Abstract
Background
Lying in a shallow continental shelf cyclically affected by oscillating sea levels since the Miocene, the Seychelles islands are particularly interesting for evolutionary studies. Recent molecular studies are generating an emerging picture of the origin of its biota, yet very little is known regarding their phylogeographic structure or on the factors promoting diversification within the archipelago. Here we aimed to obtain a detailed depiction of the genetic structure and evolution of one of the most widespread vertebrate groups in the archipelago: the day-geckos of the genus Phelsuma. In parallel, we aimed to infer divergence times between species and subspecies, testing a long-standing hypothesis that argues for different time since sympatry between species as the cause of their different morphological differentiation across the archipelago.
Results
Molecular data corroborated the existence of two main lineages, corresponding to the two currently recognized species. Divergences between species likely date back to the Mio-Pliocene, while more recent, Pleistocenic, divergences are suggested within each species. Populations from outer islands share mtDNA haplotypes with inner island populations, suggesting very recent dispersals (or introductions). We found no evidence of current gene flow between species, but results pointed to the possibility of gene flow between (now allopatric) subspecies. Time estimates suggest a synchronous divergence within each species (between island groups).
Conclusions
The geographic patterns of genetic variation agree with previous taxonomic subdivisions within each species and the origin of outer islands populations is clearly tracked. The similar intraspecific divergence time estimates obtained suggest that the differential body-size differentiation between species within each group of islands may be driven by factors other than character displacement proportional to time since sympatry, as previously suggested. These factors could include different habitats/resources available within each island group, niche differentiation and/or character displacement. We also bring again into consideration the hypothesis of body size being influenced by the distribution of native vegetation and social systems within this group, although it remains to be tested. Our results highlight not only the necessity of clarifying the role of ecology and interspecific interactions in this group’s morphological diversification and community assemblage, but also the importance of co-evolutionary mechanisms and their importance for appropriate conservation of island biodiversity. Further, we provide a detailed description of the phylogeographic structure of these taxa across these islands, which still remain poorly characterized in this respect.
Keywords: Phelsuma, Seychelles, Phylogeography, Species-trees, Diversification, Morphological evolution, Character displacement, Biogeography
Background
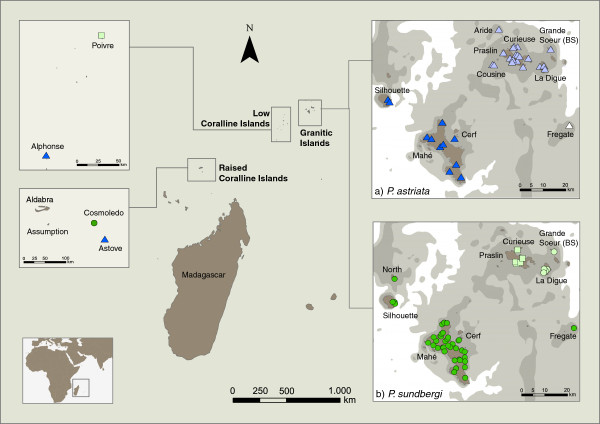
The Western Indian Ocean islands of Madagascar, Comoros, the Mascarenes, and the Seychelles are one of the 34 “hotspots” defined by Conservation International [1]. While there is an emerging picture from molecular data about the origin of their biota (see [2-7] for recent reviews), diversification mechanisms within each archipelago and island are less understood. That is particularly true for the Seychelles, where only a handful of studies have explored the genetic structure of different species within the archipelago [8-13]. The separation of the Seychelles and the Indian subcontinent is estimated to have been completed 65 Mya and the Seychelles include about 155 islands that can be divided in 3 groups: granitic, low and raised coralline islands (Figure 1). The granitics are a group of about 40, clustered together on the undersea shelf of granite, that is the Seychelles bank. These comprise the islands of North, Silhouette, Mahé, Fregate, Praslin, La Digue, Curieuse and several smaller ones encircling them. The low coralline islands are very recent, probably less than 6,000 years ago, and formed from marine sediments cemented with guano deposits [14]. The raised coralline islands (Aldabra, Assumption, Astove and Cosmoledo) are also oceanic, formed by reef-building corals acting on submerged volcanic seamounts that may have formed some 20 million years ago (Mya) [15] and were submerged and re-emerged several times since their formation.
Figure 1.
Sampling sites for Phelsuma species along the granitic and coralline Seychelles islands. Species and subspecies are coded using different symbols: dark blue triangles - P. astriata astriata; light blue triangles - P. astriata semicarinata; white triangle - P. astriata (Fregate population); dark green circles - P. sundbergi longinsulae; light green squares - P. sundbergi sundbergi; light green circles - P. sundbergi ladiguensis. Within the granitic islands (insets a and b), different shadings show areas that would be emerged at −30 m (dark grey) and −50 m (light grey) below present sea-level stands. Low-stands below 60 m would allow contact between all islands in this granitic group. Current data on Indian Ocean sea level changes support low-stands of up to 145 ± 5 m below present sea level (bpsl) at six episodes in the last 0.5 Myr, some of which persisted for up to 50,000 years at a time see [6] and geological records going back further suggest low-stands of 80–120 m bpsl at 0.64, 0.88, 1.04, 1.26 and 1.54 Mya, and eleven additional low-stand episodes of 50 m or more bpsl during the last 5 Myr [18]. Detailed locations and accession numbers are given in Additional file 1.
Sea-level oscillations occurred throughout the Pleistocene and back through the Miocene, with low stands of up to 145 m below present sea-level (bpsl) [16-18]. All the islands in the plateau would have been united with sea-levels of roughly 60 m bpsl, but even less pronounced minima greatly increased the size of each island and their connectivity (Figure 1a, b). It is probable that these cycles of allopatry and contact between islands shaped distribution patterns, organismal divergence and diversification within the archipelago. Nevertheless, intra-archipelago genetic structure of Seychellois taxa is poorly known, as well as the relative influence of isolating vs. connecting periods on species diversification. As in other archipelagos cyclically isolated and connected, cryptic diversity might be higher than currently appreciated [19,20].
Historically, Mahé and surrounding islands, plus Silhouette and North have been considered to form a biogeographic unit separated from the Praslin-La Digue (and surrounding islands) group. Fregate, located roughly in the middle of the two groups of islands, is generally regarded as an isolated or intermediate unit [8,21,22]. Distribution data for insects [21], molluscs [23], and a few studies of intraspecific variation within amphibians [24] and reptiles [8,25] are consistent with this pattern. However, this does not seem to stand for some other groups [9,26]. Most of the few existing studies on intraspecific genetic variation suggest deep and cryptic divergences among the granitic Seychelles [12,13,27,28], highlighting the need for thorough sampling and phylogeographic analyses across these islands. At the same time, it becomes important to understand also the relative influence of historical and ecological factors in the generation and maintenance of such diversity. Day-geckos of the genus Phelsuma are ideal in this respect, as its two Seychellois species are widespread across most of the islands, with both sympatric and allopatric populations. Phelsuma mostly diversified in Madagascar, from where they colonized the surrounding archipelagos and often speciated (reviewed in [28]). Within each island clade, Phelsuma species diverged ecologically and morphologically, evolving a wide variety of colour patterns and sizes and occupying a broad range of arboreal habitats (see [29] and references therein). However the relative contribution of geographic isolation, local adaptation and interspecific interactions, including hybridization, in the generation of these species assemblages is rather unclear.
In the granitic Seychelles, Phelsuma is represented by a small endemic clade [8,30]. The classification of the Seychelles “forms” of Phelsuma has been a persistent problem due to the considerable variation in colour pattern and size across islands. Cheke [31] and Gardner [25,32,33] assessed the systematics of the Seychelles forms and currently two endemic species are recognized with an unclear number of subspecies. Within P. sundbergi, up to six subspecies have been recognized: P. sundbergi longinsulae, inhabiting Mahé (and associated islands) plus Fregate; P. (sundbergi) longinsulae rubra and umbrae, respectively from North and Silhouette sensu [34] reflecting previous consideration of P. longinsulae as a separate species; P. s. sundbergi, from Praslin, Curieuse, Aride and surrounding islands, and P. s. ladiguensis from La Digue, Mariane, Grande and Petite Soeur and surrounding islands (Figure 1). Gardner [25,33] examined variation within these species based on body size, scalation and colour pattern variables and found support for only three of the groups, P. s. longinsulae, P. s. sundbergi and P. s. ladiguensis (the latter two being clearly more similar), but not for the distinction of the North and Silhouette forms, rubra and umbrae. These were synonymised with P. sundbergi longinsulae, as was also P. (sundbergi) longinsulae menaiensis sensu [35], a putative subspecies from the coralline islands of Cosmoledo described in some detail by Cheke [31]. Within P. astriata two allopatric subspecies are recognized; P. a. astriata and P. a. semicarinata, from the southern and northern group of islands respectively (Figure 1), with populations from Fregate described as “intermediate forms” and hypothesized to be hybrids between the two subspecies [31]. Individuals from Astove (raised coralline island) were considered by Cheke [31] to be closely related to the nominate astriata on Mahé.
