Background: Mechanisms underlying the cardioprotective profile of G-protein-coupled receptor kinase 2 (GRK2) inhibitors are incompletely understood.
Results: GRK2 inhibition activated the growth-promoting MAPK pathway, which contributed to cardioprotection by preventing cardiomyocyte death.
Conclusion: Cardioprotective activity of GRK2 inhibitors overlaps with enhanced tumor growth.
Significance: A promising class of kinase inhibitors for heart failure treatment shows overlapping of cardioprotective signaling with tumor growth promotion.
Keywords: Cell Proliferation, Fos, Heart Failure, MAP Kinases (MAPKs), Transgenic Mice, Tumor Cell Biology, Adrbk1, G-protein-coupled Receptor Kinase, PEBP1, Raf Kinase Inhibitor Protein
Abstract
Inhibition of G-protein-coupled receptor kinase 2 (GRK2) is an emerging treatment option for heart failure. Because GRK2 is also indispensable for growth and development, we analyzed the impact of GRK2 inhibition on cell growth and proliferation. Inhibition of GRK2 by the dominant-negative GRK2-K220R did not affect the proliferation of cultured cells. In contrast, upon xenograft transplantation of cells into immunodeficient mice, the dominant-negative GRK2-K220R or a GRK2-specific peptide inhibitor increased tumor mass. The enhanced tumor growth upon GRK2 inhibition was attributed to the growth-promoting MAPK pathway because dual inhibition of the GRK2 and RAF-MAPK axis by the Raf kinase inhibitor protein (RKIP) did not increase tumor mass. The MAPK cascade contributed to the cardioprotective profile of GRK2 inhibition by preventing cardiomyocyte death, whereas dual inhibition of RAF/MAPK and GRK2 by RKIP induced cardiomyocyte apoptosis, cardiac dysfunction, and signs of heart failure. Thus, cardioprotective signaling induced by GRK2 inhibition is overlapping with tumor growth promotion.
Introduction
G-protein-coupled receptor kinases (GRKs),3 were initially characterized as a family of kinases, which promote the desensitization of G-protein-coupled receptors (GPCRs) by receptor phosphorylation and subsequent (β-)arrestin-mediated uncoupling of the receptor from the heterotrimeric G-protein (1). Among different members of the GRK family, GRK2 has a unique role in the control of growth and development (2, 3), and individuals lacking GRK2 are not viable (3, 4). The growth-promoting activity of GRK2 in vivo was attributed to kinase-dependent and kinase-independent effects of GRK2 involving Smoothened and Patched homolog 1 (2, 3).
Apart from its essential physiological role in growth and development, exaggerated GRK2 activity is an important pathophysiological feature of cardiovascular diseases such as hypertension and heart failure (5, 6). Under such conditions of increased GRK2 activity, GRK2 inhibition confers cardioprotection (7). Therefore many research efforts focus on the development and characterization of GRK2-specific inhibitors (7–9).
A common approach of GRK2 inhibition in vivo relies on expression of the carboxyl-terminal domain of GRK2, i.e. the βARK1ct (GRK2ct), which inhibits membrane translocation and activation of GRK2 by scavenging Gβγ subunits of heterotrimeric G-proteins (10–12). However, neutralization of Gβγ subunits by the GRK2ct may also exert GRK2-independent effects, which could contribute to cardioprotection as well (13). The final “proof of concept” for a beneficial profile of GRK2 inhibition came from studies applying mice with cardiac-specific deletion of GRK2 where GRK2 deficiency protected against myocardial damage (14) and prevented adverse remodeling after myocardial infarction (15).
Although the beneficial cardiac profile of GRK2 inhibition is thus firmly established, mechanisms underlying cardioprotection are not fully understood. Because growth-regulatory pathways are essential for cardiomyocyte survival (16, 17), we considered the impact of GRK2 inhibition on cell growth and proliferation. However, the role of GRK2 in cell growth and proliferation is not clear, because in addition to the above mentioned growth-promoting activity, GRK2 can also exert growth inhibition leading to suppressed growth and proliferation of tumor cells (18, 19).
To address the role of GRK2 and GRK2 inhibition in cell growth and proliferation, we performed experiments with cultured cells, or expanded cells in vivo after xenograft transplantation into immunodeficient non-obese diabetic mice homozygous for the severe combined immune deficiency spontaneous mutation (NOD.Scid mice). Systemic effects of GRK2 inhibition were also analyzed in vivo with transgenic mice expressing a GRK2-specific peptide inhibitor under control of the human cytomegalovirus (CMV) immediate-early promoter/enhancer, which directs ubiquitous expression of a transgene. Furthermore, transgenic mice with myocardium-specific expression of GRK2 inhibitors were generated to assess their cardioprotective profile. We report here that GRK2 inhibition triggered the growth-promoting MAPK pathway, which promoted tumor growth but also conferred cardioprotection by preventing cardiomyocyte death.
EXPERIMENTAL PROCEDURES
Generation of Transgenic Mice
To generate mice with myocardium-specific overexpression of RKIP, a transgene placing the PEBP1 cDNA under control of the α-myosin heavy chain promoter (20) was constructed. For myocardium-specific expression of the GRK2-specific peptide inhibitor (GRK-Inh), a DNA fragment encoding the peptide sequence, MAKFERLQTVTNYFITSE (21, 22), was inserted into the α-myosin heavy chain plasmid. The plasmid sequences were removed by NotI digestion, and the purified linear DNA (2 ng/μl) was injected into fertilized oocytes of superovulated B6 (C57BL/6J) and FVB (FVB/N) mice. Oviduct transfer of the injected embryos into pseudopregnant CD-1 mice was performed according to standard procedures (23). Genomic DNA of the F0 generation was isolated from ear punch biopsies taken at 3–4 weeks of age and analyzed by PCR for integration of the transgene. Mice of two different transgenic lines each were born at Mendelian frequency and grew to adulthood normally.
To assess the systemic effect of GRK2 inhibition by the GRK2-specific peptide inhibitor in vivo, we generated transgenic mice with expression of the GRK2-specific peptide inhibitor under control of the CMV immediate-early promoter/enhancer, which directs ubiquitous expression of a transgene. To generate CMV-GRK-Inh transgenic mice, the DNA fragment encoding the peptide sequence MAKFERLQTVTNYFITSE was inserted into the pcDNA3.1 plasmid (Invitrogen). Plasmid sequences were removed by MluI and DraIII digestion, and the purified linear DNA (2 ng/μl) was injected into fertilized oocytes of superovulated B6 and FVB mice. Oviduct transfer of the injected embryos into pseudopregnant CD-1 mice and all subsequent steps were performed as detailed above.
Cell Culture and in Vivo Cell Expansion in NOD.Scid Mice
Culture of HEK and A431 cells, generation of cell clones stably expressing GRK2, GRK2-K220R, RKIP, or the GRK2-specific peptide inhibitor (21, 22) was performed essentially as described (24). Cells were maintained in high-glucose DMEM with 10% fetal bovine serum and 4 mm l-glutamine at 37 °C, in a humidified atmosphere with 5% CO2. The cell proliferation assay was performed essentially as detailed previously (3). As indicated, HEK cells were also cultured in growth factor-supplemented (50 ng/ml of epidermal growth factor (EGF)) DMEM and plated on mitomycin-inactivated mouse embryonic fibroblasts (MEF) as feeder cells essentially as described (25). For in vivo cell expansion, NOD.Scid mice (age 3 months) received an injection of 6–8 × 106 cells/200 μl of PBS (24). Two weeks (A431) or 4 weeks (HEK) after the injection, mice were anesthetized with ketamine/xylazine (100/10 mg/kg), perfused intracardially with physiological phosphate buffer, pH 7.2, and expanded cell clones were rapidly isolated and processed for further use. In addition, cells were also isolated from NOD.Scid mouse-expanded clones and re-cultured in DMEM as detailed above. Animal experiments were performed in accordance with the NIH guidelines, and reviewed and approved by the local committee on animal care and use (University of Zurich).
Immunohistology, Immunoblotting, and Protein Techniques
Immunohistological detection of phospho-ERK1/2 was performed with phospho-ERK1/2-specific antibodies (phosphorylated at Thr202/Tyr204 of ERK1, and Thr185/Tyr187 of ERK2; E10 mouse mAb, Cell Signaling), and GRK2 was detected with GRK2-specific antibodies (raised in rabbit against full-length recombinant GRK2 protein) on cryosections of NOD.Scid mouse-expanded HEK and A431 clones, respectively, similarly as described (24, 26). For detection of phospho-ERK1/2 in hearts of transgenic mice, we used paraffin-embedded sections. Nuclear fragmentation as a marker of apoptosis was determined in situ by TdT-mediated dUTP-biotin nick end labeling (TUNEL) technology (Roche Diagnostics) with paraffin-embedded sections prepared from transgenic hearts as detailed previously (26, 27). The TUNEL technology was also used to determine nuclear fragmentation of neonatal mouse cardiomyocytes, which were isolated from B6, Tg-RKIP, and Tg-GRK-Inh mice 1–3 days after birth essentially as described (9). All sections/cells were imaged with a Leica DMI6000 microscope equipped with a DFC420 camera. Immunoblotting was used to determine the protein levels of GRK2, GRK2-K220R, ERK1 (MAPK3), ERK2 (MAPK1), and phospho-ERK1/2 in cells, NOD.Scid mouse-expanded clones and organ tissue from transgenic mice as described (24, 26). We also used standard immunoblotting and immunohistology techniques (9, 27, 28) applying RKIP-specific antibodies (affinity-purified polyclonal antibodies raised in rabbit against full-length recombinant RKIP protein) to detect the overexpressed RKIP protein in RKIP-transgenic hearts. Co-enrichment of Raf1 and GRK2 with RKIP was performed similarly as described (9). The GRK2-specific peptide inhibitor (GRK-Inh) was detected in the immunoblot after Tricine-SDS-PAGE with anti-GRK-Inh antibodies (affinity-purified polyclonal antibodies raised in rabbit against the keyhole limpet hemocyanin-coupled GRK-Inh peptide). Nuclear phospho-ERK1/2 levels were quantified with the nuclear fraction of NOD.Scid mouse-expanded HEK and A431 clones, and organ tissue of transgenic and non-transgenic mice. The nuclear fraction was adjusted to a protein concentration of 0.2 μg/μl, solubilized by buffer (500 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% SDS, 50 mm Tris pH 7.4) supplemented with protease/phosphatase inhibitors, and assayed for phospho-ERK1/2 by immunoblotting, or a sandwich ELISA according to the instructions of the manufacturer (Pierce). Nuclear samples were normalized to histone H2B.
For subcellular fractionation, dispersed NOD.Scid mouse-expanded clones or mouse organ tissue were suspended in ice-cold buffer containing 0.25 m sucrose, 20 mm HEPES, pH 7.4, 10 mm KCl, 2 mm MgCl2, 1 mm DTT, supplemented with protease/phosphatase inhibitors, and homogenized on ice with a Dounce homogenizer. After centrifugation for 10 min at 4 °C (750 × g), the nuclear pellet was washed twice with ice-cold buffer, and stored as the nuclear fraction at −80 °C. The purity of the nuclear fraction was controlled by immunoblot detection of histone H2B and the absence of cytosolic proteins.
Nano-LC-ESI-MS/MS
To enrich proteins interacting with the GRK2-specific peptide inhibitor, GRK-Inh-expressing tumors were pulverized under liquid nitrogen. After solubilization for 30 min at 4 °C with solubilization buffer (1% sodium deoxycholate, 0.05% SDS, 0.05% Tween 20 in PBS, pH 7.4, supplemented with protease inhibitors), insoluble material was removed by centrifugation. The supernatant was diluted 1:5 in PBS (supplemented with protease inhibitors) and subjected to affinity chromatography with anti-GRK-Inh antibodies (6 mg of affinity-purified IgG/ml of Affi-Gel 10). After overnight incubation at 4 °C, unbound proteins were removed by extensive washing with PBS (20 column volumes), and bound proteins were eluted with 0.25 m NH4OH, 10% dioxane, pH 11. The pH was immediately adjusted to pH 7.4, eluted proteins were concentrated by acetone precipitation, dissolved by 8 m urea, and subjected to 8% urea-containing SDS-PAGE under reducing conditions. After Coomassie Brilliant Blue staining, the GRK2-reactive band was cut and subjected to nano-LC-ESI-MS/MS. Protein identification using nano-LC-ESI-MS/MS was performed by the Proteome Factory (Proteome Factory AG, Berlin, Germany). The MS system consisted of an Agilent 1100 nano-LC system (Agilent, Boeblingen, Germany), PicoTip emitter (New Objective, Woburn, MA), and an Esquire 3000 plus ion trap MS (Bruker, Bremen, Germany). The cut protein band was in-gel digested by trypsin (Promega, Mannheim, Germany) and applied to nano-LC-ESI-MS/MS. After trapping and desalting the peptides on an enrichment column (Zorbax SB C18, 0.3 × 5 mm, Agilent) using 1% acetonitrile, 0.5% formic acid solution for 5 min, peptides were separated on a Zorbax 300 SB C18, 75 μm × 150-mm column (Agilent) using an acetonitrile, 0.1% formic acid gradient from 5 to 40% acetonitrile within 40 min. MS spectra were automatically taken by Esquire 3000 plus according to the manufacturer's instrument settings for nano-LC-ESI-MS/MS analyses. Proteins were identified using MS/MS ion search of the Mascot search engine (Matrix Science, London, England) and nr protein database (National Center for Biotechnology Information, Bethesda, MD). Ion charge in search parameters for ions from ESI-MS/MS data acquisition were set to “1+, 2+, or 3+” according to the common charge state distribution for the instrument and method.
