Abstract
Background
As well known, both natural and synthetic steroidal compounds are powerful endocrine disrupting compounds (EDCs) which can cause reproductive toxicity and affect cellular development in mammals and thus are generally regarded as serious contributors to water pollution. Streptomyces virginiae IBL14 is an effective degradative strain for many steroidal compounds and can also catalyze the C25 hydroxylation of diosgenin, the first-ever biotransformation found on the F-ring of diosgenin.
Results
To completely elucidate the hydroxylation function of cytochrome P450 genes (CYPs) found during biotransformation of steroids by S. virginiae IBL14, the whole genome sequencing of this strain was carried out via 454 Sequencing Systems. The analytical results of BLASTP showed that the strain IBL14 contains 33 CYPs, 7 ferredoxins and 3 ferredoxin reductases in its 8.0 Mb linear chromosome. CYPs from S. virginiae IBL14 are phylogenetically closed to those of Streptomyces sp. Mg1 and Streptomyces sp. C. One new subfamily was found as per the fact that the CYP Svu001 in S. virginiae IBL14 shares 66% identity only to that (ZP_05001937, protein identifer) from Streptomyces sp. Mg1. Further analysis showed that among all of the 33 CYPs in S. virginiae IBL14, three CYPs are clustered with ferredoxins, one with ferredoxin and ferredoxin reductase and three CYPs with ATP/GTP binding proteins, four CYPs arranged with transcriptional regulatory genes and one CYP located on the upstream of an ATP-binding protein and transcriptional regulators as well as four CYPs associated with other functional genes involved in secondary metabolism and degradation.
Conclusions
These characteristics found in CYPs from S. virginiae IBL14 show that the EXXR motif in the K-helix is not absolutely conserved in CYP157 family and I-helix not absolutely essential for the CYP structure, too. Experimental results showed that both CYP Svh01 and CYP Svu022 are two hydroxylases, capable of bioconverting diosgenone into isonuatigenone and β-estradiol into estriol, respectively.
Keywords: Biotransformation, Cytochrome P450, Ferredoxin, Ferredoxin reductase, Gene sequencing, Secondary metabolism
Background
Cytochrome P450 (CYP) genes refer to such genes that encode a superfamily of iron-containing hemoproteins with a maximum absorption spectrum near 450 nm, often characterized by conserved Cys residue in hydrophobic pocket(s) [1]. Most of the ORFs of CYP have three distinct characteristics used often for their identification and analysis, i.e., the I-helix of putative CYPs (a highly conserved threonine involved in oxygen activation), the conserved EXXR motif located in the K-helix and the cytochrome P450 cysteine heme-iron ligand signature motif (GXXXCXG, there are exceptions) [2]. According to a widely-accepted taxonomy, CYPs within a family share more than 40% amino acid identity and members of subfamilies share more than 55% amino acid identity [3]. Occasionally, the decision to accept a sequence in a known family depends greatly on how it clusters on a tree, not so much on the absolute amino acid identity [4].
CYPs have been confirmed existing in all eukaryotic (human, animals, plants, fungi, etc.) and prokaryotic organisms (bacteria, archaea, and even in viruse) [5-8]. They often are monooxygenases involved in oxidation of a range of endogenous compounds, such as cholesterol, lipids and steroidal hormones, as well as xenobiotics such as drugs and toxic chemicals in environment [9-11]. CYPs catalyse diverse reactions, including C-H hydroxylation, epoxidation, hetero-atom oxidation, aromatic ring oxidation and dealkylation [11-13]. In the catalytic reaction process of P450 monooxygenase, one atom of O2 is inserted into substrate while the other is reduced to H2O. CYP genes responsible for secondary metabolism are often laid in antibiotic biosynthetic gene clusters to catalyze stereo- and region- specific reaction of substrates to related derivatives.
The biotransforming capabilities of bacterial CYPs have been widely elucidated. P450soy (CYP105D1) from Streptomyces griseus was involved in the degradation of a diverse array of complex agrochemicals and environmental pollutants [14]. CYP105C1 from Actinomycete spp. had the ability to transform benanomicin A into two derivatives, 10-hydroxybenanomicin A and 11-O-demethylbenanomicin [15]. The functions of related CYP107 family members have been reported. CYP107E from Micromonospora griseorubida was found to govern the hydroxylation and epoxidization in mycinamicin biosynthesis [16], P450 Terf (107 L) from Streptomyces platensis to catalyze hydroxylation of terfenadine [17] and hydroxylase PikC (107 L1) of Streptomyces venezuelae to convert narbomycin to picromycin [18]. CYP124 of Mycobacterium tuberculosis demonstrated omega-hydroxylase activity of relevant methyl-branched lipids [19]. YbdT (CYP152A) of Bacillus subtilis was involved in fatty acid beta-hydroxylation [20]. CYP154 of Nocardia farcinica IFM10152 had the functions of the O-dealkylation and ortho-hydroxylation of formononetin [21] and 154H1 from Clostridium acetobutylicum performed biocatalytic reactions with different aliphatic and aromatic substrates [22].
Genome sequencing is an effective way to predict and annotate all the possible CYPs genes in an organism. Streptomyces coelicolor A3 (2), a typical strain which is often used for the study of physiological function and antibiotic production, is the first Streptomyces species sequenced in 2001. Its linear chromosome is 8.7 Mb [23] which contains 7825 open reading frames (ORFs) with 18 putative CYPs [24]. S. avermitilis, known for producing the antiparasitic agent avermectin, contains 7600 ORFs with 33 putative CYPs in the 9 Mb chromosomes [25,26]. The genome of Streptomyces peucetius ATCC27592 with the size of 8.7 Mb contains 19 putative CYPs [27].
S. virginiae IBL14, isolated from activated sludge for treatment of waste from a steroidal drug factory, is an effective degradative strain of various steroidal compounds, including progesterone, isotestosterone, dihydrotestosterone, hydrocortisone, cholesterol and ostrone [28]. To comprehensively understand the function of CYPs of S. virginiae IBL14 in degradation and biotransformation of diosgenin, the whole genome sequencing of S. virginiae IBL14 isolated by our lab was carried out for the first time. Using in silico technology, we predict and annotate all of the putative CYPs of S. virginiae IBL14 and analyze these CYPs evolutionarily and functionally via comparison with those of other Streptomyces species. Furthermore, functions and characteristics of CYP genes svh01 and CYP svu022 in this strain are experimentally identified and analyzed.
Results and discussion
Genome sequencing and CYPs in S. virginiae IBL14
By in silico analysis of newly-sequenced S. virginiae IBL14 8.0 Mb genome, 8288 ORFs are identified and the total GC content exceeds 70%. The annotated results via Rpsblast display that there are a total of 33 putative CYPs in the genome of this strain IBL14, contributing to approximately 0.4% of all the coding sequences. The number of CYPs is identical to that in S. avermitili and almost two times as that in S. coelicolor A3(2) and S. peucetius ATCC27952 (18 and 19 CYPs, respectively). Such high level of CYP diversity suggests the high diversity of the secondary metabolism pathways in S. virginiae IBL14.
The 32 out of 33 putative CYPs of S. virginiae IBL14 belong to 13 previously-reported CYP families, i.e., 105 (5), 107 (11), 121 (1), 124 (1), 147 (1), 152 (1), 154 (1), 157 (2), 185 (1), 191 (3), 197 (4), 247(1) and another to an unknown family, as shown in Table 1. Among the all, the CYP121A (Svu018), CYP124 (Svu19), CYP147 (Svu020), CYP152 (Svu021), CYP154H (Svu022), CYP157 (Svu023-024), CYP185 (Svu025), CYP191 (Svu26-28), CYP197 (Svu017,029-031) and CYP247 (Svu032) are firstly reported in S. virginiae, and especially, CYP107M, CYP185A and CYP247A have been found rarely in Streptomycete spp. The Svu025, Svu026 and Svu029 have lower identity with other family members (<50%) while others show more than 63% identity to CYPs of other organisms. It’s worth noting that the Svu001 presumably belongs to a new CYP family since no close homologue is found in Genbank except that in Streptomyces sp. Mg1 with 66% identity.