The degree of morphological differentiation between and within species and subspecies varies geographically. Notably, body size variation shows no clear geographic pattern within P. astriata, while clearly increases from south to north within P. sundbergi; with P. s. sundbergi and P. s. ladiguensis individuals being significantly larger than P. s. longinsulae[8,32]. Radtkey [8] proposed allopatric speciation followed by asynchronous dispersals across island groups plus resource competition generating character displacement as the drivers of speciation and body size evolution within this group. More specifically, he proposed that after the invasion of the Seychelles archipelago by a single species of Phelsuma, sea-level changes led first to allopatric speciation, with the ancestor of P. sundbergi and P. astriata differentiating in the southern and northern groups of islands, respectively, and then to the asynchronous dispersal of these species across island groups. The differences in the relative timing of the dispersal between island groups (with the ancestor of P. s. sundbergi and P. s. ladiguensis colonizing the northern group of islands much earlier than the ancestor of P. astriata astriata colonized the southern group) would have resulted in P. sundbergi evolving an intermediate body size in the group of islands associated with Mahé (southern) and a large body size in the group of islands associated with Praslin and La Digue (northern), due to character displacement, and given its longer period of sympatry with P. astriata in the northern islands. Nevertheless, this is not the only possible explanation for the different degrees of body size disparity. Body size distributions may be governed instead by different ecological variables and interactions happening within each island group [32,36]. For example, competition and selective pressures for body size within each island or island group may be distinct and lead to distinct patterns of morphological differentiation between species within island group, independently of the time since sympatry. In fact, under character displacement, phenotypic divergence accelerates in sympatry [37] but needs not be correlated with time since sympatry [38,39].
Gardner [32] proposed instead that body size in Seychellois Phelsuma has probably been influenced by the distribution of vegetation and social systems. The use of defensible pollen and nectar food supplies, especially the male inflorescences of Lodoicea maldivica, native to (and only found in) the northern islands of Praslin and Curieuse, may have resulted in the evolution of the larger body size of P. sundbergi in the northern islands while hardwood forests, shrub and seabird islands vegetation may have led to smaller sizes in the south. The competitive relationships between the species could then be influenced by their body size differences and help to support the selective pressures within the system. Sympatric species within each island may be partitioning their habitat differently or diversifying along different axis eg. [40-43], resulting in the observed pattern, without the need to invoke asynchronous sympatry as its cause. In the Mascarenes for example, sympatric species of Phelsuma often differ across several resource axes at the same time [44]. If the time since contact between the different species in the Seychelles is similar, ecological factors shall play a much more important role in morphological diversification of Seychellois Phelsuma than currently appreciated. It is also not known if these species hybridize on any island group.
Our objectives here were first to assess the intraspecific genetic variation within species and its geographical distribution, inferring possible colonization routes and the geographic origins of outer island populations. Second, using multilocus data, to explore the possibility of gene flow across species and subspecies and infer the underlying population’s history. Ultimately, we aimed to test the asynchronous divergence within each species, simultaneously providing a time scale for the diversification of this clade. Such a phylogeographic approach will help us to better understand the relative influence of oscillating sea-levels in shaping diversification across these islands, and also should provide a valuable framework upon which the ecological determinants of diversification within this group can be further explored.
Results
Mitochondrial DNA variability and genetic structure
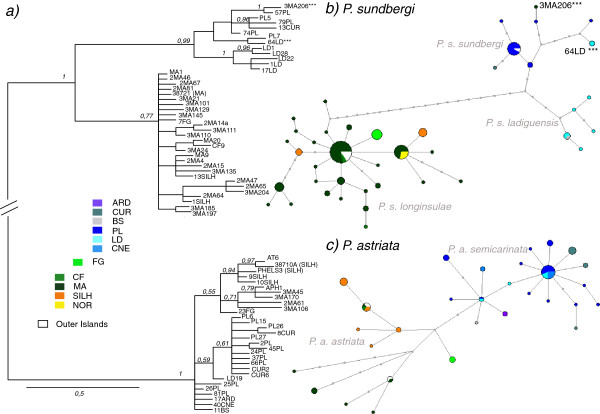
The two species, P. sundbergi and P. astriata correspond to two divergent mtDNA clades (Figure 2) (Nei’s Da = 11%). Intraspecifically, Phelsuma sundbergi longinsulae clearly differentiates from P. s. sundbergi and P. s. ladiguensis (Da = 2.5%) (Figure 2b). Within P. s. longinsulae, the North and Silhouette populations share or carry haplotypes very closely related to Mahé. Silhouette haplotypes are paraphyletic relative to Mahé, and individuals from North exhibit a single haplotype, shared with individuals from Mahé (Figure 2b). Furthermore, all individuals from Fregate carry a unique haplotype, which is just one nucleotide substitution away from the most abundant haplotype from Mahé. The haplotype of one P. sundbergi individual from Mahé (3MA206) clusters with P. sundbergi haplotypes from Praslin. We provisionally interpreted this as a possibly anthropogenic related pattern (but see Discussion) and excluded this individual for the inference of divergence times and gene flow. Concerning the P. sundbergi populations from the northern group of Praslin, La Digue and associated islands, i.e., P. sundbergi sundbergi and P. s. ladiguensis, they form two slightly divergent mtDNA clades, in agreement with their morphological variation: Praslin and Curieuse populations in one clade and La Digue and Grande Soeur populations in the other clade (Figure 2b). Yet, two individuals from La Digue exhibit haplotypes that cluster within those from Praslin.
Figure 2.
a) MtDNA haplotype phylogeny. Midpoint-rooted Bayesian 50% majority-rule consensus tree with posterior probabilities above 0.5 indicated above branches. The scale bar refers to branch lengths (expected changes per site). Median-Joining networks for P. sundbergi (b) and P. astriata (c). Islands are colour-coded and circle size is proportional to the number of individuals. Full grey dots along branches represent missing or unsampled haplotypes. Islands labelling is as follows: Aride (ARD), Curieuse (CUR), Grande Soeur (BS), Praslin (PL), La Digue (LD), Cousine (CNE), Fregate (FG), Cerf (CF), Mahé (MA), Silhouette (SILH) and North (NOR). Highlighted haplotypes (***) correspond to the individuals from Mahé and La Digue found to cluster within P. sundbergi sundbergi (from Curieuse and Praslin) (see text). The outer islands of Cosmoledo, Astove (AT), Poivre and Alphonse (APH) are identically coloured in white (see Results for detailed information).
Within P. astriata the non-overlapping geographic distribution of mitochondrial lineages agrees also with the currently recognized subspecies (Figure 2c). The divergence between these lineages is not large, with just three substitutions separating haplotypes from P. a. astriata and P. a. semicarinata, and with two and three substitutions separating both these subspecies from individuals from Fregate, where the three individuals sampled exhibit the same mtDNA haplotype.
All the outer island populations share mtDNA haplotypes with the granitic islands populations (Figures 2b and 2c, in white colour). The Cosmoledo populations of P. sundbergi longinsulae exhibit a single haplotype, the central one in the phylogenetic network and the most abundant in Mahé. Similarly, P. astriata astriata haplotypes from Alphonse are also shared with individuals from Mahé, while the Astove population of P. astriata astriata shares its (again single) haplotype with Mahé, Cerf and Silhouette individuals. Finally, individuals of P. sundbergi sundbergi from Poivre exhibit a single and same haplotype as most of the individuals from Praslin.
Nuclear DNA genealogies
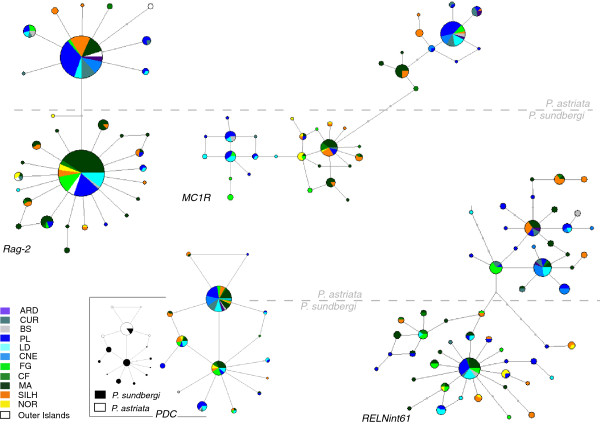
The nuclear variation is almost completely sorted between species (Figure 3). P. astriata and P. sundbergi share only a single haplotype for one of the genes (PDC), which is central in the phylogenetic network and the most abundant in P. astriata. Overall, divergence between species is not high and the conspecific subspecies in all cases share a high number of haplotypes. The only nuclear gene where intraspecific structure is noticeable is MC1R, with two subgroups within each species, with a strong geographic component.
