Abstract
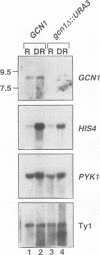
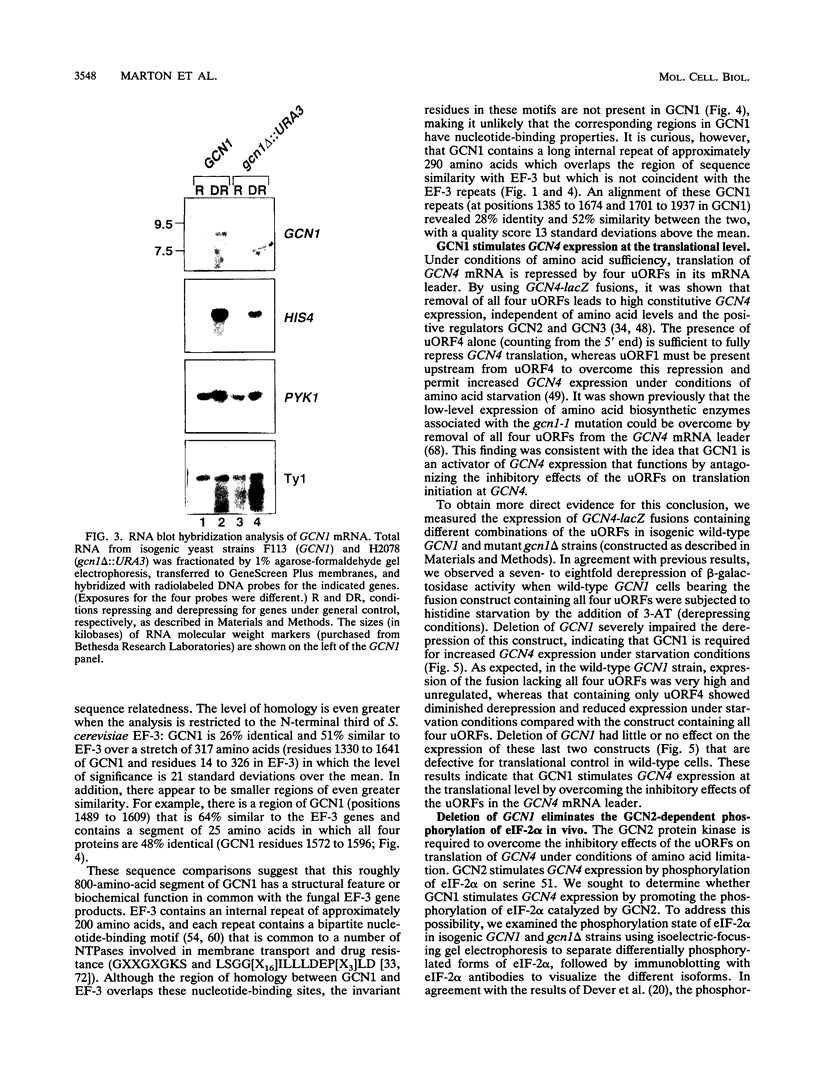
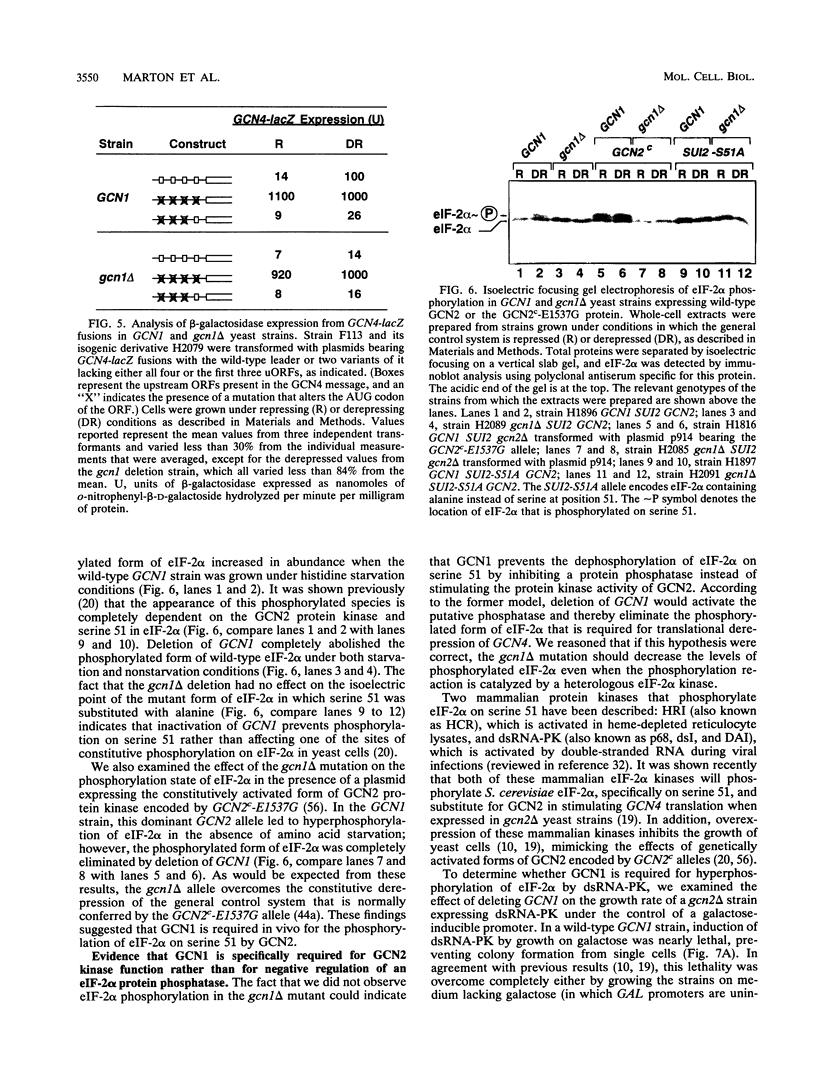
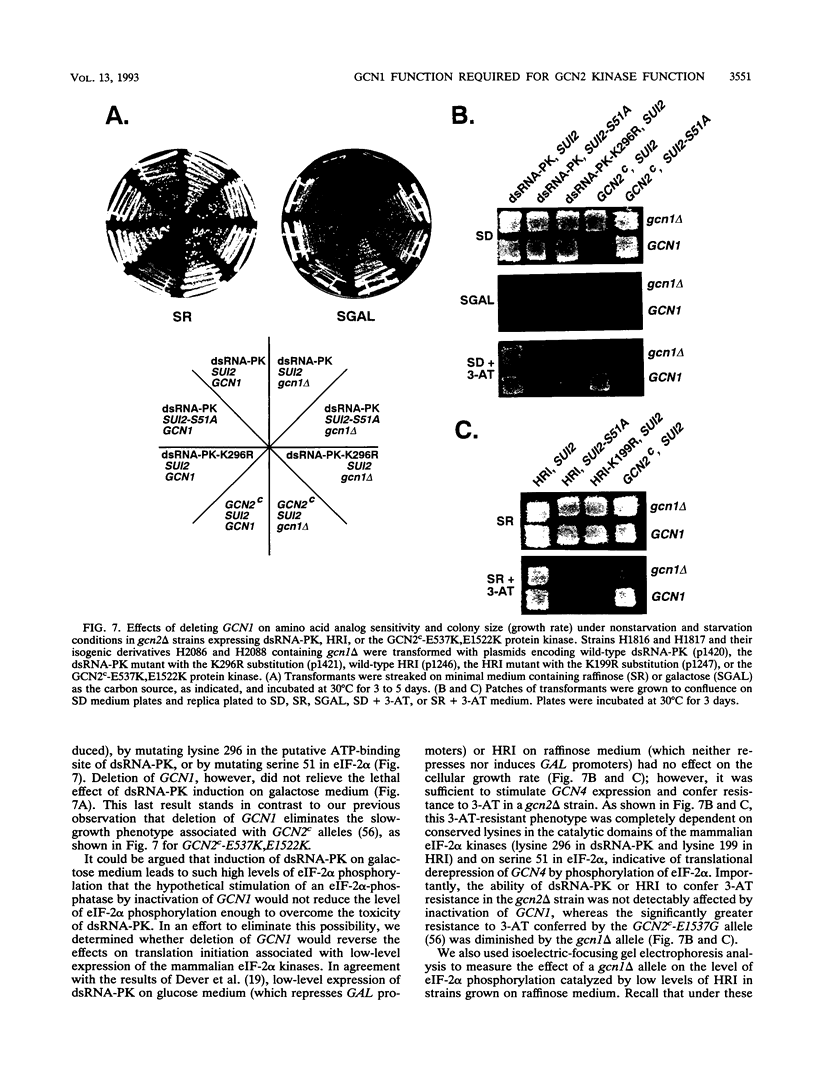
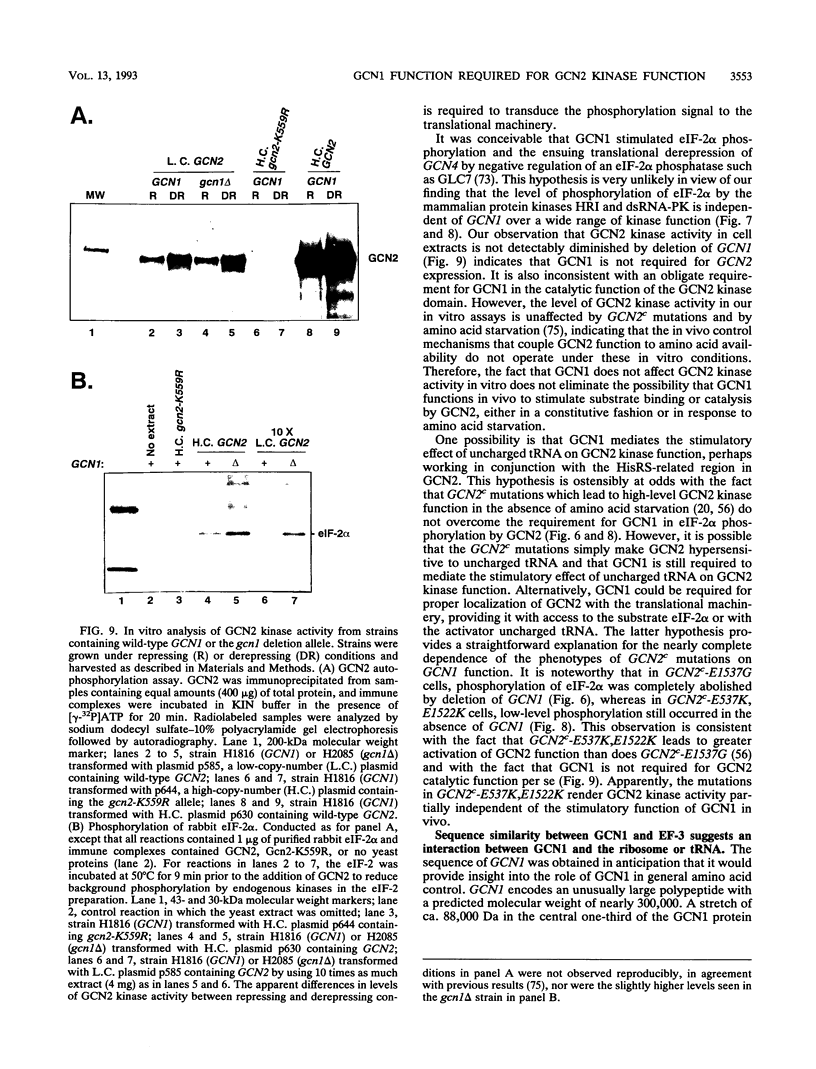
Phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2 alpha) by the protein kinase GCN2 mediates increased translation of the transcriptional activator GCN4 in amino acid-starved yeast cells. We show that this key phosphorylation event and the attendant translational induction of GCN4 are dependent on the product of a previously uncharacterized gene, GCN1. Inactivation of GCN1 did not affect the level of eIF-2 alpha phosphorylation when mammalian eIF-2 alpha kinases were expressed in yeast cells in place of GCN2, arguing against an involvement of GCN1 in dephosphorylation of eIF-2 alpha. In addition, while GCN1 is required in vivo for phosphorylation of eIF-2 alpha by GCN2, cell extracts from gcn1 delta strains contained wild-type levels of GCN2 eIF-2 alpha-kinase activity. On the basis of these results, we propose that GCN1 is not needed for GCN2 kinase activity per se but is required for in vivo activation of GCN2 in response to the starvation signal, uncharged tRNA. GCN1 encodes a protein of 297 kDa with an 88-kDa region that is highly similar in sequence to translation elongation factor 3 identified in several fungal species. This sequence similarity raises the possibility that GCN1 interacts with ribosomes or tRNA molecules and functions in conjunction with GCN2 in monitoring uncharged tRNA levels during the process of translation elongation.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Mar;13(3):1920–1932. doi: 10.1128/mcb.13.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty K., Kamath A. Protein synthesis in yeast. Int J Biochem. 1988;20(6):581–590. doi: 10.1016/0020-711x(88)90096-1. [DOI] [PubMed] [Google Scholar]