Table 1.
Putative cytochrome P450s in S. virginiae IBL14 with their closest homologs
| IDa | Sizeb |
Best matches in the database |
||||
|---|---|---|---|---|---|---|
| Species | Protein identifier | CYP family | AA overlapd | identitye% | ||
| Svu001 |
464 |
Streptomyces sp. Mg1 |
ZP_05001937 |
new |
598 |
66 |
|
Photobacterium profundum 3TCK |
ZP_01217946 |
|
113 |
25 |
||
| Svu002 |
361 |
Streptomyces virginiae |
ABR68806 |
105 L |
134 |
100 |
|
Streptomyces clavuligerus ATCC 27064 |
ZP_06769587 |
|
93.2 |
66 |
||
| Svu003 |
439 |
Streptomyces venezuelae ATCC 10712 |
CCA59424 |
105C |
608 |
77 |
|
Streptomyces cattleya NRRL 8057 |
YP_004920090 |
105C |
553 |
70 |
||
| Svu004 |
398 |
Streptomyces virginiae |
ABR68806 |
105 L |
794 |
99 |
|
Streptomyces sp. ACT50-5 |
BAG16627 |
|
529 |
69 |
||
| Svu005 |
400 |
Streptomyces sp. C |
ZP_07285089 |
105D |
595 |
74 |
|
Streptomyces avermitilis MA-4680 |
BAC75180 |
105D7 |
529 |
69 |
||
| Svh01 |
399 |
Streptomyces virginiae |
ABR68805 |
105C1 |
797 |
99 |
|
Streptomyces viridochromogenes DSM 40736 |
ZP_07307444 |
105 |
703 |
87 |
||
| Svu006 |
403 |
Streptomyces virginiae |
ABR68807 |
107 L14 |
713 |
99 |
|
Streptomyces sp. C |
ZP_07284721 |
107 L14 |
611 |
87 |
||
| Svu007 |
351 |
Streptomyces sp. C |
ZP_07290554 |
107E |
609 |
87 |
|
Streptomyces violaceusniger Tu 4113 |
YP_004815015 |
|
540 |
77 |
||
| Svu008 |
406 |
Streptomyces sp. C |
ZP_07285026 |
107 L14 |
604 |
77 |
|
Streptomyces sp. Mg1 |
ZP_04997607 |
107 L14 |
584 |
76 |
||
| Svu009 |
415 |
Streptomyces sp. C |
ZP_07286517 |
107 L |
727 |
85 |
|
Streptomyces sp. Mg1 |
ZP_04999247 |
107 L |
484 |
59 |
||
| Svu010 |
396 |
Streptomyces sp. Mg1 |
ZP_04997607 |
107 L14 |
578 |
74 |
|
Streptomyces sp. C |
ZP_07285026 |
107 L14 |
556 |
72 |
||
| Svu011 |
405 |
Streptomyces sp. C |
ZP_07287693 |
107 L |
728 |
91 |
|
Streptomyces clavuligerus ATCC 27064 |
ZP_05005324 |
107 L |
601 |
75 |
||
| Svu012 |
430 |
Streptomyces sp. C |
ZP_07287209 |
107 L |
808 |
92 |
|
Streptomyces sp. Mg1 |
ZP_05000207 |
107 L |
780 |
91 |
||
| Svu013 |
396 |
Streptomyces sp. C |
ZP_07287353 |
107 L |
578 |
75 |
|
Streptomyces hygroscopicus subsp. jinggangensis 5008 |
AEY86095 |
|
383 |
54 |
||
| Svu014 |
395 |
Streptomyces sviceus ATCC 29083 |
ZP_06921933 |
107 L |
961 |
80 |
|
Streptomyces venezuelae ATCC 10712 |
CCA53921 |
107 L |
549 |
79 |
||
| Svu015 |
406 |
Streptomyces sp. Mg1 |
ZP_05001939 |
107 L |
667 |
80 |
|
Streptomyces scabiei 87.22 |
YP_003488837 |
107 L |
640 |
77 |
||
| Svu016 |
406 |
Amycolatopsis editerranei U32 |
YP_003767608 |
107 M |
482 |
63 |
|
Actinomadura hibisc |
BAA23153 |
|
387 |
55 |
||
| Svu017 |
368 |
Streptomyces avermitilis MA-4680 |
NP_823237 |
197A1 |
436 |
64 |
|
Streptomyces scabiei 87.22 |
YP_003487606 |
|
389 |
59 |
||
| Svu018 |
393 |
Streptomyces venezuelae ATCC 10712 |
CCA55152 |
121A |
509 |
67 |
|
Mycobacterium tuberculosis 02_1987 |
ZP_06504929 |
121A |
464 |
57 |
||
| Svu019 |
421 |
Streptomyces sp. C |
ZP_07287311 |
124B |
782 |
94 |
|
Streptomyces pristinaespiralis ATCC 25486 |
ZP_06909795 |
124B |
566 |
70 |
||
| Svu020 |
416 |
Streptomyces sp. C |
ZP_07289557 |
147A |
731 |
91 |
|
Streptomyces peucetius ATCC 27952 |
CAE53704 |
147A |
667 |
79 |
||
| Svu021 |
421 |
Streptomyces sp. C |
ZP_07290439 |
152A |
515 |
71 |
|
Streptomyces sp. SirexAA-E |
YP_004806454 |
152A |
429 |
58 |
||
| Svu022 |
412 |
Streptomyces sp. Mg1 |
ZP_05002011 |
154H |
742 |
91 |
|
Streptomyces sp. SirexAA-E |
YP_004804189 |
154H |
666 |
83 |
||
| Svu023 |
409 |
Streptomyces sp. C |
ZP_07285064 |
157A |
773 |
93 |
|
Streptomyces sp. Mg1 |
ZP_05002010 |
157A |
734 |
88 |
||
| Svu024 |
450 |
Streptomyces sp. Mg1 |
ZP_05002596 |
157C |
723 |
82 |
|
Streptomyces hygroscopicus ATCC 53653 |
ZP_07300920 |
157C |
574 |
64 |
||
| Svu025 |
89 |
Streptomyces tubercidicus |
AAT45286 |
185A1 |
85.9 |
47 |
|
Actinosynnema mirum DSM 43827 |
YP_003102184 |
185A |
84.7 |
51 |
||
| Svu026 |
409 |
Streptomyces violaceusniger Tu 4113 |
YP_004813101 |
191A |
313 |
44 |
|
Rhodococcus opacus B4 |
YP_002781958 |
|
300 |
43 |
||
| Svu027 |
398 |
Streptomyces sp. C |
ZP_07286547 |
191A |
756 |
92 |
|
Streptomyces sp. Mg1 |
ZP_04998169 |
191A |
733 |
89 |
||
| Svu028 |
446 |
Streptomyces sp. Mg1 |
ZP_04997583 |
191A |
699 |
88 |
|
Streptomyces sp. C |
ZP_07290135 |
191A |
692 |
90 |
||
| Svu029 |
476 |
Singulisphaera acidiphila DSM 18658 |
ZP_09568426 |
197A |
199 |
33 |
|
Streptomyces roseosporus NRRL 11379 |
ZP_04712663 |
197A |
191 |
32 |
||
| Svu030 |
447 |
Streptomyces sp. C |
ZP_07289871 |
197B |
713 |
82 |
|
Streptomyces sp. Mg1 |
ZP_05001362 |
197B |
680 |
77 |
||
| Svu031 |
710 |
Streptomyces sp. C |
ZP_07284739 |
197B |
353 |
79 |
|
Streptomyces clavuligerus ATCC 27064 |
ZP_05006237 |
|
350 |
55 |
||
| Svu032 | 416 |
Streptomyces flavogriseus ATCC 33331 |
YP_004921083 |
247A |
693 |
81 |
| Frankia alni ACN14a | YP_712777 | 247A | 573 | 70 | ||
a The name of the putative CYPs in S. virginiae IBL14.
b Amino acid number of putative CYPs.
c Closest homologs in Genbank and the family classification of CYPs searched in CYPED.
d Number of amino acid overlap, which exceeds the protein size, is due to the introduction of gaps during BLAST comparison.
e The highest percent identity for a set of aligned segments to the same subject sequence.