Figure 3.
MJ networks representing nuclear DNA variation. Islands are colour-coded and circle size is proportional to the number of individuals. Labelling is the same as in Figure 2. Full grey dots along branches represent missing or unsampled haplotypes. Clades above and below the dashed horizontal line correspond to P. astriata and P. sundbergi, respectively (except for PDC where haplotype sharing is observed – inset network). The gene to which each network corresponds is identified on its inferior left corner.
Genetic variability and neutrality tests
Considerable variation was observed in the levels of polymorphism across loci (Table 1), with Cytb and RELNint61 being the polymorphic ones. The variability levels within each subspecies were not remarkably different, although they were slightly higher within P. sundbergi longinsulae. Tajima’s D and Fu’s Fs were usually negative, while R2 values were usually small, suggesting demographic growth across all groups, although only in a few instances (P. sundbergi longinsulae for the mitochondrial and Rag-2 markers and P. astriata semicarinata for Cytb) were values significant for all statistics.
Table 1.
Summary diversity statistics, tests of neutrality and indexes of populations growth for all subspecies of P. sundbergi and P. astriata
| Subspecies | Locus | Lenght | N |
Polymorphism |
Neutrality |
Population growth |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H | S | Hd | π | θw | D | FS | R2 | ||||
|
P. sundbergi longinsulae |
Cytb |
761 |
93 |
28 |
30 |
0.875 |
0.00362 |
0.00772 |
−1.62892* |
−17.813* |
0.0445* |
| |
MC1R |
617 |
56 |
13 |
10 |
0.758 |
0.00261 |
0.00358 |
−0.76192 |
−5.943* |
0.0759 |
| |
RELNint61 |
877 |
52 |
19 |
21 |
0.903 |
0.00401 |
0.00587 |
−1.02213 |
−6.903* |
0.0798 |
| |
Rag-2 |
796 |
136 |
17 |
15 |
0.554 |
0.00089 |
0.00345 |
−1.97263* |
−1.97263* |
0.0261* |
| |
PDC |
360 |
56 |
7 |
6 |
0.710 |
0.00255 |
0.00363 |
−0.74314 |
−2.145 |
0.0772 |
|
P. sundbergi sundbergi |
Cytb |
761 |
17 |
6 |
12 |
0.588 |
0.00234 |
0.00466 |
−1.87150* |
−0.889 |
0.0983 * |
| |
MC1R |
617 |
22 |
9 |
7 |
0.853 |
0.00391 |
0.00312 |
0.81826 |
−2.265 |
0.1645 |
| |
RELNint61 |
877 |
22 |
12 |
19 |
0.835 |
0.00364 |
0.00596 |
−1.44378 |
−4.329* |
0.0839 |
| |
Rag-2 |
796 |
34 |
6 |
5 |
0.371 |
0.00051 |
0.00154 |
−1.76914* |
−4.537* |
0.0611 |
| |
PDC |
360 |
22 |
7 |
6 |
0.762 |
0.00422 |
0.00457 |
−0.23808 |
−1.875 |
0.1186 |
|
P. sundbergi ladiguensis |
Cytb |
761 |
7 |
5 |
6 |
0.857 |
0.00300 |
0.00322 |
−0.33869 |
−1.262 |
0.1543 |
| |
MC1R |
617 |
10 |
5 |
4 |
0.844 |
0.00223 |
0.00229 |
−0.09820 |
−1.468 |
0.1652 |
| |
RELNint61 |
877 |
10 |
6 |
8 |
0.758 |
0.00211 |
0.00303 |
−1.20688 |
−1.540 |
0.1351 |
| |
Rag-2 |
796 |
14 |
2 |
1 |
0.143 |
0.00018 |
0.00040 |
−1.15524 |
−0.595 |
0.2575 |
| |
PDC |
360 |
14 |
7 |
6 |
0.890 |
0.00394 |
0.00524 |
−0.89259 |
−3.209* |
0.1071* |
|
P. astriata astriata |
Cyt b |
761 |
20 |
10 |
18 |
0.895 |
0.00541 |
0.00667 |
−0.71027 |
−1.539 |
0.1044 |
| |
MC1R |
617 |
30 |
8 |
8 |
0.660 |
0.00279 |
0.00327 |
−0.44757 |
−1.813 |
0.1059 |
| |
RELNint61 |
877 |
30 |
11 |
10 |
0.867 |
0.00356 |
0.00289 |
0,.3543 |
−2.134 |
0.1555 |
| |
Rag-2 |
796 |
40 |
3 |
2 |
0.188 |
0.00024 |
0.00059 |
−1.11593 |
−1.548 |
0.0858 |
| |
PDC |
360 |
30 |
3 |
2 |
0.439 |
0.00132 |
0.00140 |
−0.12690 |
−0.044 |
0.1184 |
|
P. astriata semicarinata |
Cyt b |
761 |
46 |
19 |
20 |
0.865 |
0.00289 |
0.00598 |
−1.67207* |
−12.687** |
0.0499* |
| |
MC1R |
617 |
64 |
6 |
4 |
0.445 |
0.00084 |
0.00137 |
−0.82812 |
−2.856* |
0.0651 |
| |
RELNint61 |
877 |
56 |
10 |
11 |
0.728 |
0.00307 |
0.00277 |
0.30059 |
−0.596 |
0.1193 |
| |
Rag-2 |
796 |
92 |
4 |
3 |
0.184 |
0.00024 |
0.00074 |
−1.23597 |
−2.748 |
0.0445 |
| PDC | 360 | 58 | 3 | 2 | 0.101 | 0.00028 | 0.00120 | −1.31498* | −2.516* | 0.0673 | |
a N, number of individuals (mtDNA)/chromossomes (nucDNA). Polymorphism indexes: H, number of haplotypes detected; S, number of segregating sites; Hd, haplotype diversity; π, nucleotide diversity; θw, population mutation parameter [62]. Neutrality test: D, Tajima’s D. Population growth tests: FS, Fu’s FS; R2, Ramos-Onsins and Rozas’ R2.* p < 0.05; ** p < 0.01.
b Statistics were not computed for P. astriata population from Fregate as only three individuals were sampled. Populations from outer islands and hypothetically introduced individuals (3, see above) were also excluded of calculations.
Time and gene flow estimates under the isolation-with-migration model
Using the IM model we obtained consistent estimates of demographic parameters across all runs (Table 2), with high effective sample size (ESS) values, convergence and a good sampling of the parameter space. Assuming a mean rate substitution of 1% per lineage per million years (Myr) at this marker, the estimated time since divergence between the two species was 6.39 Myr, with a 95% high posterior density (HPD) interval of 4.264 – 8.397. No gene flow was detected between species and the ancestral effective population size estimate was very low (Table 2, bottom row). Estimates of the splitting times between the southern and northern clades of both species were fairly coincident (~500,000 years old) as so were their posterior density intervals (Table 2 and Figure 4a), which were, nevertheless, quite large. The effective population size estimates for P. sundbergi doubled those for P. astriata, irrespectively of the island group. Accordingly, the estimated effective ancestral population sizes were also higher in P. sundbergi. Even having excluded the few individuals presumed to be the result of anthropogenic introductions (as mentioned above), significant migration rates were still recovered within both species, especially for P. sundbergi (Table 2 and Figure 4b). Neither recombination nor selection was detected at any locus.
Table 2.
Isolation-with-migration estimates
| Parameter estimates | ||||
|---|---|---|---|---|
| Group/Species |
Ancestral Effective Population Size Ne [95%HPD] |
Effective Population Size Ne [95%HPD] |
Effective Number of Migrants into the population 2 Nm [95%HPD] |
Time Since Divergence T [95%HPD] |
| |
Absolute (millions of individuals) |
Absolute (millions of individuals) |
Absolute (migrant gene copies per generation) |
Absolute (in million years) |
|
P. s. sundbergi + P. s. ladiguesis (northern) |
0.600 [0.104-2.452] |
1.826 [1.046-3.148] |
1.584 [0.3513 – 3.59]** |
0.536 [0.266-1.901] |
|
P. sundbergi longisulae (southern) |
3.584 [2.228-5.598] |
0.0075 [0 – 1.995] |
||
|
P. astriata astriata (northern) |
0.296 [0.0014 – 0.868] |
0.764 [0.424-1.280] |
0.1311 [0 – 0.692] |
0.580 [0.251-2.876] |
|
P. astriata semicarinata (southern) |
0.752 [0.390-1.436] |
0.3910 [0 – 1.503] * |
||
|
P. sundbergi |
0.002 [0 – 2.408] | 2.880 [2.143 – 3.737] |
0.0006 [0 – 0.1763] |
6.39 [4.264 – 8.297] |
| P. astriata | 0.765 [0.564 – 1.035] | 0.0003 [0 – 0.08] | ||
Estimates and 95% highest posterior densities (HPD) of population sizes (θ), effective number of migrants (2 Nm) and time since divergence (T) between groups, as inferred with IMa2 (three different comparisons). Values for migration that are significantly different from zero [LLR test: * p-value <0.05; ** p-value <0.01, 82] are indicated with asterisk. A substitution rate of 1% per site per million years and a generation time of one year were assumed to convert the estimates into absolute quantities.
