Abstract
The origin recognition complex (ORC) of Saccharomyces cerevisiae binds origin DNA and cooperates with Cdc6 and Cdt1 to load the replicative helicase MCM2–7 onto DNA. Helicase loading involves two MCM2–7 hexamers that assemble into a double hexamer around double-stranded DNA. This reaction requires ORC and Cdc6 ATPase activity, but it is unknown how these proteins control MCM2–7 double hexamer formation. We demonstrate that mutations in Cdc6 sensor-2 and Walker A motifs, which are predicted to affect ATP binding, influence the ORC–Cdc6 interaction and MCM2–7 recruitment. In contrast, a Cdc6 sensor-1 mutant affects MCM2–7 loading and Cdt1 release, similar as a Cdc6 Walker B ATPase mutant. Moreover, we show that Orc1 ATP hydrolysis is not involved in helicase loading or in releasing ORC from loaded MCM2–7. To determine whether Cdc6 regulates MCM2–7 double hexamer formation, we analysed complex assembly. We discovered that inhibition of Cdc6 ATPase restricts MCM2–7 association with origin DNA to a single hexamer, while active Cdc6 ATPase promotes recruitment of two MCM2–7 hexamer to origin DNA. Our findings illustrate how conserved Cdc6 AAA+ motifs modulate MCM2–7 recruitment, show that ATPase activity is required for MCM2–7 hexamer dimerization and demonstrate that MCM2–7 hexamers are recruited to origins in a consecutive process.
INTRODUCTION
DNA replication is initiated at origins of replication. In Saccharomyces cerevisiae, these origins are called autonomously replicating sequences and contain binding sites for the origin recognition complex (ORC) (1). ORC is a six-subunit complex (Orc1–6) that in late M-phase recruits Cdc6 to replication origins. The ORC–Cdc6 complex loads the mini-chromosome maintenance proteins 2–7 (MCM2–7) with the help of Cdt1 into a pre-replication complex (pre-RC) onto DNA (2). During this loading reaction, Cdt1 gets released from DNA in a Cdc6 ATP hydrolysis-dependent manner (3), while MCM2–7 assembles from a hexamer into a double hexamer encircling double-stranded DNA (4,5). In S-phase, MCM2–7 becomes activated and acts as the replicative helicase together with Cdc45 and GINS (6,7).
Cdc6 and Orc1–Orc5 are members of the AAA+ (ATPases associated with a variety of cellular activities) family of proteins (8,9). Cdc6, Orc1 and Orc5 are known to bind ATP, but only Cdc6 and Orc1 can hydrolyse ATP (3,10–12). AAA+ proteins contain several conserved elements that are important for ATP binding and hydrolysis: Walker A, Walker B, sensor-1, arginine finger and sensor-2 (Figure 1). The Walker A motif in Cdc6 contains a conserved GXXGXGKT sequence (13). On the basis of archaeal Cdc6 crystal structures (14,15), the Walker A motif of S. cerevisiae Cdc6 is thought to be involved in the binding of the triphosphate moiety of the nucleotide, which is important for ATP binding and hydrolysis. A mutation of the conserved lysine within the Walker A motif of human Cdc6 causes an ATP-binding and hydrolysis defect (16). A corresponding (K114E) mutant in S. cerevisiae Cdc6 has not been tested for ATP-binding or hydrolysis defects in vitro, but is lethal in vivo, fails to load MCM2–7 onto chromatin and binds weakly to ORC (9,17,18). On the other hand, the Cdc6 Walker B motif (DELD) (9) is important for ATP hydrolysis, as it coordinates a Mg2+ ion and the water molecule required for the nucleophilic attack on the β–γ bond of the ATP via a conserved glutamic acid (14,15). A Cdc6 mutant (Cdc6 E224G), carrying a glutamine instead of the conserved glutamic acid in the Walker B motif, binds ATP, but has an ATP hydrolysis defect, which causes a partial block in Cdt1 release and in MCM2–7 loading in vitro (3), as well as dominant lethality in vivo (17). ATPγS is an ATP analogue, which can be only slowly hydrolysed. It was found that ATPγS and a Cdc6 Walker B ATPase mutant lead to a similar type of pre-RC assembly arrest—reduced Cdt1 release and reduced MCM2–7 loading (3). For this reason, ATPγS is used to study the effect of Cdc6 ATP hydrolysis. The function of the sensor-1 and sensor-2 motifs is in general more difficult to predict. The sensor-1 motif is part of a hydrogen-bonding network that positions a water molecule relative to the γ-phosphate of ATP and has been implicated in ATP binding and hydrolysis (19). The sensor-2 motif contains a conserved amino acid, which contacts ATP. A mutation of this amino acid in different AAA+ proteins has been shown to result in decreased ATP binding or ATP hydrolysis activity (19). Cdc6 mutants with mutations in sensor-1 (N263A) and sensor-2 (R332E) have been shown to block MCM2–7 loading in vivo (20,21), and the sensor-1 mutant has an ATP-hydrolysis defect in the context of ORC (11), but these mutants have not been analysed in vitro for specific defects during pre-RC formation. The arginine finger motif of AAA+ proteins is important for ATP hydrolysis. Interestingly, the Orc1 ATPase becomes activated in trans by an arginine finger of the neighbouring Orc4 subunit (10). An ORC complex, containing an Orc4 arginine finger mutant ORC4R, is deficient in Orc1 ATPase activity, but is capable of MCM2–7 loading. Although ORC loads MCM2–7 in a repetitive manner, ORC4R is restricted to a single round of MCM2–7 loading. Based on these findings, it has been suggested that Orc1 ATPase may function during pre-RC disassembly by releasing ORC–Cdc6 from loaded MCM2–7 (10).
Figure 1.
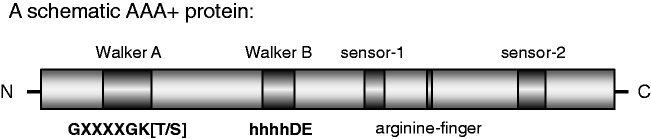
Schematic representation of conserved AAA+ ATPase motifs. A simplified AAA+ structure including the conserved Walker A, Walker B, sensor-1, arginine finger and sensor-2 is shown. Hydrophobic amino acids are abbreviated as h. The general AAA+ consensus sequence of the Walker A and Walker B motifs is shown.
Each subunit of MCM2–7 belongs to the AAA+ family of ATPases. MCM2–7 helicase activity requires ATP hydrolysis. In contrast, ATPase activity of MCM2–7 is dispensable for pre-RC formation in Xenopus (22), suggesting that Cdc6 and Orc1 are the main ATPases involved in pre-RC assembly. In summary, it is known that Cdc6 ATPase is required for MCM2–7 loading, whereas Orc1 ATPase is involved in repetitive loading of MCM2–7, thereby revealing that Cdc6 ATPase acts prior to Orc1 ATPase (3,10). However, it is not known how the conserved Cdc6 sensor-1, sensor-2 and Walker A motifs regulate pre-RC formation, why pre-RC formation is blocked in the absence of Cdc6 ATP hydrolysis and whether Orc1 ATPase is required for pre-RC disassembly upon MCM2–7 loading.