Transthoracic Echocardiography
Transthoracic echocardiography was performed with a Vivid 7 echocardiograph system (GE Healthcare) and a 12 MHz linear array transducer similarly as described (27, 29). Transthoracic echocardiography was used to characterize the cardiac function of transgenic and non-transgenic B6 and FVB mice without and with 4 weeks of chronic pressure overload imposed by abdominal aortic constriction (AAC) performed as detailed previously (27).
Microarray Gene Expression Profiling
Whole genome microarray gene expression profiling of cells, NOD.Scid mouse-expanded clones, and heart tissue of RKIP-transgenic, GRK-Inh-transgenic, and B6 mice was performed as described (24, 30). We used GeneChip Human genome U133 Plus 2.0 Arrays for human cells and Mouse genome MG430 2.0 Arrays for mouse cardiac tissue (Affymetrix). Gene ontology analyses of microarrray data were performed with GCOS/RMA processed data using GeneSpring GX software (Agilent). Data were compared between groups using the unpaired two-tailed Student's t test. p values of <0.05 were considered significant unless specified otherwise. All microarray gene expression data were deposited to the NCBI GEO database (accession numbers GSE42753 and GSE42771).
Selected gene expression data were confirmed by real-time quantitative (q) RT-PCR using a LightCycler 480 (Roche Diagnostics). Sequences of the forward and reverse primers of genes studied were as follows: FOS forward, 5′-CATAGCATTAACTAATCTATTGGGTTCA-3; FOS reverse, 5′-CACACTATTGCCAGGAACACAGTAGT-3′; PEBP1 (RKIP) forward, 5′-GGAGACCACCGTGGCAAATTCAAG-3′; PEBP1 (RKIP) reverse, 5′-GCTGCTCGTACAGTTTGGGCACAT-3′; FN forward, 5′-ATGTTAGCAGACCCAGCTTAGAGTTC-3′; FN reverse, 5′-TCAGGAAACTCCCAGGGTGATGCTTG-3′; GRK-Inh forward, 5′-CATGGCCAAGTTCGAGC-3′; GRK-Inh reverse, 5′-GTCACTCGCTGGTGATG-3′; Scd1 forward, 5′-GTTATAGACGGCAGTTGGCACTGG-3′; Scd1 reverse, 5′-CTTAGCCCAGGAGTTTCTTGGGTT-3′; Fasn forward, 5′-CCGACTCCCTTACGAGGCTGTGT-3′; Fasn reverse, 5′-CTGCACAGCGCTCTATCCACTGA-3′; Ucp1 forward, 5′-CACTGCCAAAGTCCGCCTTCAGA-3′; Ucp1 reverse, 5′-GCAGGCAGACCGCTGTACAGTT-3′; Retn forward, 5′-GTCCTGCTAAGTCCTCTGCCAC-3′; Retn reverse, 5′-GGCTGCTGTCCAGTCTATCCTTG-3′.
Confocal FRET Imaging
The GRK2-mediated interaction of enhanced yellow fluorescent protein (EYFP)-tagged β-arrestin1 with bradykinin B2 receptor (B2R)-Cerulean in transfected HEK cell clones without (control) or with expression of GRK2 or the dominant-negative GRK2-K220R, respectively, was determined in the absence or presence of bradykinin (1 μm, 8 min) and quantified by confocal FRET imaging and acceptor photobleaching with a confocal laser-scanning microscope (SP5-CLSM, Leica) similarly as described (21).
Plasmids
For transfection of HEK and A431 cells, we used the eukaryotic expression plasmid pcDNA3.1 (Invitrogen) and inserted the following cDNAs: ARRB1-EYFP (EYFP fused in-frame with the C-terminal amino acid codon of ARRB1), B2R-Cerulean (21) (Cerulean fused with a linker encoding Gly-Gly-Gly-Gly-Gly in-frame with the codon for the C-terminal amino acid 391 of BDKRB2), RKIP (cDNA encoding PEBP1 (28)), GRK2 (cDNA encoding ADRBK1), GRK2-K220R (cDNA encoding ADRBK1 with point mutation exchanging lysine 220 for arginine), GRK-Inh (DNA encoding the sequence MAKFERLQTVTNYFITSE) (21), a GRK2-specific peptide inhibitor (21, 22). Expression of GRK2 was down-regulated by RNA interference (RNAi) by POL II driven expression of a GRK2-targeting miR. The following hairpin-forming oligonucleotides were hybridized to form a 60-bp duplex and inserted into pcDNA6.2-GW(miR) (Invitrogen): 5′-TGCTGATAGAATTCCACCAAGGGCCTGTTTTGGCCACTGACTGACAGGCCCTTTGGAATTCTAT-3′, 5′-CCTGATAGAATTCCAAAGGGCCTGTCAGTCAGTGGCCAAAACAGGCCCTTGGTGGAATTCTATC-3′.
Statistics
Results are presented as mean ± S.D. unless indicated otherwise. Unpaired two-tailed Student's t test was used to calculate p values. Analysis of variance was performed with Prism (GraphPad). Statistical significance was set at a p value of <0.05.
RESULTS
Dominant-Negative Activity of the Kinase-deficient GRK2-K220R Mutant in HEK Cells
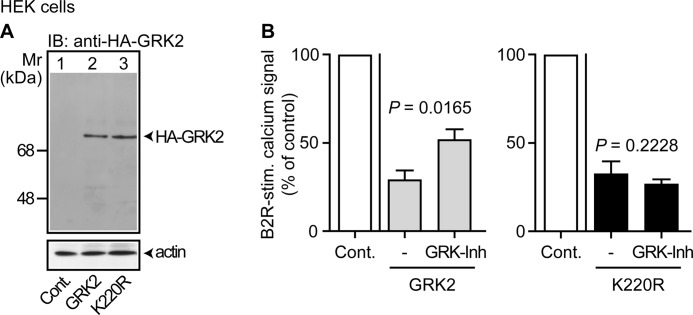
To analyze the impact of GRK2 and GRK2 inhibition on cell growth, we used the kinase-deficient GRK2-K220R, which is reported to act as a dominant-negative mutant (31). We generated HEK cell clones stably expressing GRK2 or the kinase-deficient GRK2-K220R (Fig. 1A). Protein levels were similar because GRK2 and GRK2-K220R exerted comparable kinase-independent functions such as inhibition of a Gαq/11-mediated calcium signal stimulated by the B2R (Fig. 1B). To confirm kinase activity of GRK2, we expressed a GRK2-specific peptide inhibitor (21, 22), and determined its effect on GRK2-mediated desensitization of the calcium signal stimulated by B2R, which is a kinase substrate of GRK2 (32). In agreement with kinase inhibition, GRK2-mediated desensitization of the B2R-stimulated calcium signal was partially reversed by expression of the GRK2-specific peptide inhibitor, whereas the kinase inhibitor did not affect the kinase-inactive GRK2-K220R (Fig. 1B).
FIGURE 1.
Kinase-dependent and kinase-independent activity of GRK2 in cultured HEK cells. A, immunoblot (IB) detection of HA-GRK2 in cultured HEK control cells (Cont., lane 1), and cells expressing HA-GRK2 (GRK2, lane 2) or HA-GRK2-K220R (K220R, lane 3). The lower panel is a control immunoblot detecting β-actin. B, expression of comparable levels of a GRK2-specific kinase inhibitor (GRK-Inh; MAKFERLQTVTNYFITSE) partially reversed the GRK2-mediated desensitization of a bradykinin B2R-stimulated calcium signal in GRK2-expressing cells (left panel), whereas GRK2-K220R-expressing cells were not affected (right panel). Bars represent mean ± S.E., n = 5.
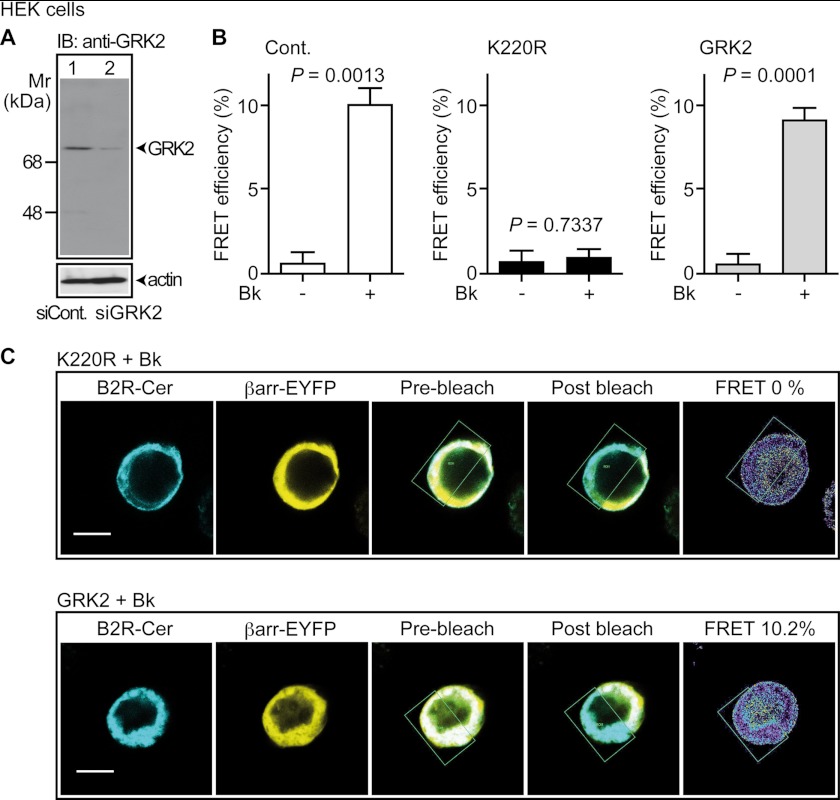
We next analyzed whether GRK2-K220R exerted dominant-negative activity in HEK cells and inhibited the endogenously expressed GRK2 (Fig. 2A). As a kinase effect of GRK2, we determined the GRK2-triggered interaction of Cerulean-tagged B2R with β-arrestin1-EYFP by fluorescence resonance energy transfer (FRET) measurement. Confocal FRET imaging revealed that the kinase-deficient GRK2-K220R completely prevented the bradykinin-stimulated interaction of B2R-Cerulean with β-arrestin1-EYFP indicating dominant-negative activity (Fig. 2, B, and C). As a control, the expression of GRK2 did not decrease the bradykinin-enhanced FRET intensity of the B2R-Cerulean interaction with β-arrestin1-EYFP (Fig. 2, B and C).
FIGURE 2.
GRK2-K220R acts as a dominant-negative mutant in HEK cells. A, immunoblot detection (IB: anti-GRK2) of the endogenously expressed GRK2 protein of HEK cells transfected with siRNA control plasmid (siCont., lane 1) or siGRK2 plasmid targeting GRK2 by RNA interference (siGRK2, lane 2). The loading control detected β-actin (lower panel). B and C, GRK2-K220R inhibited the GRK2-mediated interaction of bradykinin-stimulated B2R-Cerulean with β-arrestin1-EYFP. The interaction of B2R-Cerulean with β-arrestin1-EYFP was determined by confocal FRET imaging in the absence or presence of bradykinin (±Bk, 1 μm, 8 min) with control HEK cells (B, left panel), and HEK cells expressing GRK2-K220R (B, middle panel; C, upper panels) or GRK2 (B, right panel; C, lower panels). FRET efficiency was determined by acceptor photobleaching. Data represent mean ± S.E. (B, left panel: n = 3; middle, right panel, n = 9); bar, 10 μm.
Dominant-Negative GRK2-K220R Enhanced the Growth of NOD.Scid Mouse-expanded HEK Clones
We determined whether GRK2 or the dominant-negative GRK2-K220R affected the proliferation of HEK cells. In agreement with previous results (3), neither GRK2 nor GRK2-K220R affected the proliferation rate of cultured HEK cells (Fig. 3A).
FIGURE 3.
Dominant-negative GRK2-K220R enhanced the growth of NOD.Scid mouse-expanded HEK clones. A, cell proliferation of in vitro cultured control HEK cells (Cont.), and HEK cells expressing GRK2 (GRK2) or GRK2-K220R (K220R). The left panel presents cell counts of day 4, and the right panel shows growth curves. Data represent mean (±S.D.) of three independent experiments performed in triplicates. B, left panel: mass of HEK clones after expansion in NOD.Scid mice (± S.D., n = 5). The right panel shows a representative experiment (bar, 5 mm). C, immunoblot detection of GRK2 (IB: anti-GRK2) in lysates of the indicated NOD.Scid mouse-expanded HEK clones (upper panel). The lower panel shows a control blot detecting β-actin. D, immunohistological detection of GRK2 with GRK2-specific antibodies (anti-GRK2) in sections of a NOD.Scid mouse-expanded control HEK clone (Cont.), and HEK clones with overexpression of GRK2 (GRK2) or GRK2-K220R (K220R), respectively. The preabsorption control is a GRK2-K220R-expressing clone. Cell nuclei were stained with hematoxylin (HE), bar, 50 μm. E, immunoblot detection (IB) of phospho-ERK1/2 in total cell lysates of NOD.Scid mouse-expanded HEK clones (as indicated) with phospho-ERK1/2-specific antibodies. The left panel shows a representative immunoblot and the middle panel presents quantitative immunoblot evaluation by densitometric scanning of three different experiments. Bars represent mean ± S.D., n = 3. Right panel, total ERK1/2 protein levels were comparable between control, GRK2- and GRK2-K220R-expressing clones, respectively, as determined by immunoblotting (anti-ERK1/2). All results are representative of 2–3 different HEK cell clones each.