Figure 4.
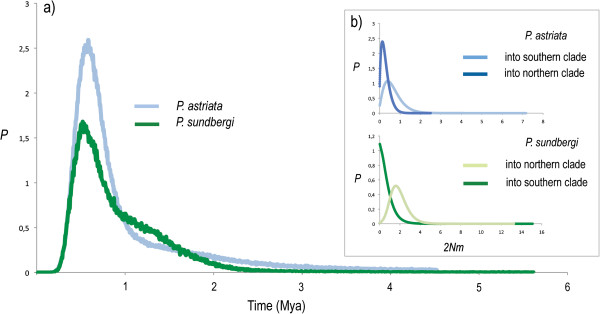
a) Probability distribution for the splitting time between northern and southern clades of P. astriata (light blue) and P. sundbergi (dark green); b) number of migrant gene copies per generation (2 Nm) between those same clades within P. astriata (blue, upper) and P. sundbergi (green, down).
Seychelles’ Phelsuma species tree
*BEAST analyses recovered consistent results across replicates, with high ESS values for all parameters of interest. Estimates without data recover completely different values and distributions, showing that the data, and not the prior, are primarily responsible for the posterior distributions. The maximum clade credibility species tree (Figure 5) was in general agreement with the inferences made under the IM model in respect to the ages of the branching events. The initial split between the two species (P. sundbergi and P. astriata) was dated to circa 3.6 Myr, although the 95% HPD age intervals were very wide (1.33 – 6.86). These estimates are slightly more recent than those obtained under the IM model (Table 2), but the HPD intervals considerably overlap. Estimates of 95% HPD intervals for the north–south split within both species again clearly overlap. The population of P. astriata from Fregate (included here though excluded from the IMa analysis so data could fit a two populations model) was recovered as sister taxa to P. astriata semicarinata, with moderate support.
Figure 5.
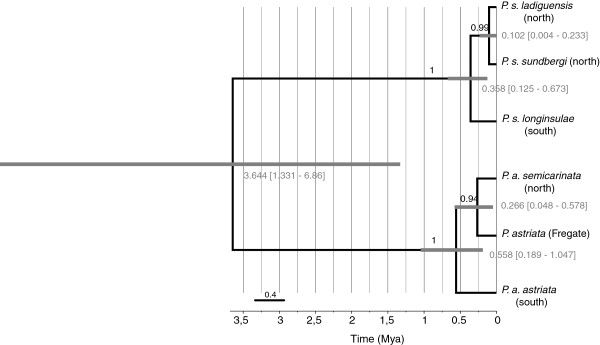
“Species” tree of the Phelsuma day-geckos endemic from the Seychelles. The tree shown corresponds to the maximum clade credibility tree with mean estimates used for node heights and it is based on the multispecies coalescent analysis of the five molecular markers and assuming a rate of evolution of 0.01 (SD = 0.0027) substitutions per lineage per My for Cytb. Clade posterior probability is indicated above each branch. Node bars correspond to the 95% high posterior density intervals for node height (age), which are given below/in front of each node. Horizontal axis corresponds to time in million years before present (Mya).
Discussion
Species divergence and intraspecific genetic structure
Here we analyzed in detail for the first time the genetic structure of the Phelsuma day-geckos in the Seychelles based on a comprehensive sampling across the archipelago. Molecular data from widespread populations across these islands corroborate the two recognized species as two single main lineages and place their divergence in the late Miocene or early Pliocene. That is earlier than the Pliocene/Pleistocene divergence estimates within the similarly distributed Seychellois crab genus Seychellum[12] but in the same range as the north–south split within the co-distributed Seychellois gecko species Urocotyledon inexpectata[13].
Very recently, Carranza et al. [45] obtained rate estimates for part of the same Cytb fragment used in this study that are higher that the one we used (approximately the double and up to three times higher, depending on the group of taxa, although with high standard deviations). Pending further confirmation of these rate estimates, we preferred to keep the use here of a more conservative (and widespread) average rate of 1% substitutions per lineage per Myr. Still, we note the need of taking these absolute age estimates with caution. We note also that using a faster rate would lead us to more recent estimates of species and subspecies diversification (although yet early Pliocenic and Pleistocenic, respectively) but that it would not change the relative timing between these events. Importantly, it would not change the inference of a simultaneous split within each species.
We did not find evidences of past or current gene flow between the two species. While the application of the IM model is debatable given the high levels of population structure observed, Strasburg and Rieseberg [46] showed that most parameter estimates in IMa are fairly robust to population structure over a wide range of parameter values, down to Nm = 0.1 among conspecific populations. It is nevertheless possible that estimates of population sizes and divergence times are slightly biased upwards in these cases [46].
Within each species, our data are congruent with previous multivariate morphometric studies from Gardner [25], with Cheke’s [31] taxonomic classification and with previous inferences from Radtkey [8]: three lineages within P. sundbergi and two lineages within P. astriata. The limited number of individuals of P. astriata sampled from Fregate, as well as the little intraspecific differentiation found between the northern and southern islands did not allow us, though, to infer whether the individuals from Fregate are in fact the result of hybridization between distinct clades (as previously proposed) or are just a distinct, equidistant, clade.
Due to the reciprocal monophyly of the main clades within each species at the mtDNA level and the low variation and lack of sorting at the nuclear markers, it seems too speculative to depict historical colonization sequences and routes within the granitic Seychelles. Several scenarios could fit the observed patterns. Moreover, extinction and demographic changes may have affected populations differently, further obscuring any historical signal to be inferred. As argued by Gardner [32], current diversity could be explained by the sectioning of pre-existing clinal variation due to rising sea levels. It seems likely therefore that divergence between and within each species in the granitic islands was mostly driven by vicariance through cycles of allopatry between populations, rather than through stepping-stone colonization events. However, if isolation was due solely to oscillating sea-levels we would expect that the main split occurred between Silhouette and North versus remaining islands, such as in [12] (see Figure 1). It seems though, that distance was also an important factor for these species, and they were likely able to perform short overwater dispersals more often than long ones, leading to the main subdivision being between northern (Praslin-group) and southern (Mahé group, including Silhouette and North) islands.
Within P. sundbergi, some observations point to the possibility of (past or ongoing) gene flow between subspecies (island groups): one individual collected in Mahé as P. s. longinsulae exhibited in fact an haplotype clustering within P. s. sundbergi (from Curieuse and Praslin), and the same happened with two individuals collected in La Digue that grouped within P. s. sundbergi. Given that we did not collect morphological measurements or photos that would allow us to reconfirm the identification of these specimens, and given previous records of occasional dispersals that do not seem to form viable populations [31] we provisionally considered these individuals as possible cases of anthropogenic or very recent dispersals, and eliminated them from further analyses when appropriate. Nevertheless, even without considering these individuals, the IMa analyses returned positive estimates of gene flow within P. sundbergi (specifically, into the northern clade of P. s. sundbergi and P. s. ladiguesis), suggesting that the possibility of gene flow should be investigated in more detail. Also between P. astriata there were signs of possible gene flow, albeit weaker. Given the estimated Pleistocenic divergence between these subspecific clades, and low sea levels assumed during this period [6,18], which could have facilitated contact between island groups, it is reasonable to expect that gene flow may have occurred. Pending further studies shedding light on gene flow estimates and possible ecological differences across island groups, we consider the current taxonomy of this group appropriate, with only two species, P. sundbergi and P. astriata, that include three and two subspecies (or lineages; we are not necessarily advocates of the subspecies rank) respectively. Phelsuma astriata population from Fregate remains unassigned as P. astriata ssp.
The phylogeographic pattern of P. sundbergi (and P. astriata to a certain extent) mirrors the patterns found within U. inexpectata, a gecko species co-distributed with Phelsuma across the granitic Seychelles [13] and may thus reflect a more general biogeographic pattern. Many similarities can also be found with a co-distributed endemic freshwater crab species, Seychellum allaudi[12] with the difference that in this case Silhouette populations split first from the remaining populations, which seems logical due to their lower dispersal capacity.