Features of CYPs from S. virginiae IBL14
Table 2 displays the three characteristic motifs of CYPs of S. virginiae IBL14. The critical residues are highlighted with bold fonts, which are threonine (T) in GXXTT motif of I-helix, glutamic acid (E) and arginine (R) in EXXR motif of K-helix and cysteine (C) in the GXXXCXG heme-binding domain signature, respectively.
Table 2.
A comparison of the conserved domain of putative CYPs in S. virginiae IBL14 with those of the same (sub)family in CYPED using ClustalW
| ID | I-helix | K-helix | Heme binding motif | Accession numbers |
|---|---|---|---|---|
| Svu001 |
Unidentified |
E335TLR338 |
F403LPFGAGPRHCVG415 |
JX119062 |
| Svu002 |
Unidentified |
Unidentified |
L297RVGVDRRLCCG308 |
|
| Svu003 |
G276LDTT280 |
E314LLR317 |
H375LGFGHGIHQCLG387 |
JX119063 |
| Svu004 |
G237HETT241 |
E275SLR278 |
H337LGFGHGIHQCLG349 |
JX119064 |
| Svu005 |
G247HETT251 |
E285LMR288 |
H346LAFGFGIHQCLG358 |
JX119065 |
| Svh01 |
G235FDTT239 |
E273LLR276 |
H334LAFSHGIHQCLG346 |
EF646279 |
| Svu006 |
G277HETT281 |
E315MLR318 |
H377IAFGHGLHYCLG389 |
JX119066 |
| Svu007 |
G238HETT342 |
E276LLR279 |
H339LGFGHGVHHCLG351 |
JX119067 |
| Svu008 |
G236HETT240 |
E275MLR278 |
H337LAFGHGLHFCIG349 |
JX119068 |
| Svu009 |
G236HKTT240 |
E274MQR277 |
H338LGFGYGAHYCLG350 |
JX119069 |
| Svu010 |
G234HETT238 |
E273MLR276 |
H335LAFGHGIHFCIG347 |
JX119070 |
| Svu011 |
G242HEAT246 |
E285LMR288 |
H346LTFGAGIHYCLG358 |
JX119071 |
| Svu012 |
G259FETT263 |
E302LLR305 |
H364LGYGHGIHYCLG376 |
JX119072 |
| Svu013 |
G237SETV241 |
E275LFR278 |
H337LALGHGVHYCLG349 |
JX119073 |
| Svu014 |
G234HETT238 |
E272LLR275 |
H334LAFGHGVHRCLG346 |
JX119074 |
| Svu015 |
F247APTT251 |
E285VVR288 |
Q347LSFGIGVHSCLG359 |
JX119075 |
| Svu016 |
G244YHTT248 |
E282ALR285 |
H345LAFGAGIHFCLG357 |
JX119076 |
| Svu017 |
G207FLTT211 |
E245GLR248 |
H307VAFGYGPHACPG319 |
JX119077 |
| Svu018 |
G231VIST235 |
E269LLR272 |
H332FSFGGGSHYCPA344 |
JX119078 |
| Svu019 |
G256VETT260 |
E295MIR298 |
H356LGFGGGGPHFCLG369 |
JX119079 |
| Svu020 |
G251HETT255 |
E289LLR292 |
H351LGLGSGIHSCFG363 |
JX119080 |
| Svu021 |
T247WFTT251 |
E281VRR284 |
E347LIAQGGGNARTGHRCPG364 |
JX119081 |
| Svu022 |
G251HETT255 |
E286TLR289 |
H349ISFGHGPHVCPG361 |
JX119082 |
| Svu023 |
G238HQPT242 |
E276VLW279 |
F337SFGHGEHRCPFPA350 |
JX119083 |
| Svu024 |
A247FETT251 |
E284QILW288 |
S344HLAFSSGPHECPG357 |
JX119084 |
| Svu025 |
Unidentified |
Unidentified |
H50LALGIGPHVCMG62 |
JX119085 |
| Svu026 |
G249NETT253 |
E287VLR290 |
H348LALGSGPHYCLG360 |
JX119086 |
| Svu027 |
G238NETT242 |
E274IVR277 |
H335LGFGGGGPHFCLG348 |
JX119087 |
| Svu028 |
G284NDTV288 |
E322LLR325 |
H383VSFGDGPHVCLG395 |
JX119088 |
| Svu029 |
A242HETT246 |
E297TLR300 |
A367FMPFGGGPRTCLG380 |
JX119089 |
| Svu030 |
G259HETT263 |
E314AMR317 |
A383WFPFGGGPRACIG396 |
JX119090 |
| Svu031 |
G499HETT503 |
E545TLR548 |
A614YLPFGIGPGPAWARSSRCGS634 |
|
| Svu032 | A252NVTT256 | E290GLR293 | R351HGAFGFGPHFCIG364 | JX119091 |
From the Table 2, we can find the I-helix is absent in Svu001(new family), and the I-helix and K-helix missing in Svu002 (105 L, often for hydroxylation activity) [29], which reflects I-helix is not absolutely essential for the CYP structure. The 2 members of CYP157 family Svu023 (E276VLW279)/157A and Svu024 (E284QILW288)/157C do not have arginine residue in K-helix like the CYP157C1 from S. coelicolor A3(2) having a motif E 297 QSLW [30] and the CYP157A2 and CYP157C2 from S. avermitilis exhibiting a 257EVLW motif and a 257EQSLW motif [26]. The CYP157 family proteins that lack consensus EXXR motifs but genetically are linked to their upstream conservons imply that they have functions linked to the upstream pathway(s) [30]. Besides, Svu002, Svu018, Svu021, Svu023 and Svu031 do not strictly follow the GXXXCXG motif of heme-binding.
Multiple alignments and phylogenetic analysis
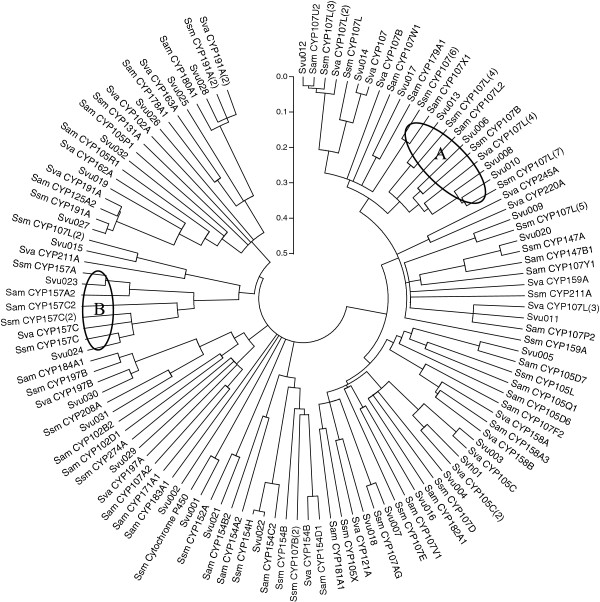
The phylogenetic tree of the combined CYPs of S. virginiae IBL14, S. avermitilis MA-4680, S. venezuelae ATCC 10712 and Streptomyces sp. Mg1 is presented in Figure 1. From Figure 1, we can find almost of all the CYPs in S. virginiae IBL14 are closely related to their homologues. More than 10 of CYPs from S. virginiae IBL14 are close to those from Streptomyces sp. Mg1 and the member (Svu001) of new CYP family found in S. virginiae IBL14 is only close to Streptomyces sp. Mg1. These results indicate that the CYPs from S. virginiae IBL14 are closer to those from Streptomyces sp. Mg1 than those from other Streptomyces spp, including S. avermitilis MA-4680 and S. venezuelae ATCC 10712. For the four species of S. virginiae IBL14, sp. Mg1, avermitilis MA-4680 and S. venezuelae ATCC 10712, the families CYP 107 and CYP157 (labeled with circle A and B in Figure 1, respectively) have more closely evolutionary relationship.