The development of a reconstituted pre-RC assay that uses purified proteins (4,5,23) allows now the analysis of these questions.
Our work, using purified proteins from yeast, identifies the function of several conserved AAA+ motifs within Cdc6 and Orc1 during pre-RC formation. We find that Cdc6 sensor-2 and Walker A mutants severely impair the ability of Cdc6 to interact with ORC and therefore fail to bind and load MCM2–7 efficiently. Based on our analysis, we predict that both mutants share an ATP-binding defect and consequently block MCM2–7 loading owing to a weak ORC–Cdc6 interaction. On the other hand, Cdc6 sensor-1 and Walker B mutants interacted efficiently with ORC and recruited MCM2–7, but led to slightly reduced Cdt1 release and poor MCM2–7 loading, highlighting how the sensor-1 and Walker B Cdc6 ATPase motifs affect pre-RC formation. Based on this work, Cdc6 ATP hydrolysis appears to be particularly important for MCM2–7 loading, but the function of the Cdc6 ATPase is only poorly understood. Here, we discovered that Cdc6 ATP hydrolysis regulates MCM2–7 double-hexamer assembly, as in the absence of ATP hydrolysis, only a single MCM2–7 hexamer associates with ORC–Cdc6–Cdt1, whereas in the presence of ATP hydrolysis, MCM2–7 double hexamers are formed. Lastly, we showed that an Orc1 ATP hydrolysis mutant did not affect pre-RC assembly or release of ORC–Cdc6 from loaded MCM2–7, suggesting that Orc1 ATPase is required for the reactivation of the ORC complex during repetitive MCM2–7 loading. In summary, our data identify the functions of Cdc6 Walker A, sensor 1 and sensor 2 motifs in pre-RC formation, show for the first time that in the absence of Cdc6 ATPase activity only a single MCM2–7 hexamer associates with origin DNA and identify that Cdc6 ATPase regulates assembly of an MCM2–7 double hexamer.
MATERIALS AND METHODS
In vitro pre-RC assembly assay
The pre-RC was assembled in a one-step reaction: 40 nM ORC or ORC4R, 80 nM Cdc6 or Cdc6 mutants, 40 nM Cdt1, 40 nM MCM2–7 and 120 U Lambda phosphatase in buffer A (50 mM HEPES–KOH pH 7.5, 100 mM KGlu, 10 mM MgAc, 50 μM ZnAc, 3 mM ATP, 5 mM DTT, 0.1% Triton X-100 and 5% glycerol) plus 2 mM MnCl2 were added to 6 nM pUC19-ARS1 plasmid beads (4) for 15 min at 24°C. Beads were washed three times with buffer A plus 1 mM EDTA or buffer B (50 mM HEPES–KOH pH 7.5, 1 mM EDTA, 500 mM NaCl, 5% glycerol, 0.1% Triton X-100 and 5 mM DTT) before digestion with 1 U of DNase I in buffer A plus 5 mM CaCl2 for 6 min at 24°C. The samples were separated by poly acrylamide gel electrophoresis (PAGE) and analysed by silver staining or by immune blotting for Cdc6 with anti-Cdc6 (9H8/5, Abcam, ab20150).
In vitro pre-RC assembly followed by crosslinking
Pre-RC assembly was performed as described. Samples were washed with buffer A containing KAc instead of KGlu. For crosslinking in solution, the samples were released from DNA by DNase I, which was followed by the addition of 2% glutaraldehyde for 10 min at 4°C. Glutaraldehyde was inactivated by addition of 1 volume of Laemmli buffer (24). Crosslinking on DNA was performed with 2% glutaraldehyde for 10 min at 4°C; samples were washed with buffer B and eluted with DNase I. The complexes were separated on a discontinuous gradient by SDS-PAGE (25), where three quarters of the gel were a 3.5–7.5% gradient and the top quarter was a continuous gel of 3.5%, both with an acrylamide/bisacrylamide ratio of 80:1; the gel was analysed by silver staining and by immune blotting for pre-RC proteins using anti-Orc3 (SB3) (26), anti-Cdc6 (9H8/5, Abcam, ab20150), anti-Cdt1 (CS1411) (unpublished; Speck and Stillman) and anti-Mcm2 (#49) (26) antibodies.
Co-immunoprecipitation assays
Two standard size pre-RC reactions were prepared as described above (In vitro pre-RC assembly assay) using MBP-tagged MCM2–7 (20 nM) and untagged MCM2–7 (20 nM). The complexes were released from magnetic beads with AluI (NEB) for 7.5 min at 24°C; the two reactions were pooled, immune precipitated with anti-MBP (NEB) antibody coupled to protein G beads for 7.5 min at 24°C, washed three times with buffer A and analysed by Western blot with anti-Mcm2.
Cloning of MBP-Mcm2
Using site-directed mutagenesis with QuickChange II XL, an XmaI site was inserted between amino acids 221 and 222 in Mcm2. Afterwards MBP was amplified from pMAL C2x (NEB) with primers (MBP-20A_linker FWD TCCCAAGCATGCTATAGAACTTTGACTGTTTTGAAAATCGAAGAAGGTAAACTGGT and MBP-20A_linker REV TCCCAAGCATGCTATAGAACTTTGACTGTTTTGTTTAGTAATTCTAGTCTGCGCGTCTTTCA), which introduced flexible linkers at both ends, and the resulting product was inserted in the XmaI site.
Complex assembly for EM analysis
The samples were prepared as described (4). Briefly, samples were prepared as described for the in vitro pre-RC assembly assay, but DNA was not coupled to the beads. The mixture was centrifuged and then gelfiltered through a Sephacryl 400 MicroSpin column. The sample was diluted 1:10 in 10 mM Tris (pH 7.5) and 10 mM MgCl2, adsorbed to freshly cleaved mica and rotary shadowed with platinum vapor (27). Negative stain with 2% uranyl acetate was performed as described (28).
Analysis of ORC4R ATPase activity
This was performed as described (11,12). Briefly, 2.5 pmol of ORC and 2.5 pmol of Cdc6 were incubated for 30 min on ice in 12 μl of buffer (25 mM HEPES, pH 7.6, 100 mM KGlu, 5 mM MgAc, 1 mM DTT, 1 mM EDTA, 1 mM EGTA, 0.1% (v/v) Triton X-100, 10% glycerol) containing 2.5 pmol of DNA (when indicated) and 100 μM ATP. After the incubation, 5 μCi of [α-32P] ATP (3000 Ci/mmol) was added. Two microlitres of aliquots were removed and stopped with 0.5 μl of 2% SDS stop solution. One microlitre of the samples were consequently spotted on Cellulose PEI TLC plates (Baker) and developed in 1 M HCOOH, 0.4 M LiCl.
Protein purification
The untagged ORC complex was expressed in insect cells and purified via SP sepharose, Mono Q sepharose and by Superdex 200-mediated gelfiltration as described (12). Cdc6 was expressed in bacteria as a GST fusion protein and purified by GST agarose, followed by Prescission protease cleavage to remove the GST tag and by hydroxyapatite resin as described (9). Cdt1 was expressed in bacteria as a GST fusion protein and purified by GST agarose, followed by Prescission protease cleavage to remove the GST tag and by SP sepharose as described (4). MCM2–7 with a single HA tag at the N-terminus of Mcm3 was expressed in yeast and purified by anti-HA agarose and Superdex 200 gelfiltration as described (4).