- Chang H. W., Watson J. C., Jacobs B. L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong K. L., Feng L., Schappert K., Meurs E., Donahue T. F., Friesen J. D., Hovanessian A. G., Williams B. R. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992 Apr;11(4):1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Foiani M., Hannig E. M., Hinnebusch A. G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991 Jun;11(6):3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Feng L., Donahue T. F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M. J., Galpine A., Austin S. A., Panniers R., Henshaw E. C., Duncan R., Hershey J. W., Pollard J. W. Regulation of polypeptide chain initiation in Chinese hamster ovary cells with a temperature-sensitive leucyl-tRNA synthetase. Changes in phosphorylation of initiation factor eIF-2 and in the activity of the guanine nucleotide exchange factor GEF. J Biol Chem. 1987 Jan 15;262(2):767–771. [PubMed] [Google Scholar]
- Colthurst D. R., Santos M., Grant C. M., Tuite M. F. Candida albicans and three other Candida species contain an elongation factor structurally and functionally analogous to elongation factor 3. FEMS Microbiol Lett. 1991 May 1;64(1):45–49. doi: 10.1016/0378-1097(91)90207-q. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra B., Chakraburtty K. Protein synthesis in yeast. I. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J Biol Chem. 1981 Oct 10;256(19):9999–10004. [PubMed] [Google Scholar]
- Davies M. V., Elroy-Stein O., Jagus R., Moss B., Kaufman R. J. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J Virol. 1992 Apr;66(4):1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992 Feb 7;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domenico B. J., Lupisella J., Sandbaken M., Chakraburtty K. Isolation and sequence analysis of the gene encoding translation elongation factor 3 from Candida albicans. Yeast. 1992 May;8(5):337–352. doi: 10.1002/yea.320080502. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M., Pabich E. K., Valavicius B. C. Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988 Aug 26;54(5):621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Goldman E., Jakubowski H. Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol Microbiol. 1990 Dec;4(12):2035–2040. doi: 10.1111/j.1365-2958.1990.tb00563.x. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Cigan A. M., Freeman B. A., Kinzy T. G. GCD11, a negative regulator of GCN4 expression, encodes the gamma subunit of eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):506–520. doi: 10.1128/mcb.13.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Hinnebusch A. G. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol Cell Biol. 1988 Nov;8(11):4808–4820. doi: 10.1128/mcb.8.11.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990 Nov;126(3):549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J. W. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., Holland I. B., Gray L., Buckel S. D., Bell A. W. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986 Oct 2;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath A., Chakraburtty K. Role of yeast elongation factor 3 in the elongation cycle. J Biol Chem. 1989 Sep 15;264(26):15423–15428. [PubMed] [Google Scholar]
- Katze M. G., Krug R. M. Translational control in influenza virus-infected cells. Enzyme. 1990;44(1-4):265–277. doi: 10.1159/000468764. [DOI] [PubMed] [Google Scholar]
- Kawasaki G., Fraenkel D. G. Cloning of yeast glycolysis genes by complementation. Biochem Biophys Res Commun. 1982 Oct 15;108(3):1107–1122. doi: 10.1016/0006-291x(82)92114-3. [DOI] [PubMed] [Google Scholar]
- Koromilas A. E., Roy S., Barber G. N., Katze M. G., Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992 Sep 18;257(5077):1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- Lee T. G., Tomita J., Hovanessian A. G., Katze M. G. Characterization and regulation of the 58,000-dalton cellular inhibitor of the interferon-induced, dsRNA-activated protein kinase. J Biol Chem. 1992 Jul 15;267(20):14238–14243. [PubMed] [Google Scholar]
