Abstract
While common in viral infections and neoplasia, spontaneous cell-cell fusion, or syncytialization, is quite restricted in healthy tissues. Such fusion is essential to human placental development, where interactions between trophoblast-specific human endogenous retroviral (HERV) envelope proteins, called syncytins, and their widely-distributed cell surface receptors are centrally involved. We have identified the first host cell-encoded protein that inhibits cell fusion in mammals. Like the syncytins, this protein, called suppressyn, is HERV-derived, placenta-specific and well-conserved over simian evolution. In vitro, suppressyn binds to the syn1 receptor and inhibits syn1-, but not syn2-mediated trophoblast syncytialization. Suppressyn knock-down promotes cell-cell fusion in trophoblast cells and cell-associated and secreted suppressyn binds to the syn1 receptor, ASCT2. Identification of the first host cell-encoded inhibitor of mammalian cell fusion may encourage improved understanding of cell fusion mechanisms, of placental morphogenesis and of diseases resulting from abnormal cell fusion.
Although virtually pathognomonic of viral infections and neoplasia, spontaneous cell-cell fusion, or syncytialization, is quite restricted in healthy tissues. Normally seen in muscle1 and in bone2, such fusion is best described in the placenta. In this organ, interactions between the human endogenous retroviral (HERV) envelope proteins, syncytin-1 (syn1)3,4 and syncytin-2 (syn2)5, and their receptors, the widely-distributed neutral amino acid transporter, ASCT-26, and the placenta-specific carbohydrate transporter, Major Facilitator Superfamily Domain Containing-2 (MFSD2)7, respectively, play key roles. We have recently identified the first reported host cell-encoded protein, named suppressyn, that inhibits cell fusion in mammals. The protein is derived from HERV-Fb1, it is transcribed only in the placenta in vivo and highly homologous gene and protein sequences can be traced to evolutionarily distant simian representatives. Inhibition of trophoblast cell fusion is syn1-specific. Implication of syn1 involvement in the pathogenesis of diseases of abnormal placental development and in neoplasia and of ASCT-2 involvement in viral transmission suggest that the discovery of a novel, syn1-specific inhibitor of cell fusion that binds to ASCT-2 may advance our understanding of cell fusion itself and of the role of these proteins in human health and disease.
Results
Amino acid sequence, genomic structure and expression of HERV-Fb1
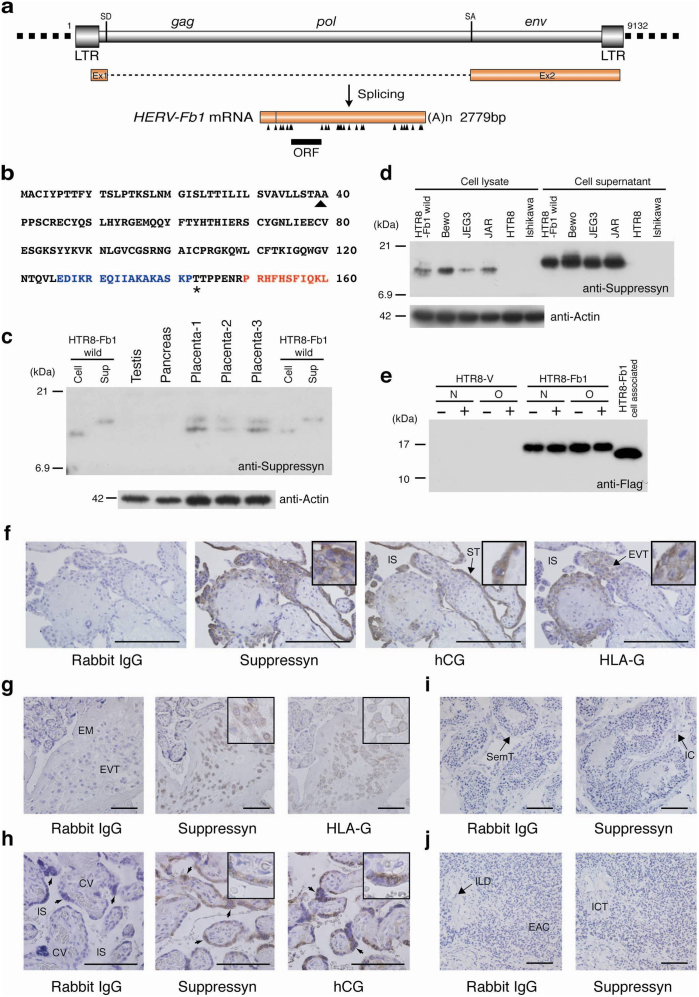
The suppressyn coding sequence is derived from the env-coding region of a HERV-F (type b) family member: HERV-Fb1 (Fb1) (Fig. 1a). Fb1 transcripts can be detected in placental tissues from early and late gestation8,9 and its protein product is a 160 aa polypeptide (Fig. 1b). The predicted 18 kDa HERV-Fb1 translation product is part of the surface subunit (SU) and contains a putative signal sequence (39aa) and a premature stop codon that truncates the protein product prior to the SU-transmembrane (TM) cleavage site. The TM subunit of other HERV envelope genes commonly contains an immunosuppressive domain (ISD)10; neither the TM nor the ISD present in suppressyn. The translation product contains no predicted N-linked glycosylation sites and a single O-linked site (Fig 1b). All of these characteristics suggest that suppressyn may function differently from other HERV envelope-derived proteins. Typical of endogenous retroviral proteins, the Fb1 sequence is found in several locations in the human genome, although its full length coding sequence is present only in a reverse orientation on chromosome 21q22.3. The Fb1 DNA and suppressyn amino acid sequences are highly conserved through simian evolution (Supplementary Fig. S1a online and unpublished observations) suggesting Fb1 is not a pseudogene. Suppressyn protein was identified by immunoprecipitation-western immunoblotting or direct immunoblotting using a combination of polyclonal and monoclonal anti-suppressyn antibodies in human placental samples (Fig. 1c) and in trophoblast cell lines (Fig. 1d, Supplementary Fig. S2 online). Both methods identify cell-associated and secreted forms of the protein. In cell lysates, suppressyn was identified at a molecular mass of 14 kDa, identical to that of recombinant suppressyn and consistent with that predicted after truncation of its putative signal peptide. Suppressyn in cell supernatants had an unexpected size of approximately 15–16 kDa. Suppressyn contains no predicted N-glycosylation sites, (Fig. 1b) and exhibits no change in migration after O-glycosidase treatment (Fig. 1e), suggesting the size difference between cell-associated and secreted suppressyn may result from other types of O-linked glycosylation or alternate protein modifications. A polyclonal antibody generated against a suppressyn-specific C-terminal 17-mer (Fig. 1b and Supplementary Fig. S2 online) detected intracellular expression of suppressyn in extravillous trophoblast (EVT) cells isolated from first and third trimester human placental samples, mirroring the expression of the EVT-specific non-classical MHC product, HLA-G11,12 (Figs. 1f, g). Additional detection of suppressyn in the human chorionic gonadotropin (hCG) positive, syncytiotrophoblast layer of placental floating villi (Figs. 1f, h) is consistent with RNA expression patterns previously observed through in situ hybridization8. Although syn1 is typically detected at the cell surface, the cell-type expression patterns of suppressyn mimic those of syn1 protein in human placenta13,14.