As GRK2-dependent growth control is active in vivo (2–4), we performed cell expansion in immunodeficient NOD.Scid mice because the xenograft transplantation model had previously been used to overcome limitations of cell culture (24). In contrast to cultured cells, GRK2 inhibition by the dominant-negative GRK2-K220R led to a strongly increased growth rate in vivo upon cell expansion in NOD.Scid mice, whereas GRK2-expressing clones showed a reduced cell mass (Fig. 3B). As a control, immunoblotting and immunohistology demonstrated comparable protein levels of GRK2-K220R and GRK2, respectively (Fig. 3, C and D).
We searched for the mechanism underlying the growth control by GRK2. The mitogen-activated protein kinase (MAPK) pathway triggered by RAS-dependent activation of RAF and MEK is a universal signal transduction cascade involved in cell survival, growth, and proliferation (33, 34). Because MAPK pathway activation is also a common feature of many GRK2 receptor substrates (35), we analyzed the activation of the MAP kinases, ERK1/2, of in vivo expanded HEK clones. The GRK2-K220R-induced increase in HEK cell mass correlated with a high level of activated phospho-ERK1/2 (Fig. 3E). Vice versa, the reduced growth of GRK2-expressing clones was accompanied by decreased ERK1/2 phosphorylation (Fig. 3E).
Up-regulation of MAPK Pathway Genes in NOD.Scid Mouse-expanded HEK Clones Expressing GRK2-K220R
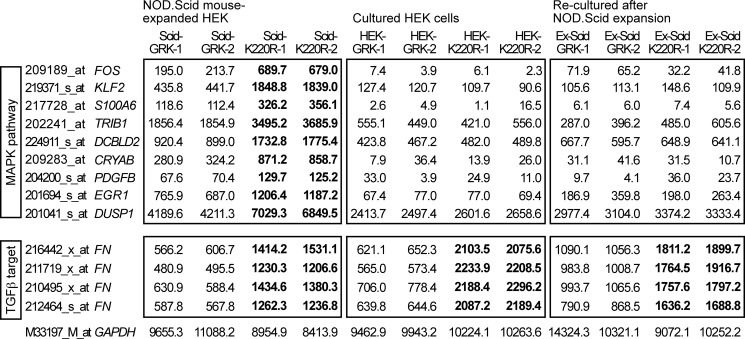
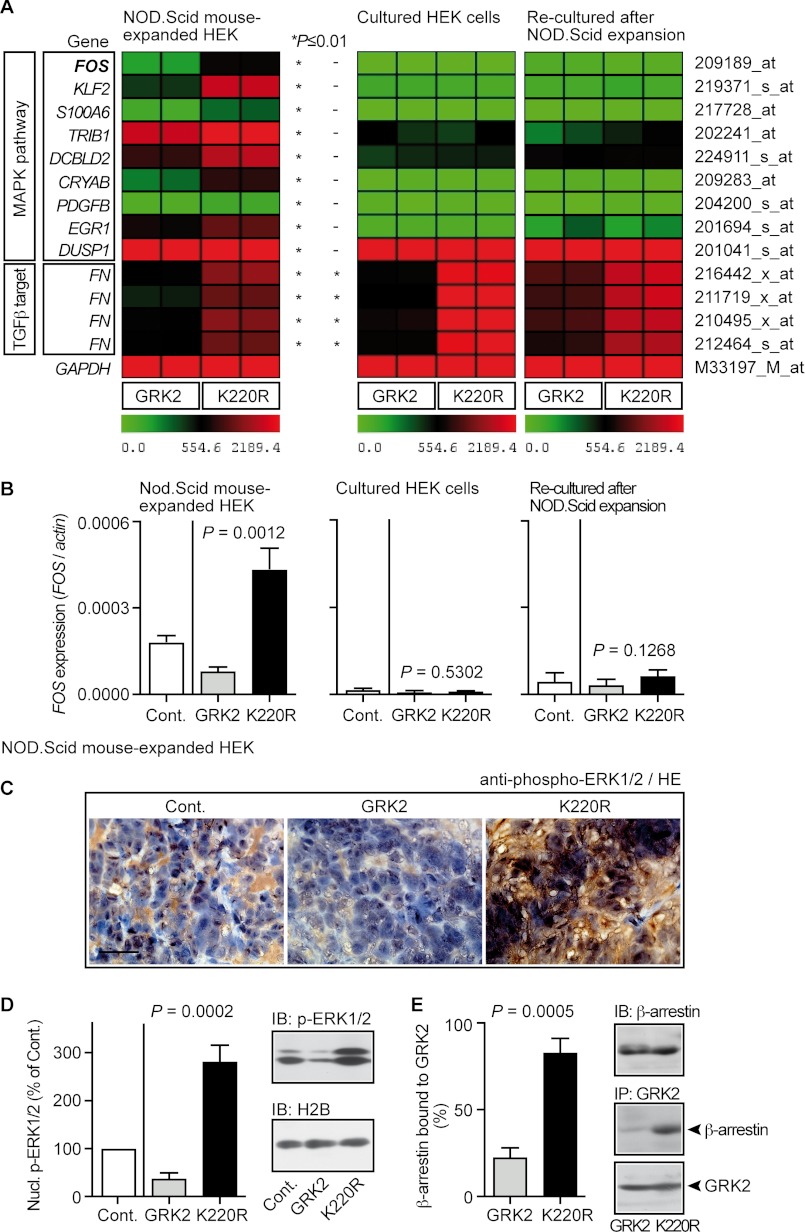
To decipher the basis of the differential growth regulation of cultured cells and NOD.Scid mouse-expanded clones, we performed microarray gene expression profiling. In agreement with sustained ERK1/2 activation (cf. Fig. 3E), microarray gene expression analysis showed that MAPK pathway target genes, such as FOS, were specifically increased in NOD.Scid mouse-expanded clones expressing dominant-negative GRK2-K220R compared with GRK2-expressing clones (Figs. 4 and 5A, left panel). In contrast to the NOD.Scid model, expression of MAPK target genes was low in cultured HEK cells and not different between GRK2- and GRK2-K220R-expressing cells, respectively (Figs. 4 and 5A, middle panel). NOD.Scid mice did not irreversibly modify the MAPK pathway because MAPK target gene induction disappeared when cells were “re-cultured” in vitro after in vivo expansion (Figs. 4 and 5A, right panel). As a control, a typical kinase-mediated effect of GRK2, i.e. inhibition of fibronectin (FN) expression depending on TGFβ (36) as a stimulus, which is active in vitro and in vivo, was similarly detected in HEK clones expanded in vivo or cultured in vitro (Figs. 4 and 5A). Taken together, in vivo cell expansion in NOD.Scid mice revealed GRK2-dependent regulation of the MAPK pathway.
FIGURE 4.
Microarray gene expression profiling revealed up-regulation of MAPK pathway genes in NOD.Scid mouse-expanded HEK clones expressing GRK2-K220R. Microarray gene expression profiling was performed with HEK clones expanded in vivo in NOD.Scid mice (Scid, left panels), of the respective in vitro-cultured HEK cell clones (HEK, middle panels), and of re-cultured cells after NOD.Scid mouse expansion (Ex-Scid, right panels). Gene expression data from GRK2-expressing (GRK-1 and GRK-2) and GRK2-K220R-expressing HEK clones (K220R-1 and K220R-2) are shown. Microarray probe sets were selected according to the following criteria: (i) significant difference (p ≤ 0.01) between NOD.Scid mouse-expanded clones expressing GRK2-K220R (Scid-K220R-1; Scid-K220R-2) and GRK2 (Scid-GRK-1; Scid-GRK-2), (ii) >1.6-fold up-regulation in GRK2-K220R-expressing clones relative to GRK2-expressing clones, and (iii) involvement in the MAPK pathway according to gene ontology analysis. Probe sets with significant differences (K220R- relative to GRK2-expressing HEK cells/clones) are marked in bold. The following genes were identified: FOS, v-fos FBJ murine osteosarcoma viral oncogene homolog; KLF2, Kruppel-like factor 2; S100A6, S100 calcium-binding protein A6, calcyclin; TRIB1, tribbles homolog 1; DCBLD2, discoidin, CUB and LCCL domain containing 2; CRYAB, crystallin, αB; PDGFB, platelet-derived growth factor β polypeptide; EGR1, early growth response 1; DUSP1, dual specificity phosphatase-1. Signal intensities of probe sets detecting the TGFβ target fibronectin (FN) were significantly lower in all GRK2-expressing HEK cells/clones indicative of TGFβ-antagonistic activity of GRK2. Selected data of two gene chips are presented for each group (two different clones/cell preparations per gene chip). The probe set detecting GAPDH is shown as a control.
FIGURE 5.
Dominant-negative GRK2-K220R triggered the nuclear MAPK pathway of NOD.Scid mouse-expanded HEK clones. A, microarray gene expression data of HEK clones after in vivo expansion in NOD.Scid mice (left panel), of cultured HEK cells (middle panel), and re-cultured cells after NOD.Scid mouse expansion (right panel) are presented as a heat map centered around the median value. Data filtering applied the following criteria: (i) significant difference (*, p ≤ 0.01) between NOD.Scid mouse-expanded HEK clones expressing GRK2-K220R and GRK2, (ii) >1.6-fold up-regulation in GRK2-K220R-expressing clones relative to GRK2-expressing clones, and (iii) involvement in the MAPK pathway according to gene ontology analysis. Signal intensities of probe sets detecting the TGFβ target fibronectin (FN) were significantly lower in all GRK2-expressing HEK clones/cells reflecting intact TGFβ-antagonistic activity of GRK2. The probe set of GAPDH is shown as control. B, left panel, real-time qRT-PCR showed up-regulated FOS expression of GRK2-K220R-expressing HEK clones expanded in NOD.Scid mice. Middle and right panels, real-time qRT-PCR analysis revealed low FOS expression of cultured HEK cells (middle panel) and cells re-cultured after in vivo expansion in NOD.Scid mice (right panel). Expression data were normalized to β-actin (FOS/actin) and represent mean ± S.D., n = 3. C, detection of phospho-ERK1/2 by immunohistology with phospho-ERK1/2-specific antibodies in sections of a NOD.Scid mouse-expanded control HEK clone (Cont.), and HEK clones expressing GRK2 (GRK2) or GRK2-K220R (K220R). Nuclei were stained with hematoxylin (HE); bar, 50 μm. D, nuclear phospho-ERK1/2 levels of NOD.Scid mouse-expanded HEK clones without (Cont., set to 100%) or with expression of GRK2 or GRK2-K220R (all samples were normalized to histone H2B). Data are mean ± S.D., n = 3. The right panels show a representative immunoblot experiment. E, increased interaction of β-arrestin with GRK2-K220R compared with GRK2 as determined by immunoenrichment of GRK2 (IP) from GRK2-expressing or GRK2-K220R-expressing HEK clones, respectively, followed by immunoblot (IB) detection of co-enriched β-arrestin and enriched GRK2. Bars represent mean ± S.D., n = 3. The right panels show a representative experiment (middle and lower panels) and a control immunoblot detecting β-arrestin (upper panel). All results are representative of 2–3 different HEK cell clones each.
Real-time qRT-PCR confirmed substantial FOS expression after NOD.Scid mouse expansion, whereas FOS was low in cultured cells (Fig. 5B). Real-time qRT-PCR also confirmed that GRK2-K220R up-regulated FOS in NOD.Scid mouse-expanded HEK clones compared with clones expressing GRK2 (Fig. 5B, left panel). In contrast to in vivo cell expansion, GRK2 and GRK2-K220R did not affect the expression of FOS in cultured HEK cells, either before or after in vivo expansion in NOD.Scid mice (Fig. 5B, middle and right panel). These experiments further support that in vivo cell expansion in NOD.Scid mice was essential to manifest MAPK target gene modulation by GRK2 and GRK2-K220R, respectively.
Dominant-Negative GRK2-K220R of NOD.Scid Mouse-expanded HEK Clones Enhanced MAPK Activation and Nuclear Translocation
The high expression of the nuclear MAPK target, FOS, pointed to sustained activation and nuclear translocation of the MAP kinases, ERK1/2 (34), upon GRK2 inhibition by GRK2-K220R. In agreement with that notion, immunohistology analysis with phospho-ERK1/2-specific antibodies revealed an increased total phospho-ERK1/2 level with a substantial amount of phospho-ERK1/2 in the nuclei of a NOD.Scid mouse-expanded GRK2-K220R-expressing HEK clone compared with the control or GRK2-expressing clone (Fig. 5C). Quantitative immunoblot evaluation confirmed those data and showed a high level of activated phospho-ERK1/2 in the nuclear fraction of GRK2-K220R-expressing clones, whereas nuclear phospho-ERK1/2 was low in GRK2-expressing clones (Fig. 5D).
Similarly as in cultured HEK cells (cf. Fig. 2), kinase-inactivated GRK2-K220R of NOD.Scid mouse-expanded HEK clones could act as a dominant-negative protein, which prevented signal desensitization by scavenging the cellular β-arrestin pool, whereas GRK2 was released from β-arrestin as determined by co-immunoenrichment (Fig. 5E). Together these findings suggest that dominant-negative GRK2-K220R could enhance receptor-stimulated (MAPK) signaling by preventing β-arrestin-dependent signal desensitization.