- Lee T. G., Tomita J., Hovanessian A. G., Katze M. G. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M. B. Control of translation in adenovirus-infected cells. Enzyme. 1990;44(1-4):250–264. doi: 10.1159/000468763. [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Kagiyama H. Soluble factor requirements for the Tetrahymena peptide elongation system and the ribosomal ATPase as a counterpart of yeast elongation factor 3 (EF-3). J Biochem. 1990 Dec;108(6):1001–1008. doi: 10.1093/oxfordjournals.jbchem.a123298. [DOI] [PubMed] [Google Scholar]
- Moehle C. M., Hinnebusch A. G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991 May;11(5):2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Harashima S., Hinnebusch A. G. A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc Natl Acad Sci U S A. 1987 May;84(9):2863–2867. doi: 10.1073/pnas.84.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Myers K. K., Fonzi W. A., Sypherd P. S. Isolation and sequence analysis of the gene for translation elongation factor 3 from Candida albicans. Nucleic Acids Res. 1992 Apr 11;20(7):1705–1710. doi: 10.1093/nar/20.7.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S. L., Xie A. G., Bonato M. C., McLaughlin C. S. Sequence analysis of the translational elongation factor 3 from Saccharomyces cerevisiae. J Biol Chem. 1990 Feb 5;265(4):1903–1912. [PubMed] [Google Scholar]
- Ramirez M., Wek R. C., Hinnebusch A. G. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M., Wek R. C., Vazquez de Aldana C. R., Jackson B. M., Freeman B., Hinnebusch A. G. Mutations activating the yeast eIF-2 alpha kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol Cell Biol. 1992 Dec;12(12):5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987 Mar 27;48(6):1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Sandbaken M. G., Lupisella J. A., DiDomenico B., Chakraburtty K. Protein synthesis in yeast. Structural and functional analysis of the gene encoding elongation factor 3. J Biol Chem. 1990 Sep 15;265(26):15838–15844. [PubMed] [Google Scholar]