Similarity calculation
The similarity was calculated using a Blosum62 matrix and with these gap penalties: 10 for creating a gap and 0.2 every time the gap is extended one amino acid. The alignment was done with Clustal X (2.0) iterating each alignment step.
Structural prediction
The S. cerevisiae Cdc6p sequence from S73 to H513 was submitted to I-TASSER online modelling platform, which generates three-dimensional structure models by multiple-threading alignments and iterative structural assembly simulations. The program identified Sulfolobus solfataricus Cdc6 (PDB ID 2QBY) as the top template for structure prediction. Accuracy of the I-TASSER model was estimated based on C-score, TM-score, RMSD and cluster density (C-score: −60; Exp. TM-Score: 0.64 ± 0.13; Exp. RMSD: 8.4 ± 4.5; Cluster density: 0.1711). Then, the model was refined using ModRefiner. Improvement by ModRefiner was judged considering the RMSD and TM-score to initial model (1.057 and 0.9829, respectively) (29–31). Finally, the protein model was compared with the S. solfataricus Cdc6 crystal structure using the program DaliLite (32), and their alignment revealed two homologous structures (Z-score 34.7, RMSD 2.4 Å). The images were rendered using Pymol 1.5 (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
RESULTS
Cdc6 sensor-1 and sensor-2 mutants cause an MCM2–7-loading defect
Cdc6 is a central protein in the MCM2–7-loading reaction (15,17,18,33–37). Previous studies have shown in vivo that Cdc6 sensor-1 (N263A) and sensor-2 (R332E) mutants block MCM2–7 loading (20,21), indicating that these motifs are essential for Cdc6 function. However, it is unknown how these motifs contribute to Cdc6 function in helicase loading. Here, we used an in vitro pre-RC assay to determine directly the functionality of these mutants (4). Because the sensor-1 and sensor-2 mutants could affect either ATP binding or ATP hydrolysis (19), we performed in parallel experiments with a Cdc6 ATP binding (Walker A—K114E) (18) and a Cdc6 ATPase (Walker B—E224G) (3) mutants. The pre-RC assay can distinguish between MCM2–7 associated with DNA and MCM2–7 loaded onto DNA. While associated MCM2–7 is connected to DNA via ORC–Cdc6–Cdt1 and is salt sensitive, loaded MCM2–7 encircles double-stranded DNA and is salt resistant. Pre-RC reactions were incubated for 5 min or 15 min and then washed with low salt buffer to analyse MCM2–7 association with origin DNA in a time-dependent manner. An alternative reaction was incubated for 15 min and washed with high salt buffer to identify loaded MCM2–7. With wt proteins and ATP(gamma),S we detected that ORC, Cdc6, Cdt1 and MCM2-7 associated efficiently with DNA (Figure 2A, lane 7). While using ATP, we observed ORC, Cdc6 and MCM2-7 in complex with DNA (Figure 2A, lane 8 and 9). Cdt1 instead was released during the MCM2–7-loading reaction in a time-dependent manner (Figure 2A, lane 8 and 9) (3). A high salt wash removed ORC–Cdc6 and identified the presence of loaded MCM2–7 (Figure 2A, lane 10). The Cdc6 sensor-2 mutant R332E interacted weakly with ORC, promoted weak association with MCM2–7 (Figure 2A, compare lanes 14 and 15) and did not support MCM2–7 loading (Figure 2A, lane 16). Interestingly, a Cdc6 Walker A ATP-binding mutant (K114E) interacted even less efficiently with ORC, but otherwise behaved nearly identically to the Cdc6 sensor-2 mutant (Figure 2A, lanes 11–13), suggesting a similar defect. On the other hand, the Cdc6 sensor-1 mutant N263A interacted efficiently with ORC on DNA, recruited MCM2–7 efficiently, but Cdt1 release was slightly reduced and MCM2–7 loading was absent (Figure 2B, lanes 14–16). Interestingly, a Cdc6 E224G ATP hydrolysis mutant behaved similar to Cdc6 N263A, although the Walker B mutant led to some MCM2–7 loading (Figure 2B, lanes 11–13).
Figure 2.
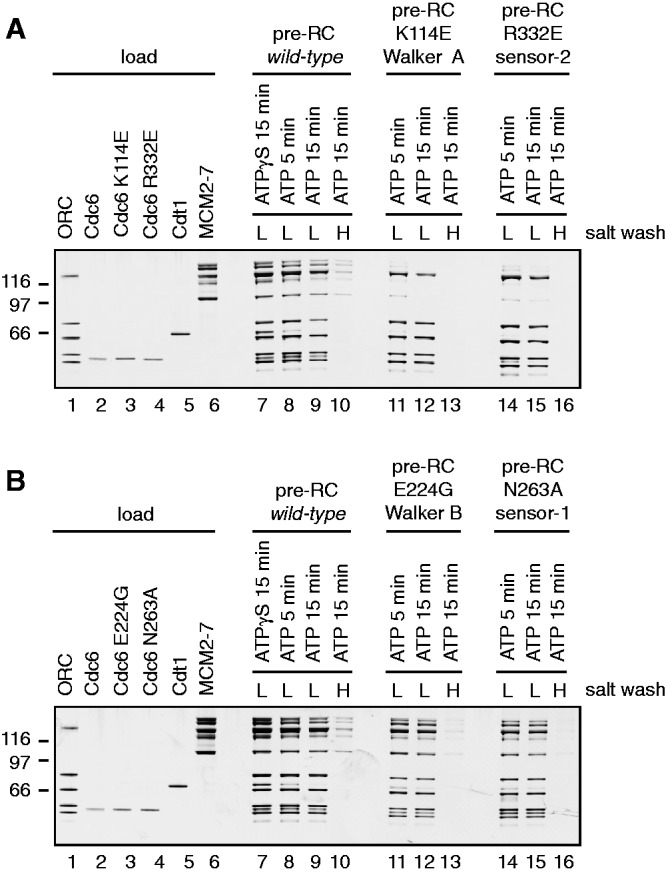
The influence of Cdc6 mutants on pre-RC formation. Pre-RC assembly was performed as described in the methods section. A 30% load of the pre-RC proteins is shown in lanes 1–6. The use of the mutants is indicated in the figures. The gel was silver stained to visualize the proteins; however, the smallest subunit of the Orc1–6 complex stains only weakly. (A) Cdc6, Cdc6 K114E (Walker A) and Cdc6 R332E (sensor-2) were used in pre-RC assays in the presence of ATP (lanes 8–16) or ATPγS (lane 7). Pre-RCs were washed with a low salt buffer (lanes 7–9, 11, 12, 14 and 15) or a high salt buffer (lanes 10, 13 and 16). (B) Cdc6, Cdc6 E224G (Walker B) and Cdc6 N263A (sensor-1) were used in pre-RC assays in the presence of ATP (lanes 8–16) or ATPγS (lane 7). Pre-RCs were washed with a low salt buffer (lanes 7–9, 11, 12, 14 and 15) or a high salt buffer (lanes 10, 13 and 16).
