Duplications of FOXG1 gene at 14q12 have been reported in patients with infantile spasms and developmental delay of variable severity.1, 2, 3 FOXG1 encodes the forkhead protein G1, a brain-specific transcriptional repressor, regulating corticogenesis in the developing brain and neuronal stem cell self-renewal in the postnatal brain.4 Recently, Amor et al.5 reported on this journal an interstitial duplication of ∼88 kb at 14q12 in a father–son pair with hemifacial microsomia and normal neurocognitive phenotype. The duplication contains only two polypeptide-encoding genes, FOXG1 and C14orf23, suggesting that FOXG1 duplication may be benign or at least incompletely penetrant. That makes the involvement of FOXG1 duplication in the pathogenesis of the neurocognitive impairment and epilepsy controversial. As also discussed by Brunetti-Pierri et al,6 we feel that this statement needs special caution.
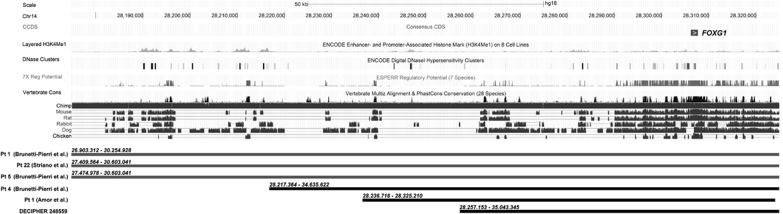
Functional consequences of chromosomal microduplication and microdeletion rely on the final gene dosage, which is strongly influenced by the location of the breakpoint. In this context, the understanding of the contribution of regulatory sequences in gene transcription is critical to understand the relationship between CNVs and human diseases. With this purpose, the Encyclopedia of DNA Elements (ENCODE) project has recently performed a systematic analysis of transcriptional regulation in different human cell lines, providing new understanding about transcription start sites, including their relationship with specific regulatory sequences and histone modification and features of chromatin accessibility.7, 8 Interestingly, analysis of histone modifications from the ENCODE project revealed the presence of a putative regulatory element upstream FOXG1 gene between 28 188 and 28 217 kb (UCSC genome browser, NCBI Build 36/hg18) (Figure 1). This conserved region localizes about 130 kb upstream FOXG1 gene and contains histone modifications typical of enhancers of gene transcription (eg, histone H3 and Lysine 4 monomethylation) in eight different human cells lines. Analysis of regulatory potential scores, comparing frequencies of short alignment patterns between known regulatory elements and neutral DNA,9 also disclose two additional putative elements typical of cis-regulatory modules within this region (Figure 1). Moreover, it contains a DNaseI hypersensitive site (DHS). DHSs reflect genomic regions thought to be enriched for regulatory information and many DHSs reside at or near transcription start site. Notably, no other polypeptide-encoding genes or non-coding RNAs and pseudogenes are present in the region, suggesting that this regulatory element might regulate FOXG1 transcription. Analysis of duplication breakpoints previously reported on 14q12 revealed that duplications associated with an epileptic phenotype localizes uniquely upstream this regulatory element, whereas downstream duplications were identified only in the cases without seizures (Figure 1). On the basis of this finding, we suggest that FOXG1 duplication including this putative regulatory region allows the efficient transcription of the supernumerary copy of FOXG1 gene, resulting in an effective increase in FOXG1 expression and, thereby, in brain hyperexcitability. In contrast, duplications starting downstream this putative regulatory site do not allow efficient transcription of FOXG1, which may underlie the lack of neurological phenotype in the case reported by Amor et al5.
Figure 1.
UCSC genome browser (NCBI36/hg18) schematic view of reported 14q12 duplications encompassing FOXG1 gene. Histone modifications from the ENCODE project7 in eight cells lines (GM12878, H1-hESC, HMEC, HSMM, HUVEC, K562, NHEK and NHLF) indicate the presence of regulatory elements upstream FOXG1 gene. Monomethylation of histone 3 at lysine1 (H3K4me1) is often found near regulatory elements, whereas the presence of DNaseI hypersensitive clusters reflect genomic regions containing regulatory informations. Potential regulatory elements identified with the ESPERR (Evolutionary and Sequence Pattern Extraction through Reduced Representations) computational method and evolutionary conservation are also shown. Grey bars represent duplications associated with an epileptic phenotype; black bars represent duplication identified in non-epileptic patients.
Even if the functional relevance of this putative long-range regulatory element on FOXG1 transcription deserves to be experimentally verified, it provides an interesting clue to dissect genotype–phenotype correlation in FOXG1 microduplication and to uncover the real actual contribution of FOXG1 in the neurodevelopmental phenotype associated with 14q12 duplication.
Notably, chromosome rearrangements disrupting or displacing putative cis-regulatory elements distal to FOXG1 gene in patients with severe cognitive disabilities has been also reported,10, 11 pointing out the relevance of regulatory sequences in the expression of FOXG1 gene.
The authors declare no conflict of interest.
References
- Yeung A, Bruno D, Scheffer IE, et al. 4.45 Mb microduplication in chromosome band 14q12 including FOXG1 in a girl with refractory epilepsy and intellectual impairment. Eur J Med Genet. 2009;52:440–442. doi: 10.1016/j.ejmg.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Striano P, Paravidino R, Sicca F, et al. West syndrome associated with 14q12 duplications harboring FOXG1. Neurology. 2011;76:1600–1602. doi: 10.1212/WNL.0b013e3182194bbf. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Paciorkowski AR, Ciccone R, et al. Duplications of FOXG1 in 14q12 are associated with developmental epilepsy, mental retardation, and severe speech impairment. Eur J Hum Genet. 2011;19:102–107. doi: 10.1038/ejhg.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Pivetta C, Granzotto M, et al. Emx2 and Foxg1 inhibit gliogenesis and promote neuronogenesis. Stem Cells. 2010;28:1206–1218. doi: 10.1002/stem.443. [DOI] [PubMed] [Google Scholar]
- Amor DJ, Burgess T, Tan TY, et al. Questionable pathogenicity of FOXG1 duplication. Eur J Hum Genet. 2012;20:595–596. doi: 10.1038/ejhg.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Cheung SW, Stankiewicz P. Reply to Amor et al. Eur J Hum Genet. 2012;20:597. [Google Scholar]
- ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Kolbe D, Taylor J, Elnitski L, et al. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 2004;14:700–707. doi: 10.1101/gr.1976004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet SA, Kunde SA, Viertel P, et al. Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations and microcephaly. Hum Genet. 2005;117:536–544. doi: 10.1007/s00439-005-1310-3. [DOI] [PubMed] [Google Scholar]
- Kortüm F, Das S, Flindt M, et al. The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet. 2011;48:396–406. doi: 10.1136/jmg.2010.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]