Figure 1. Human suppressyn structure, coding sequence and protein expression.
(a) Splicing pattern of the HERV-Fb1 gene and its transcriptional product. Arrowheads, stop codons; LTR, long terminal repeat; SD, splice donor; SA, splice acceptor. (b) Primary human suppressyn sequence. Arrowhead, predicted signal sequence cleavage site; Asterisk, predicted O-glycosylation site. The peptide sequence used to raise the polyclonal antibody is indicated in blue, that for the monoclonal in red. (c) Northern analysis has demonstrated placental specificity for Fb19; here western analysis using a monoclonal antibody against suppressyn confirms protein expression and demonstrates cell-associated and secreted (sup) forms in cell lines and in human tissues. Lanes 5–7: three independent human placental samples from term gestations; Lane 1 and 8, 2 and 9: cell-associated and secreted suppressyn from HTR8 cells stably-transfected with a suppressyn-expressing vector (positive controls). (d) Suppressyn protein expression in cell lines. Positive controls as in (c). (e) Soluble, tagged suppressyn was purified over Flag-M2 agarose and treated with or without N- and O-glycosidase. No differences in migration were seen between treated and untreated preparations. (f) Immunohistochemical detection of suppressyn using a polyclonal antibody demonstrates suppressyn localization to hCG-expressing syncytiotrophoblast (ST) and to the HLA-G positive extravillous trophoblast (EVT) in early gestation placenta. All negative tissue controls use rabbit IgG as primary antibody; HLA-G detection in EVT used a monoclonal anti-human MEMG/1 antibody; hCG detection in syncytiotrophoblast used a polyclonal antibody against hCG. (g–j) Localization of suppressyn protein in human third trimester placental tissues using a polyclonal antibody; (g) extravillous trophoblast and (h) syncytiotrophoblast. Negative control tissues exposed to the polyclonal anti-suppressyn antibody are depicted in (i) testis and (j) pancreas. EVT, extravillous trophoblast; EM, extracellular matrix; IS, intervillous space; CV, chorionic villi (fetal tissue); syncytiotrophoblast (arrow heads); EAC, exocrine acinar cells; ICT, interlobular connective tissue; ILD, interlobular duct; IC, interstitial cells; SemT, seminiferous tubule. Magnifications of positively–stained sections in the upper right corners of select images are to aid in discrimination of subcellular localization. Scale bar indicates 100 μm.
Fb1 gene knock-down induces cell fusion
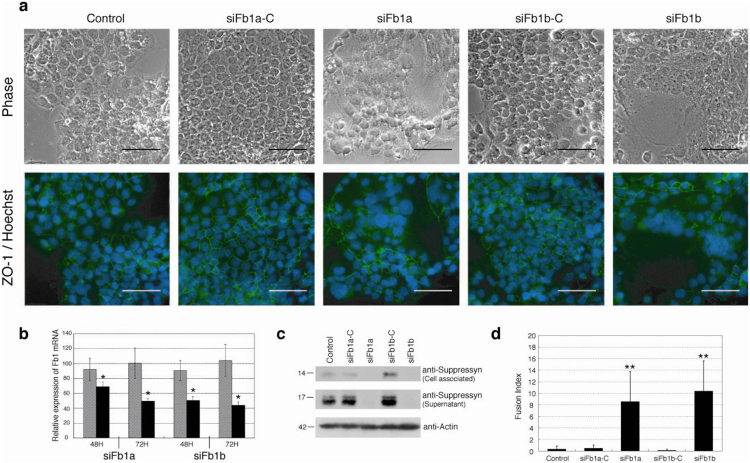
To identify a potential function for this novel HERV-derived human placental protein, we used siRNA knock-down and the BeWo cell line. Suppressyn-specific mRNA and protein knock-down (50–60% mRNA knock-down; Figs. 2b, c) in BeWo cells using distinct siRNA (siFb1a and siFb1b) or their respective scrambled siRNA controls induced a significant increase in cell fusion when compared to parental cells or to those treated with control siRNA (Figs. 2a, d). hCG transcripts and secreted protein were increased 2.5-fold (mRNA) and 1.5-fold (protein) 72 hours after suppressyn knock-down (Supplementary Fig. S3a, b online). Human placental lactogen (hPL) mRNA expression and progesterone secretion were likewise increased at 72 hours; estriol was undetectable at all time points (Supplementary Fig. S3c, d online). These data show that, in addition to inhibiting syn1-mediated fusion, suppressyn may also promote differentiation of BeWo cells. Trophoblast fusion and trophoblast differentiation have been shown in cell lines (BeWo) and in ex vivo models to be linked, but distinct processes15,16. Inhibition of fusion using Fb1 siRNAs was also visualized immunocytochemically using antibodies against the intercellular tight junction protein, zona occludens-1 (ZO-1, Fig. 2a, lower panels). Unlike most human trophoblast cell lines, BeWo cells syncytialize poorly when grown under standard culture conditions but differentiate into multinucleate, syncytiotrophoblast-like cells and secrete the syncytiotrophoblast cell differentiation marker, hCG, in response to forskolin17. Although some controversy remains, this secretory product may help to distinguish these cells from trophoblast giant cells11,18,19,20. BeWo cells spontaneously translate cell-associated and soluble suppressyn (Fig. 1d) and cell-associated syn1 (Supplementary Fig. S2d online). Although forskolin exposure increases suppressyn transcription in BeWo cells, it also decreases ASCT2 levels (data not shown). Still, suppressyn knock-down promotes cell fusion in the absence of forskolin. We therefore hypothesize that endogenous suppressyn exerts tonic control of syn1-mediated BeWo cell fusion.