The origins of the outer island populations
Cheke [31] discussed the status of the populations from Cosmoledo and Astove, keeping their designation as separate subspecies (P. longinsulae menaiensis and P. astriata astovei) based on slight colour variations. Gardner [25,33] found no morphological differentiation between P. sundbergi populations from the coralline and granitic islands and argued for their probable recent colonization or introductions by man (in the coralline islands). Accordingly, the fact that all individuals analysed from each of these populations possess the same mtDNA haplotype, shared with populations from the granitic islands, supports their very recent origin, either by natural dispersion or as a result of human-driven introductions. Specifically, based on mtDNA variation, the population from Cosmoledo could be assigned to P. sundbergi longinsulae, Poivre to P. sundbergi sundbergi, and the populations from Astove and Alphonse to P. astriata astriata.
The timing and mechanisms driving diversification of Phelsuma in the Seychelles
Perhaps the most unexpected result of this study was the estimate of simultaneous divergence of the northern and southern lineages within both species. In the biogeographic scenario previously proposed by Radtkey [8], the asynchronous divergence between these clades within each species would be responsible for the different degrees of morphological differentiation observed between the two species within each island group (the degree of character displacement being proportional to time since contact between the different species). Our results contradict this hypothesis, implying that other factors should be responsible for the morphological differentiation between species within each island group. The biogeographic scenario proposed by Radtkey [8] was largely based on his estimate of the mtDNA phylogeny, and on the different pairwise genetic distances between the different mtDNA clades within each species. However, this variance in gene divergence could also be due to stochasticity and different levels of ancestral polymorphism within each ancestral population [47,48]. The degree to which gene divergence predates population divergence can be substantial when the effective population size (Ne) of the common ancestor is large relative to the divergence time [47]. In fact, recent studies emphasize the need for model-based approaches to test the effects of shared historical factors in species/clades divergence [49,50]. Moreover, with just a single non-recombining locus, and especially if reciprocal monophyly has been achieved, it is not possible to differentiate between variance in sequence divergence due to divergence times and that resulting from coalescence. Given the different levels of intraspecific genetic divergence seen at the mtDNA, very different long-term population sizes would have to be assumed for each species to make them compatible with a synchronous north–south split. Multiple unlinked loci are extremely useful in addressing this question [47,51], as the expected variance in pairwise genetic distances from unlinked nuclear loci is directly proportional to the ancestral Ne, thereby providing information on coalescent processes in ancestral populations and reducing confidence intervals surrounding estimated divergence times eg. [49]. Accordingly, we estimated higher current and ancestral population sizes within P. sundbergi than within P. astriata. Interestingly, these estimates oppose current census sizes for both species, especially in the case of P. sundbergi populations of the northern islands [43,52], possibly indicating wide demographic changes across the evolutionary history of these species.
Eco-morphological divergence
Assuming a simultaneous divergence between both the northern and southern clades in each species leads us to hypothesize that the different ecological/habitat conditions in the different island groups had a predominant role on the different degrees of body-size divergence within each island group. As shown with body size measurements from Gardner [32] and Radtkey [8], the mean difference in snout-vent length (SVL) between P. sundbergi and P. astriata within the northern group (Praslin, La Digue, and associated islands) is significantly higher than the corresponding difference in the southern group. Gardner [32] demonstrated the importance of body size in resource use with observations of the differential use of palm tree pollen and nectar. Palms from the genera Lodoicea, Deckenia and Cocos all produce male inflorescences that are actively defended for highly nutritious resources, and in all reported cases the largest individual displaced smaller individuals, regardless of the intruding species [32]. This author further suggested that the large body size of P. sundbergi in the northern group likely evolved through interference competition associated with the differential distribution of palm trees. More specifically, he argued that because Lodoicea maldivica (the larger palm species) occurs only in the northern islands of Praslin and Curieuse, the selection pressure necessary to drive the evolution of larger body size was absent in the southern group. Radtkey [8] discarded this hypothesis arguing that competition in Cocos nucifera plantations was equally intense, and that this palm, along with Deckenia, was distributed throughout all the Seychelles. Nevertheless, L. maldivica (native only to the northern islands of Praslin and Curieuse), was the dominant tree on these islands, occurring across a broad range of habitats from the coast to the uplands, prior to human colonization [11] and recent data highlights the clear association of P. sundbergi with adult L. maldivica males in Praslin [43]. Harmon et al. [44] investigating the competition and community structure within Phelsuma from the Mascarene islands also found that one of their unique characteristics compared to other diurnal arboreal lizards is the fact that they partition the habitat between palms and non-palm trees. They also reported that interspecific interactions exist and that species rapidly react to other species presence or removal shifting their habitat use. As argued by Gardner [32], it is therefore possible that in palm forests selection will favour adults with large body-size as they will be able to defend inflorescences successfully, ensuring a constant food supply. In fact, all large species of this genus in the Mascarenes are also associated with potentially defensible items, in which large size may be advantageous [32]. In other habitats small size is probably more effective.
All these aspects become relevant given our divergence time estimates and warrant further investigation on the ecological determinants of eco-morphological diversification within Phelsuma in the Seychelles. According to our results, the intermediate size of P. sundbergi in the southern group is not the result of its evolution as a solitary species to use a wider resource distribution; rather other variables should be considered. Species are likely partitioning their habitat differently within each island, which leads to different outcomes in terms of selection and body size evolution. The low overlap of structural niche categories shows for example that P. astriata from Praslin and Silhouette use different parts of the tree [32]. For example, on Silhouette P. astriata are more abundant than P. sundbergi in coconut plantations, where they use a similar microhabitat to P. sundbergi on Praslin. In Silhouette habitat segregation is considerable (with P. astriata reaching larger sizes than P. sundbergi and dominating nectiferous resources) while in Mahé species seem to co-exist without obvious resource partitioning, although some segregation is possible (P. astriata seems to be much less abundant at the medium perch height where P. sundbergi is most frequently observed; S. Rocha, pers. obs). Further investigation on habitat use and niche partitioning within both species across island groups will be determinant for the understanding of the role of the habitat and interspecific competition in the microevolutionary patterns within this group.
Conclusions
Geographic patterns of genetic variation within Phelsuma species from the Seychelles support current taxonomy, with the taxonomic status of P. astriata population from Fregate remaining undetermined. The data are also congruent with the very recent origin of all outer islands populations. Despite extensive sympatric distributions, no gene flow seems to occur between the two species, whose divergence most probably pre-dates the Pleistocene. The genetic structure within each species is well correlated with geography, with the main mtDNA clades (subspecies) being distributed exclusively across northern and southern islands. Such differentiation is consistent with isolation caused by oscillating sea levels, which also seems to have allowed post-divergence gene flow within both species. The divergence between the main (northern and southern) clades within each species seems to have been approximately simultaneous, and highlights the importance of studying the potential role of habitat and ecological factors in microevolutionary patterns within this group.
Methods
Sampling and molecular data collection
Tissue samples (tail tips) of Phelsuma sundbergi and P. astriata, covering almost their entire distribution were collected and stored in 100% ethanol. Sampling spanned a 4-year period (2005–2008) with sampling locations not repeated, thus the likelihood of having sampled the same individual more than once is minimal. No vouchers were collected. DNA extraction followed standard salt or phenol-chloroform protocols [53,54]. Cytochrome-b (Cyt-b) was amplified in 126 P. sundbergi individuals and 73 P. astriata individuals, comprising all recognized morphological variation within each species (Figure 1). Between one and ten individuals per island per species were genotyped for four additional nuclear markers (see Additional file 1).
Primers for Cyt-b were CBL14753 [55] and CB3H [56] with amplification conditions as given by Rocha et al. [30]. Phosducin (PDC) amplification was made with primers and conditions from Gamble et al. [57], recombination-activating gene 2 (Rag-2) as in Rocha et al. [30], melanocortin-1 receptor (MC1R) and intron 61 of the reelin gene (RELNint61) using primers and conditions from Pinho et al. [58]. No evidences of multiple copies (size variation, modifications of reading frame) were found for any fragment. Sequences were determined by a commercial facility (Macrogen, Seoul, Korea). For most individuals we sequenced both strands of nuclear fragments to assure double peaks were real and not base calling errors.
Sequences were aligned manually using BioEdit [59]. For the nuclear genes, the haplotype phases were resolved using two complementary methods: first, for sequences that were heterozygous for insertions or deletions (some individuals for the RELNint61 fragment), we used the method described by Flot et al. [60], after which we applied the Bayesian algorithm implemented in PHASE software [61] using the known phases of haplotypes determined by the previous method, when available. For the remaining genes PHASE was applied directly to the datasets. We considered all positions for which phases were resolved with a posterior probability higher than 0.8 (though the majority of them were actually resolved with 1 or > 0.9 probability). Remaining positions were coded as missing data (N) or with ambiguity codes for subsequent sequence analysis as appropriate, but complete individuals were never excluded. We performed a minimum of three runs of PHASE for each dataset and checked phase calls for consistency. Sequence accession numbers are KC347737-KC348370. All sequence files used in downstream analyses can also be downloaded from the DRYAD database (doi:http://10.5061/dryad.6q3t1).
