Abstract
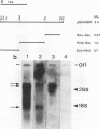
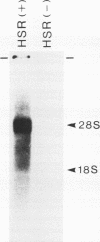
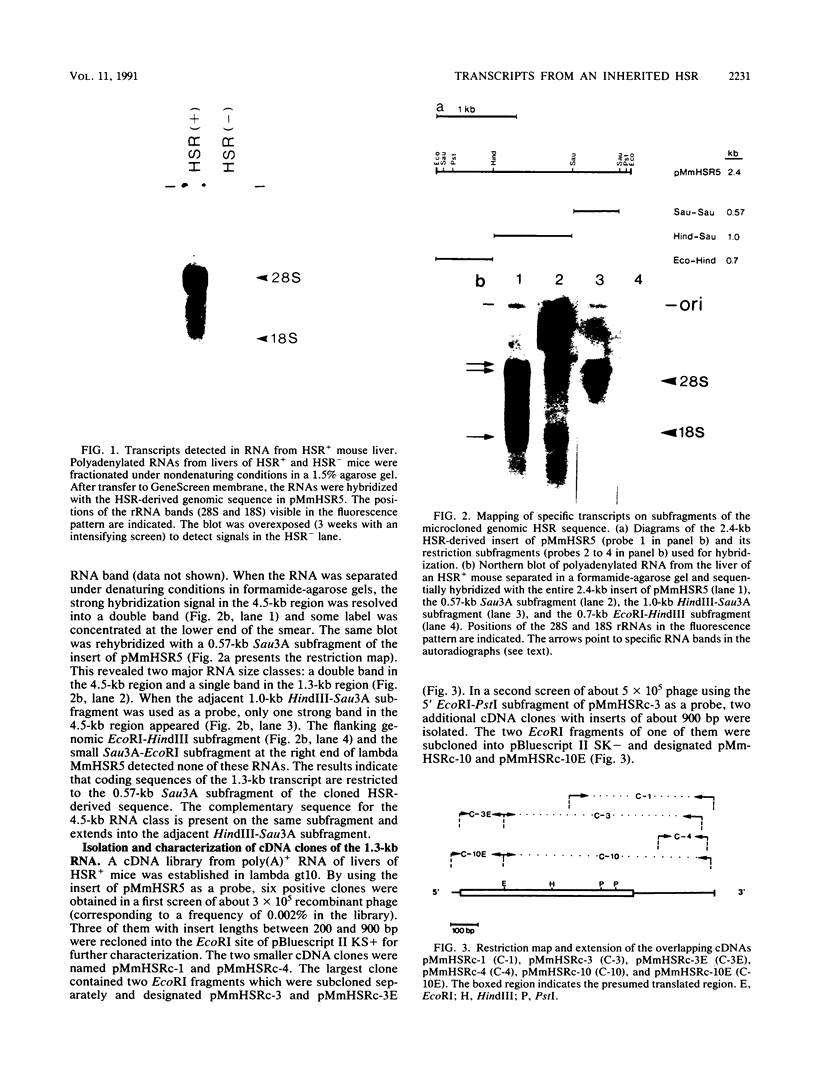
Several populations of the house mouse, Mus musculus, are polymorphic for the presence or absence of an inherited homogeneously staining region (HSR) in chromosome 1. The HSR consists of highly amplified DNA sequences, present in low copy numbers in the HSR- genome. A cloned HSR-derived genomic sequence detected transcripts of about 1.3 and 4.5 kb on blots of poly(A)+ RNA from liver of HSR+ mice but not from that of HSR- mice. A cDNA library was established from RNA of HSR+ mice and screened with the HSR-derived genomic clone. Positive clones were isolated and shown to be complementary to the 1.3-kb RNA species and to amplified DNA sequences in the HSR+ genome. The combined sequence of four overlapping cloned cDNAs is 959 nucleotides long and includes an open reading frame encoding a putative protein of 208 amino acids. The pertinent gene is unidentified. No homologous sequence is stored in the EMBL data base. A stretch of 109 nucleotides at the 3' end of the 1.3-kb RNA homology region in the same genomic fragment, as indicated by hybridization data and sequence motifs resembling promoter elements. Thus, our data suggest that at least two genes or gene families are encoded in the HSR.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Schwab M. Oncogene amplification in tumor cells. Adv Cancer Res. 1986;47:235–281. doi: 10.1016/s0065-230x(08)60201-8. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biedler J. L., Spengler B. A. Metaphase chromosome anomaly: association with drug resistance and cell-specific products. Science. 1976 Jan 16;191(4223):185–187. doi: 10.1126/science.942798. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. M. The molecular genetics of cancer. Science. 1987 Jan 16;235(4786):305–311. doi: 10.1126/science.3541204. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldyreff B., Winking H., Weith A., Traut W. Evidence for in situ amplification of a germ line homogeneously staining region in the mouse. Cytogenet Cell Genet. 1988;47(1-2):84–85. doi: 10.1159/000132512. [DOI] [PubMed] [Google Scholar]
- Borst P., Greaves D. R. Programmed gene rearrangements altering gene expression. Science. 1987 Feb 6;235(4789):658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- Bradley G., Juranka P. F., Ling V. Mechanism of multidrug resistance. Biochim Biophys Acta. 1988 Aug 3;948(1):87–128. doi: 10.1016/0304-419x(88)90006-6. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cowell J. K. Double minutes and homogeneously staining regions: gene amplification in mammalian cells. Annu Rev Genet. 1982;16:21–59. doi: 10.1146/annurev.ge.16.120182.000321. [DOI] [PubMed] [Google Scholar]