Figure 2. Fb1 knock-down increases cell fusion in syn1 expressing cells.
Fb1-specific mRNA knock-down in BeWo choriocarcinoma cells using distinct siRNA (siFb1a and siFb1b) or their respective scrambled siRNA controls. Fb1 knock down induced a significant increase in cell fusion (a, d). (a) ZO-1-FITC and Hoechst 33342 immunocytochemistry was performed 72 hours after siRNA exposure and images are depicted in the lower row. The upper row shows matched, phase-contrast images. Scale bar represents 100 μm. Quantitative RT-PCR, immunoprecipitation and immunoblotting confirm decreases in (b) Fb1 mRNA (50–60% knock-down) and (c) protein (cell-associated and secreted) in siRNA treated Bewo cells. Expression (b) is normalized to unexposed samples cultured for similar time periods (48 or 72H). (d) Mean fusion indices for siRNA-exposed and control cells (error bars represent standard deviations; n = 4 × 5 fields). Cell fusion was assessed using phase contrast microscopy and quantitated using cell fusion indices. All observations were performed at a final magnification of 200× and total nuclei were counted per field using Leica MetaMorph image analyzing software. The number of fused syncytial aggregates and the number of nuclei in each aggregate was counted manually and fusion indices were defined as [(N-S)/T] × 100. N is the number of nuclei in syncytia, S is the number of syncytia, and T is the total number of nuclei counted. The fusion index quantitates the percentage of fusion events in a cell population. Black bars- Fb1 siRNA exposed; hatched bars-control siRNA exposed. Data in (b, d) are representative of three independent experiments performed in duplicate. *p < 0.05 and **p < 0.01 compared to matched siRNA control. Statistical comparisons used Kruskal-Wallis and Mann-Whitney U-testing with (d) or without (b) Bonferroni corrections.
Suppressyn inhibits syn1-induced cell fusion and binds to the syn1 receptor, ASCT2
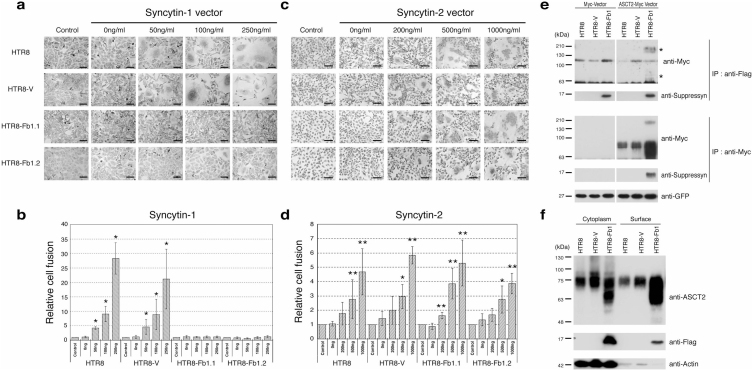
A trophoblast cell line, HTR8/SVneo21, that does not spontaneously syncytialize and exhibits low to undetectable endogenous expression of suppressyn and syn1 (Fig. 1d, Supplementary Fig. S2 online) was stably transfected with a vector driving Fb1 expression (HTR8-Fb1). Control cells and two distinct HTR8-Fb1 clones (HTR8-Fb1.1 and HTR8-Fb1.2) were then transiently-transfected with vectors driving the expression of syn1 or syn2 and analyzed for cell size (FSC) and granularity (SSC) using flow cytometry. The presence of large, multinucleated (more granular) syncytialized cells increased in direct proportion to the amount of transfected syn1 and syn2 in HTR8 (parent) and HTR8-V (vector only control) cells and continuous expression of suppressyn in stably-transfected HTR8-Fb1.1 and HTR8-Fb1.2 cells abrogated syn1- but not syn2-induced cell fusion (Figs. 3a–d). Transient expression of syncytins in transfected cells was verified using western immunoblotting, excluding the possibility of inter-vector inhibition of transcription in doubly-transfected cells, supporting the concept that suppressyn modulates trophoblast fusion, and suggesting that suppressyn does not inhibit syn1 production or maturation22 (Supplementary Fig. S4a, b online).
Figure 3. Suppressyn inhibits syn1- but not syn2-induced trophoblast cell fusion and suppressyn binds to the syn1 receptor ASCT2.
