR2/DBD), which significantly weakens the avidity of DNA binding (23), abolished chromatin decondensation, whereas array decondensation was induced by mCherry-LacR fused to the juxtaposed GR1 and GR2 (Figure 4b). It is noteworthy that the failure to achieve decondensation in cells expressing mCherry-LacR-EBNA1-GR2/DBD excludes the involvement of proteins binding to GR2, such as EBP2 and HMGB2 that were proposed to play a role in the loading of EBNA1 to cellular chromatin (44,45).
Figure 4.
The bipartite Gly-Arg rich domain of EBNA1 is required for chromatin decondensation. (a) Schematic representation of the EBNA1 protein: N terminus, amino acid 1–39; Gly-Arg repeat-1 (GR1), amino acid 39–89; Gly-Ala repeat (GA), amino acid 89–324; Gly-Arg repeat-2 (GR2), amino acid 324–379; Nuclear Localization Signal (NLS), amino acid 379–386; viral DBD, amino acid 459–598. (b) The GR1 and GR2 domains are necessary and sufficient for the chromatin decondensation effect. A03-1 cells were transfected with mCherry-LacR or mCherry-LacR fused to EBNA1 or the indicated EBNA1 subdomains. Decondensation of the array was observed on tethering of the full length EBNA1, EBNA1 deletion mutants containing both the GR1 and GR2 domains or the juxtaposed GR1 and GR2 domains alone. The relative size ±SE of the array in >30 cells analyzed in three independent experiments is shown, ***P < 0.001.
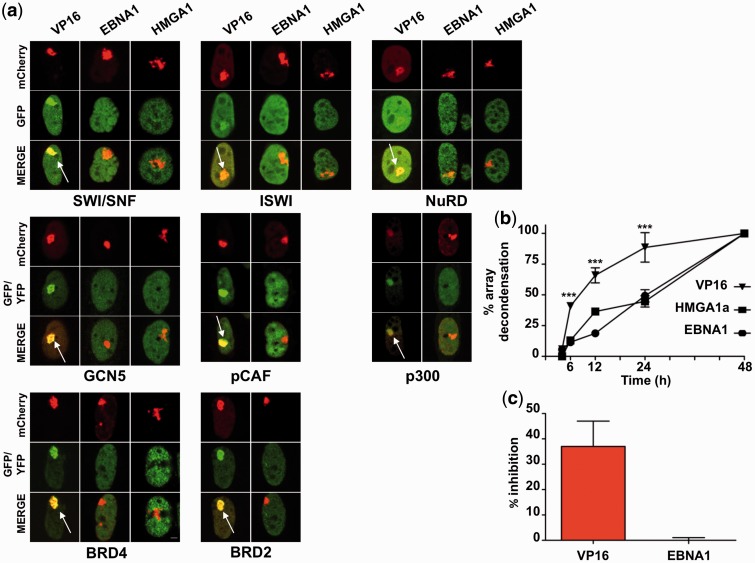
Chromatin remodeling is a multistep process by which high-order chromatin structures are relaxed to allow the binding of large protein complexes that regulate transcription (46). Powerful transactivators, such as VP16, promote chromatin decondensation through recruitment of histone acetyltransferases and ATP-dependent chromatin remodelers, whereas members of the HMG protein family are believed to exert their remodeling function via displacement of linker histones (47,48). To investigate the mechanisms by which EBNA1 promotes chromatin remodeling, mCherry-LacR tagged VP16, EBNA1 and HMGA1a (that is equally efficient in promoting decondensation of the LacO array in A03-1 cells, Supplementary Figure S5) were compared for their ability to recruit a panel of GFP-tagged ATPase subunits of known ATP-dependent remodeling complexes, including the BRG1 subunit of SWI/SNF complexes, the SNF2H subunit of ISWI complexes and the CHD4 subunit of NuRD complexes (49), histone acetyltransferases, such as GCN5 (50), pCAF (51) and p300 (52), and two bromodomain containing proteins, BRD2 and BRD4, that bind to acetylated histones and are often hijacked by viruses to promote transcription (53). As expected, all the tested proteins were recruited to the site of chromatin decondensation in cells expressing LacR-VP16, whereas recruitment was not observed in cells expressing LacR-EBNA1 or LacR-HMGA1a (Figure 5a).
Figure 5.