This analysis allows us to clearly categorize the Cdc6 sensor-1 and sensor-2 motif and understand their functional roles in pre-RC formation. A Cdc6 sensor-2 mutant blocks pre-RC formation owing to an inefficient ORC–Cdc6 interaction and a reduced ability to recruit Cdt1–MCM2–7. A Walker A ATP-binding mutant behaved identically, suggesting that both the sensor-2 and Walker A motifs influence the Cdc6–ATP interaction. Moreover, the Cdc6 sensor-1 mutant allows efficient pre-RC assembly, but led to a partial block in Cdt1 release and reduced MCM2–7 loading. A Cdc6 Walker B ATPase mutant displays the same features in a pre-RC assay (Figure 2B) (3) and therefore we suggest that the sensor-1 and Walker B motifs are both important for Cdc6 ATPase activity in the context of pre-RC assembly.
An ATPγS-arrested pre-RC complex contains a single MCM2–7 hexamer
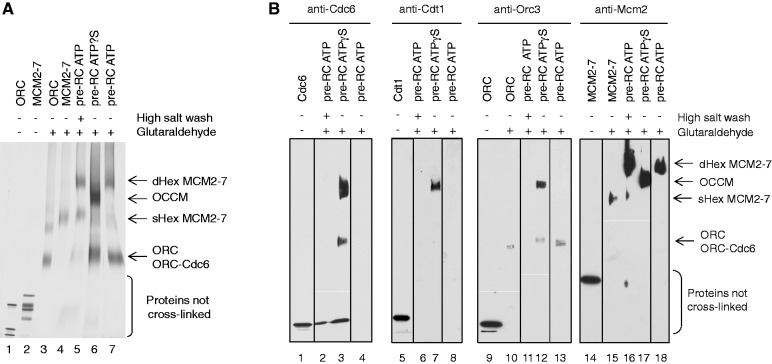
As we have just described, the sensor-1 motif is important for Cdc6 ATP hydrolysis-dependent pre-RC formation. However, the function of Cdc6 ATP hydrolysis remains unclear. To address a potential role of the Cdc6 ATPase activity in pre-RC assembly, we aimed to determine the size and composition of the protein–DNA complexes that form in the presence or absence of Cdc6 ATP hydrolysis. Thus, we assembled pre-RCs onto origin DNA linked to magnetic beads in the presence of ATPγS or ATP. Then we released protein complexes from the magnetic beads by DNase I treatment and immediately crosslinked the complexes with glutaraldehyde to obtain stable-fixed complexes. Subsequently, the samples were denatured and separated by size using PAGE. The resulting gels were stained with silver (Figure 3A) or processed for Western blotting (Figure 3B). As size standards, we used ORC and MCM2–7 proteins in the absence and presence of glutaraldehyde (Figure 3A, lanes 1–4). In the presence of ORC, Cdc6, Cdt1 and ATP, MCM2–7 assembled into a large complex (Figure 3A, lane 7), which was salt stable (Figure 3A, lane 5), consistent with the formation of a salt-resistant MCM2–7 double hexamer. Western blotting confirmed that the low salt washed (Figure 3B, lane 4, 8, 13 and 18) and high salt washed (Figure 3B, lanes 2, 6, 11 and 16) complex contained only Mcm2. In the presence of ATPγS, we observed a complex that migrated in the gel slower than the MCM2–7 hexamer, but faster than the MCM2–7 double hexamer (Figure 3A, lane 6). Western blotting identified the presence of Cdc6 (Figure 3B, lane 3), Cdt1 (Figure 3B, lane 7), Orc3 (Figure 3B, lane 12) and Mcm2 (Figure 3B, lane 17), suggesting that all pre-RC proteins are part of this complex. A complex containing one subunit of ORC, Cdc6, Cdt1 and MCM2–7 has a theoretical molecular weight of 1142 kDa, while an MCM2–7 double hexamer has a molecular weight of 1211 kDa. As a pre-RC complex formed in the presence of ATPγS contains all pre-RC components in roughly equimolar amounts (Figure 2A, lane 7—compare with inputs) and the crosslinked ATPγS complex migrated just slightly smaller than the MCM2–7 double hexamer, we conclude that this complex contains besides Cdc6 and Cdt1 most likely only one ORC and one MCM2–7 hexamer.
Figure 3.
Pre-RC complexes crosslinked in solution. During pre-RC formation with ATPγS, an ORC–Cdc6–Cdt1–MCM2–7 complex is formed that is labelled OCCM. During pre-RC formation with ATP, an MCM2–7 double hexamer is formed that is labelled dHex MCM2–7. The single MCM2–7 hexamer is labelled sHex MCM2–7. A small amount of single hexameric MCM2–7 can be detected in the high salt-washed pre-RC ATP owing to destabilization of the complex. (A) Analysis of crosslinked pre-RC reactions using silver staining. Purified ORC and MCM2–7 (lanes 1 and 2), crosslinked purified ORC and MCM2–7 (lanes 3 and 4), crosslinked high salt-washed pre-RC ATP (lane 5), crosslinked pre-RC ATPγS (lane 6) and low salt-washed crosslinked pre-RC ATP (lane 7) are shown. (B) Purified Cdc6, Cdt1, ORC and MCM2–7 (lanes 1, 5, 9 and 14); purified crosslinked ORC and MCM2–7 (lanes 10 and 15); crosslinked pre-RC ATP (lanes 2, 4, 6, 8, 11, 13, 16 and 18); and crosslinked pre-RC ATPγS (lanes 3, 7, 12 and 17) were analysed by Cdc6, Cdt1, Orc3 and Mcm2 Western blot.