HTR8 trophoblast cells were stably transfected with vectors driving the expression of Fb1 (HTR8-Fb1.1 and 1.2) or vector alone (HTR-V). Cells were then transiently transfected with increasing amounts of a vector driving expression of syn1 (a) or syn2 (c), counterstained with hematoxylin and analyzed using phase contrast microscopy. To calculate fusion indices, cells were resuspended and analyzed by flow cytometry (b and d). Relative cell fusion was defined as the number of fused syn1- or syn2-transfected cells (gated by flow cytometry) divided by the number of fused cells in parent (HTR8) and in stable vector- or Fb1-transfected (HTR8-V, HTR8-Fb1.1, HTR8-Fb1.2), but syncytin non-transfected, cells cultured under identical conditions (indicated as controls for each cell line, *p < 0.05 **p < 0.01 when compared to non-transfected controls). Statistical comparisons were made using Kruskal-Wallis and Mann-Whitney U-testing with Bonferroni corrections. Data in (b) and (d) are representative of three independent experiments performed in duplicate. (e) HTR8 parental cells were stably transfected with vectors driving expression of flag-tagged Fb1 and then transiently transfected with a myc-vector or with a vector driving the expression of myc-tagged ASCT2. For the transfection internal control, the pAcGFP1-C1 vector was co-transfected with the myc tag vector and GFP protein expression quantitated using a monoclonal anti-GFP antibody. Lysates were immunoprecipitated with an anti-myc tag antibody and protein-G agarose or anti-flag-M2 agarose. Immunoprecipitants were separated by PAGE and immunoblotted with a myc-specific antibody. *Co-precipitated high and low molecular weight ASCT2. (f) HTR8, HTR8-V and HTR8-Fb1 cells were surface biotinylated and proteins purified using the Pierce cell surface protein isolation kit. Equal amounts of cytoplasmic and biotinylated surface protein were subsequently analyzed with standard SDS-PAGE and immune detection. Data in (e) and (f) are representative of three independent experiments.
To assure we were not documenting endoreduplication, a common characteristic of end-differentiated EVT, particularly trophoblast giant cells23,24,25,26, we transfected mixed cultures of equal numbers of CSFE (green)- and Cell Vue Claret (red)-stained HTR8, HTR8-V, HTR8-Fb1.1 and HTR8-Fb1.2 with a vector driving syn1 expression (Supplementary Fig. S4c online). Non-fused cells in all cultures continued to display distinct red or green fluorescence at 72 hours post-transfection; fused cells in the HTR8 and HTR8-V (Fb1-negative) controls displayed a mixture of green and red fluorescence, indicating fusion rather than endoreduplication.
Although suppressyn does not contain the SDGGGX2DX2R conserved motif (Supplementary Fig. S1b online) thought to be essential for syn1/hASCT-2 interactions27 nor the heptad repeat sequences that characterize previously described synthetic inhibitors of syn1-mediated fusion28, syn1 inhibition is still most likely to occur at the level of the receptor. Immunoprecipitation of suppressyn co-preciptiates ASCT2 and immunoprecipitation of ASCT-2 co-precipitates suppressyn in doubly transfected cells (Fig. 3e), indicating direct binding of suppressyn to ASCT-2. The high molecular weight form of ASCT bound by Fb1 is likely a glycosylated multimer29. Purification of surface biotinylated proteins from HTR8, HTR8-V and HTR8-Fb1 revealed ASCT-2 on the cell surface of all tested cells, as expected29, and surface-expressed suppressyn only on HTR-Fb1 cells (Fig. 3f). Since suppressyn lacks a transmembrane domain, the detection of this ASCT-2 binding HERV product at the cell surface suggests that it is ligand-bound and promotes a potential mechanism for syn1 antagonism.
Secreted suppressyn binds ASCT2 and inhibits cell fusion
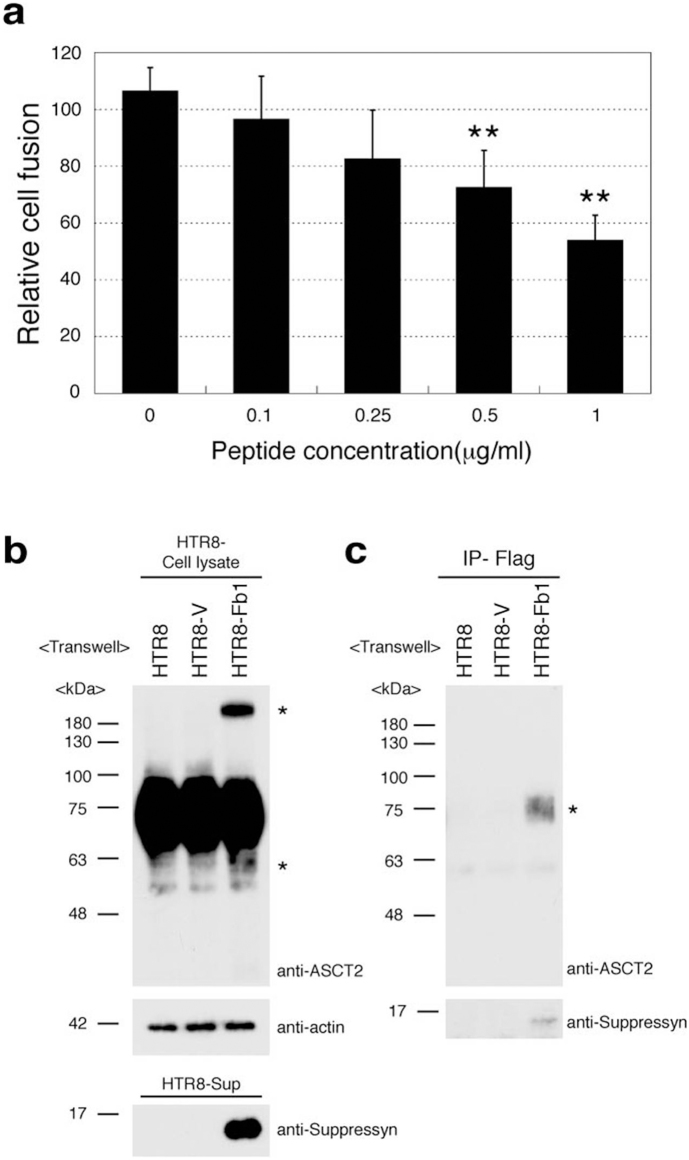
We hypothesized that secreted suppressyn may exert effects on nearby trophoblast cells as well as on the secreting cell itself. To test the former hypothesis, syn1-transfected HTR8 parent cells were exposed for 24 hours in culture to suppressyn protein purified from JEG3-Fb1-flag supernatants and cell fusion was assessed as in Fig. 3b. Exposure to exogenous suppressyn reduced syn1-induced fusion in a dose-dependent fashion, reaching a level of nearly 50% suppression at a suppressyn concentration of 1 μg/ml (Fig. 4a).
Figure 4. Secreted suppressyn binds ASCT2 and inhibits syn1- induced trophoblast cell fusion.