EBNA1 promotes large-scale chromatin decondensation without recruitment of ATP-dependent chromatin remodeling complexes. (a) A03-1 cells transfected with either mCherry-LacR-VP16, mCherry-LacR-EBNA1 or mCherry-LacR-HMGA1a were co-transfected with the ATPase subunits of the SWI/SNF, ISWI or NuRD ATP-dependent remodelers, GFP-BRG1, SNF2H-GFP and GFP-CHD4, respectively, the histone acetyltransferases YFP-GCN5, YFP-pCAF, YFP-p300 or the bromodomain containing proteins YFP-BRD2 and YFP-BRD4 that bind to acetylated histone tails. The VP16-dependent recruitment to the array is visualized in the GFP/YFP channel and in the merged images as an accumulation of green fluorescence overlapping with the site of the decondensed array (indicted by arrows). (b) Time kinetics of array decondensation. To achieve synchronous initiation of array decondensation, A03-1 cells were transfected with the indicated plasmids in the presence of IPTG, which allows the accumulation of unbound LacR-fusion proteins by preventing the interaction of LacR with the LacO sequence. After 24 h, the IPTG was removed and the cells were incubated in complete medium for the indicated time before fixation and determination of the array size in 25 cells for each condition. The percentage of array decondensation was calculated relative to the size of the condensed array in cells expressing the LacR alone and the maximal decondensation induced by the indicated LacR-fusion protein at 48 h according to the formula [(LacR-Ptx − LacR)/(LacR-Pt48 − LacR)] × 100, where P is either VP16, EBNA1 or HMGA1a. One representative experiment out of three is shown. (c) To assess the requirement of ATP for array decondensation, the cells were incubated for 30 min before fixation with ATP depletion medium containing 60 mM deoxyglucose, 30 mM NaAz. The assay was performed when the array decondensation had reached ∼50% of the maximum (6 h for VP16 and 24 h for EBNA1). The percentage inhibition was calculated according to the following formula: (1 − sizeATP−/sizeATP+) × 100, ***P < 0.001.
To further characterize the remodeling process, the kinetics of chromatin decondensation was investigated in A03-1 cells transfected with the LacR-fusion proteins in the presence of IPTG, which allows the accumulation of unbound products by preventing the interaction of LacR with LacO sequences (54). After 24 h, the IPTG was removed to allow synchronous binding of the accumulated products to DNA and the size of the array was monitored over time in cells chased in complete medium. As illustrated in Figure 5b, the decondensation process was significantly slower in cells expressing LacR-EBNA1 or LacR-HMGA1a compared with cells expressing LacR-VP16, with half maximal effect achieved in <6 h in VP16-expressing cells, whereas >24 h were needed in cells expressing HMGA1a or EBNA1. Furthermore, while depletion of ATP by culturing cells for 30 min in the presence of 60 mM deoxyglucose and 30 mM NaAz halted VP16-induced chromatin decondensation, the treatment did not affect chromatin unfolding in EBNA1-expressing cells (Figure 5c). Thus, EBNA1 resembles HMGA1a in the capacity to initiate chromatin decondensation with slow kinetic, without the help of ATP-dependent chromatin remodelers and in the absence of histone tail acetylation.