Figure 1.
Phylogenetic tree of the CYPs from S. virginiae IBL14 and three related bacteria. Sequences were aligned using Clustal W and the tree was calculated and constructed using MEGA 5.0. (Streptomyces sp. Mg1, Ssm; S. avermitilis MA-4680, Sam; S. venezuelae ATCC 10712, Sva).
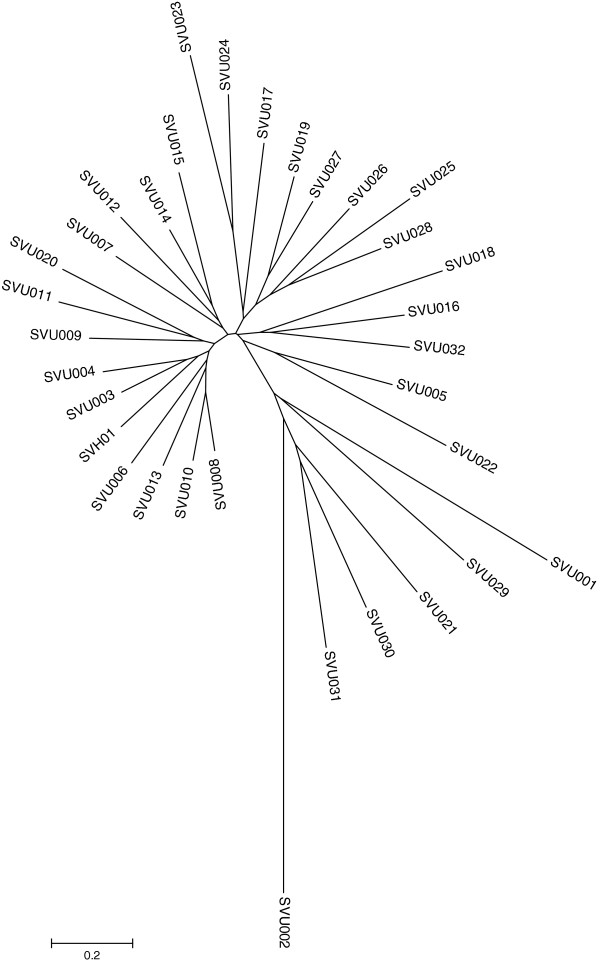
Further, the paralogous relationship of the 33 CYPs in S. virginiae IBL14 was generated with the neighbor-joining methods (Clustal W and MEGA 5.0). From Figure 2, we can find that svh01 and svu03 and svu04 as well as svu022 and svu005 in S. virginiae IBL14 have the closest homologous evolutionary relationship, respectively. It’s worth noting that most members belonging to the same CYP family are clustered together as expected, e.g., the 11 members of CYP107 family.
Figure 2.
A paralogous tree of all CYP sequences from S. virginae IBL14.
The prediction of functions of CYPs in S. virginiae IBL14
A high identity over 70% among different protein sequences reasonably suggests that they may hold similar function [26]. As shown in the Table 1, we can find a sum of 26 CYP sequences of S. virginiae IBL14 have best matches to those of other Streptomyces, which are helpful in function prediction.
CYP105 and CYP107 are the most studied bacterial cytochromes which are associated with the degradation and biotransformation of a diverse array of xenobiotics and antibiotic biosynthesis. Analysis of CYPs sequence of S. virginiae IBL14 shows that there are 11 CYPs belonging to CYP107, five to CYP105, four to CYP197, three to CYP191, two to CYP157 and one to each other family, which indicates the diversity and importance of the two groups CYP105 and CYP107. The predicted functions of several putative CYPs in S. virginiae IBL14, combined with reported experimental evidences, were listed in Table 3.
Table 3.
Prediction of functions of several putative CYPs in S. virginiae IBL14
| ID | Functions | Reference |
|---|---|---|
| Svu003 |
Hydroxylation & O-demethylation |
[15] |
| Svu005 |
N-demethylation & Hydroxylation |
[31] |
| Svh01 |
Hydroxylation |
[32] and this study |
| Svu006 |
Hydroxylation |
[17] |
| Svu007 |
Hydroxylation |
[16] |
| Svu019 |
Hydroxylation |
[19] |
| Svu021 |
Hydroxylation& Decarboxylation |
[33] |
| Svu022 | Hydroxylation& O-dealkylation | This study |
CYPs in S. virginiae IBL14 and their ferredoxin reductase and ferredoxin
The catalytic activity of CYPs depends greatly on individual ferredoxin or/and ferredoxin reductase associated with. It was reported that there are three, six and four ferredoxin reductase genes and six, nine and two ferredoxin genes in S. coelicolor A3 (2), S. avermitilis and S. peucetius, respectively. In S. coelicolor A3 (2) only CYP105D5 is arranged in an operon with a ferredoxin gene [24]. In S. peucetius CYP147F is clustered with ferredoxin reductase [27]. In S. avermitilis both CYP105P1 and CYP105D6 are clustered with ferredoxin, CYP147B1 is arranged in an operon with a ferredoxin and ferredoxin reductase, CYP105Q1 is associated in an operon containing both a ferredoxin and ferredoxin reductase, and CYP102 is fused to a P450 reductase [26].
Three ferredoxin reductase genes and seven ferredoxin genes are found in S. virginiae IBL14 after annotation of S. virginiae IBL14 genome. That is, the activities of many of the CYPs in S. virginiae IBL14 are supported by different combinations with the three ferredoxin reductases and seven ferredoxins. Also in S. virginiae IBL14, svu005 (CYP105D), svh01 (CYP105C) and svu019 (CYP124B) is found to cluster with ferredoxin svf03, svf09 and svf07, respectively and svu020 (CYP147A) clustered with ferredoxin reductase svfr03 and ferredoxins svf06. The facts suggest that the functional realization of CYPs Svu005, Svh01, Svu019 and Svu020 needs the participation of electron transfer. The result of homology analysis by Blast-searching the Genbank are listed in the Table 4.
Table 4.
Putative ferredoxin reductases and ferredoxins in S. viginiae IBL14 with their closest homologs
| IDa | Accession numbers | NO. nucleic acids |
Match in the databasesb |
||
|---|---|---|---|---|---|
| Species | Accession | Identity% | |||
| Putative ferredoxin reductases | |||||
|
svfr01 |
JX119052 |
453 |
Streptomyces sp. C |
ZP_07290734 |
94 |
|
Streptomyces pristinaespiralis ATCC 25486 |
ZP_06911868 |
92 |
|||
|
svfr02 |
JX119053 |
463 |
Streptomyces sp. C |
ZP_07285271 |
87 |
|
Streptomyces sp. Mg1 |
ZP_05002250 |
84 |
|||
|
svfr03 |
JX119054 |
464 |
Streptomyces sp. C |
ZP_07289558 |
94 |
|
Streptomyces peucetius ATCC 27952 |
CAF33360 |
84 |
|||
| Putative ferredoxins | |||||
|
svf03 |
JX119055 |
219 |
Streptomyces sp. C |
ZP_07285090 |
84 |
|
Streptomyces viridochromogenes DSM 40736 |
ZP_07308348 |
79 |
|||
|
svf04 |
JX119056 |
1143 |
Streptomyces sp. Mg1 |
ZP_04996989 |
83 |
|
Streptomyces griseoflavus Tu4000 |
ZP_07315146 |
83 |
|||
|
svf05 |
JX119057 |
234 |
Streptomyces sp. C |
ZP_07286537 |
88 |
|
Streptomyces sp. Mg1 |
ZP_05002165 |
79 |
|||
|
svf06 |
JX119058 |
231 |
Streptomyces peucetius ATCC 27952 |
ACE73829 |
62 |
|
Streptomyces hygroscopicus subsp |
AEY87986 |
61 |
|||
|
svf07 |
JX119059 |
600 |
Streptomyces sp. C |
ZP_07287304 |
98 |
|
Streptomyces venezuelae ATCC 10712 |
CCA56325 |
94 |
|||
|
svf08 |
JX119060 |
315 |
Streptomyces sp. C |
ZP_07285869 |
89 |
|
Streptomyces peucetius ATCC 27952 |
ACE73824 |
88 |
|||
| svf09 | JX119061 | 243 |
Streptomyces cattleya NRRL 8057 |
YP_004920089 |
63 |
| Streptomyces diastaticus | AAR16520 | 61 | |||
a The name of gene in S. virginiae IBL14.
b Homologues searched in Genbank.