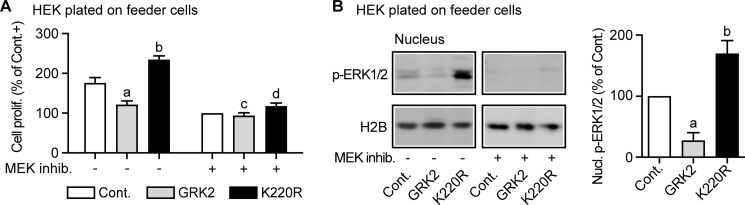
Trophic Effects of MEF Feeder Cells Reconstituted GRK2-dependent Growth Control in Vitro
To reconstitute the trophic effect of NOD.Scid mice on HEK growth in vitro, growth factor-supplemented HEK cells were plated on MEFs as feeder cells (25). Culture of HEK cells with MEF feeder cells revealed a GRK2-mediated reduction of HEK cell proliferation, whereas dominant-negative GRK2-K220R promoted cell proliferation (Fig. 6A). The MEK inhibitor PD0325901 prevented GRK2-mediated growth regulation indicating dependence on the MAPK pathway (Fig. 6A). In agreement with involvement of the MAPK pathway, cell proliferation control by GRK2 correlated with the regulation of the nuclear phospho-ERK1/2 level (Fig. 6B). Similarly as in the NOD.Scid mouse-expanded clones, GRK2 reduced the nuclear phospho-ERK1/2 level, whereas GRK2 inhibition by GRK2-K220R significantly increased the amount of nuclear phospho-ERK1/2 of HEK cells plated on MEF feeder cells (Fig. 6B). These experiments strongly suggest that the apparent difference between in vivo and in vitro experiments could be due to the lack of sustained MAPK-activating growth-promoting stimuli under standard cell culture conditions.
FIGURE 6.
Trophic effects of MEF feeder cells reconstituted GRK2-dependent growth control in vitro. A, cell proliferation analysis of HEK cells plated on mitomycin-inactivated MEF feeder cells showed decreased cell proliferation of GRK2-expressing HEK cells compared with HEK control cells and enhanced cell proliferation of dominant-negative GRK2-K220R-expressing cells. As indicated, EGF-supplemented cells were treated without or with the MEK inhibitor PD0325901. Cell proliferation is expressed as percentage of control, i.e. MEK inhibitor-treated control cells (set to 100%). Bars represent mean ± S.D., n = 3 (a, p = 0.0022; and b, p = 0.0018 versus Cont. without MEK inhibitor; c, p = 0.1515; and d, p = 0.1029 versus Cont. with MEK inhibitor). B, nuclear phospho-ERK1/2 levels of HEK cells plated on mitomycin-inactivated MEF feeder cells. The left and middle panels show a representative experiment and the right panel presents the quantitative evaluation of three different experiments without MEK inhibitor (± S.D., n = 3; a, p = 0.0113, and b, p = 0.0312 versus Cont.).
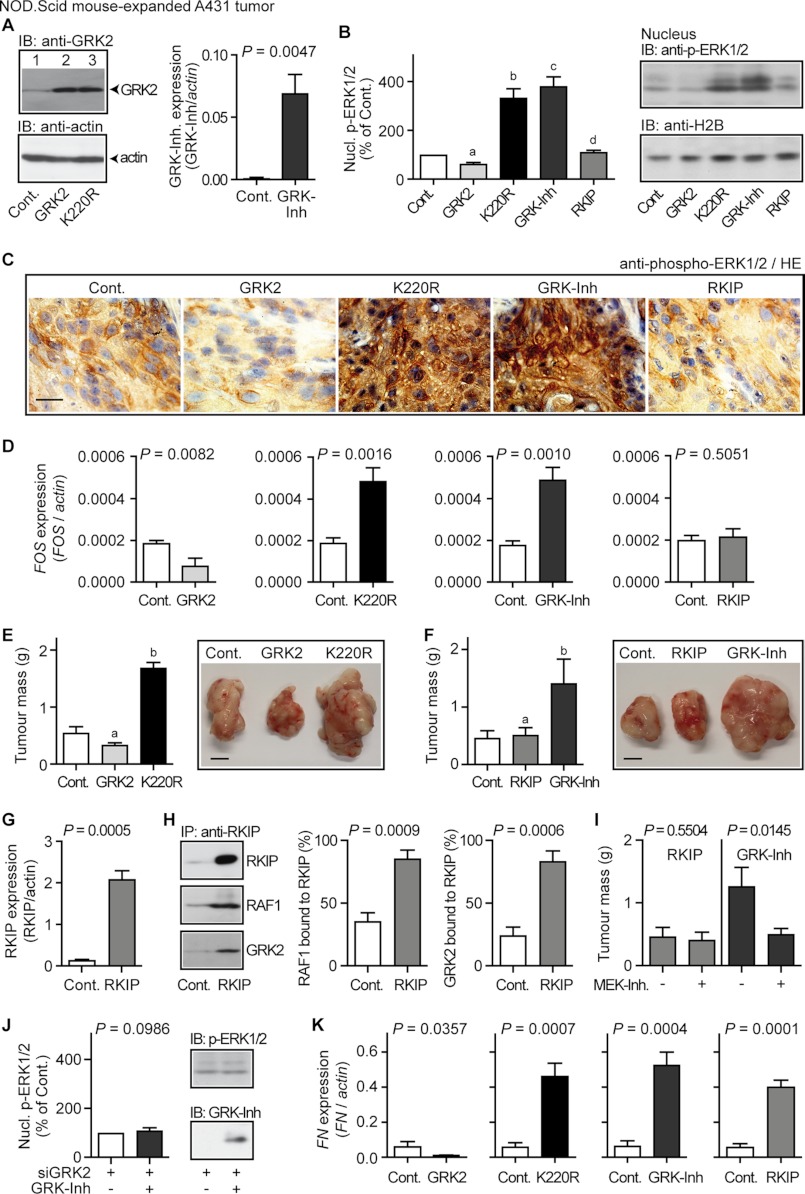
The RAF-MAPK Axis Triggered by GRK2 Inhibition Promotes Tumor Growth
Sustained MAPK pathway activation induces and/or promotes growth of malignant tumors such as squamous cell carcinoma (37). Therefore we used squamous cell carcinoma A431 cells to analyze whether GRK2-K220R-dependent enhancement of the MAPK pathway affected tumor growth. A431 clones with comparable protein levels of GRK2 and GRK2-K220R, respectively, were used (Fig. 7A, left panel). Analogous to NOD.Scid mouse-expanded HEK clones, immunotechniques revealed a high protein level of activated phospho-ERK1/2 in the nuclei of NOD.Scid-expanded A431 tumors expressing dominant-negative GRK2-K220R, whereas phospho-ERK-1/2 was low in GRK2-expressing tumors (Fig. 7, B and C). In agreement with growth-promoting MAPK activation, expression of the MAPK target, FOS, was induced in GRK2-K220R-expressing tumors (Fig. 7D), and A431 tumor mass was increased (Fig. 7E).
FIGURE 7.
The RAF-MAPK pathway triggered by GRK2 inhibition promotes tumor growth. A, left panel, immunoblot (IB) detection of GRK2 with GRK2-specific antibodies (anti-GRK2) in lysates of a NOD.Scid mouse-expanded control A431 tumor (Cont., lane 1), and A431 tumors expressing GRK2 (lane 2) or GRK2-K220R (lane 3). The loading control detected β-actin (lower panel). Right panel, expression level of the GRK2-specific peptide inhibitor (GRK-Inh, MAKFERLQTVTNYFITSE) in NOD.Scid mouse expanded A431 tumors was determined by real-time qRT-PCR relative to mock-transfected A431 control tumors (Cont.). Bars represent mean ± S.D., n = 4. B, left panel, nuclear phospho-ERK1/2 levels of NOD.Scid mouse expanded A431 tumors without (Cont., set to 100%) or with expression of GRK2 (GRK2), GRK2-K220R (K220R), the GRK2-specific peptide inhibitor (GRK-Inh) and RKIP (RKIP), respectively (all samples were normalized to histone H2B). Data represent mean ± S.D., n = 3 (a, p = 0.0150; b, p = 0.0087; c, p = 0.0067; d, p = 0.1537 versus Cont). The right panels show a representative immunoblot experiment. C, detection of phospho-ERK1/2 by immunohistology with phospho-ERK1/2-specific antibodies in a NOD.Scid mouse expanded control A431 tumor (Cont.), and A431 tumors expressing GRK2, GRK2-K220R (K220R), the GRK2-specific peptide inhibitor (GRK-Inh), or RKIP. Nuclei were stained with hematoxylin (HE); bar, 20 μm. D, real-time qRT-PCR analysis of FOS expression of different NOD.Scid mouse expanded A431 tumors without (Cont.) or with expression of GRK2, GRK2-K220R (K220R), the GRK2-specific peptide inhibitor (GRK-Inh), and RKIP. Data represent mean ± S.D. (n = 3). E, tumor mass of NOD.Scid mouse expanded A431 tumors without (Cont.) or with expression of GRK2 or the dominant-negative GRK2-K220R. Data represent mean ± S.D.; n = 5 (a, p = 0.0029; b, p = 0.0001 versus Cont.); bar, 5 mm. F, GRK2 inhibition by the GRK2-specific peptide inhibitor (GRK-Inh) promoted A431 tumor growth, whereas dual inhibition of the GRK2 and RAF-MAPK axis by RKIP did not. Data represent mean ± S.D.; n = 5 (a, p = 0.5439; b, p = 0.0014 versus Cont.); bar, 5 mm. G, the expression of RKIP was determined by real-time qRT-PCR in NOD.Scid mouse expanded A431 tumors expressing RKIP relative to A431 control tumors (Cont.). Bars represent mean ± S.D. (n = 4). H, overexpressed RKIP scavenged the cellular RAF1 and GRK2 pools. Enrichment of RKIP was performed with RKIP-specific antibodies (IP, anti-RKIP) from control A431 tumors (Cont.) and RKIP-expressing A431 tumors (RKIP) followed by immunoblot detection of enriched RKIP (upper panel), co-enriched RAF1 (middle panel) or co-enriched GRK2 (lower panel). The left panel shows a representative experiment and middle and right panels present quantitative evaluation of three different experiments (mean ± S.D.; n = 3). RKIP-bound RAF1 is expressed as % of total RAF1 (set to 100%) and RKIP-bound GRK2 is expressed as % of total GRK2 (set to 100%). Total RAF1 and total GRK2 protein was determined by quantitative immunoblotting. I, tumor mass of RKIP-expressing and GRK-Inh-expressing A431 tumors, respectively, expanded in NOD.Scid mice treated without or with the MEK inhibitor PD0325901 (20 mg/kg/day). Data represent mean ± S.D., n = 3. J, GRK-Inh expression (+GRK-Inh) did not significantly increase the nuclear phospho-ERK1/2 level of A431 tumors with down-regulated GRK2 (+siGRK2). Nuclear phospho-ERK1/2 is expressed as percentage of control, i.e. siGRK2-expressing A431 tumors without GRK-Inh (set to 100%). GRK2 down-regulation by siGRK2 was controlled by immunoblotting. Data represent mean ± S.D., n = 3. K, expression analysis of the GRK2-regulated TGFβ target, fibronectin (FN) was performed by real-time qRT-PCR with the NOD.Scid mouse expanded A431 clones used in D. Data represent mean ± S.D. (n = 3). All results are representative of 2–5 different A431 cell clones each.
To further investigate whether GRK2 inhibition triggered the growth-promoting MAPK pathway, we used a GRK2-specific peptide inhibitor (cf. Fig. 1B and Refs. 21 and 22). Expression of the GRK2-specific peptide inhibitor (Fig. 7A, right panel) significantly increased the level of activated phospho-ERK1/2 (Fig. 7, B and C), and induced the nuclear MAPK target FOS similarly as did GRK2-K220R (Fig. 7D, third versus second panel). Concomitantly, tumor growth was enhanced (Fig. 7F). Thus, GRK2 inhibition either by a dominant-negative mutant or a peptide inhibitor activated the growth-promoting MAPK pathway, induced the expression of the MAPK target FOS, and enhanced tumor growth.
The MAPK pathway is triggered by the proto-oncogene RAF. To analyze the involvement of RAF, we applied the RAF kinase inhibitor protein (RKIP) (Fig. 7G), which is a dual-specific GRK2 and RAF-MAPK inhibitor (9, 28, 38). In contrast to the sole GRK2 inhibition, RKIP prevented the increase in nuclear phospho-ERK1/2 (Fig. 7, B and C), and FOS was not induced in A431 clones expressing RKIP (Fig. 7D, fourth panel versus first to third panels). Concomitantly, GRK2 inhibition by RKIP did not enhance tumor growth (Fig. 7F). As a control, the expressed RKIP protein level was sufficient to scavenge the cellular RAF1 and GRK2 protein pools as assessed by co-immunoenrichment (Fig. 7H), confirming dual-specific GRK2 and RAF1 inhibition by RKIP (9, 28, 38).
In agreement with the requirement of the RAF/MEK/ERK pathway for enhanced tumor growth upon GRK2 inhibition, the MEK inhibitor PD0325901 significantly reduced the tumor mass of the GRK2 inhibitor-expressing A431 tumors, whereas the mass of RKIP-expressing A431 tumors was not affected (Fig. 7I). Moreover, MAPK activation induced by the GRK2-specific inhibitor was dependent on GRK2 because the GRK2-specific peptide inhibitor did not significantly increase the nuclear phospho-ERK1/2 level upon down-regulation of GRK2 by RNA interference (Fig. 7J). As an indicator for comparable GRK2 inhibition, the target gene FN (cf. Fig. 5A) was similarly up-regulated by the three different GRK2 inhibitors, whereas wild-type GRK2 reduced the expression of FN (Fig. 7K). Together the experiments strongly suggest that the RAF-MAPK axis triggered by GRK2 inhibition promoted growth of malignant A431 tumors.