EBNA1 enhances the mobility of histone H1
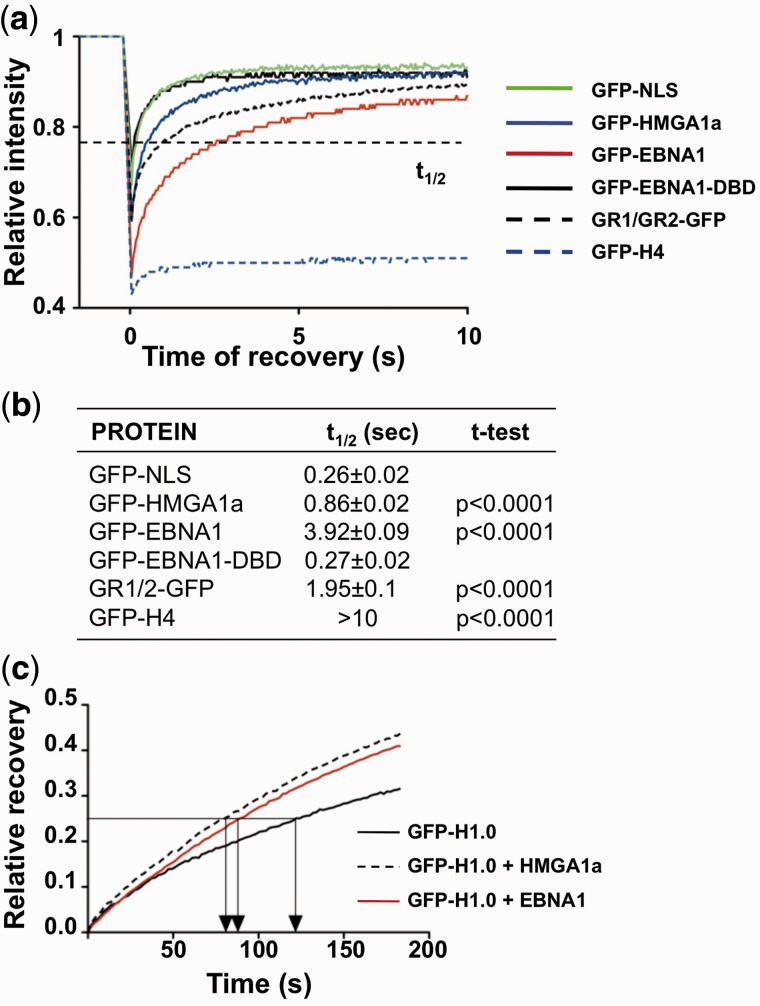
To reveal additional features of the interaction of EBNA1 with cellular DNA that may be relevant for the chromatin remodeling function, the mobility of EBNA1 on interphase chromatin was measured in live cells by FRAP. The FRAP recovery time of GFP-EBNA1 was compared with that of a nuclear GFP (GFP-NLS) that does not bind to chromatin, to the architectural protein HMGA1a (GFP-HMGA1a) and to the nucleosome-resident protein histone H4 (GFP-H4) (Figure 6a). The recovery time of GFP-EBNA1 was similar to that of HMGA1a although the slope of the curve was less steep, suggesting a longer chromatin residence time. The requirement of the GR domains for chromatin targeting was confirmed by the rapid recovery time of GFP-EBNA1-DBD that resembled that of soluble GFP-NLS. Furthermore, the mobility of GFP fused to the juxtaposed GR1/GR2 was comparable with that of full-length EBNA1, confirming that the bipartite domain is essential and sufficient to mediate efficient chromatin retention (Figure 6b).
Figure 6.
EBNA1 is highly mobile on interphase chromatin and displaces histone H1. (a) The GR1 and GR2 domains mediate a highly dynamic interaction of EBNA1 with cellular chromatin. FRAP recovery curves of GFP-tagged proteins in transfected U2OS cells. The fast recovery of the FRAP signal for GFP-NLS indicates that the protein does not bind to chromatin, whereas the failure to recover the signal of GFP-H4 confirms the tight association with DNA. The FRAP recovery curve of EBNA1 and GR1/GR2 containing constructs was comparable with that of HMGA1a. (b) Compilation of the time required of 50% recovery of fluorescence (t1/2) in 20 cells. The curves were fitted with the Prism5 software and significance was calculated relative to GFP-NLS. (c) FRAP recovery curves of GFP-H1.0 in NIH2/4 cells co-transfected with GFP-H1.0 and either mCherry-LacR, mCherry-LacR-EBNA1 or mCherry-LacR-HMG1a. The faster recovery time of GFP-H1.0 in cells expressing mCherry-LacR-EBNA1 or mCherry-LacR-HMGA1a indicates that the proteins reduce the chromatin-binding of histone H1.
Given that EBNA1 shares the rapid diffusion property of HMGs, we then tested whether it may also interfere with the binding of linker histone H1. Thus, we used FRAP to measure the relative mobility of transfected GFP-H1.0 at the LacO array in NIH2/4 cells expressing mCherry-LacR, mCherry-LacR-EBNA1 or mCherry-LacR-HMGA1a. The FRAP recovery of H1.0 in cells expressing mCherry-LacR was relatively slow, with a t1/2 value of ∼130 s (Figure 6c). Targeting of HMGA1a to the region reduced the t1/2 to ∼82 s, as expected. A comparable acceleration of recovery was observed in cells expressing EBNA1 (t1/2 90 s), indicating that EBNA1 efficiently competes with GFP-H1.0 and reduces its chromatin-retention time.