A single MCM2–7 hexamer associates on DNA with ORC–Cdc6–Cdt1
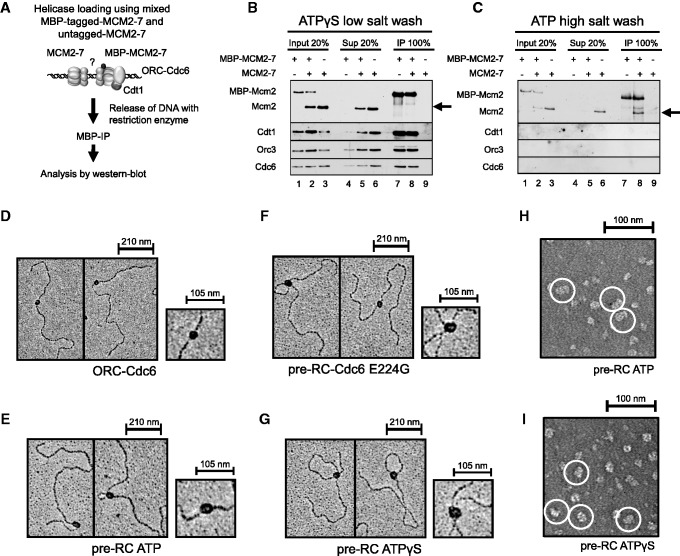
As just discussed, the complex formed in the presence of ATPγS most likely contained only one MCM2–7 hexamer, but a loosely associated second hexamer could have been lost owing to DNase I treatment prior to crosslinking. To distinguish between these two scenarios, we used a co-IP approach to measure MCM2–7 dimerization directly (Figure 4A). For this assay, both tagged and untagged MCM2–7 were combined with ORC, Cdc6, Cdt1 and origin DNA. After complex formation, the protein complexes were released from the magnetic beads by restriction digestion. Importantly, the restriction enzyme cuts outside of the DNA replication origin to maintain complex integrity. The tagged MCM2–7 was then immunoprecipitated and analysed for co-precipitation of untagged MCM2–7, which can identify dimerization of MCM2–7. We did not observe co-precipitation of untagged MCM2–7 in the presence of ATPγS (Figure 4B), while reactions prepared with ATP led to efficient co-precipitation of untagged MCM2–7 (Figure 4C). This experiment shows that in the presence of ATPγS, only one MCM2–7 hexamer associates with ORC, Cdc6 and Cdt1, while ATP leads to high salt-resistant MCM2–7 double hexamer formation, and these results are also consistent with our crosslinking analysis. In an independent approach, we visualized the ATPγS and pre-RC–Cdc6 E224G complex on DNA by electron microscopy and metal shadowing; while using ORC–Cdc6 and the pre-RC–ATP (MCM2–7 double hexamer) as controls (Figure 4D–G). The ORC–Cdc6 complex appeared fairly small and round, in contrast the MCM2–7 double hexamer was elongated and larger (Figure 4D and E). The ATPγS and E224G complex appeared slightly smaller than the MCM2–7 double hexamer (Figure 4; compare F and G with E). As metal shadowing does not allow for exact size measurements, we used negative staining with uranyl acetate to visualize ATP and ATPγS complexes (Figure 4H and I). With ATP, we observed double hexamers of 23.6 nm (±1.66, n = 24) in length as reported (4,5), while the ATPγS complex was 18.0 nm (±1.32, n = 48) long. This work establishes that in the absence of ATP hydrolysis, only a single MCM2–7 hexamer associates with ORC–Cdc6–Cdt1–DNA. This indicates that Cdc6 ATPase activity becomes activated in the context of the single MCM2–7 hexamer. Furthermore, this finding reveals that MCM2-7 hexamers associate in a consecutive manner with replication origins, as initially only one MCM2–7 hexamer binds to ORC–Cdc6–Cdt1, and Cdc6 ATPase is then required to allow the recruitment of a second MCM2–7 hexamer prior to stable MCM2–7 double hexamer formation.
Figure 4.
Pre-RC complexes on DNA. (A) Experimental outline for (B) and (C). Pre-RC assays were assembled in the presence of ATPγS and ATP. When tagged and untagged MCM2–7 were used, equimolar amounts of each complex were combined in pre-RC reactions. Complexes were released from DNA via restriction digest, and DNA-bound complexes were immune precipitated (IP) and together with input and supernatant (Sup) analysed by Western blotting with anti-Mcm2, anti-Cdt1, anti-Orc3 and anti-Cdc6 antibodies. (B) Co-immunoprecipitation of MBP-tagged and untagged MCM2–7 in the presence of ATPγS. (C) Co-immunoprecipitation of high salt-washed MBP-tagged and untagged MCM2–7 in the presence of ATP. Electron micrographs of metal-shadowed protein–DNA complexes with (D) ORC–Cdc6, (E) pre-RC ATP, (F) pre-RC Cdc6 E224G and (G) pre-RC ATPγS. Electron micrographs of negative-stained samples of (H) pre-RC ATP—(double hexamers are circled in white) and (I) pre-RC ATPγS (ORC–Cdc6–Cdt1–MCM2–7 complexes are circled in white).
Orc1 ATPase is not required for pre-RC assembly and disassembly
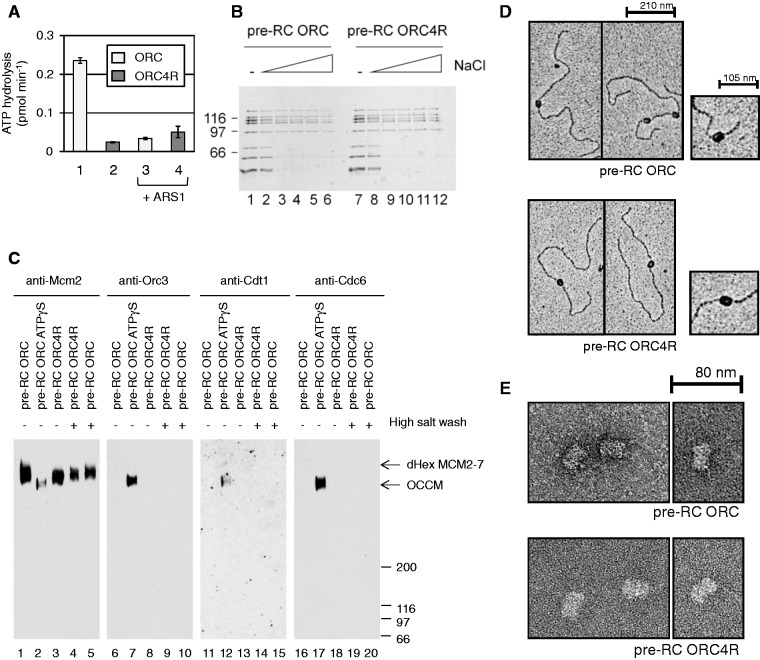
Besides Cdc6, Orc1 is the other main ATPase involved in pre-RC formation (10). It has been shown that Orc1 binds ATP, but requires an arginine finger in the neighbouring subunit Orc4 to function as an ATPase. An ORC complex containing the Orc4R mutant protein (ORC4R) is lethal in vivo, loads salt-stable MCM2–7 on origin DNA, but does not function in repetitive MCM2–7 loading (10). Currently it is not clear why the Orc1 ATPase mutant ORC4R cannot promote repetitive loading. One possibility is that MCM2–7 forms a stable intermediate with ORC4R that precludes ORC4R from repetitive MCM2–7 loading. To address this question, we compared MCM2–7 loading with wild-type ORC or ORC4R. An ORC complex containing the mutant Orc4R subunit (ORC4R) was purified and we verified the ATP hydrolysis defect of this mutant in vitro (Figure 5A). As expected, ORC4R hydrolyses ATP less efficiently than ORC (10). The addition of origin DNA leads to a suppression of ATPase activity in wild-type ORC and in the case of ORC4R, the ATPase activity remains low after addition of ARS1 DNA. This result verifies that the ORC4R protein we used has an ATPase defect. Then we analysed the ability of the mutant to load MCM2–7 (Figure 5B), by testing the association of MCM2–7 with origin DNA in low salt or high salt buffers (100–500 mM sodium chloride). We found both ORC4R and ORC formed a pre-RC complex containing ORC/ORC4R, Cdc6 and MCM2–7. Furthermore, both ORC versions promoted efficient and salt-stable MCM2–7 loading.