(a) Syn1 transfected HTR8 cells were cultured in the presence of increasing amounts of recombinant secreted suppressyn for 24 hours prior to fusion assessment. Relative cell fusion was assessed by flow cytometry as in Fig 3b but was defined as the number of fused cells (gated by flow cytometry) in suppressyn peptide-exposed cells normalized to the number of fused cells in syn1-transfected HTR8 cells exposed to a control peptide. **p < 0.01 compared to 0 μg/ml sample. Statistical comparisons were made using Kruskal-Wallis and Mann-Whitney U-testing with Bonferroni corrections. Data are representative of three independent experiments performed in duplicate. (b, c) Transwell (0.4 μm; Becton Dickenson 353493) cell culture models (upper chamber; HTR8-V or HTR8-Fb1, lower chamber; HTR8 parent cell) were used to assess the effects of secreted suppressyn on non-transfected ASCT2-expressing target cells. Structural changes (*high and low molecular weight suppressyn forms) were observed in HTR8 cells after exogenous exposure to suppressyn (b). Coprecipitation experiments demonstrate direct binding of ASCT2 and suppressyn (c). Data in (b) and (c) are representative of three independent experiments. *Co-precipitated ASCT2.
Transwell cell culture models (upper chamber: HTR8-V or HTR8-Fb1; lower chamber: Fb1 negative HTR8 parent cells) were used to assess the effects of secreted suppressyn on non-transfected ASCT2-expressing target cells. HTR8 target cells were lysed for protein isolation after 3 days of physically-separated co-culture with Fb1-expressing or control cells. In cultures exposed to exogenous suppressyn, co-precipitation experiments demonstrated direct binding of ASCT2 and suppressyn and bound ASCT2 exhibited the same structural changes (degradation and multimer formation) observed in cells endogenously expressing ASCT2 (Figs. 4b, c). That the secreted and cell-associated suppressyn forms in primary placental tissues and in trophoblast cell lines exert similar effects on ASCT2 and inhibit cell fusion suggests that this novel suppressor of cell fusion may utilize intracellular, autocrine and paracrine pathways.
Discussion
We are aware of no previous reports of a protein encoded by a mammalian genome that specifically inhibits cell fusion. The implications of this finding to human health and disease may be far-reaching. In the placenta, cytotrophoblast progenitor cells syncytialize into a continuous, multinucleated layer of terminally-differentiated fused cells that line the intervillous space; this layer is called the syncytiotrophoblast. The syncytiotrophoblast sheds cell-derived microparticles into the maternal circulation, likely via activation or apoptosis30, but certainly through processes that appear to be tightly regulated31,32. While the exact control mechanisms for syncytiotrophobalst turnover remain undefined, it is clear that syn1 is involved in formation of syncytiotrophoblast and that several diseases of pregnancy, including intrauterine growth retardation (IUGR) and preeclampsia, are characterized by abnormal placental development and/or villous turnover32,33. The possibility of central involvement of a placental inhibitor of syn1-induced syncytialization (i.e., suppressyn) in these disorders is enticing and indicates an important pathway for future investigations. While we have here described the cell fusion effects of suppressyn in syncytiotrophoblast models, syn1 and suppressyn are also detected in invasive EVT. Like abnormal syncytialization, shallow placental invasion is also characteristic of preeclampsia and IUGR, but also of spontaneous pregnancy loss33,34. While the role of syn1 in placental invasion remains incompletely defined, the co-expression of syn1 and suppressyn at this maternal-fetal also promotes intriguing hypotheses. For example, the presence of suppressyn in EVT could be involved in the inhibition of EVT end-differentiation into non-invasive, HLA-G positive, non-secretory trophoblast giant cells20,23,25,35.
In a remarkable example of convergent evolution, several mammalian species, including rodents, lagomorphs, Carnivora, and humans have co-opted endogenous retroviral envelope proteins for use in normal placental development36,37,38,39. Also remarkable is the finding that, despite widely varying sequences and independent acquisition, human syn1 and rabbit syn-Ory1 can use the same neutral amino acid transporter, ASCT-2, as a receptor mediating cell fusion6,38; the receptors for the murine synA and Carnivoral syncytin (syncytin-Car1) have not been identified. Our demonstration of direct binding of suppressyn to ASCT-2 supports an important role for suppressyn in human fusion events and predicts that a search for similar retrovirally-derived fusion-control proteins in other species is likely to be fruitful.
While a recent exciting description of syn1 expression and function in osteoclast fusion2 adds to the very limited evidence for tissue expression of syn1 among healthy organs other than the placenta, in diseased tissues, syn1 has been implicated in neoplastic cell fusion40 and ASCT2 appears to play roles in cancer aggression41 and in viral fusion. ASCT2 has been identified as the receptor for a diverse family of retroviruses, including baboon endogenous retrovirus, avian reticuloendothelial virus, feline endogenous virus (RD114), and type D simian retroviruses29. Syn1 can pseudotype HIV-1 viral cores42, likely through interactions with RD114 (ASCT2). We hypothesize that suppressyn-mediated inhibition of signaling through ASCT2 could help to explain the fairly robust ability of the human placenta to delimit vertical viral transmission, at least for a subset of exogenous pathogens.
Finally, it is intriguing to speculate on the possible function of Fb1 in its ancestral exogenous retrovirus. Several retroviruses have well-described mechanisms by which they downregulate the host cell surface receptors mediating their entry after infection has occurred43,44. Like those that share ASCT2, many other retroviruses share alternate but common host surface receptors that mediate entry by any of the viruses in that particular family. The subset of viruses sharing a given receptor make up an interference group; those that utilize ASCT2 for entry into human cells comprise one of the largest of these groups: the RD114/type D interference group44. In this context, surface receptor downregulation after infection may function to inhibit superinfection by a second virus from the same interference group. This mechanism has been called superinfection resistance or viral interference. The Fb1 protein may have originally been part of a HERV-Fb virus-encoded mechanism to inhibit superinfection by other interference group members, including HERV-W, that shared preference for ASCT2-mediated entry into host cells. A similar role has been described for a murine gene called Fv4 that is derived from a murine leukemia virus (MuLV) Env-like sequence and can function in cellular resistance to MuLV infection45. Like our description of the effects of suppressyn on the syn1 receptor, ASCT2, interference by Fv4 has been hypothesized occur at the level of the MuLV receptor.