The transcriptional effect of EBNA1 is similar to that of HMGAs
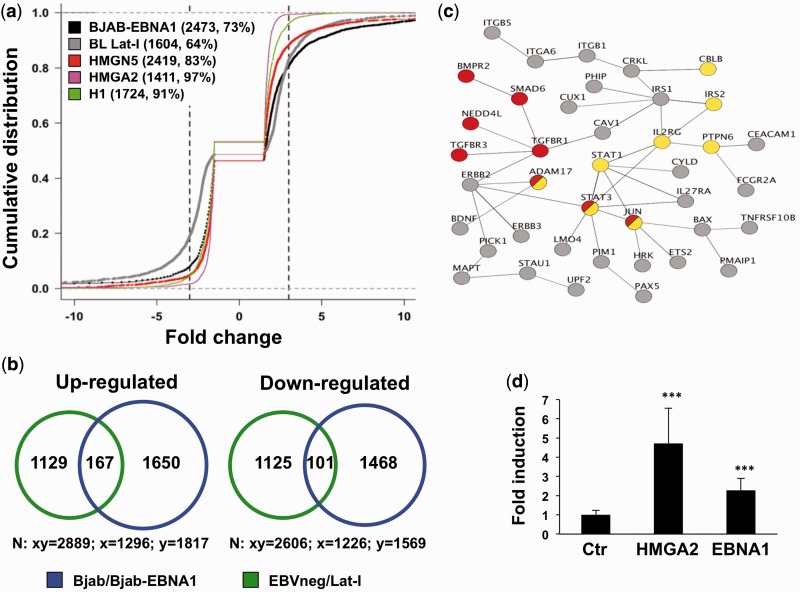
Treatments that induce global changes in chromatin architecture, such as modifications of histone acetylation (55) or alterations in the expression levels of MeCP2 [methyl-CpG binding protein-2 (56)], the genome organizing protein SATB1 [special AT-rich sequence-binding protein-1 (57)], histone H1 (34) or architectural transcription factors (58,59) promote a broad rearrangement of the cellular transcription profile featuring low average changes and both up- or down-regulation of a large number of genes. The chromatin remodeling capacity of EBNA1 raise the possibility that the viral protein may have a similar effect on transcription. To address this possibility, we took advantage of published gene expression microarray data to compare the transcription profiles of (i) the BJAB/BJAB-EBNA1 cell pair; (ii) a panel of EBV negative and EBV positive BL cell lines expressing physiological levels of EBNA1 in the absence of other latency proteins (EBV Lat I); (iii) cell pairs differing in the expressing of HMGN5 (32), HMGA2 (33) or histone H1 (34) following transfection or gene knockdown. This analysis revealed that, similar to the effect of ectopic expression of HMGN5 or HMGA2 and diminished expression of H1, the expression of EBNA1 is associated with differential regulation of a large number of genes (3386 in the BJAB/BJAB-EBNA1 pair and 2522 in the BL EBVneg/Lat-I pairs, Figure 7a), with approximately equal numbers of genes being up- or down-regulated (Figure 7b). Although statistically significant, the changes were relatively small for the majority of the genes, falling in a narrow range between 1.5- and 3-fold (Figure 7a).
Figure 7.
The transcriptional effect of EBNA1 is similar to that of HMGA proteins. (a) Empirical cumulative distribution function plots illustrating the distribution of genes differentially expressed in BJAB/BJAB-EBNA1 (black), BL EBVneg/Lat-I pairs (gray), HMGN5 (pink) or HMGA2 (red) transfected cells and histone H1 knockdown cells (green). The number of differentially regulated genes falling in the −3- to +3-fold interval (dotted lines) and the % relative to the total number of regulated genes are indicated. (b) Venn diagram depicting the overlap of genes whose expression is regulated in BJAB-EBNA1 and Lat-I cells compared with EBV negative BLs. A total of 268 genes, 167 upregulated and 101 downregulated, were commonly affected. (c) Protein–protein interaction networks of 66 genes concordantly regulated that are predicted by the STRING database to have one or more interacting partner with the highest confidence score (>0.9). The Panther and KEGG databases were used to identify signaling pathways in the network. Several components of the TGFβ (red circles) and JAK/STAT (yellow circles) signaling pathways were concordantly regulated. (d) Luciferase reporter assay of Twist promoter activity in HepG2 cells. The bars represent the mean ± SD of normalized luciferase activity from four independent experiments, each performed in triplicate.