Figure 5.
Analysis of the role of ORC4R in pre-RC formation. (A) ATPase activity of ORC and ORC4R in the absence (columns 1 and 2) and presence of DNA (columns 3 and 4). (B) The stability of pre-RCs formed in the presence of ORC (lanes 1–6) or ORC4R (lanes 7–12). Pre-RCs were formed in the presence of low salt buffer (lanes 1 and 7) or with buffer containing 100, 200, 300, 400, 500 mM sodium chloride, respectively (lanes 2–6 and 8–12). (C) Pre-RC complexes were crosslinked in solution. Pre-RC ATP (lanes 1, 5, 6, 10, 11, 15, 16 and 20), pre-RC ATPγS (lanes 2, 7, 12 and 17) and pre-RC with ORC4R (lanes 3, 4, 8, 9, 13, 14, 18 and 19). (D) Electron micrographs of metal-shadowed pre-RCs formed with ORC (upper picture) and ORC4R (lower picture). (E) Electron micrographs of uranyl acetate-stained pre-RC samples prepared with ORC (upper picture) and ORC4R (lower picture).
To address the organization of the pre-RC complex formed by ORC4R, we used once more glutaraldehyde crosslinking to study the size and composition of the complex. We assembled pre-RCs in the presence of low salt or high salt using ORC or ORC4R. In addition, we assembled a pre-RC in the presence of ATPγS as a positive control for a salt-unstable pre-RC intermediate containing ORC, Cdc6, Cdt1 and MCM2–7. Consequently, we released the complexes from DNA using DNase I and immediately crosslinked the proteins with glutaraldehyde. The size and composition of the complexes were analysed by PAGE and Western blotting. We found as expected that ORC promoted the assembly of a large double hexameric complex, which was salt stable (Figure 5C, lane 1 and 5). The complex assembled in the presence of ATPγS was smaller, but contained besides MCM2–7 also ORC, Cdc6 and Cdt1 (Figure 5C, lanes 2, 7, 12 and 17). ORC4R also produced an MCM2–7 double hexamer-sized complex that was salt resistant (Figure 5C, lanes 3 and 4). Moreover, this complex did not contain ORC, Cdc6 or Cdt1 (Figure 5C, lanes 8, 13 and 18). These data show that ORC4R promotes assembly of an MCM2–7 double hexamer.
However, as the complex was released from DNA prior to crosslinking, we also wanted to verify the structure of the complex on DNA. The complexes were crosslinked on DNA, then metal-shadowed or negative stained and visualized by electron microscopy (Figure 5D and E). We found that the length of MCM2–7 complexes loaded by ORC (24.5 nm ± 1.2 nm; n = 24) and by ORC4R (24.5 nm ± 1.3 nm; n = 24) were identical (Figure 5E). Furthermore, our work shows no evidence for an ORC4R–MCM2–7 intermediate that could interfere with repetitive MCM2–7 loading. Based on this analysis, Orc1 ATPase is not required for pre-RC assembly (loading of the MCM2–7 double hexamer) or pre-RC disassembly (release of ORC from loaded MCM2–7), but may function to reset the ORC complex, a process that could reactivate ORC for another round of MCM2–7 loading.
DISCUSSION
Although it is clear that Orc1 and Cdc6 are essential for helicase loading (2,3,17,18,21,36–40), little is known about how Cdc6 regulates MCM2–7 loading, whether Cdc6 ATPase is coordinated with MCM2–7 double hexamer assembly and whether Orc1 ATPase is required for release of ORC–Cdc6 upon MCM2–7 loading. Our work identified how several conserved AAA+ ATP-binding and hydrolysis motifs in Cdc6 and in Orc1 function during specific steps of the MCM2–7-loading process. Importantly, our work revealed that inhibition of Cdc6 ATPase blocks MCM2–7 dimerization and highlights that MCM2–7 hexamers are recruited in a consecutive manner to replication origins.
AAA+ proteins contain a Walker A motif, which is essential for ATP binding (19). A point mutation of a conserved lysine within the Walker A motif blocks the interaction of human Cdc6 with ATP (16). A point mutation of the same lysine in budding yeast Cdc6 blocks MCM2–7 loading in vivo and is causing lethality (17,18). Although less is known about the role of the sensor-2 motif, it is predicted to affect either nucleotide binding or ATP hydrolysis (15). In vivo analysis of Cdc6 sensor-2 mutants have shown a slow growth phenotype and an MCM2–7 loading defect (20,21), while Walker A K114E mutant is lethal and fails to load MCM2–7 (17,18). Here, we have shown in vitro that the Cdc6 Walker A mutant interacted weakly with ORC, promoted only weak MCM2–7 association and blocked MCM2–7 loading completely. The sensor-2 mutant exhibited a nearly identical phenotype in vitro, but interacted with ORC slightly better than the Walker A mutant, potentially explaining why the sensor-2 mutant displayed a less severe in vivo phenotype than the Walker A mutant (17,18,20,21). These results show that both mutants act in a similar fashion on pre-RC formation in vitro.
To understand how these two sequence motifs, which are located in the primary sequence far away from each other (Figure 1), are arranged in the 3D structure of Cdc6, we have modelled the S. cerevisiae Cdc6 3D structure using the I-Tasser server (29). The resulting structure is overall similar to the crystal structure of an S. solfataricus Cdc6 orthologue (41) (Figure 6A and B). Indeed, the similarity of both proteins is high (72.2%). Both, the crystal structure of an S. solfataricus Cdc6 and the modelled structure show that the Walker A and sensor-2 motifs are embracing ATP from two different sides (Figure 6C–D). This structural information helps us to explain why both motifs affect the Cdc6 function in an almost identical way. For these reasons, we strongly suggest that the Cdc6 sensor-2 mutant primarily fails to load MCM2–7 owing to a Cdc6 ATP-binding defect, which causes a weak ORC–Cdc6 interaction and reduced MCM2–7 recruitment. These are features of a typical allosteric regulation—where ATP acts as an allosteric activator of Cdc6. The binding of Cdc6 to ATP involves likely a structural change in Cdc6 that in turn allows its interaction with ORC.
Figure 6.
