Abstract
Amide hydrogen/deuterium exchange detected by mass spectrometry (HXMS) is seeing wider use for the identification of intrinsically disordered parts of proteins. In this review, we discuss examples of how discovery of intrinsically disordered regions and their removal can aid in structure determination, biopharmaceutical quality control, the characterization of how posttranslational modifications affect weak structuring of disordered regions, the study of coupled folding and binding, and the characterization of amyloid formation.
Keywords: amide exchange, intrinsically disordered proteins, flow quench, amyloid, coupled folding and binding, biopharmaceuticals
1. INTRODUCTION
1.1 Intrinsically Disordered Proteins in health and disease
At the time the complete human genome was sequenced, Dunker‘s group, attempting to predict the structures of all the new protein coding sequences, noted that the published genomes contained sequences that did not look like they would code for structured proteins 1. Remarkably, some 30% of bacterial proteins, nearly 40% of archaeal proteins and perhaps as many as 50% of eukaryotic proteins were predicted to contain stretches of at least 40 consecutive disordered amino acids 2. Around the same time, protein biophysicists discovered several proteins that appeared to not have a stable, unique structure, at least until they interacted with their physiological partners 3; 4; 5; 6. The first review describing these early experimental results was published in 1999 7. Many algorithms for prediction of disordered sequences in proteins have since been developed 8; 9; 10; 11; 12; 13; 14; 15.
Intrinsically disordered proteins (IDPs) are found in functional niches that include transcriptional and translational control, cell cycle control and signaling. IDPs were found to be enriched among cancer related and signaling proteins 16, all processes that are in some way involved in physiological control. IDPs are frequently associated with diseases. Increases in the number of disordered repeat sequences increases susceptibility to the neurodegenerative diseases Huntington’s and Parkinson’s diseases 17; 18; 19. Mutations in disordered regions can also result in the loss of important post-transcriptional modification sites, leading to disease 20. Diseases such as cancer, cardiovascular disease, amyloidosis, neurodegenerative diseases and diabetes often involve disordered proteins 17; 21 probably because these often involve failures in protein signaling, structure, posttranslational modification and inability to interact correctly with physiological partners 21. The more virulent strains of disease-causing viruses may be characterized by their increased levels of intrinsic disorder 22, and viruses in general appear to have a high proportion of genes potentially coding for disordered proteins 23.
1.2 Experimental Characterization of Intrinsic Disorder
Intrinsically disordered regions of proteins can range from surface loops in otherwise structured proteins that are not observed in x-ray crystal structures to long stretches of polypeptide with low sequence complexity and no predicted structure. It is probably most helpful to think of intrinsic disorder as a continuum from slightly less than uniquely structured all the way to completely unfolded. Given this viewpoint, it is easy to see how different experimental measures will weigh-in differently on whether or not a particular region of a protein qualifies as intrinsically disordered. NMR spectroscopy is often a good experimental approach to detecting intrinsic disorder, as it provides a highly sensitive read-out of resonances that change their chemical environment and this is very useful for monitoring changes in an IDP upon interaction with a binding partner 24; 25. NMR is also very useful for detecting protein dynamics, however there is a blind spot for dynamic regions that move on the NMR timescale because resonances that exchange between different chemical environments will have broadened signals that may disappear completely. If the disordered region is so dynamic that the atoms move very quickly between different environments, the resonances will have averaged chemical shifts and will appear as strong signals. Other more global indicators of secondary structure, such as circular dichroism (CD) and/or packing of the hydrophobic cores, 1-anilino 8-naphthalene sulfonate (ANS) binding and fluorescence changes are also useful for detecting large regions of disorder and changes in disorder upon binding.