In spite of largely non-overlapping sets of genes being affected in different cell lines, several previously identified EBNA1 targets (11) were found in a small group of genes that are concordantly regulated in transfected and endogenously expressing cells (Figure 7b), including components of the TGFβ signaling pathways such as TGFBR1 and TGFBR3, SMAD4, STAT3 and c-Jun (Figure 7c). The TGFβ signaling cascade acts via intracellular SMAD transducers to regulate a variety of cellular events, including epithelial-to-mesenchymal transition (EMT) that is active during embryogenesis and in invasive carcinomas (60). The transcriptional network of the EMT program is orchestrated by the AT-hook protein HMGA2 (61) that binds to SMADs and to the promoter of the transcription factors SNAIL (62) and TWIST (63). Thus, in the final set of experiments, we asked whether EBNA1 might regulate the expression of an HMGA2 target gene such as Twist. To this end, the expression of luciferase was compared in HepG2 hepatocellular carcinoma cells co-transfected with a murine Twist promoter-luciferase reporter plasmid and either EBNA1 or HMGA2. We found that, similar to the effect of HMGA2, expression of EBNA1 was accompanied by a robust activation of the promoter (Figure 7d).
DISCUSSION
Dynamic changes in chromatin structure play a key role in the orderly progression of DNA transcription, replication, recombination and repair. The access of transcription factors to gene regulatory elements is controlled by the local chromatin architecture, which contributes to their tissue-specific action. We have found that EBNA1 promotes a widespread remodeling of chromatin organization and a broad rearrangement of transcription, featuring both up- and down-regulation of a large number of genes. This effect is reminiscent of that of ‘architectural’ or ‘pioneer’ transcription factors that confer competence for gene expression by opening the chromatin for binding by remodelers, transcription factors and co-repressors (18), and play thereby critical roles in cell programming and in the responsiveness to environmental cues.
Here we have shown that EBNA1 induces chromatin decompaction with slow kinetics and independent of the recruitment of ATP-dependent remodelers, sharing these two properties with HMGA1a. The HMG ‘architectural factors’ modulate nucleosome and chromatin structures by weakening the chromatin binding of linker histones (47,48), which are themselves mobile proteins (64). It is surmised that by rapid diffusion through the nucleus, HMGs gain access to temporarily vacated nucleosomal sites, and can thereby counteract the ability of linker histones to stabilize higher order chromatin structures and restrict the access of regulatory factors, nucleosome remodeling complexes and histone modifiers (65). We have found that, similar to HMGAs, EBNA1 enhances the mobility of histone H1, which correlates with global chromatin decompaction, increased accessibility to MNase digestion and reduced length of the nucleosome repeat. The importance of H1 displacement for these effects is supported by the induction of a similar phenotype in mouse embryonic stem cells on diminished occupancy of the linker region following knockdown of multiple H1 variants (34,66).
The HMG proteins bind to partially overlapping sites in the linker region (47). Proteins of the HMGA family are targeted to the region via the AT-hook that mediates sequence-independent binding to the minor groove of AT-rich DNA and promotes DNA bending and the interaction with high-order DNA and RNA structures such as cruciform DNA and G-quadruplex (67). Our findings demonstrate that the AT-hook–like chromatin-targeting module of EBNA1 is necessary and sufficient for a highly dynamic interaction with chromatin. This module is also involved in two additional properties shared by EBNA1 and HMGAs, the capacity to bend DNA and the interaction with cruciform DNA and G-quadruplex that are particularly frequent in genomic regions flanking transcription start sites (68). The functional similarity is further substantiated by the finding that a chimeric protein containing the C-terminal viral-DBD of EBNA1 fused to HMGA1 retains most of the functional properties of EBNA1, including the capacity to regulated the replication and correct partitioning of the viral episome (5,7,8). It is noteworthy that while the capacity to bend DNA may play a role in the regulation of transcription, for example, by favoring long-range enhancer-promoter interactions, this property in unlikely to contribute to the chromatin remodeling effect because the latter requires the displacement of histones and other inhibitory proteins.