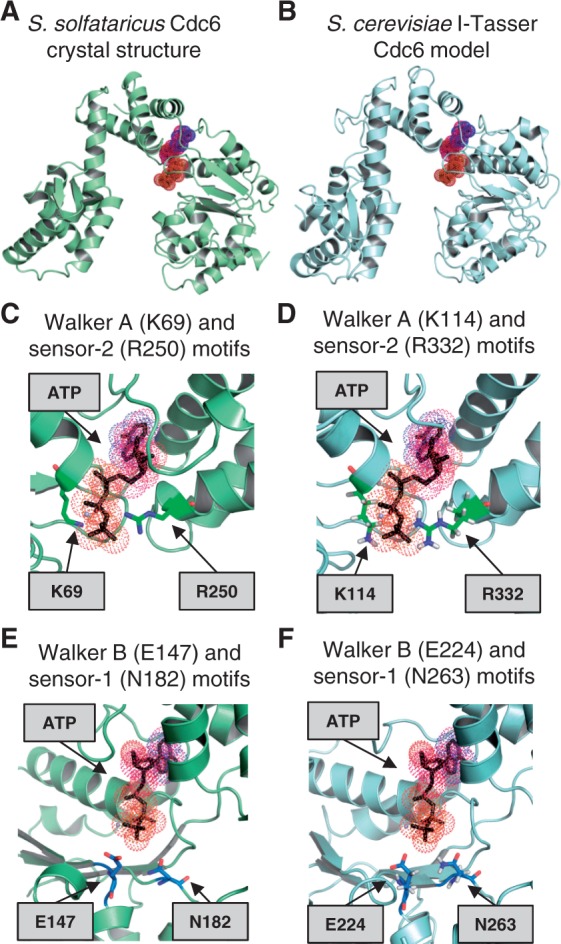
Structural analysis of conserved Cdc6 ATP-binding and hydrolysis motifs. (A) Crystal structure of S. solfataricus Cdc6 and (B) an I-Tasser-predicted S. cerevisiae structure of Cdc6. The sequence of S. cerevisiae Cdc6 (amino acids 73–513) was submitted to the I-TASSER server for protein structure prediction. A close-up view of the of S. solfataricus (C) and the predicted S. cerevisiae Cdc6 (D) ATP-binding motif showing the conserved Walker A (K114) and sensor-2 (R332) motifs. The S. solfataricus (E) and the predicted S. cerevisiae (F) Walker B (E224) and sensor-1 (N263) motifs of Cdc6 are shown.
Another key part of AAA+ proteins is the Walker B motif, which is important for ATP hydrolysis (19). Consistent with that, it was found that a Cdc6 Walker B mutant is dominant lethal in vivo, reduces MCM2–7 loading in vitro (3,17) and has an ATP hydrolysis defect (3,11). In contrast, the sensor-1 motif of AAA+ proteins is known to influence either ATP binding or ATP hydrolysis (19). Our analysis of a Cdc6 sensor-1 mutant identified efficient MCM2–7 recruitment, slightly reduced Cdt1 release and strongly reduced MCM2–7 loading. These are trademarks of an ATP hydrolysis defect (3), which is consistent with the finding that a Cdc6 sensor-1 mutant has an ATPase defect in the context of ORC (11). Our structural prediction suggests that the Walker B and sensor-1 motifs are located close to each other near the reactive γ-phosphate of ATP (Figure 6E and F). This spatial proximity fits with the nearly identical phenotypes that we observed for these two mutants. We observed that Cdc6 sensor-1 and Walker B mutants have only a weak defect in Cdt1 release and MCM2–7 loading when compared with ATPγS. One possibility is that both motifs do synergize during ATP hydrolysis and that a mutation of a single motif on its own has only a mild defect. Another possibility is that the point mutation used here is not completely blocking Cdc6 ATPase and that further amino acids in each motif must be mutated to block the ATPase activity completely. Based on this analysis, we predict that the Cdc6 sensor-1 motif primarily affects ATP hydrolysis.
The function of Cdc6 ATP hydrolysis during pre-RC formation has been a mystery for the longest time. It was noted that in the absence of Cdc6 ATP hydrolysis, the generation of a salt-stable MCM2–7 complex is blocked (3). However, the reason for this block in pre-RC formation was unknown. Our data demonstrate now that only one MCM2–7 hexamer is recruited in the absence of ATP hydrolysis to replication origins. This indicates that Cdc6 ATPase activity becomes activated in the context of the single MCM2–7 hexamer. One important question is now: What could Cdc6 ATP hydrolysis do? It has been proposed that Cdc6 ATPase is involved in opening the hexameric ring to allow loading of MCM2–7 onto DNA (3,9). If that is true, then Cdc6 ATPase would load each MCM2–7 hexamer individually on DNA. Another interesting question is: How is a Cdc6 ATPase-induced structural change transmitted to the rest of the complex? Structural data of the archaeal Cdc6 homologue have shown markedly different conformations between ATP and ADP–Cdc6 (14), suggesting that Cdc6 ATP hydrolysis could promote a structural change in an interaction partner. This structural change could occur owing to the negative free-energy change accompanying the release of Pi (44). Based on our work, we suggest that Cdc6 ATP hydrolysis induces a structural change in MCM2–7 that makes it competent to recruit a second MCM2–7 hexamer. We note that this structural change could release also Cdt1, as ATP hydrolysis and Cdt1 release appear to be coupled (3). Furthermore, multiple Cdt1 molecules have been shown to act in conjunction with ORC–Cdc6 during pre-RC formation (43). Based on this finding and our work, it would be interesting to see if indeed two Cdt1 molecules transmit a Cdc6 ATPase-mediated structural change in MCM2–7. One intriguing possibility is that Cdc6 ATPase promotes several events simultaneously: inducing dimerization competence in MCM2–7, opening of MCM2–7 ring and releasing Cdt1 in a single Cdc6 ATP hydrolysis-controlled step. It will be exciting to discover in the future how Cdc6 ATPase in conjunction with Cdt1 assembles the MCM2–7 double hexamer onto DNA.
It has been shown that Cdc6 ATPase activity precedes Orc1 ATPase activity and that Orc1 ATPase is only required for repetitive MCM2–7 loading reactions (10). Electron microscopy analysis of crosslinked complexes show now that ORC4R is able to promote MCM2–7 double hexamer formation. This is consistent with the earlier finding that ORC4R allows loading of a salt-resistant MCM2–7 complex (10), but extends this concept by showing directly the presence of an MCM2–7 double hexamer. Moreover, it was suggested that Orc1 ATP hydrolysis could facilitate the release of ORC–Cdc6 from MCM2–7 (3,10). However, crosslinking and electron microscopy analysis revealed that ORC4R is efficiently released from the MCM2–7 double hexamer, highlighting that Orc1 ATPase is not required for pre-RC assembly or disassembly. Therefore, the question is: What is the function of Orc1 ATPase? We propose a model in which ORC adopts upon MCM2–7 loading an altered conformation that is inhibitory for pre-RC formation. This altered conformation could be a result of Cdc6 ATP hydrolysis, as Cdc6 alters the structure of Orc6 in an ATP hydrolysis-dependent manner (40). Once MCM2–7 loading is finished, ORC could be arrested in a conformation that hinders interaction with Cdc6, potentially reflecting negative allosteric regulation. An Orc1 ATP hydrolysis cycle could be required to reinstate the active ORC conformation, as it has been observed in case of an ABC transporter or the F1-ATPase (44,45). It will be interesting to understand how the Orc1 ATPase is activated. One possibility is that Orc1 ATPase is connected to MCM2–7 release. Another possibility is that an additional factor could induce Orc1 ATPase, which could add another layer of regulation towards pre-RC formation.