Regulatory elements and functional genes clustered with CYPs
The CYPs in S. peucetius ATCC27952 clustered with regulatory elements were reported [27]. In the annotations of gene arrangement around the putative CYPs on the S. virginiae IBL14 chromosome, svu022, svu023 and svu024 were found to cluster with the genes of ATP/GTP binding proteins (having a phosphate-binding loop for energy requiring metabolic reactions) [34], svu001, svu015 to cluster with LysR-family transcriptional regulator (regulating a diverse set of genes, including those involved in virulence, metabolism, quorum sensing and motility) [35], svu011 to cluster with two component transcriptional regulators and LuxR family (quorum sensing signals in Gram-negative bacteria often regulated by acylated homoserine lactones) [36], svu018 to cluster with a transcriptional regulator, AraC family (transcriptional regulators having diverse functions ranging from carbon metabolism to stress responses to virulence) [37] and two component transcriptional regulators, LuxR family and svu020 to cluster with the ATP-binding protein fbpC and TetR-family transcriptional regulators (among bacteria with an HTH DNA-binding motif for the transcriptional control of multidrug efflux pumps, pathways for the biosynthesis of antibiotics, response to osmotic stress and toxic chemicals, control of catabolic pathways, differentiation processes, and pathogenicity) [38].
As described above, the CYPs in S. virginiae IBL14 chromosome are responsible for the transcriptional regulation of many functional genes related with primary, secondary metabolism, as well as the responses to environmental factors as expected. Besides, CYPs are clustered with other functional genes. svh01 is adjacent to the genes of MdlB, ABC-type multidrug transport system, ATPase and permease components, which may be involved in the transportation of substrates [39]. svu009 lies next to alcohol dehydrogenase, suggesting that svu009 may take part in alcohol bioconversion and biodegradation. svu013 is next to 4, 5-DOPA dioxygenase which is a member of the class III extradiol dioxygenase family (a group of enzymes which use a non-heme Fe (II) to cleave aromatic rings between a hydroxylated carbon and an adjacent non-hydroxylated carbon), suggesting that the combination of svu013 and 4, 5-DOPA dioxygenase may be responsible in biodegradation of substrates with aromatic rings. svu026 is adjacent to MbtH-like protein which is found in known antibiotic synthesis gene clusters [40]. The cholesterol oxidase ChoL from S. virginiae IBL14 in the bioconversion and biodegradation of diosgenin responsible for the conversion of diosgenin to diosgenone (a 4-ene-3-keto steroid) via a couple of C3-dehydrogenation and C4-5-isomerization was reported [41]. In S. virginiae IBL14 the gene encoding Svu004 (CYP105L) clusters with the genes of putative ferredoxin Svfr2 and cholesterol oxidase (ChoL), suggesting that the cytochrome P450 joins with the cholesterol oxidase ChoL to catalyze the oxidation of cholesterol and its structural analogs. In conclusion, CYPs from S. virginiae IBL14 may have multiple functions in secondary metabolism, including hydroxylation, dehydrogenation, ring-cleavage, transportation, etc.
Functional identification and characteristics of svh01 and svu022
To elucidate all putative CYPs’ functions in S. virginiae IBL14, four CYP genes of the strain IBL14 were firstly selected. Among them, the functional identities of CYP genes Svh01(105C1) and svu022 (154H) has been finished.
The cytochrome P450 Svh01 (responsible for the C25-hydroxylation of diosgenin) [32] belongs to the class I (prokaryotic/mitochondrial) P450 system based on a taxonomic split, in which electrons are transferred from NADPH or NADH to ferredoxin reductase and ferredoxin. Sequence analysis revealed the complete sequence of svh01 with ATG as the start codon has 70% G + C content. The sequence of possible ribosome-binding site is located on the upstream of svf09 (a coenzyme of Svh01).
Both svh01 and svf09 contain 1200 bp and 243 bp, respectively, based on sequence analysis. To obtain the expressed products of them, both svh01 and svf09 sequences were first ligated into a pET22b vector in a cluster to generate the expression plasmid pET22b-svh01-svf09 that was then cloned into E. coli JM109 (DE3) to form a recombinant strain E. coli IBL161 [JM109 (DE3)/pET22b-svh01-svf09]. The PCR results of svh01 and svf09 from the recombinant strain E. coli IBL161 were analyzed by gel electrophoresis (Figure 3A and B) and also confirmed by gene sequencing.
Figure 3.
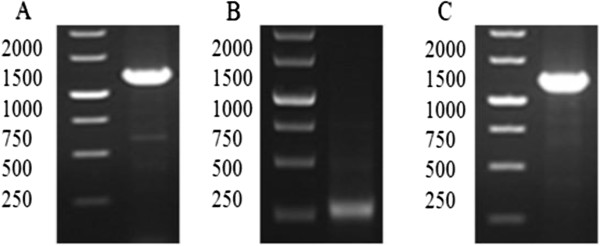
DNA analysis of svh01, svf09 and svu022 cloned from S. virginiae IBL14. A, B and C are the PCR results of svh01, svf09 and svu022, respectively.
The svu022 with a G + C content of 73% (clustering with the gene of ATP/GTP binding protein) consists of 1239 nucleotides. Similarly, the complete sequence of svu022 was first inserted to the shuttle plasmid pHCMC05 to form the recombinant plasmid pHCMC05-svu022, and then cloned in B. subtilis WB800N (improving the extracellular expression level of Svu022 for the analysis of enzymatic biotransformation) to produce the recombinant strain B. subtilis IBL 241 [WB800N/pHCMC05-svu022]. The PCR result of svu022 from the recombinant strain B. subtilis IBL 241 is shown in Figure 3C.
Svh01 (105C1) is a peptide of 399 amino acids, with a molecular weight of 44.04 kDa and a pI value of 4.97 estimated by the ExPASy (a computing pI/MW tool). To obtain its expressed product and study product characteristics, the recombinant strain E. coli IBL161 was incubated and induced. The expression of Svh01 was shown in Figure 4A. From the SDS-PAGE, we can find that the two distinctly additional protein bands should be Svh01 with an about MW of 44 kDa and Svf09 with an about MW of 8.0 kDa, respectively. The further functional identification of the Svh01/FcpC of S. virginiae IBL14, hydroxylating the C25-tertiary carbon of diosgenin to form isonuatigenone, was experimentally confirmed [32].
Figure 4.