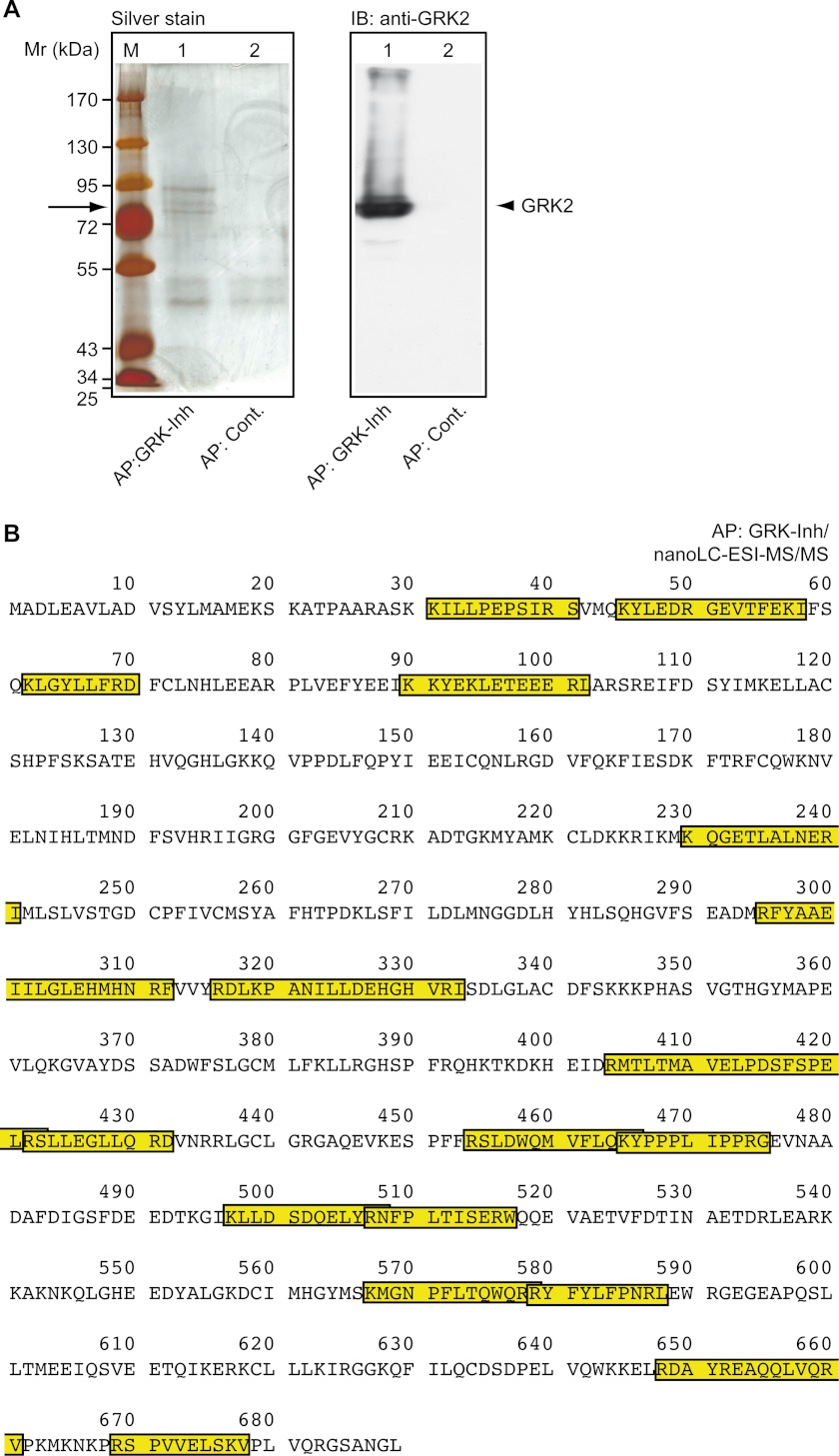
Enrichment of GRK2 by GRK-Inh-specific Immunoaffinity Chromatography and Identification by Nano-LC-ESI-MS/MS
To analyze interaction of the GRK2-specific peptide inhibitor (GRK-Inh) with GRK2 in vivo, we performed affinity chromatography with GRK-Inh-reactive antibodies. Proteins were immunoaffinity enriched from GRK-Inh-expressing tumor tissue and separated by SDS-PAGE. Silver staining of enriched proteins revealed a predominant protein band with an apparent molecular mass of 79 ± 3 kDa, which was not enriched by the control column (Fig. 8A). Immunoblotting demonstrated that the enriched protein co-migrated with the GRK2-immunoreactive protein band (Fig. 8A). The protein band was excised from the gel, and subjected to identification by nano-LC-ESI-MS/MS. With 27 matching peptides, the Mascot search engine identified the human β-adrenergic receptor kinase 1 (GRK2) with the highest probability score (Fig. 8B and supplemental Data). Regions of identified peptides matching with the human GRK2 protein sequence are shown (Fig. 8B). Together these experiments provide strong evidence that the GRK2-specific peptide inhibitor interacts with the GRK2 protein in vivo.
FIGURE 8.
Enrichment of GRK2 by GRK2-specific peptide inhibitor immunoaffinity chromatography and identification by nano-LC-ESI-MS/MS. A, proteins were enriched from GRK-Inh-expressing tumors by immunoaffinity chromatography with antibodies specific for GRK-Inh. The left panel shows a silver-stained gel of proteins enriched by immunoaffinity purification with GRK2-inhibitor-specific antibodies (AP:GRK-Inh; lane 1) compared with a control column (AP:Cont., lane 2). The right panel shows the immunoblot (IB) detection of GRK2 in the specific eluate (lane 1) compared with the control eluate (lane 2). B, after immunoaffinity chromatography with GRK-Inh-specific antibodies (AP:GRK-Inh), the GRK2-reactive band was cut and subjected to nano-LC-ESI-MS/MS. With 27 matching peptides, the Mascot search engine identified the human β-adrenergic receptor kinase 1 (GRK2) with the highest probability score. Regions of identified peptides matching with the human β-adrenergic receptor kinase 1 (GRK2) protein sequence are marked.
MAPK Pathway Activation in Transgenic Mice with Systemic Expression of the GRK2-specific Peptide Inhibitor
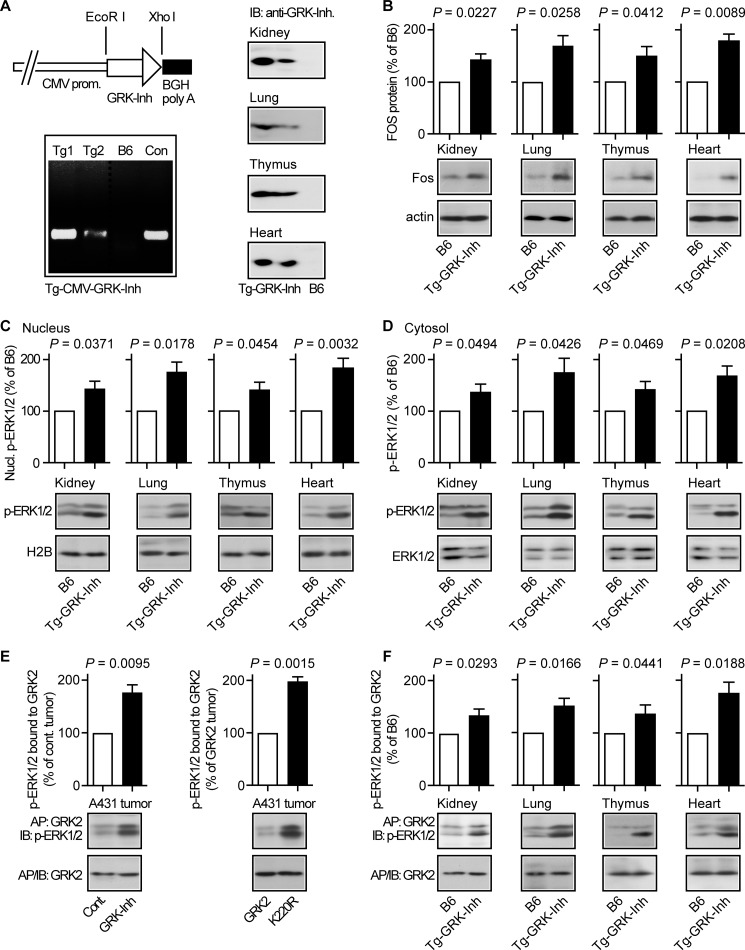
To investigate whether GRK2 inhibition by the GRK2-specific peptide inhibitor also regulated the MAPK pathway in non-tumor tissue, we generated transgenic mice expressing the GRK2-specific peptide inhibitor under control of the CMV immediate-early promoter/enhancer, which directs ubiquitous expression in transgenic mice (39). Two different transgenic mouse lines were identified, which showed high and low levels, respectively, of the GRK2-specific peptide inhibitor, in different organs, i.e. kidney, lung, thymus, and heart (Fig. 9A).
FIGURE 9.
MAPK pathway activation in transgenic mice with systemic expression of the GRK2-specific peptide inhibitor. A, left, generation of transgenic mouse lines (Tg1 and Tg2) with expression of the GRK2-specific peptide inhibitor under control of the CMV immediate-early promoter/enhancer (upper panel, left, diagram of the transgene; lower panel, left, identification of transgenic lines with integration of Tg-CMV-GRK-Inh into the mouse genome). The right panels present immunoblot detection of the GRK2-specific peptide inhibitor in different organs isolated from Tg-CMV-GRK-Inh-transgenic mice. B, immunoblot detection showed increased Fos protein levels in different organs from transgenic mice with systemic GRK2-specific peptide inhibitor expression under control of the CMV promoter (Tg-GRK-Inh) compared with B6 mice. The lower panels present control blots detecting β-actin. Bars represent mean ± S.D., n = 3 mice/group. C and D, nuclear (C) and cytosolic (D) phospho-ERK1/2 protein levels were determined by immunoblot analysis of different organs from transgenic mice with GRK2-specific peptide inhibitor expression under control of the CMV promoter. Nuclear proteins were normalized to H2B (C). Total cytosolic ERK1/2 protein was comparable between Tg-GRK-Inh mice and non-transgenic B6 mice (D). Bars represent mean ± S.D., n = 3 mice/group. E, stabilization of activated phospho-ERK1/2 by kinase-inhibited GRK2 (inhibited by GRK-Inh) or kinase-deficient GRK2-K220R. GRK2 protein was enriched by immunoaffinity purification (AP: GRK2) from A431 control tumors (Cont.) or A431 tumors expressing the GRK2-specific peptide inhibitor (GRK-Inh, left panel), and from GRK2-expressing or GRK2-K220R-expressing A431 tumors (right panel). Enriched GRK2 (IB: GRK2) and co-enriched phospho-ERK1/2 (IB: p-ERK1/2) were detected in the immunoblot. Quantitative assessment of co-enriched phospho-ERK1/2 was performed and is presented as percentage of the control A431 tumor (left panel) or the GRK2-expressing A431 tumor (right panel). Bars represent mean ± S.D., n = 3. F, immunoaffinity enrichment of GRK2 (AP: GRK2) from different organs isolated from transgenic mice with systemic GRK2-specific peptide inhibitor expression (Tg-GRK-Inh) or from non-transgenic B6 mice followed by immunoblot detection of enriched GRK2 (IB: GRK2) and co-enriched phospho-ERK1/2 (IB: p-ERK1/2). Quantitative assessment of co-enriched phospho-ERK1/2 is presented as percentage of B6 (±S.D., n = 3 mice/group).
Organs with transgenic expression of the GRK2-specific peptide inhibitor also displayed an increased Fos protein level compared with non-transgenic B6 controls, suggesting enhanced activation of the MAPK pathway upon GRK2-specific peptide inhibitor expression (Fig. 9B). In agreement with MAPK activation, nuclear and cytosolic protein levels of activated phospho-ERK1/2 were significantly higher in organs with GRK2-specific peptide inhibitor expression compared with B6 control tissue, whereas total cytosolic ERK1/2 protein was not different (Fig. 9, C and D). These experiments show that the GRK2-specific peptide inhibitor promoted activation of the MAPK pathway and Fos induction in vivo, not only in xenografted tumor tissue, but also in different organs of transgenic mice.
Kinase-inhibited GRK2 Stabilized Activated Phospho-ERK1/2 in Vivo
We analyzed the interaction of GRK2 with activated phospho-ERK1/2. Co-enrichment studies revealed that kinase-inhibited GRK2 (inhibited by the GRK2-specific peptide inhibitor) or kinase-deficient GRK2-K220R showed an increased interaction with activated phospho-ERK1/2 compared with kinase-active GRK2 of A431 tumor tissue (Fig. 9E, left and right panel). The increased interaction of kinase-inhibited GRK2 with phospho-ERK1/2 was also detected in different organs of mice with transgenic expression of the GRK2-specific peptide inhibitor (Fig. 9F). These findings suggest that kinase-inhibited GRK2 stabilized phospho-ERK1/2. The phospho-ERK1/2-stabilizing activity of kinase-inhibited GRK2 could contribute to sustained phospho-ERK1/2 activation required for MAPK target gene induction of A431 tumors and organs from transgenic mice.