- Croce C. M. Role of chromosome translocations in human neoplasia. Cell. 1987 Apr 24;49(2):155–156. doi: 10.1016/0092-8674(87)90552-6. [DOI] [PubMed] [Google Scholar]
- Debatisse M., Hyrien O., Petit-Koskas E., de Saint-Vincent B. R., Buttin G. Segregation and rearrangement of coamplified genes in different lineages of mutant cells that overproduce adenylate deaminase. Mol Cell Biol. 1986 May;6(5):1776–1781. doi: 10.1128/mcb.6.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M., Saito I., Buttin G., Stark G. R. Preferential amplification of rearranged sequences near amplified adenylate deaminase genes. Mol Cell Biol. 1988 Jan;8(1):17–24. doi: 10.1128/mcb.8.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988 Oct 25;16(20):9878–9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott J. A., Ling V. The biochemistry of P-glycoprotein-mediated multidrug resistance. Annu Rev Biochem. 1989;58:137–171. doi: 10.1146/annurev.bi.58.070189.001033. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Foreman P. K., Hamlin J. L. Identification and characterization of a gene that is coamplified with dihydrofolate reductase in a methotrexate-resistant CHO cell line. Mol Cell Biol. 1989 Mar;9(3):1137–1147. doi: 10.1128/mcb.9.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L., Milbrandt J. D., Heintz N. H., Azizkhan J. C. DNA sequence amplification in mammalian cells. Int Rev Cytol. 1984;90:31–82. doi: 10.1016/s0074-7696(08)61487-4. [DOI] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapler G. M., Beverley S. M. Transcriptional mapping of the amplified region encoding the dihydrofolate reductase-thymidylate synthase of Leishmania major reveals a high density of transcripts, including overlapping and antisense RNAs. Mol Cell Biol. 1989 Sep;9(9):3959–3972. doi: 10.1128/mcb.9.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kister K. P., Eckert W. A. Characterization of an authentic intermediate in the self-splicing process of ribosomal precursor RNA in macronuclei of Tetrahymena thermophila. Nucleic Acids Res. 1987 Mar 11;15(5):1905–1920. doi: 10.1093/nar/15.5.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Mock B., Krall M., Blackwell J., O'Brien A., Schurr E., Gros P., Skamene E., Potter M. A genetic map of mouse chromosome 1 near the Lsh-Ity-Bcg disease resistance locus. Genomics. 1990 May;7(1):57–64. doi: 10.1016/0888-7543(90)90518-y. [DOI] [PubMed] [Google Scholar]
- Moriwaki K., Miyashita N., Suzuki H., Kurihara Y., Yonekawa H. Genetic features of major geographical isolates of Mus musculus. Curr Top Microbiol Immunol. 1986;127:55–61. doi: 10.1007/978-3-642-71304-0_6. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Owens J., Maizel J. V., Jr Sequence signals in eukaryotic upstream regions. Biochim Biophys Acta. 1986 Mar 26;866(2-3):109–119. doi: 10.1016/0167-4781(86)90107-7. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer G., Telford J., Baldari C., Pirrotta V. Isolation of cloned genes differentially expressed at early and late stages of Drosophila embryonic development. Dev Biol. 1981 Sep;86(2):438–447. doi: 10.1016/0012-1606(81)90202-5. [DOI] [PubMed] [Google Scholar]