Methods
Cell lines and human tissues
The human choriocarcinoma cell lines JEG3, JAR and BeWo were obtained from ATCC. The endometrial Ishikawa cell line was generously supplied by Dr Susan Nagel at The University of Missouri. The human trophoblast cell line, HTR8/SVneo, was kindly provided by Dr. Charles H. Graham, Department of Anatomy & Cell Biology, Queen's University, Toronto, Canada. With the exception of BeWo cells, which were maintained in Kaighn's Modification medium (F-12K, Invitrogen) supplemented with 15% fetal calf serum (FCS), all cells were grown in D-MEM containing 10% fetal bovine serum (Invitrogen). Early and term placental tissues for immunohistochemical analyses were obtained from women at 7–12 weeks of gestation undergoing dilation and evacuation or those undergoing an uncomplicated delivery at 37–41 weeks of gestation. Normal paraffin-embedded human pancreas (T2234188) and testis (T2234260) tissue sections were purchased from Biochain. All human tissue specimens were obtained under University of Missouri or University of the Ryukyus IRB-approved protocols.
Immunohistochemistry and antibodies
The anti-suppressyn polyclonal antibody used in immunohistochemistry, immunoprecipitation and western immunoblotting was generated in rabbits against a synthetic C-terminal KLH-conjugated 17 aa polypeptide (EDIKRAQIIAKAKASKP; Fig. 1b) and purified over a peptide affinity column loaded with the immunizing peptide (Genemed Synthesis, Inc.). The monoclonal antibody was raised against a C-terminal 10-mer (PRHFHSFIQKL; Fig. 1b; AbMart). The commercially available antibodies for immunohistochemistry included: 1) monoclonal anti-FLAG M2 (F3165; Sigma-Aldrich), 2) monoclonal anti-Myc tag (05-724; Millipore) 3) polyclonal anti-human chorionic gonadotropin (AB936; Chemicon), 4) monoclonal anti-human HLA-G (ab52455; Abcam), 5) polyclonal anti-human syncytin-1 (sc-50369; Santa Cruz Biotechnology), 6) monoclonal anti-beta actin (MAB1501; Millipore), 7) monoclonal anti-GFP (04363-24; Nakalai tesque), 8) monoclonal anti-ZO-1-FITC (33-9111; Zymed), 9) polyclonal anti-GAPDH (905-734-100; Assay Designs) 10) monoclonal anti-ASCT2 (#8057S; CST). Human first trimester and term placental tissues, testis and pancreas were paraffin-embedded, sectioned at 5 μm and deparaffinized according to standard procedures. Exposure to primary polyclonal anti-suppressyn (1:300), polyclonal anti-human hCG (1:2500), negative control rabbit IgG (identical concentration, 731642; DAKO), anti-human HLA-G (1:500), and negative control mouse IgG (identical concentration, 015-000-003; Jackson ImmunoResearch Laboratories) antibodies occurred overnight at 4°C. Sections were washed and processed using a Histofine SAB peroxidase kit (424021, 424031; Nichirei) or diaminobenzidine (DAB). Samples were visualized using standard microscopy. Specificity of the primary polyclonal anti-suppressyn antibody used in immunohistochemistry was confirmed using a blocking peptide competition assay (BPCA). Suppressyn polyclonal antibody was incubated with 0- or 500-fold molar excess peptide (EDIKRAQIIAKAKASKP, the sequence used for antibody generation) over night at 4°C; both antibodies were then used for standard immunohistochemistry as described above (Supplementary Fig. S2c online).
Immunoprecipitation and western immunoblot analyses
Whole cell extracts from cell lines or placental tissues were prepared in cell lysis buffer (50 mM Tris HCl pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, supplemented with a protease inhibitor cocktail (11-873-580-001; Roche). Twenty to five hundred micrograms of extract were used for immunoprecipitation with 2 microgram of polyclonal anti-suppressyn antibody or anti-Myc-tag antibody. For purifications, antibodies were captured with 50 μl of protein G-agarose (11-243-233-001; Roche) or 50 μl of anti-FLAG M2 agarose (A-2220; Sigma) according to the manufacturer's recommendations. Captured proteins were separated by standard SDS–PAGE and analyzed by immunoblotting with specific primary antibodies (above) and peroxidase conjugated goat anti-rabbit Ig (#32460; Pierce) or anti-mouse IgG (#32430; Pierce) secondary antibodies. Antibodies were detected using chemoluminescence (ECL Prime, RPN2232; GE Healthcare).
Vectors and constructs
Fb1, syn1, syn2 and ASCT2 were reverse transcribed and amplified using term placental mRNA or RNA from trophoblast cell lines. Further amplification used restriction enzyme site-linked primers and fragments were cloned into the pGEM-T Easy vector (Promega) for sequencing. Clones with 100% sequence identity to c21orf105, syn1 (NM_014590.3), syn2 (BC068585) or ASCT2 (BC000062) were inserted into a pFlag-EF1 vector that replaced the CMV promoter of the pFlag-CMV expression vector (E6908, Sigma Aldrich) with an elongation factor 1 (EF1) promoter. A separate ASCT2 vector replaced its flag tag with a myc tag. Flag tag-fused or -unfused suppressyn fragments were transferred to a mammalian expression vector with a CAG promoter to obtain stable suppressyn-expressing cell lines. Cloning primer sequences and 5′ sequence positions are listed in Supplementary Table S1 online.
siRNA gene knock-down
BeWo cells at 60% confluency were transfected using 1.5 μl of lipofectamine 2000 (Invitrogen) and 20 pmol siRNA in 500 μl of Opti-MEM medium. After 6 hours, medium was changed and incubation continued for 72 hours. siRNA sequences are listed in Supplementary Table S1 online.