Based on our findings, we propose the following model of pre-RC formation (Figure 7): ORC binds in an ATP-dependent way to the replication origin (1) (Figure 7A). Cdc6 needs to interact with ATP to interact with ORC (9,11,33–35) (Figure 7B), as mutations in the ATP binding Walker A and sensor-2 motifs interfere with the ORC–Cdc6 interaction (Figure 2). It is known that Cdt1 forms a complex with MCM2–7 and that this complex is recruited by ORC–Cdc6 to replication origins (4,5,46,47). Our data show now that a Cdc6 sensor-1 mutant, which is deficient in ATP hydrolysis (12), blocks pre-RC formation at that stage. Interestingly, we discovered that in the absence of ATP hydrolysis, only a single MCM2–7 hexamer associates with a single ORC complex (Figure 7C). This result demonstrates that Cdc6 ATP hydrolysis acts on a single MCM2–7 hexamer, shows that Cdc6 ATPase is required for the recruitment of the second MCM2–7 hexamer and identifies that the two MCM2–7 hexamers are assembled on origins in a consecutive manner. It remains an open question, whether a second Cdt1–MCM2–7 heptamer could be recruited by the existing ORC–Cdc6–Cdt1–MCM2–7 complex or whether two ORC–Cdc6–Cdt1–MCM2–7 complexes dimerize in a cooperative manner. Orc1 ATPase functions after Cdc6 ATP hydrolysis and is essential for the cell (10). We found that even in the absence of Orc1 ATP hydrolysis, MCM2–7 double hexamer formation occurs in vitro and that ORC separates from the loaded MCM2–7 complex (Figure 7D). For this reason, we suggest that Orc1 ATP hydrolysis is not involved in assembly or disassembly of the pre-RC, but reactivates the ORC complex for another round of MCM2–7 loading.
Figure 7.
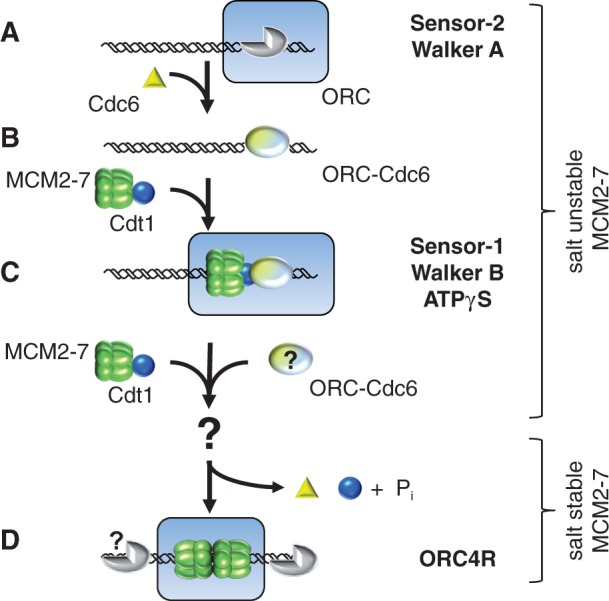
Blocked Cdc6 ATP hydrolysis leads to formation of an ORC–Cdc6–Cdt1–MCM2–7 complex, and ORC4R promotes double hexamer formation. A model of the MCM2–7-loading process (A–D). (A) ATP–ORC is bound to DNA. (B) ATP–Cdc6 associates with ORC. (C) We show that in the absence of ATP hydrolysis, only one MCM2–7 hexamer associates with ORC–Cdc6. This complex could contain multiple Cdt1 molecules. (D) ATP hydrolysis promotes the recruitment of a second Cdt1–MCM2–7 complex potentially with the help of a second ORC–Cdc6 complex. In consequence, MCM2–7 becomes loaded, forms a stable double hexamer encircling DNA and ORC, Cdc6 and Cdt1 become released from MCM2–7. Orc1 ATPase may function to reactivate ORC for a new round of pre-RC assembly.
FUNDING
Medical Research Council UK (to C.S.). Funding for open access charge: Medical Research Council UK.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Bruce Stillman for antibodies (Orc3, Mcm2, Cdt1).
REFERENCES
- 1.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 2.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the MCM2–7 helicase. Mol. Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl Acad. Sci. USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of MCM2–7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/MCM2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 9.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative MCM2–7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Speck C, Stillman B. Cdc6 ATPase activity regulates ORC x Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J. Biol. Chem. 2007;282:11705–11714. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 13.Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc. Natl Acad. Sci. USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol. 2004;343:547–557. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 16.Herbig U, Marlar CA, Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkins G, Diffley JF. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 18.Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl Acad. Sci. USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendler P, Ciniawsky S, Kock M, Kube S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Schepers A, Diffley JF. Mutational analysis of conserved sequence motifs in the budding yeast Cdc6 protein. J. Mol. Biol. 2001;308:597–608. doi: 10.1006/jmbi.2001.4637. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi N, Tsutsumi S, Tsuchiya T, Stillman B, Mizushima T. Functions of sensor 1 and sensor 2 regions of Saccharomyces cerevisiae Cdc6p in vivo and in vitro. J. Biol. Chem. 2002;277:16033–16040. doi: 10.1074/jbc.M108615200. [DOI] [PubMed] [Google Scholar]
- 22.Ying CY, Gautier J. The ATPase activity of MCM2–7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J. 2005;24:4334–4344. doi: 10.1038/sj.emboj.7600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Casas-Terradellas E, Garcia-Gonzalo FR, Hadjebi O, Bartrons R, Ventura F, Rosa JL. Simultaneous electrophoretic analysis of proteins of very high and low molecular weights using low-percentage acrylamide gel and a gradient SDS-PAGE gel. Electrophoresis. 2006;27:3935–3938. doi: 10.1002/elps.200600141. [DOI] [PubMed] [Google Scholar]
- 26.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiess E, Lurz R. Electron-microscopic analysis of nucleic-acids and nucleic acid-protein complexes. Method Microbiol. 1988;20:293–323. [Google Scholar]
- 28.Steven AC, Trus BL, Maizel JV, Unser M, Parry DAD, Wall JS, Hainfeld JF, Studier FW. Molecular substructure of a viral receptor-recognition protein - the Gp17 tail-fiber of bacteriophage-T7. J. Mol. Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 29.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011;101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 33.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 34.Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 35.Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 36.Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoeber K, Mills AD, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey RA, Williams GH. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seki T, Diffley JF. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl Acad. Sci. USA. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J. Cell. Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizushima T, Takahashi N, Stillman B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 41.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 42.Bray D, Duke T. Conformational spread: the propagation of allosteric states in large multiprotein complexes. Annu. Rev. Biophys. Biomol. Struct. 2004;33:53–73. doi: 10.1146/annurev.biophys.33.110502.132703. [DOI] [PubMed] [Google Scholar]
- 43.Takara TJ, Bell SP. Multiple Cdt1 molecules act at each origin to load replication-competent MCM2–7 helicases. EMBO J. 2011;30:4885–4896. doi: 10.1038/emboj.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu G, Westbrooks JM, Davidson AL, Chen J. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc. Natl Acad. Sci. USA. 2005;102:17969–17974. doi: 10.1073/pnas.0506039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe R, Iino R, Noji H. Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 2010;6:814–820. doi: 10.1038/nchembio.443. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka S, Diffley JF. Interdependent nuclear accumulation of budding yeast Cdt1 and MCM2–7 during G1 phase. Nat. Cell. Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- 47.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated MCM2–7 loading. Genes Dev. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]