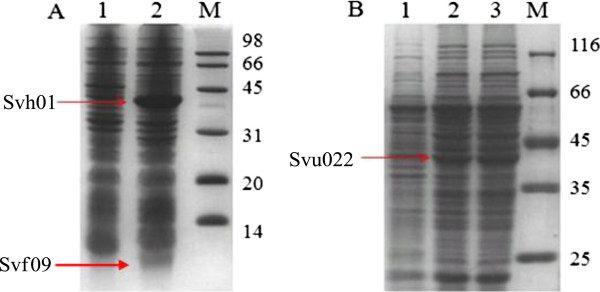
SDS-PAGE analysis of Svh01 and Svu022. (A) Lane 1, sample from JM109 (DE3)/pET22b cells; Lane 2, sample from E. coli IBL161. (B) Lane 1, WB800N/pHCMC05 cells; Lane 2 and 3, B. subtilis IBL 241.
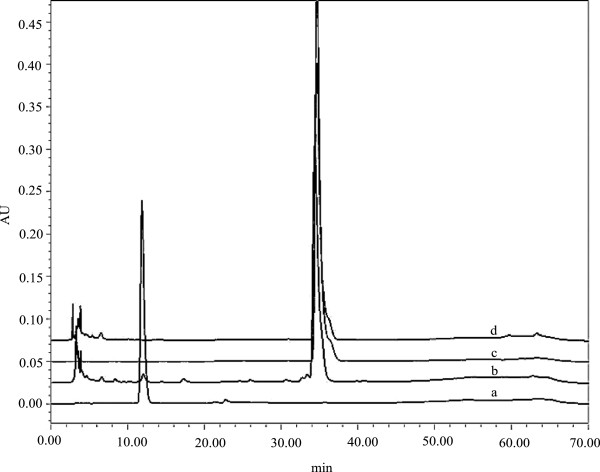
Svu022 (154H) is a deduced protein of 412 amino acids which shares 91% identity with that in Streptomyces sp. Mg1. The estimation of MW and pI of SVU022 are 44.59 kDa and 5.00, respectively. Similarly, the recombinant strain B. subtilis IBL 241 was incubated and induced to study the product expression and its characteristics. The expressed result of Svu022 from the recombinant strain B. subtilis IBL 241 was shown in Figure 4B. The SDS-PAGE displays a distinct protein band with about MW of 45.0 kDa as expected. The further experimental results from TLC, HPLC and LC/MS indicated that the CYP Svu022 enables to biotransform β-estradiol into estriol. Figure 5 shows the profiles of the biotransformation of β-estradiol by strains B. subtilis WB800N and B. subtilis IBL 241 in HPLC. The functional identification of the Svu005 (CYP105D) and Svu019 (CYP124B) is in progress.
Figure 5.
The profiles of the transformation of β-estradiol by strain B. subtilis IBL 241 in HPLC. a: standard estriol; b: sample from B. subtilis IBL 241; c: standard β-estradiol; d: sample from B. subtilis WB800N.
Conclusion
S. virginiae IBL14 contains 33 putative CYPs, 7 ferredoxins and 3 ferredoxin reductases in its 8.0 Mb linear chromosome. Most of the CYPs in S. virginiae IBL14 belong to the CYP107 (11 members) family and CYP105 (5 members) family. Compared phylogenetically with CYPs from 3 typical Streptomycete spp., S. virginiae IBL14 appears to be closest to those of Streptomyces sp. Mg1.
Further analysis showed that among all of the 33 CYPs in S. virginiae IBL14, three CYPs are clustered with ferredoxins, one with ferredoxin and ferredoxin reductase and three CYPs with ATP/GTP binding proteins, four CYPs arranged with transcriptional regulatory genes and one CYP locates on the upper of ATP-binding protein and transcriptional regulators as well as four CYPs associated with other functional genes involved in secondary metabolism and degradation.
The new characteristics found in CYPs from S. virginiae IBL14 suggest that the EXXR motif in the K-helix is not absolutely conserved in CYP157 family as reported [30] and I-helix not absolutely essential for the CYP structure. Particularly, one new family was found based on the CYP svu001 in S. virginiae IBL14 which shares 66% identity only to that from Streptomyces sp. Mg1.
Two recombinant strains E. coli IBL161 [JM109 (DE3)/pET22b-svh01-svf09] and B. subtilis IBL 241 [WB800N/pHCMC05-svu022] were constructed and subsequently their functions were identified, respectively. Experimental results showed that both CYP Svh01 and CYP Svu022 are two hydroxylases, capable of bioconverting diosgenone into isonuatigenone and β-estradiol into estriol, respectively.
Methods
Strains and plasmids
S. virginiae IBL-14 (CCTCCM 206045) [42] as the strain of interest was used for the Cytochrome P450 gene identification and functional analysis. E. coli JM109, JM109 (DE3) and B. subtilis WB800N were used as the host for plasmid construction and target protein expression in the functional identification of the CYPs, respectively. The vector pET22b was used for cloning and expression of genes of interest in E. coli. The shuttle plasmid pHCMC05 was used for the expression of target proteins in B. subtilis (a GRAS strain by FDA). The features of the bacterial strains and plasmids used in this study are listed in Table 5.
Table 5.
Microorganisms and plasmids used in this study
| Strains | Relevant properties | Source |
|---|---|---|
|
Escherichia coli | ||
| JM109 |
Cloning host, genotype:endA1, recA1, gyrA96, thi, hsdR17 (rk–, mk+), relA1, supE44, (lac-proAB), [F’ traD36, proAB, laqIqZΔM15] |
Promega |
| JM109 (DE3) |
Expression host, genotype:endA1, recA1, gyrA96, thi, hsdR17 (rk–, mk+), relA1, supE44, λ–, Δ(lac-proAB), [F’, traD36, proAB, lacIqZΔM15], lDE3 |
Promega |
|
Bacillus subtilis | ||
| WB800N |
Secretion host with resistance to neomycin, genotype: nprE aprE epr bpr mpr :: ble nprB :: bsr vpr wprA :: hyg cm :: neo; NeoR |
Mo Bi Tec |
|
Streptomyces virginiae | ||
| IBL14 |
Wild type |
Our lab |
| Plasmids | ||
| pET22b |
Expression vector in E. coli |
Novagen |
| pHCMC05 |
Shuttle plasmid |
BGSC |
| pET22b-svh01-svf09 |
The fragment of svh01 and svf09 were digested with NdeI/EcoRIand EcoRI/Hind Ш, respectively, and ligated into the NdeIand Hind Ш sites of pET22b |
This study |
| pHCMC05-svu022 | The gene of svu022 digested with BamHI/SmaIligated into BamHI/SmaIdigested pHCMC05 | This study |
Media and cultivation
Luria-Bertani (LB) medium was used for plasmid construction and protein expression. A final concentration of 70 μg/ml ampicillin was supplemented into the medium when E. coli IBL161 [JM109 (DE3)/pET22b-svh01-svf09] and E. coli IBL152 [JM109/pHCMC05-svu022] were cultivated. A final concentrations of 25 μg/ml chloramphenicol was added to the medium when B. subtilis IBL 241 [WB800N/pHCMC05-svu022] was cultivated. The cultivating procedure of S. virginiae IBL-14 has been described previously [42]. Diosgenin in 95% purity (J&K Chemical Ltd, China) and β-estradiol in 98% purity (J&K Chemical Ltd, China) were dissolved in anhydrous ethanol before adding into medium.
Sequencing and in-silico identification analyses of CYPs
The S. virginiae IBL14 genome sequencing was performed at 454 platform (Encode Genomics Co. Ltd., Suzhou, China) for the first time (sequence data will be published step by step). All of the ORFs of this genome were predicted using glimmer3.0 and prodigal, respectively. To dig out all possible CYP gene function information in S. virginiae IBL14, the genome sequence of the strain was compared with the SWISSPROT, TrEMBL, KEGG databases by using Blastp and the CDD and COG databases by using Rpsblast, respectively.
The deduced amino acid sequences of the putative CYPs of S. virginiae IBL14 were aligned with the CYPs from S. avermitilis MA-4680, S. venezuelae ATCC 10712 and Streptomyces sp. Mg1 by using ClustalW [43]. Then the molecular evolution and phylogenetic analyses by neighbor-joining methods were carried out using MEGA5.0 [44]. To forecast the possible functions involved in secondary metabolism, comparison between all putative CYPs of S. virginiae IBL14 with those in other organisms based on homologues was done by using Blastp too.