Immunocytochemistry
siRNA treated cells were grown on 24 well plates for 72 h. Cells were then fixed in PBS-4% paraformaldehyde. After blocking in PBS-1% BSA, cells were immunostained with a mouse anti-ZO-1-FITC conjugated antibody at 4°C overnight. Cells were washed and counterstained for 5 min at room temperature with Hoechst33342 (0.5 μg/ml). Exposed cells were examined with an EVOSfl (AMF-4302) fluorescence microscope. Matched phase-contrast images were also obtained.
Fusion assays
HTR8 cells were transiently transfected with syn1 or syn2 (as above), incubated for the stated times, trypsinized for 10 min using TrypLE Express (Invitrogen) and collected as a confirmed single cell suspension in PBS. In co-transfection assays, HTR8 cells stably–expressing suppressyn or control vectors were transiently co-transfected with a vector driving syncytin expression using lipofectamine (Invitrogen) prior to trypsin exposure and collection as above. Standard flow cytometry (BD FACSCalibur) generated dot plots of FSC vs. SSC and a gate was created based on untreated HTR8 cells. Fused, syncytialized cells were defined by larger size and increased granularity (more nuclei) by flow cytometry.
Conventional and real-time quantitative PCR
Total RNA was extracted using QIAshredder (79654; Qiagen) and an RNeasy Plus Mini Kit (74134; Qiagen) and single-strand cDNA was synthesized using TaqMan Reverse Transcription Reagents (N8080234; Life Technologies). A Nano drop (Thermo Scientific) analyzer was used to titer mRNA and amplification was performed using the Power Sybr Green PCR Master Mix (4368577; Life Technologies) or conventional ampli Taq polymerase (10342046; Life Technologies). Primers used for quantitative PCR are listed in Supplementary Table S1.
ELISA
hCG, progesterone and estriol were measured using standard ELISA kits (hCG- Calbiotech EKCB10057; progesterone and estriol-ENZO Life Sciences ADI-900-011 and ADI-900-100, respectively).
N-, O-glycosidase treatment
Secreted suppressyn protein was purified from HTR8-V and HTR8-Fb1 cells using Flag-M2 agarose beads. Equal amounts of protein were treated with N-glycosydase (P0704S; New England Biolabs) and O-glycosydase (P0720L; New England Biolabs) according to the manufactures recommendations. Proteins were then analyzed using standard PAGE and western immunoblotting.
Fluorescence activated flow cytometry
Syn1 transfected cells were stained with the polyclonalanti-syncytin-1 antibody (1:50) for 60 min. Cells were washed and exposed to a secondary DyLight™ 488 Donkey anti-rabbit IgG antibody (1:200, #406404; Biolegend) at room temperature for 60 min and then analyzed by flow cytometry (BD FACSCalibur).
Surface Biotinylation
Cell surface biotinylation was performed using a cell surface protein isolation kit (Pierce; 89881).
Statistical analyses
Data are expressed and plotted as means +/- standard deviations. Means were compared using Mann-Whitney U-tests or Kruskal-Wallis testing followed by Mann-Whitney U-tests with Bonferroni corrections as appropriate (described in figure legends). Statistical significance was defined as P < 0.05 or P < 0.01, as indicated.
HERV-Fb1 gene and protein accession number: AB610407
Additional experimental and statistical details can be found online in the supplemental materials.
Author Contributions
Project Design: JS, DS; Experimental (design and performance): JS, MS, DS; Data Interpretation: JS, HB, DS; Manuscript Preparation: JS, YJ, DS.
Supplementary Material
Supplementary Information
Acknowledgments
The authors would like to thank Dr. Linda Schust for her expert editing. We are indebted to Dr. Toshio Nikaido, Department of Regenerative Medicine and Dr. Atsushi Hayashi, Department of Ophthalmology at The University of Toyama for their wise advice. No author on this manuscript declares a conflict of interest.
References
- Abmayr S. M. & Pavlath G. K. Myoblast fusion: lessons from flies and mice. Development 139 (4), 641–656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soe K. et al. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone 48 (4), 837–846 (2011). [DOI] [PubMed] [Google Scholar]
- Frendo J. L. et al. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol Cell Biol 23 (10), 3566–3574 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S. et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403 (6771), 785–789 (2000). [DOI] [PubMed] [Google Scholar]
- Blaise S., de Parseval N., Benit L. & Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A 100 (22), 13013–13018 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond J. L. et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. Journal of virology 74 (7), 3321–3329 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C. et al. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc Natl Acad Sci U S A 105 (45), 17532–17537 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudaka W., Oda T., Jinno Y., Yoshimi N. & Aoki Y. Cellular localization of placenta-specific human endogenous retrovirus (HERV) transcripts and their possible implication in pregnancy-induced hypertension. Placenta 29 (3), 282–289 (2008). [DOI] [PubMed] [Google Scholar]
- Okahara G. et al. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84 (6), 982–990 (2004). [DOI] [PubMed] [Google Scholar]
- Benit L., Dessen P. & Heidmann T. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. Journal of virology 75 (23), 11709–11719 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke Y. W., Burland K. & Butterworth B. Antigen bearing trophoblast in human placental implantation site. in Reproductive Immunology, edited by Clark Croy D. A. B. A. (Elsevier Science Publishers, Amsterdam, 1986), pp. 53–59. [Google Scholar]
- Patel R. N., Quack K. C., Hill J. A. & Schust D. J. Expression of membrane-bound HLA-G at the maternal-fetal interface is not associated with pregnancy maintenance among patients with idiopathic recurrent pregnancy loss. Mol Hum Reprod 9 (9), 551–557 (2003). [DOI] [PubMed] [Google Scholar]
- Malassine A. et al. Expression of HERV-W Env glycoprotein (syncytin) in the extravillous trophoblast of first trimester human placenta. Placenta 26 (7), 556–562 (2005). [DOI] [PubMed] [Google Scholar]