- Sandbaken M., Lupisella J. A., DiDomenico B., Chakraburtty K. Isolation and characterization of the structural gene encoding elongation factor 3. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):230–234. doi: 10.1016/0167-4781(90)90172-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogerson L. Separation and characterization of yeast elongation factors. Methods Enzymol. 1979;60:676–685. doi: 10.1016/s0076-6879(79)60063-0. [DOI] [PubMed] [Google Scholar]
- Skogerson L., Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci U S A. 1976 Jan;73(1):73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritani M., Miyazaki M. Characterization of the ATPase and GTPase activities of elongation factor 3 (EF-3) purified from yeasts. J Biochem. 1988 Mar;103(3):522–530. doi: 10.1093/oxfordjournals.jbchem.a122302. [DOI] [PubMed] [Google Scholar]
- Uritani M., Miyazaki M. Role of yeast peptide elongation factor 3 (EF-3) at the AA-tRNA binding step. J Biochem. 1988 Jul;104(1):118–126. doi: 10.1093/oxfordjournals.jbchem.a122405. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Cannon J. F., Dever T. E., Hinnebusch A. G. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2 alpha kinase GCN2. Mol Cell Biol. 1992 Dec;12(12):5700–5710. doi: 10.1128/mcb.12.12.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Jackson B. M., Hinnebusch A. G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Ramirez M., Jackson B. M., Hinnebusch A. G. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990 Jun;10(6):2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. P., Hinnebusch A. G., Donahue T. F. Mutations in the structural genes for eukaryotic initiation factors 2 alpha and 2 beta of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7515–7519. doi: 10.1073/pnas.86.19.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Fonzi W. A., Sypherd P. S. Fungus-specific translation elongation factor 3 gene present in Pneumocystis carinii. Infect Immun. 1992 Oct;60(10):4140–4145. doi: 10.1128/iai.60.10.4140-4145.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]