The AT-hook of EBNA1 is similar but not identical to the AT-hook of HMGAs (Supplementary Figure S4). The GR1 and GR2 domains contain multiple highly conserved Arg-Gly-Arg motifs, whereas HMGAs contain three independent AT-hooks, each carrying a single Arg-Gly-Arg motif. The NMR structure of the HMGA1-DNA complex shows that the Arg-Gly-Arg peptide forms a narrow concave surface perfectly inserted into the minor groove with the extended Arg side chains binding to opposite A-T base pairs (67). The conserved Arg and Lys residues that flank the core motif establish electrostatic interactions with DNA, while the Pro-Lys-Gly-Ser residues form a type II β-turn that bridges the edges of the minor groove, and the Pro residues direct the peptide backbone away from the groove. The lack of Pro residues in the GR1 and GR2 of EBNA1 is likely to be compensated by the presence of longer repeats in a random coil conformation. The extended positively charged surface formed by the multiple Arg residues could mediate electrostatic interactions with DNA. Indeed, bacterially expressed GR1 and GR2, and baculovirus-expressed EBNA1 were shown to selectively bind to AT-rich DNA in vitro, and the binding of GR1 and GR2 was competed by the synthetic AT-hook analogs Netropsin and Distamycin A (5). It remains to be seen whether the structural differences may account for the somewhat slower mobility of EBNA1 and less-efficient displacement of H1, and whether other features, such as the intervening GA repeat or the capacity to form dimers, may contribute to fine tune the activity of the viral protein.
HMGA proteins are physiologically expressed at high levels during embryogenesis and have important roles in development (19,69,70). Downregulation of HMGA1 and HMGA2 promotes chromatin compaction during neocortical development and reduces the neurogenic potential of neuronal cell precursors, while overexpression in late-stage NCPs is associated with chromatin opening and increased neurogenesis (71). Notably, both chromatin opening and neurogenesis are antagonized by overexpression of histone H1. The oncogenic potential of HMGAs is highlighted by their ectopic expression in a broad spectrum of human malignancies, and is confirmed by the induction of different types of neoplasia in transgenic mouse models (72). Several mechanisms have been proposed to account for the transforming ability of HMGAs, but a common theme is the capacity to cooperate with oncogenes in the regulation of genes that control cell proliferation and apoptosis. In addition, HMGAs play a key role in tumor cell migration and metastasis by controlling the cellular reprogramming associated with EMT (62,63,73). Although EBNA1 is regularly expressed in all EBV-associated malignancies, its role in oncogenesis is debated and conflicting data have been reported on the capacity to serve as an oncogene in transgenic mouse models (74,75). Our findings provide a possible explanation for these discrepancies by highlighting a scenario where the chromatin remodeling function of EBNA1, although insufficient to promote malignancy in the absence of co-factors, may contribute to malignant transformation by sensitizing the infected cells to the activity of cellular or viral oncogenes. Indeed, EBNA1 was shown to synergize with c-Myc in a mouse model of lymphomagenesis (40). It is also noteworthy that the regulation of genes involved in EMT suggests that, in combination with appropriate environmental cues, EBNA1 may promote metaplasia. It is tempting to speculate that this effect could underlie the association of the virus with malignancies, such as smooth muscle lejomyosarcoma (76) and gastric carcinoma (77) that express EBNA1-only latency programs and arise in cells and tissues that are normally not susceptible to EBV infection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Swedish Cancer Society, the Swedish Medical Research Council, the Karolinska Institutet, Stockholm, Sweden, and by the European Community Network of Excellence RUBICON, Contract no. LSG-CT-2005-018683 and STREP project VITAL, Contract no. LSG-CT-2006-037874. N.M. was supported by a fellowship awarded by the government of Pakistan. Funding for open access charge: Swedish Cancer Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Kieff, J. Middeldorp, H. van Hattikum, I. Rafalska-Metcalf, T. Misteli, A. Belmont and R.D. Everett for the kind gift of reagents, plasmids and cell lines.