- Muir A., Lever A. M. & Moffett A. Human endogenous retrovirus-W envelope (syncytin) is expressed in both villous and extravillous trophoblast populations. J Gen Virol 87 (Pt 7), 2067–2071 (2006). [DOI] [PubMed] [Google Scholar]
- Huppertz B. & Gauster M. Mechanisms regulating human trophoblast fusion. in Cell Fusions, edited by Larsson L. I. (Springer Science, Dordrecht, 2011), pp. 201–217. [Google Scholar]
- Pidoux G. et al. Review: Human trophoblast fusion and differentiation: lessons from trisomy 21 placenta. Placenta 33 Suppl, S81–86 (2012). [DOI] [PubMed] [Google Scholar]
- Wice B., Menton D., Geuze H. & Schwartz A. L. Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp Cell Res 186 (2), 306–316 (1990). [DOI] [PubMed] [Google Scholar]
- al-Lamki R. S., Skepper J. N. & Burton G. J. Are human placental bed giant cells merely aggregates of small mononuclear trophoblast cells? An ultrastructural and immunocytochemical study. Hum Reprod 14 (2), 496–504 (1999). [DOI] [PubMed] [Google Scholar]
- Hoffman L. H. & Wooding F. B. Giant and binucleate trophoblast cells of mammals. J Exp Zool 266 (6), 559–577 (1993). [DOI] [PubMed] [Google Scholar]
- Quenby S. et al. Antiphospholipid antibodies prevent extravillous trophoblast differentiation. Fertility and sterility 83 (3), 691–698 (2005). [DOI] [PubMed] [Google Scholar]
- Graham C. H. et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res 206 (2), 204–211 (1993). [DOI] [PubMed] [Google Scholar]
- Cheynet V. et al. Synthesis, assembly, and processing of the Env ERVWE1/syncytin human endogenous retroviral envelope. Journal of virology 79 (9), 5585–5593 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B. et al. Invasive depth of extravillous trophoblast correlates with cellular phenotype: a comparison of intra- and extrauterine implantation sites. Histochemistry and cell biology 117 (5), 401–414 (2002). [DOI] [PubMed] [Google Scholar]
- Nakayama H., Scott I. C. & Cross J. C. The transition to endoreduplication in trophoblast giant cells is regulated by the mSNA zinc finger transcription factor. Dev Biol 199 (1), 150–163 (1998). [DOI] [PubMed] [Google Scholar]
- Stanek J. & Biesiada J. Sensitivity and specificity of finding of multinucleate trophoblastic giant cells in decidua in placentas from high-risk pregnancies. Human pathology 43 (2), 261–268 (2012). [DOI] [PubMed] [Google Scholar]
- Zybina T. G., Frank H. G., Biesterfeld S. & Kaufmann P. Genome multiplication of extravillous trophoblast cells in human placenta in the course of differentiation and invasion into endometrium and myometrium. II. Mechanisms of polyploidization. Tsitologiia 46 (7), 640–648 (2004). [PubMed] [Google Scholar]
- Cheynet V., Oriol G. & Mallet F. Identification of the hASCT2-binding domain of the Env ERVWE1/syncytin-1 fusogenic glycoprotein. Retrovirology 3, 41 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Chen P. T., Chang G. D., Huang C. J. & Chen H. Functional characterization of the placental fusogenic membrane protein syncytin. Biology of reproduction 71 (6), 1956–1962 (2004). [DOI] [PubMed] [Google Scholar]
- Tailor C. S., Nouri A., Zhao Y., Takeuchi Y. & Kabat D. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. Journal of virology 73 (5), 4470–4474 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharon A. & Brenner B. Microparticles and pregnancy complications. Thromb Res 127 Suppl 3, S67–71 (2011). [DOI] [PubMed] [Google Scholar]
- Rajakumar A. et al. Transcriptionally active syncytial aggregates in the maternal circulation may contribute to circulating soluble fms-like tyrosine kinase 1 in preeclampsia. Hypertension 59 (2), 256–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman C. W. et al. Review: Does size matter? Placental debris and the pathophysiology of pre-eclampsia. Placenta 33 Suppl, S48–54 (2012). [DOI] [PubMed] [Google Scholar]
- Roberts J. M. & Escudero C. The placenta in preeclampsia. Pregnancy Hypertens 2 (2), 72–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauniaux E., Poston L. & Burton G. J. Placental-related diseases of pregnancy: Involvement of oxidative stress and implications in human evolution. Hum Reprod Update 12 (6), 747–755 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnenborg R., Vercruysse L. & Brosens I. Deep placentation. Best practice & research 25 (3), 273–285 (2011). [DOI] [PubMed] [Google Scholar]
- Cornelis G. et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci U S A 109 (7), E432–441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A. et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A 108 (46), E1164–1173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann O., Vernochet C., Dupressoir A. & Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new "syncytin" in a third order of mammals. Retrovirology 6, 107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernochet C. et al. A syncytin-like endogenous retrovirus envelope gene of the guinea pig specifically expressed in the placenta junctional zone and conserved in Caviomorpha. Placenta 32 (11), 885–892 (2011). [DOI] [PubMed] [Google Scholar]
- Larsson L. I., Bjerregaard B., Wulf-Andersen L. & Talts J. F. Syncytin and cancer cell fusions. ScientificWorldJournal 7, 1193–1197 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte D., Ali N., Carlson N. & Younes M. Overexpression of the neutral amino acid transporter ASCT2 in human colorectal adenocarcinoma. Anticancer Res 22 (5), 2555–2557 (2002). [PubMed] [Google Scholar]
- An D. S., Xie Y. & Chen I. S. Envelope gene of the human endogenous retrovirus HERV-W encodes a functional retrovirus envelope. Journal of virology 75 (7), 3488–3489 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiner K. M., Urban S., Glass B. & Schaller H. Envelope protein-mediated down-regulation of hepatitis B virus receptor in infected hepatocytes. Journal of virology 75 (1), 143–150 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerfelt M. A. & Weiss R. A. Receptor interference groups of 20 retroviruses plating on human cells. Virology 176 (1), 58–69 (1990). [DOI] [PubMed] [Google Scholar]
- Nethe M., Berkhout B. & van der Kuyl A. C. Retroviral superinfection resistance. Retrovirology 2, 52 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information