GRK2 Inhibition by Transgenic RKIP or GRK-Inh Expression in Hearts of Transgenic Mice
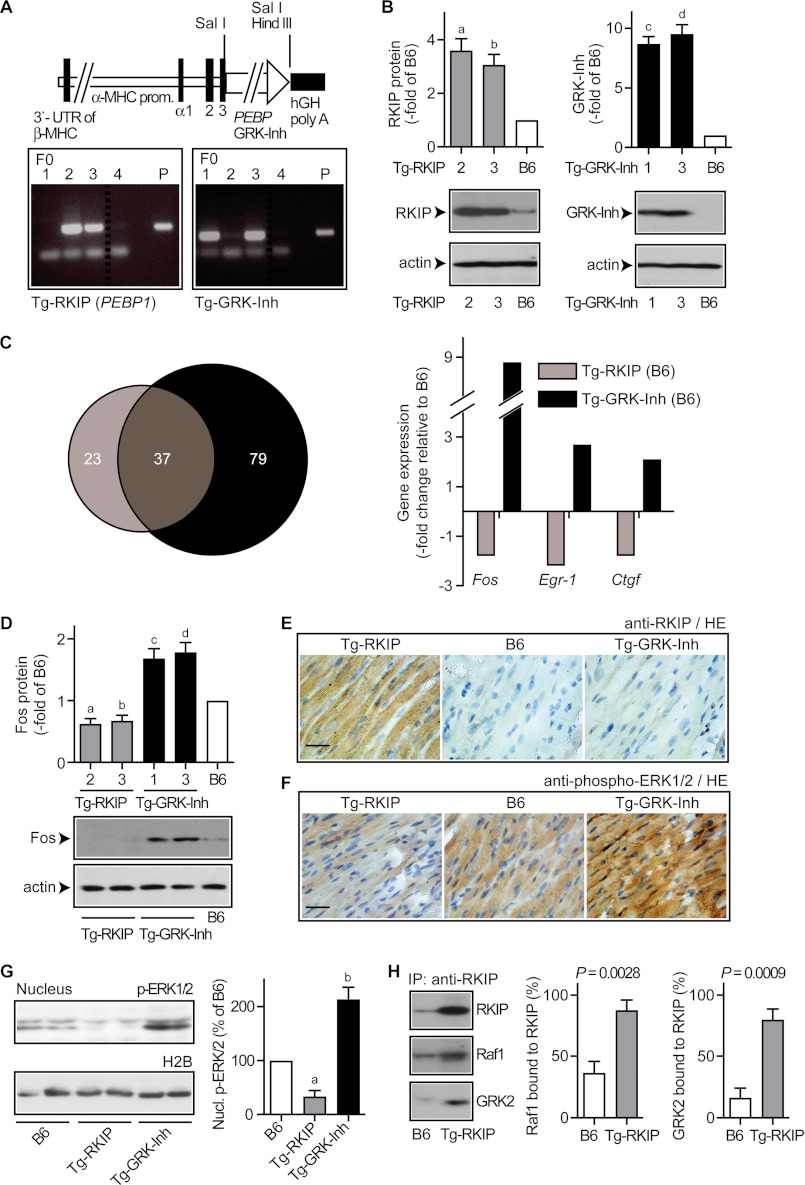
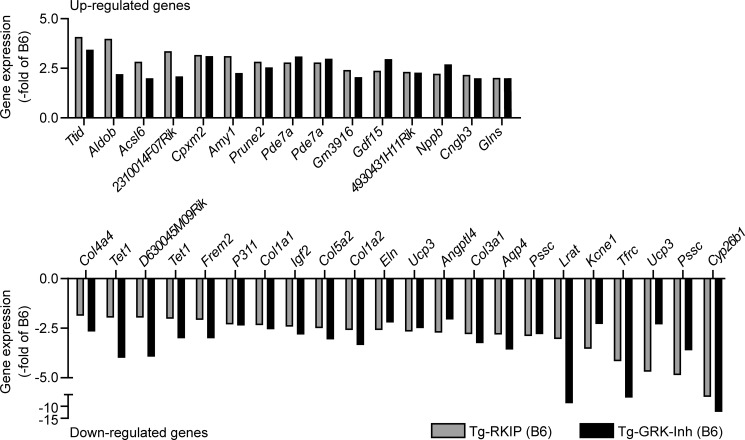
Enhanced activation of the MAPK pathway can exert cardioprotection (16). Because heart tissue from transgenic mice with GRK2-specific peptide inhibitor expression under control of the CMV promoter showed increased MAPK pathway activation (cf. Fig. 9), we investigated the cardiac profile of the GRK2-specific peptide inhibitor relative to RKIP, which is a dual-specific GRK2 and RAF/MAPK inhibitor. To this end mice with myocardium-specific expression of RKIP or the GRK2-specific peptide inhibitor, respectively, were generated (Fig. 10A). Hearts of transgenic founder lines (two different lines for each transgene) showed increased levels of the RKIP protein and the GRK2-specific peptide inhibitor, respectively (Fig. 10B). GRK2 inhibition was effective in both models as evidenced by microarray gene expression profiling: more than 60% of regulated probe sets of RKIP-transgenic hearts showed concordant regulation with hearts expressing the GRK2-specific peptide inhibitor (Fig. 10C, left panel, and Fig. 11). Moreover, the extent of gene regulation was comparable between RKIP-transgenic and GRK-Inh-transgenic hearts (Fig. 11). These in vivo observations are compatible with equivalent in vitro potencies of RKIP and GRK-Inh in inhibiting GRK2 (9, 22).
FIGURE 10.
GRK2 inhibition induced MAPK signaling in hearts of transgenic mice. A, diagram of the α-myosin heavy chain (MHC) plasmid used for the generation of transgenic mice with myocardium-specific RKIP or GRK2-specific peptide inhibitor (GRK-Inh) expression. Lower panels show the identification of founder mice (F0) with stable integration of the respective plasmid (P) encoding the RKIP transgene (left) or the GRK-Inh transgene (right) into genomic DNA. B, immunoblot analysis revealed increased RKIP protein in cardiac tissue of transgenic Tg-RKIP mice (left), and the GRK-specific peptide inhibitor was detected in cardiac tissue of transgenic Tg-GRK-Inh mice (right). Bars represent mean ± S.D., n = 3 mice/group (a, p = 0.0102; b, p = 0.0125; c, p = 0.0028; d, p = 0.0035 versus B6, set to 1), and lower panels show representative immunoblots. C, microarray gene expression profiling identified significantly regulated probe sets (p ≤ 0.01, fold-change relative to B6 mice ≥2 or ≤ −2, and signal intensity ≥100) in hearts of 2-month-old Tg-RKIP and Tg-GRK-Inh mice, respectively, relative to age-matched non-transgenic B6 control mice. The Venn diagram (left) illustrates the number of significantly regulated probe sets for each model. Thirty-seven probe sets (i.e. 61.7%) of Tg-RKIP hearts showed concordant regulation with Tg-GRK-Inh mice. Right panel, data filtering identified probe sets with significant up-regulation in Tg-GRK-Inh hearts (≥2-fold over the B6 control; p ≤ 0.01), and reduced expression in Tg-RKIP hearts relative to B6. D, the Fos protein was increased in Tg-GRK-Inh hearts relative to B6 controls, whereas Fos was decreased in Tg-RKIP hearts. Bars represent mean ± S.D., n = 3 mice/group (a, p = 0.0184; b, p = 0.0284; c, p = 0.0179; d, p = 0.0137 versus B6, set to 1), and lower panels show representative immunoblots. E, immunohistology detection revealed a high RKIP protein level in a heart section of a Tg-RKIP mouse relative to an age-matched B6 control mouse or a GRK2-specific peptide inhibitor-expressing mouse (Tg-GRK-Inh). Nuclei were stained with hematoxylin (HE), bar, 40 μm. F, immunohistology detection of phospho-ERK1/2 in heart sections of Tg-RKIP, B6 control, and Tg-GRK-Inh mice (bar, 40 μm). Histology data are representative of three different mice each (E and F). G, increased nuclear phospho-ERK1/2 level of transgenic hearts with GRK2-specific peptide inhibitor expression. The left panels show a representative immunoblot experiment, and the right panel presents quantitative data (±S.D., n = 3 mice/group; a, p = 0.0099; b, p = 0.0132; versus B6, set to 100%). H, transgenic RKIP scavenged the cardiac Raf1 and GRK2 protein pools as determined by immunoenrichment of RKIP with RKIP-specific antibodies (IP: RKIP) from B6 hearts (B6) and RKIP-transgenic hearts (Tg-RKIP) followed by immunoblot detection of enriched RKIP (upper panel), co-enriched Raf1 (middle panel), and co-enriched GRK2 (lower panel). The left panels show a representative experiment and the middle and right panels represent quantitative evaluation of three different experiments. Raf1 bound to RKIP is expressed as % of total Raf1, and GRK2 bound to RKIP is expressed as % of total GRK2 (±S.D., n = 3).
FIGURE 11.
Concordant regulation of gene expression by RKIP and the GRK2-specific peptide inhibitor in hearts of transgenic mice. Whole genome microarray gene expression profiling of heart tissue from 2-month-old mice identified probe sets of RKIP-transgenic hearts (Tg-RKIP) and GRK-Inh-transgenic hearts (Tg-GRK-Inh) with concordant up-regulation (upper panel) or down-regulation (lower panel) compared with B6 control mice (fold-change relative to B6: ≥2 or ≤ −2, and p ≤ 0.01). Bars represent mean of two different gene chips (three mice/gene chip). All data were normalized to β-actin.
The GRK2-specific Peptide Inhibitor Enhanced, Whereas RKIP Inhibited the MAPK Pathway in Vivo
Only 3 probe sets were up-regulated by the GRK2-specific peptide inhibitor and down-regulated by RKIP, i.e. MAPK target genes Fos, Egr-1, and Ctgf (Fig. 10C, right panel). Altered Fos expression was confirmed by immunoblotting, which showed increased cardiac Fos protein in transgenic hearts expressing the GRK2-specific peptide inhibitor, whereas RKIP decreased Fos (Fig. 10D). The induction of Fos correlated with increased MAPK activation as demonstrated by increased (nuclear) phospho-ERK1/2 levels in transgenic hearts expressing the GRK2-specific peptide inhibitor, whereas phospho-ERK1/2 was reduced by RKIP (Fig. 10, E–G). As a control, the protein level of overexpressed RKIP was sufficient to scavenge the entire cardiac GRK2 and Raf1 pools (Fig. 10H). Together the experiments show that transgenic RKIP overexpression induced Raf1-MAPK axis inhibition, whereas the GRK2-specific peptide inhibitor enhanced the MAPK pathway.
Inhibition of the MAPK Pathway by Transgenic RKIP Overexpression Triggered Signs of Heart Failure in B6 Mice
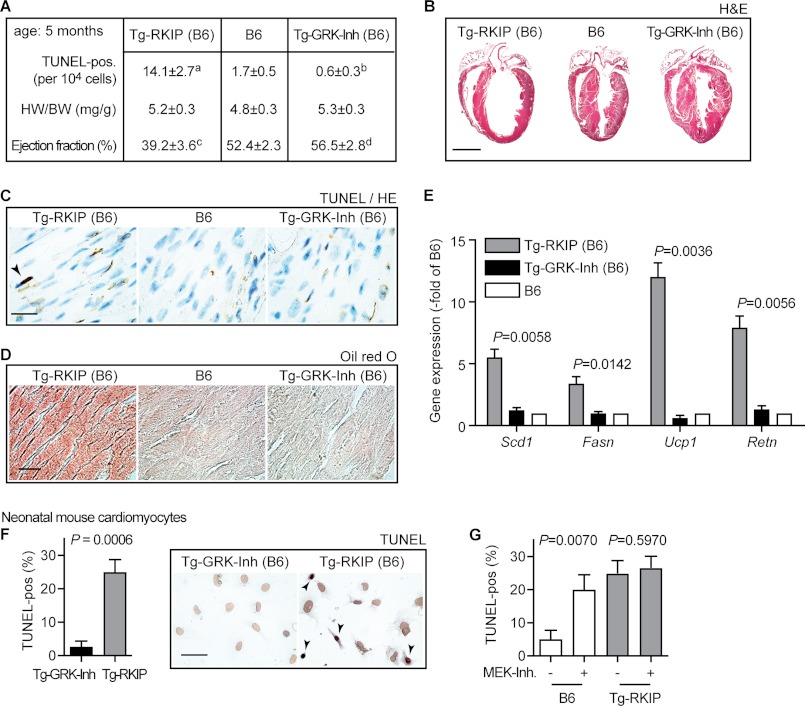
As the RAF-MAPK axis is essential for cardiomyocyte survival (16, 17, 40), cardiomyocyte death was assessed by TUNEL staining. There was a significant increase in the number of TUNEL-positive nuclei in RKIP-transgenic hearts, whereas signs of cardiomyocyte apoptosis were low in GRK2 inhibitor-expressing hearts (Fig. 12, A and C).
FIGURE 12.
Inhibition of the MAPK pathway by transgenic RKIP overexpression triggered signs of heart failure in B6 mice. A, quantitative assessment of TUNEL-positive nuclei, determination of heart weight to body weight ratio (HW/BW, mg/g), and measurement of left ventricular ejection fraction (%) by transthoracic echocardiography of Tg-RKIP, B6 control, and Tg-GRK-Inh mice (age: 5 months). Data represent mean ± S.D., n = 5 mice/group (a, p = 0.0001; b, p = 0.0018; c, p = 0.0001; d, p = 0.0352 versus B6). B, representative hematoxylin-eosin (H&E)-stained heart sections of 5-month-old Tg-RKIP, B6, and Tg-GRK-Inh mice (bar, 2 mm). C, TUNEL staining of heart sections from Tg-RKIP, B6 control, and Tg-GRK-Inh mice (bar, 20 μm). D, 5-month-old mice with myocardium-specific RKIP overexpression showed cardiac lipid overload as detected by Oil Red O staining (bar, 40 μm). E, gene expression analysis by real-time qRT-PCR detected increased expression of lipid metabolism genes (Scd1, Fasn, Ucp1, Retn) in hearts of 5-month-old RKIP-transgenic mice (all samples were normalized to Gapdh). Bars represent mean ± S.D. (n = 3). p values of significantly different gene expression data (versus B6) are indicated. F, quantitative assessment of TUNEL-positive nuclei of neonatal mouse cardiomyocytes isolated from transgenic hearts with GRK2-specific peptide inhibitor expression (Tg-GRK-Inh) or from RKIP-transgenic hearts. The left panel shows the percentage of TUNEL-positive cardiomyocytes (±S.D., n = 3), and the middle and right panels show representative images. G, treatment of neonatal mouse cardiomyocytes with the MEK-specific inhibitor PD0325901 (MEK-Inh) increased the number of TUNEL-positive neonatal mouse cardiomyocytes from B6 mice, whereas RKIP-transgenic cardiomyocytes were not substantially affected (±S.D., n = 3). Histology experiments are representative of three different mice each (B–D).