Genetic variability, neutrality tests and gene genealogies estimation
To assess how genetic variability was distributed across and within species we calculated a series of summary statistics for each marker and putative subspecies. As only three individuals of P. astriata from the Fregate population were collected (the “intermediate form” sensu [31]), summary statistics were not computed in this case.
For each locus, we calculated the number of haplotypes, the number of segregating sites, haplotype diversity, nucleotide diversity π, and population mutation parameter θ [62]. We tested for non-neutral evolution by computing Tajima’s D [63] and further tested each population for signals of demographic expansions by calculating Fu’s Fs [64] and R2[65]. We tested significance of the results of the three tests using 10,000 coalescent simulations. All these analyses were conducted in DnaSP v5 [66].
We used MrBayes v3.1.2 [67] to obtain an estimate of the Cytb phylogeny (BI). The dataset was reduced to single haplotypes using ALTER [68] and the best fitting model estimated using jModeltest [69] and PhyML [70], under the AICc criteria [71,72]. Two runs of 11 million generations with default heating parameters were performed. Convergence and congruence across runs were assessed using AWTY [73]. MEGA v4 [74] was used to estimate the mean distance between clades (Nei’s Da). Median-joining networks [75] with MP posterior optimization [76] were constructed using NETWORK v4.510 [77] and used to illustrate the relationships between nuclear and mitochondrial haplotypes (applied separately for each species in the latter).
Testing the asynchronous colonization hypothesis: estimates of divergence times and gene flow under the isolation-with-migration model
Based on the results on population structure, we fitted a subset of each species data to a two-populations model (the northern and southern clades of each species), and used the isolation-with-migration (IM) model as implemented in the program IMa2 [78,79] to estimate the six parameters involved: ancestral and current population sizes (θA, θ1, and θ2), migration in both directions (m1 and m2) and time since the population split (T). One individual of P. sundbergi longinsulae from Mahé island that carried a haplotype that clustered within P. sundbergi sundbergi haplotypes from Praslin was excluded from the analysis, as it could possibly reflect an anthropogenic introduction (see results and discussion). Within P. astriata, the Fregate population (N = 3) was also excluded due to its intermediate position relative to the other two groups of haplotypes.
The HKY [80] model was used for the mtDNA dataset and also for Rag-2, RELNint61 and MC1R nuclear gene fragments, where homoplasic positions were detected, and the IS model [81] for PDC. Prior bounds were first selected based on the summary statistics and optimized and adjusted in a few preliminary runs, to ensure that prior distributions covered the whole range of possible values for that parameter. Multiple subsequent runs of each dataset were then performed and effective sample size of parameters (ESS) and trend plots examined to assure proper mixing and convergence. The significance of the estimated migration rates was measured by the LLR test of Nielsen and Wakeley [82]. A substitution rate of 1% per lineage per Myr (mtDNA, see below) and a generation time of one year, as approximately estimated by Gardner [32] for P. astriata (9 months to attain maturity and approximately 2.5 months egg incubation time) were used to convert parameter estimates into biologically meaningful quantities.
Additionally, to obtain an estimate of the divergence time between P. sundbergi and P. astriata, and to test for the existence of post-divergence gene flow across species, we also fitted the IM model to the whole dataset, treating each species as a separate population.
Because IMa2 assumes no intragenic recombination or selection we tested each locus for recombination using the pairwise homoplasy index [83] implemented in PhiPack [84], and for selection performing multiple loci HKA tests [85] using SITES and HKA [86].
Species-tree inference
To obtain a multilocus perspective of the diversification within the whole clade, we employed the Bayesian MCMC method from Heled and Drummond [87], *BEAST, implemented in BEASTv1.6.1 [88]. This method makes use of the multispecies coalescent model to co-estimate the multiple gene trees and the “species-tree” they are embedded within, together with divergence times and effective population sizes of extant and ancestral species. Especially at this shallow level of divergence, it is likely that multiple processes such as demography and incomplete lineage sorting influence the gene genealogies and the implementation of a method that properly accommodates this variation, rather than a concatenation approach, becomes especially relevant [89,90] and references therein. A minimum of two individuals per “species” (the term species not necessarily referring to the taxonomic rank but to any diverging “population structure” – in our case the five subspecies) per loci is needed, while additional individuals increase accuracy [87]. We therefore constructed a dataset with a subsample of our individuals, using some of those that were genotyped for the five genes: 19 P. sundbergi longinsulae, 8 P. sundbergi sundbergi, 5 P. sundbergi ladiguensis, 20 P. astriata astriata, 22 P. astriata semicarinata and finally 2 individuals from P. astriata from Fregate (Additional file 1).
We estimated the appropriate model of evolution for each fragment as previously and implemented it choosing the most similar within the ones available in BEASTv1.6.1, with all related parameters (gamma; proportion of invariant sites, transition/transversion rates) being co-estimated along the runs. The multispecies coalescent allows for gene-specific mutation rates. Throughout, the substitution rate of the mitochondrial locus was set to a normal distribution prior of mean 0.01 (~1% per lineage per Myr) and standard deviation of 0.0027, as in [91]. Similar rates have been consistently recovered for mtDNA fragments of other reptile and amphibian groups at the intrageneric level, ranging from 0.99% in Hydromantes[92], to 1.35% in lacertids [93] and references therein], estimates available from calibrated trees for gekkonid falling within this range [94]; 1, 15% for Hemidactylus). Rate estimates around 1% per lineage per million year were also obtained by Chapple et al. [95] (mean 1.01%; David Chapple pers. comm) and Cox et al. [96] (S. Carranza pers comm) and the range 1.15-1.35% used also by Miralles and Carranza [97]. In the absence of any available external calibration, we believe that our rate distribution prior, which encompasses these values, is appropriate to obtain an approximate estimate of the time-scale of the diversification of this group (although see Discussion for the possibility of faster substitution rates). Nuclear rates were co-estimated along the run, relative to the mitochondrial one. An uncorrelated relaxed clock model was assumed for the Cytb dataset, whereas a strict clock was assumed for the nuclear gene fragments given their low variability (causing runs under relaxed clock models to fail to converge). We ran the dataset under both a Yule and a birth-death prior for the species-tree and a population size model of continuous growth and constant root (as recommended by the authors; J. Heled, pers. comm.). Default values were used for all other parameters. Multiple runs of 100–500 million generations were performed and sampled at appropriate intervals to obtain 10,000 final samples (trees). To test the influence of the priors on the posterior estimates, an additional run of identical length was made without data, sampling only from the prior. Tracer v1.5 was used to visualize the results of each run, to check the effective sample size of each parameter and chose appropriate burnin values [98]. After discarding the burnin samples, a consensus species tree (Maximum Clade Credibility tree), mean node heights and supports were obtained using TreeAnnotator v1.6.1 [88] and visualized using FigTree v1.3.1 [99].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SR and DJH collected the samples in the field. SR performed the laboratorial work, analysed the data and drafted the manuscript. SR, DJH and DP devised the analytical strategy. DJH and DP provided critical corrections and comments on the paper. All authors read and approved the final manuscript.
Supplementary Material
List of all analysed specimens with details on geographic origin and Genbank numbers.
Contributor Information
Sara Rocha, Email: sprocha@gmail.com.
David Posada, Email: dposada@uvigo.es.
D James Harris, Email: james@cibio.up.pt.
Acknowledgements
This research was supported by grants from Fundação para a Ciência e Tecnologia SFRH/BD/1745/2004 (to SR), PTDC/BIA-BDE/65745/2006 and the Gulbenkian Society. We are grateful to SBS of Seychelles for permits for fieldwork and tissue collection and to the Ministry of Environment and Natural Resources for logistic support during fieldwork. We thank NPTS, ICS Seychelles, SIF and several entities and persons that made fieldwork across all islands possible. A special acknowledgement is due to Raquel Vasconcelos, Ana Perera and Andreia Silva for help during fieldwork and also to Justin Gerlach from NPTS for support and information in earlier phases of the study. Thanks to Ana Perera for Figure 1 and to Ricardo Pereira for insightful discussions. We further thank Nancy Bunbury, Anthony Cheke, Miguel Carretero, Luke Harmon and Ricardo Pereira for helpful discussions and comments on previous versions of the manuscript. Cover picture from Christopher K. Bunbury.