Using the three motifs as described above as criteria, the CYP gene candidates of S. virginiae IBL14 were blast searched against GenBank non-redundant protein database to identify their closest bacterial homologues and tentatively distribute all of the CYPs of S. virginiae IBL14 into the corresponding family or subfamily [26]. Similar procedure was performed to the putative ferredoxin and ferredoxin reductase genes to identify their closest bacterial homologues.
Construction and cloning of expression plasmids
The genes of svh01, svf09 and svu022 from the genomic DNA of S. virginiae IBL14 were amplified by using PCR method (Pfu DNA Polymerase, Fermentas, Thermo Fisher Scientific Inc.) and the primers used are listed in Table 6. The PCR products of svh01 and svf09 were digested with NdeI/EcoRIand EcoRI/Hind III, respectively, then ligated into a pET22b vector, and finally transformed to the host bacterium E. coli JM109 (DE3). Similarly, the PCR product of svu022 was digested with BamHI/Sma I, then ligated into a shuttle plasmid pHCMC05 and finally transformed to B. subtilis WB800N.
Table 6.
The PCR primers used in this study
| Primer | Primer sequencea (5′ to3′) | Restriction site |
|---|---|---|
| pSVH01F |
GCCCCCCATATGAGTGAGTCCCTCCACACCGTC |
NdeI |
| pSVH01R |
GGAGGAATTCACTTCGCGTCCCAGGTG |
EcoRI |
| pSVF09F |
CCGGAATTCGGGACGCGAAGTGAGCGCGG |
EcoRI |
| pSVF09R |
CCCAAGCTTTCAGGCGGAGGGTGGGCGG |
Hind III |
| pSVU022F |
CTGGATCCATGAGCTGCCCGATCGACC |
BamHI |
| pSVU022R | CCTAAGCTTTCAGGGGTGCAGGCGTACCG | SmaI |
aThe underlined sequence are recognition sites of restriction enzymes and the nucleotides before it are the protected bases. All primers are designed by Primer Premier 5.0 and verified by Oligo 7.0.
Expression and analysis of target proteins
0.3 ml (inoculation ratio of 1%) of the overnight culture of E. coli IBL161 as seed was inoculated in 30 ml LB medium (containing 70 μg/ml ampicillin) and then cultivated at a shaking speed of 200 rpm at 37°C. The expression of target protein was induced by adding 0.2 mM IPTG when the OD value reached 0.5 ~ 0.6 at 600 nm. Then the culture was continuously cultivated for another 24 h at 25°C at a speed of 200 rpm in a rotary shaker. Similarly, the overnight culture of B. subtilis IBL 241 was inoculated with 1% ratio in 30 ml LB medium (25 μg/ml chloramphenicol, 200 rpm at 30°C). After adding 0.2 mM IPTG in logarithmic growth phase, the culture was continuously cultivated for another 48 h at the same conditions. The harvested recombinant cells were resuspended and subjected to ultrasonication in 50 mM PBS (pH 7.4), and then centrifuged at 6000 rpm for 5 min. The supernatant was analysed by SDS-PAGE.
Biotransformation and product extraction
One milliliter of β-estradiol/diosgenin (a final concentration of 0.2 mg/ml) for each flask was added for biotransformation analysis after E. coli IBL161 was induced by IPTG at 25°C for 2 h. After cultivated for another 24 h under the same conditions, the cultures were extracted two times with a half volume of 100% ethyl acetate (Sinopharm Chemical Reagent Co., Ltd). The extracts were evaporated to dryness, then re-dissolved in 1 ml anhydrous ethanol, and finally detected and analyzed (thin layer chromatography/TLC, high performance liquid chromatography/HPLC and liquid chromatography–mass spectrometry LC-MS).
DNA and protein analytical methods
DNA electrophoresis for recombinant plasmid analysis was carried out in agarose gels at 110 V for 30 min [45]. SDS-PAGE with a 15% (w/v) acrylamide gel for expressed protein analysis was run at 110 V for 2 h according to Schagger’s publication [46]. The bands were visualized by Coomassie R-250 staining.
HPLC analysis of biotransformation products
To identify and analyze the metabolites, high performance liquid chromatography (HPLC) was carried out. Simply, the sample of 10 μl was first loaded onto 250 mm Symmetry C 18 (4.6 mm × 250 mm, Waters Co., USA) and eluted with ethanol/water (60/40, v/v). The flow rate, the wavelength for UV-detection and the temperature of the column on the HPLC system (Breeze 1525 series, Waters Co., USA) were set at 1 ml/min, 245 nm and 35°C, respectively. The products after biotransformation were qualitatively and quantitatively analyzed by comparing with corresponding standard material.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Z-ZL carried out the bioinformatic and genomic analyses and experiments and drafted the manuscript. X-FL carried out the experiments and bioinformatic and genomic analyses, WY, XD, JY, S-LZ, ML, LX participated in the experiments or interpretation of results. W-YT contributed to study design, data analysis and writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Zhi-Zhen Li, Email: kungfuyang@126.com.
Xiao-Fei Li, Email: lxfqd99@163.com.
Wei Yang, Email: bukeneng_100@163.com.
Xiang Dong, Email: dongx61@126.com.
Jie Yu, Email: jieyu@mcmaster.ca.
Shu-Liang Zhu, Email: zhu_shu_liang@126.com.
Man Li, Email: lme123654@sina.com.
Li Xie, Email: xieli6860131@163.com.
Wang-Yu Tong, Email: tongwy@ahu.edu.cn.
Acknowledgments
Preparation of this manuscript was supported by the National Natural Science Foundation of China (20976001) and by the Key Scientific Research Projects of Department of Education of Anhui Province of China (KJ2009A161) and by the Academic Leader Foundation of 211 Project (phase-3) of Anhui University of China (AU02303154). Also, we are grateful to Hui Peng and Ya-Zhong Xiao, School of Life Science, Anhui University, Hefei, China, for generous gift of plasmids and to Hong Yu, Encode Genomics Co. Ltd., Suzhou, China, for the genome sequencing of the strain IBL14.