In agreement with enhanced cardiomyocyte death, RKIP-transgenic B6 mice developed heart dilatation and cardiac dysfunction at advanced age as determined by transthoracic echocardiography and histology analysis, respectively (Fig. 12, A and B). Cardiac dysfunction of RKIP-transgenic B6 mice was accompanied by cardiac lipid overload and up-regulation of cardiac lipid metabolism genes (Fig. 12, D and E), indicating the transition to heart failure (27). As a control, there was a trend toward improved cardiac function of GRK2-specific peptide inhibitor-expressing B6 mice (Fig. 12A), confirming positive inotropy induced by GRK2 inhibition (7, 10–15). Moreover, GRK-Inh-transgenic hearts did not develop cardiac dilatation or cardiac lipid overload (Fig. 12, B–E). Thus, GRK2 inhibition seemed to confer cardioprotection, whereas dual inhibition of the GRK2 and Raf/MAPK axis by RKIP produced cardiotoxic side effects.
The cardiotoxic effect of RKIP was confirmed with neonatal cardiomyocytes isolated from RKIP-transgenic hearts (Fig. 12F). RKIP overexpression induced a 9-fold increase in TUNEL-positive cardiomyocytes compared with neonatal cardiomyocytes isolated from GRK-Inh-transgenic mice (Fig. 12F). RKIP-dependent cardiomyocyte apoptosis was attributed to MAPK pathway inhibition, because a MEK inhibitor strongly increased cardiomyocyte death of B6 control cardiomyocytes but had no substantial effect on RKIP-transgenic cardiomyocytes (Fig. 12G).
Inhibition of GRK2 in FVB Mice by Transgenic Expression of RKIP or GRK-Inh
We asked whether the genetic background of B6 mice was linked to the different phenotype of the two GRK2 inhibitors. Transgenic FVB mice with myocardium-specific RKIP expression were generated (Fig. 13, A–C). Analogous to RKIP-transgenic B6 mice, the TUNEL assay revealed enhanced cardiomyocyte apoptosis of RKIP-transgenic FVB mice (Fig. 13A). Concomitantly, cardiac hypertrophy with dilatation and cardiac dysfunction developed in RKIP-transgenic FVB mice (Fig. 13, A and B). Taken together these data show that dual inhibition of the GRK2-RAF/MAPK axis by RKIP could be cardiotoxic in B6 and FVB mice, independent of the genetic background of the mouse.
FIGURE 13.
Inhibition of GRK2 in transgenic FVB mice. A, quantitative assessment of TUNEL-positive nuclei, determination of heart weight to body weight ratio (HW/BW, mg/g), and measurement of left ventricular ejection fraction (%) by transthoracic echocardiography of FVB mice with myocardium-specific RKIP expression, and non-transgenic FVB control mice (age: 10 weeks). Data represent mean ± S.D., n = 5 mice/group (a, p = 0.0001; b, p = 0.0004; c, p < 0.0001). B, representative hematoxylin-eosin (H&E)-stained heart sections of a 10-week-old FVB mouse with transgenic RKIP expression (Tg-RKIP, FVB) and a non-transgenic FVB control mouse show cardiac hypertrophy with dilatation of the RKIP-transgenic FVB mouse (bar, 1.8 mm). C, increased cardiac RKIP protein levels of two different FVB mouse lines with myocardium-specific RKIP expression (±S.D., n = 5 mice/group; a, p = 0.0025; b, p = 0.0003 versus FVB). D, cardiac GRK2-specific peptide inhibitor detection in FVB mice with transgenic GRK-Inh expression (±S.D., n = 5 mice/group; a, p = 0.0031; b, p = 0.0004 versus FVB). E, representative hematoxylin-eosin (H&E)-stained heart sections of 4-month-old FVB mice with transgenic GRK2-specific peptide inhibitor expression (Tg-GRK-Inh, FVB; left panel) and non-transgenic FVB control mice (right panel) without (Sham) or with 4 weeks of AAC (bar, 1.8 mm). F, determination of heart weight to body weight ratio (HW/BW) and left ventricular ejection fraction (%) by transthoracic echocardiography of Tg-GRK-Inh (FVB) mice and non-transgenic FVB mice without (Sham) and with pressure overload imposed by 4 weeks of AAC. Data represent mean ± S.D., n = 4 mice/group (a, p = 0.0201 versus Sham-Tg-GRK-Inh; b, p = 0.0033 versus AAC-FVB; c, p = 0.0002 versus Sham FVB; d, p = 0.0001 versus Sham-Tg-GRK-Inh; e, p = 0.0004 versus AAC-FVB; f, p < 0.0001 versus Sham FVB). Histology experiments are representative of three different mice each (B and E).
On the other hand, hearts of transgenic FVB mice expressing the GRK2-specific peptide inhibitor (Fig. 13D) were not dilated and showed a high left ventricular ejection fraction similarly as B6 mice with transgenic GRK-Inh expression (Fig. 13, E and F, versus 12). In agreement with cardioprotective GRK2 inhibition (10–15), the GRK2-specific peptide inhibitor retarded the development of chronic pressure overload-induced cardiac hypertrophy with dilatation and cardiac dysfunction imposed by 4 weeks of abdominal aortic constriction (Fig. 13, E and F). Thus, the GRK2-specific peptide inhibitor had a comparable cardiac profile in FVB and B6 mice and was protective against pressure overload-induced cardiac damage.
DISCUSSION
Our study revealed a previously unrecognized role of the desensitizing kinase, GRK2, in restraining the MAPK pathway. GRK2-mediated desensitization of MAPK-activating GPCRs and non-GPCR substrates is well established (1, 35, 41). In addition, prevention of nuclear MAPK signaling could be mediated by the GRK2-dependent recruitment of β-arrestin, which retains MAPK in the cytosol (42–44). This mechanism is not only valid for GPCRs (42, 43) but also reduces nuclear translocation of ERK1/2 upon receptor tyrosine kinases co-stimulation such as the epidermal growth factor receptor (44).
Although GRK2 suppressed MAPK activation and nuclear translocation, GRK2 inhibitors enhanced the MAPK pathway, induced nuclear ERK1/2 targets, notably the proto-oncogene FOS, and promoted tumor growth. Activation of the nuclear MAPK pathway upon GRK2 inhibition was not only observed in two different xenograft cell transplantation models but also occurred in vivo, in different organs of transgenic mice upon systemic GRK2 inhibition by transgenic expression of a GRK2-specific peptide inhibitor.
Increased MAPK activation and target gene induction upon GRK2 inhibition could be due to β-arrestin scavenging/neutralization by kinase-inhibited GRK2. Thereby the formation of receptor·β-arrestin complexes is prevented as well as subsequent receptor/MAPK desensitization. In this respect, kinase-inhibited GRK2 acts as a dominant-negative protein, which could differ from GRK2 knockdown because siGRK2 may trigger a shift to GRK5/6-β-arrestin-mediated MAPK activation (45). Nevertheless, kinase-inhibited GRK2 and GRK2 down-regulation both enhance MAPK activity, and confer cardioprotection (10–15). In addition to β-arrestin neutralization, kinase-inhibited GRK2 could lead to sustained MAPK pathway activation essential for target gene induction (34), by stabilization of activated phospho-ERK1/2, which was detected by co-enrichment studies in A431 tumor tissue and various organs of transgenic mice with systemic expression of the GRK2-specific peptide inhibitor.
The tumor growth-promoting MAPK activity triggered by GRK2 inhibition overlapped with the cardioprotective activity of the MAPK cascade, which protects the myocardium against death-promoting stimuli (16, 46). Notably, activation of ERK1/2 protects the heart against pressure overload-induced hypertrophic cardiomyopathy by stimulating cardiomyocyte survival and proliferation (46). Vice versa, genetic inhibition of ERK1/2 enhanced cardiomyocyte apoptosis and predisposed to the development of pressure overload-induced heart failure (47). In agreement with those data, the pro-survival function of the MAPK cascade could contribute to the beneficial cardiac profile of GRK2 inhibition because dual inhibition of the GRK2 and RAF/MAPK axis by RKIP was detrimental and induced signs of heart failure in transgenic mouse lines with two different genetic backgrounds, most likely due to cardiotoxic RAF inhibition (17, 40) and lack of cardioprotective MAPK activation (16, 46, 48).
On the other hand, GRK2 inhibition without MAPK inhibition, by transgenic GRK2-specific peptide inhibitor expression, was beneficial and retarded the development of chronic pressure overload-induced cardiac hypertrophy with dilatation and cardiac dysfunction. The cardioprotective activity of the GRK2-specific peptide inhibitor was comparable with the profile of GRK2 inhibition induced by the GRK2ct. Analogous to the GRK2-specific peptide inhibitor, transgenic expression of the GRK2ct also prevented the development of left ventricular dilatation and hypertrophy triggered by long-term chronic pressure overload (49). Moreover, inhibition of GRK2 by the Gβγ-scavenging GRK2ct, also preserved the activated MAPK pathway under aortic constriction (50).
In addition to the cardioprotective MAPK pathway (48), GRK2 inhibition could enhance cardiomyocyte survival by increasing AKT-mediated induction of nitric oxide (51). Such a mechanism is active in vivo and contributes to cardioprotection of GRK2ct-transgenic mice against acute myocardial ischemia/reperfusion injury (51). Because pro-survival signaling by MAPK and AKT is integrated in the heart by specific scaffold proteins (52), future studies will have to determine the impact of the GRK2-specific peptide inhibitor on synergistic enhancement of MAPK- and AKT-mediated pro-survival pathways.
Although we found analogous results of the GRK2-specific peptide inhibitor and the dominant-negative GRK2-K220R mutant regarding MAPK activation and tumor cell proliferation, the cardiac phenotype of GRK2-K220R still needs to be determined. Because GRK2-K220R is capable of inhibiting GRK2 as a dominant-negative protein and by scavenging Gβγ subunits similarly as GRK2ct, GRK2-K220R is also expected to exert cardioprotection.
Taken together, our study investigated the in vivo profile of GRK2 inhibition by a GRK2-specific peptide inhibitor and the dominant-negative GRK2-K220R mutant. Our study provides strong evidence that GRK2 inhibition induces activation of the growth-promoting MAPK pathway in vivo, which seems critical for cardiomyocyte survival. In view of the well established cell proliferation and cell growth-stimulating activity of the MAPK pathway, future efforts will have to balance the cardiomyocyte survival profile of GRK2 inhibitors with the inherent risk of tumor cell growth. New approaches could focus on the development of dual-specific inhibitors, which stimulate GRK2-dependent cardiomyocyte survival but inhibit tumor cell proliferation.
Acknowledgments
We thank Said AbdAlla for establishment of transgenic mouse techniques and James Gulick for the α-myosin heavy chain (MyHC) plasmid. Confocal FRET imaging was performed at the Center for Microscopy and Image Analysis, University of Zurich.
This work was supported in part by Swiss National Science Foundation Grant 31-140679 (to U. Q.).

This article contains supplemental data.