References
- Mittermeier RA, Robles Gil P, Hoffman J, Pilgrim J, Brooks T, Mittermeier CG, Lamoreaux J, da Fonseca GAB. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. Chicago: University of Chicago Press; 2005. [Google Scholar]
- Cheke AS, Hume JP. Lost Land of the Dodo. An Ecological History of Mauritius, Réunion & Rodrigues. London: Yale University Press; 2008. [Google Scholar]
- Gerlach J. Terrestrial and freshwater vertebrates of the Seychelles islands. Leiden: Backhuys Publishers; 2007. [Google Scholar]
- Hawlitschek O, Brückmann B, Berger J, Green K, Glaw F. Integrating field surveys and remote sensing data to study distribution, habitat use and conservation status of the herpetofauna of the Comoro Islands. Zookeys. 2011;144:21–78. doi: 10.3897/zookeys.144.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in madagascar? only time will tell. Annu Rev Ecol Evol Syst. 2006;37:405–431. doi: 10.1146/annurev.ecolsys.37.091305.110239. [DOI] [Google Scholar]
- Warren BH, Strasberg D, Bruggemann JH, Prys-jones RP. Cladistics Why does the biota of the madagascar region have such a strong asiatic flavour? Cladistics. 2010;26:526–538. doi: 10.1111/j.1096-0031.2009.00300.x. [DOI] [PubMed] [Google Scholar]
- Agnarsson I, Kuntner M. In: Current topics in phylogenetics and phylogeography of terrestrial and aquatic systems. Anamthawat-Jónsson K, editor. Rijeka: In Tech Publishers; 2012. The generation of a biodiversity hotspot: biogeography and phylogeography of the western indian ocean islands; pp. 33–82. ISBN 978-953-51-0217-5. [Google Scholar]
- Radtkey RR. Adaptive radiation of Day-geckos (Phelsuma) in the seychelles archipelago: a phylogenetic analysis. Evolution. 1996;50:604–623. doi: 10.2307/2410835. [DOI] [PubMed] [Google Scholar]
- Gerlach J. Snails of the genus Pachnodus (Mollusca; Gastropoda; Enidae): their origins and evolution. J Biogeogr. 1999;26:251–255. doi: 10.1046/j.1365-2699.1999.00259.x. [DOI] [Google Scholar]
- Silva A, Harris DJ, Rocamora G, Dufrenne A, Gerlach J, Rocha S. Assessment of mtDNA genetic diversity within the terrapins Pelusios subniger and Pelusios castanoides across the Seychelles islands. Amphibia-Reptilia. 2010;31:583–588. doi: 10.1163/017353710X524723. [DOI] [Google Scholar]
- Fleischer-Dogley F, Kettle CJ, Edwards PJ, Ghazoul J, Määttänen K, Kaiser-Bunbury CN. Morphological and genetic differentiation in populations of the dispersal-limited coco de mer (Lodoicea maldivica): implications for management and conservation. Divers Distrib. 2011;17:235–243. doi: 10.1111/j.1472-4642.2010.00732.x. [DOI] [Google Scholar]
- Daniels SR. Reconstructing the colonisation and diversification history of the endemic freshwater crab (Seychellum alluaudi) in the granitic and volcanic Seychelles Archipelago. Mol Phylogenet Evol. 2011;61:534–542. doi: 10.1016/j.ympev.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Rocha S, Harris DJ, Posada D. Cryptic diversity within the endemic prehensile-tailed gecko Urocotyledon inexpectata across the Seychelles Islands: patterns of phylogeographical structure and isolation at the multilocus level. Biol J Linn Soc. 2011;104:177–191. doi: 10.1111/j.1095-8312.2011.01710.x. [DOI] [Google Scholar]
- Braithwaite CJR. In: Biogeography of the Seychelles Islands. Stoddart DR, editor. The Hague: Dr W Junk; 1984. Scientific studies in the Seychelles; pp. 1–16. [Google Scholar]
- Plummer PS. In: Aldabra world heritage site. Amin M, Willetts D, Skerett A, editor. Nairobi: Camerapix Publishers International; 1995. Planet Aldabra; pp. 49–70. [Google Scholar]
- Colonna MJ, Casanova J, Dullo WC, Camoin G. Sea level changes and ∂18O record for the past 34,000 yr from Mayotte reef, Indian Ocean. Quaternary Res. 1996;46:335–339. doi: 10.1006/qres.1996.0071. [DOI] [Google Scholar]
- Siddall M, Rohling EJ, Almogi-Labin A, Hemleben C, Meischner D, Schmelzer I, Smeed DA. Sea-level fluctuations during the last glacial cycle. Nature. 2003;423:19–24. doi: 10.1038/nature01690. [DOI] [PubMed] [Google Scholar]
- Miller KG, Kominz MA, Browning JV, Wright JD, Mountain GS, Katz ME, Sugarman PJ, Cramer BS, Christie-blick N, Pekar SF. The phanerozoic record of global Sea-level change. Science. 2005;310:1293–1298. doi: 10.1126/science.1116412. [DOI] [PubMed] [Google Scholar]
- Inger RF, Stuart BL, Iskandar DT. Systematics of a widespread Southeast Asian frog, Rana chalconota (Amphibia: Anura: Ranidae) Zool J Linn Soc-London. 2009;155:123–147. doi: 10.1111/j.1096-3642.2008.00440.x. [DOI] [Google Scholar]
- Siler CD, Oaks JR, Esselstyn JA, Diesmos AC, Brown RM. Phylogeny and biogeography of Philippine bent-toed geckos (Gekkonidae: Cyrtodactylus) contradict a prevailing model of Pleistocene diversification. Mol Phylogenet Evol. 2010;55:699–710. doi: 10.1016/j.ympev.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Scott H. General conclusions regarding the insect fauna of the Seychelles and adjacent islands. Trans Linn Soc Lond 2nd series (Zoology) 1933;19:307–391. doi: 10.1111/j.1096-3642.1933.tb00131.x. [DOI] [Google Scholar]
- Cheke AS. In: Biogeography and ecology of the Seychelles Islands. Stoddart DR, editor. The Hague: Dr. W. Junk; 1984. Lizards of the Seychelles; pp. 245–258. [Google Scholar]
- Gerlach J, van Bruggen AC. Streptaxidae (Mollusca: Gastropoda: Pulmonata) of the Seychelles Islands, Western Indian Ocean. Zool Verh Leiden. 1999;328:1–60. [Google Scholar]
- Nussbaum RA, Wu SH. Distribution, variation, and systematics of the Seychelles treefrog, Tachycnemis seychellensis (Amphibia: Anura: Hyperoliidae) J Zool. 1995;236:383–406. doi: 10.1111/j.1469-7998.1995.tb02720.x. [DOI] [Google Scholar]
- Gardner AS. The systematics of the Phelsuma madagascariensis species group of day geckos (Reptilia: Gekkonidae) in the Seychelles. Zool J Linn Soc-London. 1987;91:93–105. doi: 10.1111/j.1096-3642.1987.tb01724.x. [DOI] [Google Scholar]
- Nussbaum RA. In: Biogeography and ecology of the Seychelles Islands. Stoddart DR, editor. The Hague: Dr. W. Junk; 1984. The amphibians of the Seychelles; pp. 378–415. [Google Scholar]
- Van Der Meijden A, Boistel R, Gerlach J, Ohler A, Vences M, Meyer A. Molecular phylogenetic evidence for paraphyly of the genus Sooglossus, with the description of a new genus of Seychellean frogs. Biol J Linn Soc. 2007;91:347–359. doi: 10.1111/j.1095-8312.2007.00800.x. [DOI] [Google Scholar]
- Rocha S, Rösler H, Gehring P, Glaw F, Posada D, Harris DJ, Vences M. Phylogenetic systematics of day geckos, genus Phelsuma, based on molecular and morphological data (Squamata: Gekkonidae) Zootaxa. 2010;28:1–28. [Google Scholar]
- Harmon LJ, Melville J, Larson A, Losos JB. The role of geography and ecological opportunity in the diversification of day geckos (Phelsuma) Syst Biol. 2008;57:562–573. doi: 10.1080/10635150802304779. [DOI] [PubMed] [Google Scholar]
- Rocha S, Vences M, Glaw F, Posada D, Harris DJ. Multigene phylogeny of Malagasy day geckos of the genus Phelsuma. Mol Phylogenet Evol. 2009;52:530–537. doi: 10.1016/j.ympev.2009.03.032. [DOI] [PubMed] [Google Scholar]
- Cheke AS. Phelsuma GRAY 1825 in the Seychelles and neighbouring islands: a re-appraisal of their taxonomy and description of two new forms (Reptilia: Sauria: Gekkonidae) Senckenberg biol. 1982;62:181–198. [Google Scholar]
- Gardner AS. The evolutionary ecology and population systematics of day-geckos genus Phelsuma in the Seychelles. Aberdeen: University of Aberdeen; 1984. (PhD thesis). [Google Scholar]
- Gardner AS. Morphological evolution in the day gecko Phelsuma sundbergi in the Seychelles: a multivariate study. Biol J Linn Soc. 1986;29:223–244. doi: 10.1111/j.1095-8312.1986.tb01774.x. [DOI] [Google Scholar]
- Borner AR, Minuth W. On the taxonomy of the Indian Ocean lizards of the Phelsuma madagascariensis species group (Reptilia: Geckonidae) J Bomb Nat Hist Soc. 1985;81:243–281. [Google Scholar]
- Mertens R. Die nichmadagassischen arten and unterarten der geckonengattung Phelsuma. Senckenberg biol. 1966;47:85–110. [Google Scholar]
- Meiri S. Size evolution in island lizards. Global Ecol Biogeogr. 2007;16:702–708. doi: 10.1111/j.1466-8238.2007.00327.x. [DOI] [Google Scholar]
- Dayan T, Simberloff D. Ecological and community-wide character displacement: the next generation. Ecol Lett. 2005;8:875–894. doi: 10.1111/j.1461-0248.2005.00791.x. [DOI] [Google Scholar]