References
- Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure (London, England: 1993) 1995;3(1):41–62. doi: 10.1016/S0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6(1):1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Cytochrome P450 nomenclature, 2004. Methods Mol Biol (Clifton, NJ) 2006;320:1–10. doi: 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Cytochrome P450 nomenclature. Methods Mol Biol. 1998;107:15–24. doi: 10.1385/0-89603-519-0:15. [DOI] [PubMed] [Google Scholar]
- Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Current drug metabolism. 2002;3(6):561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- Mogi T, Kita K. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol Int. 2010;59(3):305–312. doi: 10.1016/j.parint.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Fu C, Xiong J, Miao W. Genome-wide identification and characterization of cytochrome P450 monooxygenase genes in the ciliate Tetrahymena thermophila. BMC Genomics. 2009;10(1):208. doi: 10.1186/1471-2164-10-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb DC, Lei L, Warrilow AGS, Lepesheva GI, Mullins JGL, Waterman MR, Kelly SL. The first virally encoded cytochrome p450. J Virol. 2009;83(16):8266–8269. doi: 10.1128/JVI.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21(1):70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- Pryor WA. Cytochrome P450: Structure, mechanism, and biochemistry. New York: Montellano Plenum Press; 1996. [Google Scholar]
- McLean KJ, Hans M, Munro AW. Cholesterol, an essential molecule: diverse roles involving cytochrome P450 enzymes. Biochem Soc Trans. 2012;40(3):587–593. doi: 10.1042/BST20120077. [DOI] [PubMed] [Google Scholar]
- Ouellet H, Johnston JB, Ortiz de Montellano PR. The Mycobacterium tuberculosis cytochrome P450 system. Arch Biochem Biophys. 2010;493(1):82–95. doi: 10.1016/j.abb.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JB, Ouellet H, Podust LM, Ortiz de Montellano PR. Structural control of cytochrome P450-catalyzed ω-hydroxylation. Arch Biochem Biophys. 2011;507(1):86–94. doi: 10.1016/j.abb.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trower MK, Lenstra R, Omer C, Buchholz SE, Sariaslani FS. Cloning, nucleotide sequence determination and expression of the genes encoding cytochrome P-450soy (soyC) and ferredoxinsoy (soyB) from Streptomyces griseus. Mol Microbiol. 1992;6(15):2125–2134. doi: 10.1111/j.1365-2958.1992.tb01386.x. [DOI] [PubMed] [Google Scholar]
- Kumagai H, Umekita M, Sawa R, Takahashi Y, Arisawa A, Isshiki K, Nishimura Y, Akamatsu Y. Generation of new benanomicin analogues by biotransformation using Escherichia coli expressing actinomycete cytochrome P450. J antibiotics. 2008;61(6):394–399. doi: 10.1038/ja.2008.55. [DOI] [PubMed] [Google Scholar]
- Inouye M, Takada Y, Muto N, Beppu T, Horinouchi S. Characterization and expression of a P-450-like mycinamicin biosynthesis gene using a novel micromonospora-Escherichia coli shuttle cosmid vector. Mol Gen Genet MGG. 1994;245(4):456–464. doi: 10.1007/BF00302258. [DOI] [PubMed] [Google Scholar]
- Lombard M, Salard I, Sari M-A, Mansuy D, Buisson D. A new cytochrome P450 belonging to the 107 L subfamily is responsible for the efficient hydroxylation of the drug terfenadine by Streptomyces platensis. Arch Biochem Biophys. 2011;508(1):54–63. doi: 10.1016/j.abb.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Betlach MC, Kealey JT, Ashley GW, McDaniel R. Characterization of the macrolide P-450 hydroxylase from Streptomyces venezuelae which converts narbomycin to picromycin. Biochemist. 1998;37(42):14937–14942. doi: 10.1021/bi981699c. [DOI] [PubMed] [Google Scholar]
- Johnston JB, Kells PM, Podust LM, Ortiz de Montellano PR. Biochemical and structural characterization of CYP124: a methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis. Proc Nat Academy Sci United StatesAm. 2009;106(49):20687–20692. doi: 10.1073/pnas.0907398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef NH, Wofford N, McInerney MJ. Importance of the long-chain fatty acid beta-hydroxylating cytochrome P450 enzyme YbdT for lipopeptide biosynthesis in Bacillus subtilis strain OKB105. Inter J Mol Sci. 2011;12(3):1767–1786. doi: 10.3390/ijms12031767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-Y, Park H-Y, Kim B-G. Characterization of bi-functional CYP154 from Nocardia farcinica IFM10152 in the O-dealkylation and ortho-hydroxylation of formononetin. Enzyme Microbial Technol. 2010;47(7):327–334. doi: 10.1016/j.enzmictec.2010.08.006. [DOI] [Google Scholar]
- Schallmey A, den Besten G, Teune IGP, Kembaren RF, Janssen DB. Characterization of cytochrome P450 monooxygenase CYP154H1 from the thermophilic soil bacterium Thermobifida fusca. Appl Microbiol Biotechnol. 2011;89(5):1475–1485. doi: 10.1007/s00253-010-2965-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Lamb DC, Skaug T, Song H-L, Jackson CJ, Podust LM, Waterman MR, Kell DB, Kelly DE, Kelly SL. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3(2) J biol chemist. 2002;277(27):24000–24005. doi: 10.1074/jbc.M111109200. [DOI] [PubMed] [Google Scholar]
- Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979;15(3):361–367. doi: 10.1128/AAC.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL. Cytochrome p450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2) Biochem Biophysical Res Commun. 2003;307(3):610–619. doi: 10.1016/S0006-291X(03)01231-2. [DOI] [PubMed] [Google Scholar]
- Parajuli N, Basnet DB, Chan Lee H, Sohng JK, Liou K. Genome analyses of Streptomyces peucetius ATCC 27952 for the identification and comparison of cytochrome P450 complement with other Streptomyces. Archives Biochemist Biophys. 2004;425(2):233–241. doi: 10.1016/j.abb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Wang F-Q, Zhang C-G, Li B, Wei D-Z, Tong W-Y. New microbiological transformations of steroids by Streptomyces virginiae IBL-14. Environ Sci Technol. 2009;43(15):5967–5974. doi: 10.1021/es900585w. [DOI] [PubMed] [Google Scholar]
- Yasutake Y, Imoto N, Fujii Y, Fujii T, Arisawa A, Tamura T. Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) Biochem Biophys Res Commun. 2007;361(4):876–882. doi: 10.1016/j.bbrc.2007.07.062. [DOI] [PubMed] [Google Scholar]
- Rupasinghe S, Schuler MA, Kagawa N, Yuan H, Lei L, Zhao B, Kelly SL, Waterman MR, Lamb DC. The cytochrome P450 gene family CYP157 does not contain EXXR in the K-helix reducing the absolute conserved P450 residues to a single cysteine. FEBS Lett. 2006;580(27):6338–6342. doi: 10.1016/j.febslet.2006.10.043. [DOI] [PubMed] [Google Scholar]
- Taylor M, Lamb DC, Cannell R, Dawson M, Kelly SL. Cytochrome P450105D1 (CYP105D1) from Streptomyces griseus: heterologous expression, activity, and activation effects of multiple xenobiotics. Biochem Biophys Res Commun. 1999;263(3):838–842. doi: 10.1006/bbrc.1999.1427. [DOI] [PubMed] [Google Scholar]
- Wang W, Wang F-Q, Wei D-Z. Characterization of P450 FcpC, the enzyme responsible for bioconversion of diosgenone to isonuatigenone in Streptomyces virginiae IBL-14. Applied environ microbiol. 2009;75(12):4202–4205. doi: 10.1128/AEM.02606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A. Terminal olefin (1-alkene) biosynthesis by a novel p450 fatty acid decarboxylase from Jeotgalicoccus species. Appl Environ Microbiol. 2011;77(5):1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15(11):430–434. doi: 10.1016/0968-0004(90)90281-F. [DOI] [PubMed] [Google Scholar]
- Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiol (Reading, England) 2008;154(Pt 12):3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- Nasser W, Reverchon S. New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Analytical Bioanalytical Chemist. 2007;387(2):381–390. doi: 10.1007/s00216-006-0702-0. [DOI] [PubMed] [Google Scholar]
- Frota CC, Papavinasasundaram KG, Davis EO, Colston MJ. The AraC family transcriptional regulator Rv1931c plays a role in the virulence of Mycobacterium tuberculosis. Infect Immun. 2004;72(9):5483–5486. doi: 10.1128/IAI.72.9.5483-5486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69(2):326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72(2):317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri LEN, Sello J, Keating TA, Weinreb PH, Walsh CT. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chemist Biol. 1998;5(11):631–645. doi: 10.1016/S1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- Li B, Wang W, Wang F-Q, Wei D-Z. Cholesterol oxidase ChoL is a critical enzyme that catalyzes the conversion of diosgenin to 4-ene-3-keto steroids in Streptomyces virginiae IBL-14. Applied Microbiol Biotechnol. 2010;85(6):1831–1838. doi: 10.1007/s00253-009-2188-0. [DOI] [PubMed] [Google Scholar]
- Wang F-Q, Li B, Wang W, Zhang C-G, Wei D-Z. Biotransformation of diosgenin to nuatigenin-type steroid by a newly isolated strain, Streptomyces virginiae IBL-14. Applied microbiol biotechnol. 2007;77(4):771–777. doi: 10.1007/s00253-007-1216-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mole Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel D. Molecular cloning: a Laboratory manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochem. 1987;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]