- GRK2
- G-protein-coupled receptor kinase-2
- RKIP
- Raf kinase inhibitor protein
- GPCR
- G-protein-coupled receptor
- GRK2ct
- carboxyl-terminal domain (last 194 amino acids) of GRK2
- GRK-Inh
- GRK2-specific peptide inhibitor with the peptide sequence MAKFERLQTVTNYFITSE
- NOD.Scid mice
- non-obese diabetic mice homozygous for the severe combined immune deficiency spontaneous mutation
- AAC
- abdominal aortic constriction
- TUNEL
- TdT-mediated dUTP-biotin nick end labeling
- FOS
- FBJ osteosarcoma oncogene
- FN
- fibronectin
- MEF
- mouse embryonic fibroblast
- qRT
- quantitative RT
- B2R
- bradykinin B2 receptor
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Gurevich E. V., Tesmer J. J., Mushegian A., Gurevich V. V. (2012) G protein-coupled receptor kinases. More than just kinases and not only for GPCRs. Pharmacol. Ther. 133, 40–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Philipp M., Fralish G. B., Meloni A. R., Chen W., MacInnes A. W., Barak L. S., Caron M. G. (2008) Smoothened signaling in vertebrates is facilitated by a G protein-coupled receptor kinase. Mol. Biol. Cell 19, 5478–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang X., Yang P., Ma L. (2009) Kinase activity-independent regulation of cyclin pathway by GRK2 is essential for zebrafish early development. Proc. Natl. Acad. Sci. U.S.A. 106, 10183–10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaber M., Koch W. J., Rockman H., Smith B., Bond R. A., Sulik K. K., Ross J., Jr., Lefkowitz R. J., Caron M. G., Giros B. (1996) Essential role of β-adrenergic receptor kinase 1 in cardiac development and function. Proc. Natl. Acad. Sci. U.S.A. 93, 12974–12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gros R., Benovic J. L., Tan C. M., Feldman R. D. (1997) G-protein-coupled receptor kinase activity is increased in hypertension. J. Clin. Invest. 99, 2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ungerer M., Böhm M., Elce J. S., Erdmann E., Lohse M. J. (1993) Altered expression of β-adrenergic receptor kinase and β1-adrenergic receptors in the failing human heart. Circulation 87, 454–463 [DOI] [PubMed] [Google Scholar]
- 7. Brinks H., Koch W. J. (2010) Targeting G protein-coupled receptor kinases (GRKs) in heart failure. Drug Discov. Today Dis. Mech. 7, e129–e134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thal D. M., Yeow R. Y., Schoenau C., Huber J., Tesmer J. J. (2011) Molecular mechanism of selectivity among G protein-coupled receptor kinase 2 inhibitors. Mol. Pharmacol. 80, 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lorenz K., Lohse M. J., Quitterer U. (2003) Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 426, 574–579 [DOI] [PubMed] [Google Scholar]
- 10. Koch W. J., Rockman H. A., Samama P., Hamilton R. A., Bond R. A., Milano C. A., Lefkowitz R. J. (1995) Cardiac function in mice overexpressing the β-adrenergic receptor kinase or a β-ARK inhibitor. Science 268, 1350–1353 [DOI] [PubMed] [Google Scholar]
- 11. Rockman H. A., Chien K. R., Choi D. J., Iaccarino G., Hunter J. J., Ross J., Jr., Lefkowitz R. J., Koch W. J. (1998) Expression of a β-adrenergic receptor kinase 1 inhibitor prevents the development of myocardial failure in gene-targeted mice. Proc. Natl. Acad. Sci. U.S.A. 95, 7000–7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harding V. B., Jones L. R., Lefkowitz R. J., Koch W. J., Rockman H. A. (2001) Cardiac β-ARK1 inhibition prolongs survival and augments β blocker therapy in a mouse model of severe heart failure. Proc. Natl. Acad. Sci. U.S.A. 98, 5809–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Völkers M., Weidenhammer C., Herzog N., Qiu G., Spaich K., von Wegner F., Peppel K., Müller O. J., Schinkel S., Rabinowitz J. E., Hippe H. J., Brinks H., Katus H. A., Koch W. J., Eckhart A. D., Friedrich O., Most P. (2011) The inotropic peptide βARKct improves βAR responsiveness in normal and failing cardiomyocytes through G(βγ)-mediated L-type calcium current disinhibition. Circ. Res. 108, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raake P. W., Vinge L. E., Gao E., Boucher M., Rengo G., Chen X., DeGeorge B. R., Jr., Matkovich S., Houser S. R., Most P., Eckhart A. D., Dorn G. W., 2nd, Koch W. J. (2008) G protein-coupled receptor kinase 2 ablation in cardiac myocytes before or after myocardial infarction prevents heart failure. Circ. Res. 103, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raake P. W., Zhang X., Vinge L. E., Brinks H., Gao E., Jaleel N., Li Y., Tang M., Most P., Dorn G. W., 2nd, Houser S. R., Katus H. A., Chen X., Koch W. J. (2012) Cardiac G-protein-coupled receptor kinase 2 ablation induces a novel Ca2+ handling phenotype resistant to adverse alterations and remodeling after myocardial infarction. Circulation 125, 2108–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kehat I., Molkentin J. D. (2010) Extracellular signal-regulated kinase 1/2 (ERK1/2) signaling in cardiac hypertrophy. Ann. N.Y. Acad. Sci. 1188, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng H., Kari G., Dicker A. P., Rodeck U., Koch W. J., Force T. (2011) A novel preclinical strategy for identifying cardiotoxic kinase inhibitors and mechanisms of cardiotoxicity. Circ. Res. 109, 1401–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Métayé T., Levillain P., Kraimps J. L., Perdrisot R. (2008) Immunohistochemical detection, regulation and antiproliferative function of G-protein-coupled receptor kinase 2 in thyroid carcinomas. J. Endocrinol. 198, 101–110 [DOI] [PubMed] [Google Scholar]
- 19. Wei Z., Hurtt R., Ciccarelli M., Koch W. J., Doria C. (2012) Growth inhibition of human hepatocellular carcinoma cells by overexpression of G-protein-coupled receptor kinase 2. J. Cell Physiol. 227, 2371–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gulick J., Subramaniam A., Neumann J., Robbins J. (1991) Isolation and characterization of the mouse cardiac myosin heavy chain genes. J. Biol. Chem. 266, 9180–9185 [PubMed] [Google Scholar]
- 21. Quitterer U., Pohl A., Langer A., Koller S., Abdalla S. (2011) A cleavable signal peptide enhances cell surface delivery and heterodimerization of Cerulean-tagged angiotensin II AT1 and bradykinin B2 receptor. Biochem. Biophys. Res. Commun. 409, 544–549 [DOI] [PubMed] [Google Scholar]
- 22. Winstel R., Ihlenfeldt H. G., Jung G., Krasel C., Lohse M. J. (2005) Peptide inhibitors of G protein-coupled receptor kinases. Biochem. Pharmacol. 70, 1001–1008 [DOI] [PubMed] [Google Scholar]
- 23. Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003) Manipulating the Mouse Embryo. A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 24. Abd Alla J., Pohl A., Reeck K., Streichert T., Quitterer U. (2010) Establishment of an in vivo model facilitates B2 receptor protein maturation and heterodimerization. Integr. Biol. (Camb.) 2, 209–217 [DOI] [PubMed] [Google Scholar]
- 25. Oka Y., Nakajima K., Nagao K., Miura K., Ishii N., Kobayashi H. (2010) 293FT cells transduced with four transcription factors (OCT4, SOX2, NANOG, and LIN38) generate aberrant ES-like cells. J. Stem Cells Regen. Med. 6, 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. AbdAlla S., Lother H., el Missiry A., Langer A., Sergeev P., el Faramawy Y., Quitterer U. (2009) Angiotensin II AT2 receptor oligomers mediate G-protein dysfunction in an animal model of Alzheimer disease. J. Biol. Chem. 284, 6554–6565 [DOI] [PubMed] [Google Scholar]
- 27. Abdalla S., Fu X., Elzahwy S. S., Klaetschke K., Streichert T., Quitterer U. (2011) Up-regulation of the cardiac lipid metabolism at the onset of heart failure. Cardiovasc. Hematol. Agents Med. Chem. 9, 190–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. el Faramawy Y., Fu X., Agwa S. H., Langer A., Elzahwy S. S., Quitterer U. (2011) RKIP enhances angiotensin II-stimulated signaling. Forum Immunopathol. Dis. Therapeutics 2, 71–78 [Google Scholar]
- 29. Conner D. A., Mathier M. A., Mortensen R. M., Christe M., Vatner S. F., Seidman C. E., Seidman J. G. (1997) β-Arrestin1 knock-out mice appear normal but demonstrate altered cardiac responses to β-adrenergic stimulation. Circ. Res. 81, 1021–1026 [DOI] [PubMed] [Google Scholar]
- 30. Abd Alla J., Langer A., Elzahwy S. S., Arman-Kalcek G., Streichert T., Quitterer U. (2010) Angiotensin-converting enzyme inhibition down-regulates the pro-atherogenic chemokine receptor 9 (CCR9)-chemokine ligand 25 (CCL25) axis. J. Biol. Chem. 285, 23496–23505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kong G., Penn R., Benovic J. L. (1994) A β-adrenergic receptor kinase dominant-negative mutant attenuates desensitization of the β2-adrenergic receptor. J. Biol. Chem. 269, 13084–13087 [PubMed] [Google Scholar]
- 32. AbdAlla S., Zaki E., Lother H., Quitterer U. (1999) Involvement of the amino terminus of the B2 receptor in agonist-induced receptor dimerization. J. Biol. Chem. 274, 26079–26084 [DOI] [PubMed] [Google Scholar]
- 33. Osborne J. K., Zaganjor E., Cobb M. H. (2012) Signal control through Raf. In sickness and in health. Cell Res. 22, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murphy L. O., Smith S., Chen R. H., Fingar D. C., Blenis J. (2002) Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4, 556–564 [DOI] [PubMed] [Google Scholar]
- 35. Lappano R., Maggiolini M. (2011) G protein-coupled receptors. Novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 10, 47–60 [DOI] [PubMed] [Google Scholar]
- 36. Ho J., Cocolakis E., Dumas V. M., Posner B. I., Laporte S. A., Lebrun J. J. (2005) The G protein-coupled receptor kinase-2 is a TGFβ-inducible antagonist of TGFβ signal transduction. EMBO J. 24, 3247–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hatzivassiliou G., Song K., Yen I., Brandhuber B. J., Anderson D. J., Alvarado R., Ludlam M. J., Stokoe D., Gloor S. L., Vigers G., Morales T., Aliagas I., Liu B., Sideris S., Hoeflich K. P., Jaiswal B. S., Seshagiri S., Koeppen H., Belvin M., Friedman L. S., Malek S. (2010) Raf inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 [DOI] [PubMed] [Google Scholar]
- 38. Yeung K., Seitz T., Li S., Janosch P., McFerran B., Kaiser C., Fee F., Katsanakis K. D., Rose D. W., Mischak H., Sedivy J. M., Kolch W. (1999) Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401, 173–177 [DOI] [PubMed] [Google Scholar]
- 39. Furth P. A., Hennighausen L., Baker C., Beatty B., Woychick R. (1991) The variability in activity of the universally expressed human cytomegalovirus immediate early gene 1 enhancer/promoter in transgenic mice. Nucleic Acids Res. 19, 6205–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamaguchi O., Watanabe T., Nishida K., Kashiwase K., Higuchi Y., Takeda T., Hikoso S., Hirotani S., Asahi M., Taniike M., Nakai A., Tsujimoto I., Matsumura Y., Miyazaki J., Chien K. R., Matsuzawa A., Sadamitsu C., Ichijo H., Baccarini M., Hori M., Otsu K. (2004) Cardiac-specific disruption of the c-raf-1 gene induces cardiac dysfunction and apoptosis. J. Clin. Invest. 114, 937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Freedman N. J., Kim L. K., Murray J. P., Exum S. T., Brian L., Wu J. H., Peppel K. (2002) Phosphorylation of the platelet-derived growth factor receptor-β and epidermal growth factor receptor by G protein-coupled receptor kinase-2. Mechanisms for selectivity of desensitization. J. Biol. Chem. 277, 48261–48269 [DOI] [PubMed] [Google Scholar]
- 42. Tohgo A., Pierce K. L., Choy E. W., Lefkowitz R. J., Luttrell L. M. (2002) β-Arrestin scaffolding of the ERK cascade enhances cytosolic ERK activity but inhibits ERK-mediated transcription following angiotensin AT1a receptor stimulation. J. Biol. Chem. 277, 9429–9436 [DOI] [PubMed] [Google Scholar]
- 43. DeFea K. A., Zalevsky J., Thoma M. S., Déry O., Mullins R. D., Bunnett N. W. (2000) β-Arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 148, 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tilley D. G., Kim I. M., Patel P. A., Violin J. D., Rockman H. A. (2009) β-Arrestin mediates β1-adrenergic receptor-epidermal growth factor receptor interaction and downstream signaling. J. Biol. Chem. 284, 20375–20386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng H., Worrall C., Shen H., Issad T., Seregard S., Girnita A., Girnita L. (2012) Selective recruitment of G protein-coupled receptor kinases (GRKs) controls signaling of the insulin-like growth factor 1 receptor. Proc. Natl. Acad. Sci. U.S.A. 109, 7055–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maillet M., Purcell N. H., Sargent M. A., York A. J., Bueno O. F., Molkentin J. D. (2008) DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J. Biol. Chem. 283, 31246–31255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Purcell N. H., Wilkins B. J., York A., Saba-El-Leil M. K., Meloche S., Robbins J., Molkentin J. D. (2007) Genetic inhibition of cardiac ERK1/2 promotes stress-induced apoptosis and heart failure but has no effect on hypertrophy in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 14074–14079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lips D. J., Bueno O. F., Wilkins B. J., Purcell N. H., Kaiser R. A., Lorenz J. N., Voisin L., Saba-El-Leil M. K., Meloche S., Pouysségur J., Pagès G., De Windt L. J., Doevendans P. A., Molkentin J. D. (2004) MEK1-ERK2 signaling pathway protects myocardium from ischemic injury in vivo. Circulation 109, 1938–1941 [DOI] [PubMed] [Google Scholar]
- 49. Tachibana H., Naga Prasad S. V., Lefkowitz R. J., Koch W. J., Rockman H. A. (2005) Level of β-adrenergic receptor kinase 1 inhibition determines degree of cardiac dysfunction after chronic pressure overload-induced heart failure. Circulation 111, 591–597 [DOI] [PubMed] [Google Scholar]
- 50. Esposito G., Prasad S. V., Rapacciuolo A., Mao L., Koch W. J., Rockman H. A. (2001) Cardiac overexpression of a Gq inhibitor blocks induction of extracellular signal-regulated kinase and c-Jun NH2-terminal kinase activity in in vivo pressure overload. Circulation 103, 1453–1458 [DOI] [PubMed] [Google Scholar]
- 51. Brinks H., Boucher M., Gao E., Chuprun J. K., Pesant S., Raake P. W., Huang Z. M., Wang X., Qiu G., Gumpert A., Harris D. M., Eckhart A. D., Most P., Koch W. J. (2010) Level of G protein-coupled receptor kinase-2 determines myocardial ischemia/reperfusion injury via pro- and anti-apoptotic mechanisms. Circ. Res. 107, 1140–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sbroggiò M., Carnevale D., Bertero A., Cifelli G., De Blasio E., Mascio G., Hirsch E., Bahou W. F., Turco E., Silengo L., Brancaccio M., Lembo G., Tarone G. (2011) IQGAP1 regulates ERK1/2 and AKT signaling in the heart and sustains functional remodeling upon pressure overload. Cardiovasc. Res. 91, 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]