- Martin PR, Montgomerie R, Lougheed SC. Rapid sympatry explains greater color pattern divergence in high latitude birds. Evolution. 2010;64:336–347. doi: 10.1111/j.1558-5646.2009.00831.x. [DOI] [PubMed] [Google Scholar]
- Melo M, Warren BH, Jones PJ. Rapid parallel evolution of aberrant traits in the diversification of the Gulf of Guinea white-eyes (Aves, Zosteropidae) Mol Ecol. 2011;20:4953–4967. doi: 10.1111/j.1365-294X.2011.05099.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. New York: Oxford University Press; 2000. [Google Scholar]
- Ackerly DD, Schwilk DW, Webb CO. Niche evolution and adaptive radiation: testing the order of trait divergence. Ecology. 2006;87:S50–S61. doi: 10.1890/0012-9658(2006)87[50:NEAART]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Losos JB. Adaptive radiation, ecological opportunity, and evolutionary determinism. Am Nat. 2010;175:623–639. doi: 10.1086/652433. [DOI] [PubMed] [Google Scholar]
- Noble T, Bunbury N, Kaiser-Bunbury CN, Bell DJ. Ecology and co-existence of two endemic day gecko (Phelsuma) species in Seychelles native palm forest. J Zool. 2011;283:73–80. doi: 10.1111/j.1469-7998.2010.00751.x. [DOI] [Google Scholar]
- Harmon JL, Harmon LL, Jones CG. Competition and community structure in diurnal arboreal geckos (genus Phelsuma) in the Indian Ocean. Oikos. 2007;116:1863–1878. [Google Scholar]
- Carranza S, Arnold EN. A review of the geckos of the genus Hemidactylus (Squamata: Gekkonidae) from Oman based on morphology, mitochondrial and nuclear data, with descriptions of eight new species. Zootaxa. 2012;3378:1–95. [Google Scholar]
- Strasburg JL, Rieseberg LH. How robust are “isolation with migration” analyses to violations of the im model? A simulation study. Mol Biol Evol. 2010;27:297–310. doi: 10.1093/molbev/msp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Beerli P. Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Gene. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Hickerson MJ, Dolman G, Moritz C. Comparative phylogeographic summary statistics for testing simultaneous vicariance. Mol Ecol. 2006;15:209–223. doi: 10.1111/j.1365-294X.2005.02718.x. [DOI] [PubMed] [Google Scholar]
- Hurt C, Anker A, Knowlton N. A multilocus test of simoultaneous divergence across the isthmus of Panama using snapping shrimp in the genus Alpheus. Evolution. 2009;63:514–530. doi: 10.1111/j.1558-5646.2008.00566.x. [DOI] [PubMed] [Google Scholar]
- Knowles LL. Statistical phylogeography. Annu Rev Ecol Evol Syst. 2009;40:593–612. doi: 10.1146/annurev.ecolsys.38.091206.095702. [DOI] [Google Scholar]
- Wakeley J, Hey J. Estimating ancestral population parameters. Genetics. 1997;145:847–855. doi: 10.1093/genetics/145.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach J. Population and conservation status of the reptiles of the Seychelles islands. Phelsuma. 2008;16:31–48. [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Paäbo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals- amplification and sequencing with conserved primers. P Natl Acad Sci USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JE, Fritsh EF, Maniatis T. Molecular cloning: a laboratory manual. 2. New York: Cold Spring Harbour Press; 1989. [Google Scholar]
- Austin JJ, Arnold EN, Jones CG. Reconstructing an island radiation using ancient and recent DNA: the extinct and living day geckos (Phelsuma) of the Mascarene islands. Mol Phylogenet Evol. 2004;31:109–122. doi: 10.1016/j.ympev.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Palumbi SR, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The simple Fool’s guide to PCR, version 2.0. Hawaii: Privately published; 1991. [Google Scholar]
- Gamble T, Bauer AM, Greenbaum E, Jackman TR. Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J Biogeogr. 2008;35:88–104. [Google Scholar]
- Pinho C, Rocha S, Carvalho BM, Lopes S, Mourão S, Vallinoto M, Brunes TO, Haddad CFB, Gonçalves H, Sequeira F, Ferrand N. New primers for the amplification and sequencing of nuclear loci in a taxonomically wide set of reptiles and amphibians. Conserv Genet Resour. 2010;2:181–185. [Google Scholar]
- Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- Flot J-F, Tillier J, Samadi S, Tillier S. Phase determination from direct sequencing of length-variable DNA regions. Mol Ecol Notes. 2006;6:627–630. doi: 10.1111/j.1471-8286.2006.01355.x. [DOI] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D. ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:W14–W18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Sugiura N. Further analysis of the data by akaike’ s information criterion and the finite corrections. Commun Stat A - Theor. 1978;7:13–26. doi: 10.1080/03610927808827599. [DOI] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Polzin T, Daneshmand SV. On steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20. doi: 10.1016/S0167-6377(02)00185-2. [DOI] [Google Scholar]
- NETWORK v4.510. Available from http://fluxus-engineering.com.
- Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. P Natl Acad Sci USA. 2007;104:2785–2790. doi: 10.1073/pnas.0611164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. Isolation with migration models for more than two populations. Mol Biol Evol. 2010;27:905–920. doi: 10.1093/molbev/msp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Kimura M. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics. 1969;61:893–903. doi: 10.1093/genetics/61.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen T, Bruen T. PhiPack: PHI test and other tests of recombination. available at http://www.maths.otago.ac.nz/~dbryant/software.html.
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. SITES and HKA. available at http://genfaculty.rutgers.edu/hey/software.
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Leaché AD. Species trees for spiny lizards (genus Sceloporus): identifying points of concordance and conflict between nuclear and mitochondrial data. Mol Phylogenet Evol. 2010;54:162–171. doi: 10.1016/j.ympev.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Paulo OS, Pinheiro J, Miraldo A, Bruford MW, Jordan WC, Nichols RA. The role of vicariance vs. dispersal in shaping genetic patterns in ocellated lizard species in the western Mediterranean. Mol Ecol. 2008;17:1535–1551. doi: 10.1111/j.1365-294X.2008.03706.x. [DOI] [PubMed] [Google Scholar]
- Carranza S, Romano A, Arnold EN, Sotgiu G. Biogeography and evolution of European cave salamanders, Hydromantes (Urodela: Plethodontidae), inferred from mtDNA sequences. J Biogeogr. 2008;35:724–738. doi: 10.1111/j.1365-2699.2007.01817.x. [DOI] [Google Scholar]
- Carranza S, Arnold EN, Geniez P, Roca J, Mateo JA. Radiation, multiple dispersal and parallelism in the skinks, Chalcides and Sphenops (Squamata: Scincidae), with comments on Scincus and Scincopus and the age of the Sahara Desert. Mol Phylogenet Evol. 2008;46:1071–1094. doi: 10.1016/j.ympev.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Arnold EN, Vasconcelos R, Harris DJ, Mateo JA, Carranza S. Systematics, biogeography and evolution of the endemic Hemidactylus geckos (Reptilia, Squamata, Gekkonidae) of the Cape Verde Islands: based on morphology and mitochondrial and nuclear DNA sequences. Zool Scr. 2008;37:619–636. doi: 10.1111/j.1463-6409.2008.00351.x. [DOI] [Google Scholar]
- Chapple DG, Ritchie PA, Daugherty CH. Origin, diversification, and systematics of the New Zealand skink fauna (Reptilia: Scincidae) Mol Phylogenet Evol. 2009;52:470–487. doi: 10.1016/j.ympev.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Cox SC, Carranza S, Brown RP. Divergence times and colonization of the Canary Islands by Gallotia lizards. Mol Phylogenet Evol. 2010;56:747–757. doi: 10.1016/j.ympev.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Miralles A, Carranza S. Systematics and biogeography of the Neotropical genus Mabuya, with special emphasis on the Amazonian skink Mabuya nigropunctata (Reptilia, Scincidae) Mol Phylogenet Evol. 2010;54:857–869. doi: 10.1016/j.ympev.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracerv1.5. 2007. Available from http://beast.bio.ed.ac.uk/Trace.
- Rambaut A. FigTreev1.3.1. 2009. Available from http://tree.bio.ed.ac.uk/software/figtree/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all analysed specimens with details on geographic origin and Genbank numbers.