REFERENCES
- 1.Rickinson AB, Kieff E. In: Virology. Fields BN, Knipe DM, Howley PM, editors. Philadelphia, PA: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 2.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 3.Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy G, Sugden B. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 2003;23:6901–6908. doi: 10.1128/MCB.23.19.6901-6908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 2004;78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sears J, Kolman J, Wahl GM, Aiyar A. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein-Barr nuclear antigen 1. J. Virol. 2003;77:11767–11780. doi: 10.1128/JVI.77.21.11767-11780.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung SC, Kang MS, Kieff E. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl Acad. Sci. USA. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canaan A, Haviv I, Urban AE, Schulz VP, Hartman S, Zhang Z, Palejev D, Deisseroth AB, Lacy J, Snyder M, et al. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc. Natl Acad. Sci. USA. 2009;106:22421–22426. doi: 10.1073/pnas.0911676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sompallae R, Callegari S, Kamranvar SA, Masucci MG. Transcription profiling of Epstein-Barr virus nuclear antigen (EBNA)-1 expressing cells suggests targeting of chromatin remodeling complexes. PLoS One. 2010;5:e12052. doi: 10.1371/journal.pone.0012052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood VH, O'Neil JD, Wei W, Stewart SE, Dawson CW, Young LS. Epstein-Barr virus-encoded EBNA1 regulates cellular gene transcription and modulates the STAT1 and TGFbeta signaling pathways. Oncogene. 2007;26:4135–4147. doi: 10.1038/sj.onc.1210496. [DOI] [PubMed] [Google Scholar]
- 12.Dresang LR, Vereide DT, Sugden B. Identifying sites bound by Epstein-Barr virus nuclear antigen 1 (EBNA1) in the human genome: defining a position-weighted matrix to predict sites bound by EBNA1 in viral genomes. J. Virol. 2009;83:2930–2940. doi: 10.1128/JVI.01974-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu F, Wikramasinghe P, Norseen J, Tsai K, Wang P, Showe L, Davuluri RV, Lieberman PM. Genome-wide analysis of host-chromosome binding sites for Epstein-Barr virus nuclear antigen 1 (EBNA1) Virol. J. 2010;7:262–279. doi: 10.1186/1743-422X-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan T, Li XY, Sabo PJ, Thomas S, Stamatoyannopoulos JA, Biggin MD, Eisen MB. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 2011;7:e1001290. doi: 10.1371/journal.pgen.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Hirai H, Tani T, Kikyo N. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. Biol. 2010;54:1589–1596. doi: 10.1387/ijdb.103194hh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeves R. Nuclear functions of the HMG proteins. Biochim. Biophys. Acta. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc. Natl Acad. Sci. USA. 2009;106:2313–2318. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 22.Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coppotelli G, Mughal N, Marescotti D, Masucci MG. High avidity binding to DNA protects ubiquitylated substrates from proteasomal degradation. J. Biol. Chem. 2011;286:19565–19575. doi: 10.1074/jbc.M111.224782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luijsterburg MS, von Bornstaedt G, Gourdin AM, Politi AZ, Mone MJ, Warmerdam DO, Goedhart J, Vermeulen W, van Driel R, Hofer T. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell. Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rafalska-Metcalf IU, Powers SL, Joo LM, LeRoy G, Janicki SM. Single cell analysis of transcriptional activation dynamics. PLoS One. 2010;5:e10272. doi: 10.1371/journal.pone.0010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canning M, Boutell C, Parkinson J, Everett RD. A Ring finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 2004;279:38160–38168. doi: 10.1074/jbc.M402885200. [DOI] [PubMed] [Google Scholar]
- 31.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 32.Rochman M, Taher L, Kurahashi T, Cherukuri S, Uversky VN, Landsman D, Ovcharenko I, Bustin M. Effects of HMGN variants on the cellular transcription profile. Nucleic Acids Res. 2011;39:4076–4087. doi: 10.1093/nar/gkq1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Liu Z, Shao C, Gong Y, Hernando E, Lee P, Narita M, Muller W, Liu J, Wei JJ. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 2011;71:349–359. doi: 10.1158/0008-5472.CAN-10-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 35.Reedy EA, Heatfield BM, Trump BF, Resau JH. Correlation of cytokeratin patterns with histopathology during neoplastic progression in the rat urinary bladder. Pathobiology. 1990;58:15–27. doi: 10.1159/000163561. [DOI] [PubMed] [Google Scholar]
- 36.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drotar ME, Silva S, Barone E, Campbell D, Tsimbouri P, Jurvansu J, Bhatia P, Klein G, Wilson JB. Epstein-Barr virus nuclear antigen-1 and Myc cooperate in lymphomagenesis. Int. J. Cancer. 2003;106:388–395. doi: 10.1002/ijc.11224. [DOI] [PubMed] [Google Scholar]