- Schurr E., Skamene E., Forget A., Gros P. Linkage analysis of the Bcg gene on mouse chromosome 1. Identification of a tightly linked marker. J Immunol. 1989 Jun 15;142(12):4507–4513. [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E., Bishop J. M. The PVT gene frequently amplifies with MYC in tumor cells. Mol Cell Biol. 1989 Mar;9(3):1148–1154. doi: 10.1128/mcb.9.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skow L. C., Adkison L., Womack J. E., Beamer W. G., Taylor B. A. Mapping of the mouse fibronectin gene (Fn-1) to chromosome 1: conservation of the Idh-1-Cryg-Fn-1 synteny group in mammals. Genomics. 1987 Nov;1(3):283–286. doi: 10.1016/0888-7543(87)90057-7. [DOI] [PubMed] [Google Scholar]
- Slamon D. J., deKernion J. B., Verma I. M., Cline M. J. Expression of cellular oncogenes in human malignancies. Science. 1984 Apr 20;224(4646):256–262. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Srinivasan P. R., Tonin P. N., Wensing E. J., Lewis W. H. The gene for ornithine decarboxylase is co-amplified in hydroxyurea-resistant hamster cells. J Biol Chem. 1987 Sep 15;262(26):12871–12878. [PubMed] [Google Scholar]
- Traut W., Winking H., Adolph S. An extra segment in chromosome 1 of wild Mus musculus: a C-band positive homogeneously staining region. Cytogenet Cell Genet. 1984;38(4):290–297. doi: 10.1159/000132077. [DOI] [PubMed] [Google Scholar]
- Van der Bliek A. M., Van der Velde-Koerts T., Ling V., Borst P. Overexpression and amplification of five genes in a multidrug-resistant Chinese hamster ovary cell line. Mol Cell Biol. 1986 May;6(5):1671–1678. doi: 10.1128/mcb.6.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Vitto L., Rubnitz J. Co-amplification of rRNA genes with CAD genes in N-(phosphonacetyl)-L-aspartate-resistant Syrian hamster cells. Mol Cell Biol. 1983 Nov;3(11):2066–2075. doi: 10.1128/mcb.3.11.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weith A., Winking H., Brackmann B., Boldyreff B., Traut W. Microclones from a mouse germ line HSR detect amplification and complex rearrangements of DNA sequences. EMBO J. 1987 May;6(5):1295–1300. doi: 10.1002/j.1460-2075.1987.tb02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]
- de Bruijn M. H., Van der Bliek A. M., Biedler J. L., Borst P. Differential amplification and disproportionate expression of five genes in three multidrug-resistant Chinese hamster lung cell lines. Mol Cell Biol. 1986 Dec;6(12):4717–4722. doi: 10.1128/mcb.6.12.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]