- 41.Bryant GO. Measuring nucleosome occupancy in vivo by micrococcal nuclease. Methods Mol. Biol. 2012;833:47–61. doi: 10.1007/978-1-61779-477-3_4. [DOI] [PubMed] [Google Scholar]
- 42.Belmont AS, Li G, Sudlow G, Robinett C. Visualization of large-scale chromatin structure and dynamics using the lac operator/lac repressor reporter system. Methods Cell. Biol. 1999;58:203–222. doi: 10.1016/s0091-679x(08)61957-3. [DOI] [PubMed] [Google Scholar]
- 43.Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog. 2009;5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shire K, Ceccarelli DF, Avolio-Hunter TM, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 1999;73:2587–2595. doi: 10.1128/jvi.73.4.2587-2595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jourdan N, Jobart-Malfait A, Dos Reis G, Quignon F, Piolot T, Klein C, Tramier M, Coppey-Moisan M, Marechal V. Live-cell imaging reveals multiple interactions between Epstein-Barr virus nuclear antigen 1 and cellular chromatin during interphase and mitosis. J. Virol. 2012;86:5314–5329. doi: 10.1128/JVI.06303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 47.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lange M, Demajo S, Jain P, Di Croce L. Combinatorial assembly and function of chromatin regulatory complexes. Epigenomics. 2011;3:567–580. doi: 10.2217/epi.11.83. [DOI] [PubMed] [Google Scholar]
- 50.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 51.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 52.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 53.Weidner-Glunde M, Ottinger M, Schulz TF. WHAT do viruses BET on? Front. Biosci. 2010;15:537–549. doi: 10.2741/3632. [DOI] [PubMed] [Google Scholar]
- 54.Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, Dantuma NP. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell. Biol. 2012;197:267–281. doi: 10.1083/jcb.201106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J. Biol. Chem. 2005;280:796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- 56.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 58.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, et al. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez Hoyos J, Fedele M, Battista S, Pentimalli F, Kruhoffer M, Arra C, Orntoft TF, Croce CM, Fusco A. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: tissue specificity of the HMGA1-dependent gene regulation. Cancer Res. 2004;64:5728–5735. doi: 10.1158/0008-5472.CAN-04-1410. [DOI] [PubMed] [Google Scholar]
- 60.Fuxe J, Vincent T, Garcia de Herreros A. Transcriptional crosstalk between TGF-beta and stem cell pathways in tumor cell invasion: role of EMT promoting Smad complexes. Cell Cycle. 2010;9:2363–2374. doi: 10.4161/cc.9.12.12050. [DOI] [PubMed] [Google Scholar]
- 61.Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol. Cell. Biol. 2001;21:575–594. doi: 10.1128/MCB.21.2.575-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 2008;283:33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan EJ, Thuault S, Caja L, Carletti T, Heldin CH, Moustakas A. Regulation of transcription factor twist expression by the DNA architectural protein high mobility group A2 during epithelial-to-mesenchymal transition. J. Biol. Chem. 2012;287:7134–7145. doi: 10.1074/jbc.M111.291385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 65.Catez F, Hock R. Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim. Biophys. Acta. 2010;1799:15–27. doi: 10.1016/j.bbagrm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 68.Norseen J, Johnson FB, Lieberman PM. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009;83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reeves R. Structure and function of the HMGI(Y) family of architectural transcription factors. Environ. Health Perspect. 2000;108(Suppl 5):803–809. doi: 10.1289/ehp.00108s5803. [DOI] [PubMed] [Google Scholar]
- 70.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 71.Kishi Y, Fujii Y, Hirabayashi Y, Gotoh Y. HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat. Neurosci. 2012;15:1127–1133. doi: 10.1038/nn.3165. [DOI] [PubMed] [Google Scholar]
- 72.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 73.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson JB, Bell JL, Levine AJ. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. Embo J. 1996;15:3117–3126. [PMC free article] [PubMed] [Google Scholar]
- 75.Kang MS, Lu H, Yasui T, Sharpe A, Warren H, Cahir-McFarland E, Bronson R, Hung SC, Kieff E. Epstein-Barr virus nuclear antigen 1 does not induce lymphoma in transgenic FVB mice. Proc. Natl Acad. Sci. USA. 2005;102:820–825. doi: 10.1073/pnas.0408774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McClain KL, Leach CT, Jenson HB, Joshi VV, Pollock BH, Parmley RT, DiCarlo FJ, Chadwick EG, Murphy SB. Association of Epstein-Barr virus with leiomyosarcomas in children with AIDS. N. Engl. J. Med. 1995;332:12–18. doi: 10.1056/NEJM199501053320103. [DOI] [PubMed] [Google Scholar]
- 77.Osato T, Imai S. Epstein-Barr virus and gastric carcinoma. Semin. Cancer Biol. 1996;7:175–182. doi: 10.1006/scbi.1996.0024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.