1.3 Amide Exchange
The use of mass spectrometry to detect amide hydrogen/deuterium exchange (HDX) has found increasing applicability since its inception in the early 1990’s 26. Both electrospray ionization (ESI) 26 and MALDI-TOF 27 are suitable approaches for analyzing whether an amide group has exchanged with solvent deuterium when the sample is incubated in 2H2O (D2O). H/D exchange analysis can be performed on intact proteins 28; 29, however it is most often useful to perform the exchange reaction and then digest the protein with an acid-tolerant protease to localize the sites of exchange 30.
Mass spectrometry-detected amide exchange (HXMS) experiments were traditionally performed to study protein folding and unfolding and the measured amide proton exchange rates were used to interpret the amount of hydrogen bond 31, however, HXMS can also be a useful indicator of solvent accessibility changes at protein surfaces due to ligand binding 32; 33; 34 and for epitope mapping of antibodies 35. The rates of amide exchange in small peptides have been accurately measured by NMR, and these reveal subtle differences in the intrinsic rates of exchange due to sequence context 36. NMR has the advantage that the rate of exchange of single amides is measured, but the disadvantage is that the protein must be of a size that is tractable by NMR, and it must be able to be expressed and isotopically labeled with 15N so that the resonance assignments for the amide protons can be made. Mass spectrometric measurements avoid the need for isotopic labeling of the protein and can be performed on proteins of any size 37. The mass spectrometry measurements rely on proteolytic digestion of the protein and analysis of the peptide fragments. Thus, the resolution is limited by the sizes of the peptides although, if many overlapping peptides are obtained, this can improve the resolution. Top-down approaches as well as various alternative ionization schemes have recently been demonstrated to yield accurate single amide resolution as well 38; 39.
In folded proteins, the rates of exchange vary widely and depend on exposure to solvent as well as folding and/or dynamics. If the rate of exchange of a single amide can be measured, it is possible to compute a protection factor, which is simply the ratio of the intrinsic exchange rate to the observed rate. Protection factors vary by 10–12 orders of magnitude from amides in completely unfolded proteins to amides in very well-folded regions of proteins. Teasing apart the contributions of dynamics, solvent accessibility, and foldedness remains an unsolved problem. Computer simulations can reproduce the HDX results 40; 41 and theoretical analyses of amide exchange in proteins are now allowing insights into the mechanisms of exchange 42.
This review will discuss what can be learned about IDPs and intrinsically disordered regions of proteins from HXMS experiments. In the first part, we will discuss the utility of HXMS for detecting intrinsically disordered regions in proteins. In the second part, we will discuss how HXMS can be used to monitor coupled folding and binding, which is a phenomenon that is extremely common among IDP sequences, as they often are only disordered prior to finding their binding partner. In the third part, we will discuss the use of HXMS to characterize aggregates and oligomers. Another property of IDPs is that they are prone to aggregation, and this often leads to diseases such as Parkinson’s Alzheimers’ and Creutzfeld-Jacob disease among others. HXMS is extremely useful in characterizing the oligomers that form from IDPs. With evidence mounting that the disease-causing forms of these aggregation-prone proteins may not be the final fibrillar form but an intermediate oligomer, HXMS is becoming critical for characterizing such intermediates. In the final part, we will discuss where developments could still be made to expand the information that can be gained from analyzing IDPs by HXMS.
2.1 IDENTIFICATION OF DISORDERED REGIONS IN PROTEINS BY HXMS
2.1.1 Identification and removal of intrinsically disordered regions for structure determination
Unstructured or disordered regions often pose practical problems when it comes to structure determination by X-ray crystallography and NMR. In X-ray crystallography, flexible or disordered regions often interfere with crystal formation and result in poor quality crystals. Even if crystals can be obtained, the electron density from disordered regions often cannot be resolved and appropriate structural models of these regions cannot be built. NMR resonances for disordered regions usually are highly overlapped making them hard to assign and sometimes mask the signal from the structured residues. Disordered regions often also weakly associate, and at the high protein concentrations necessary for structure determination, weak association can cause line broadening. It is preferable, therefore, to remove these unstructured regions prior to structure determination, as long as the structural integrity is not lost.
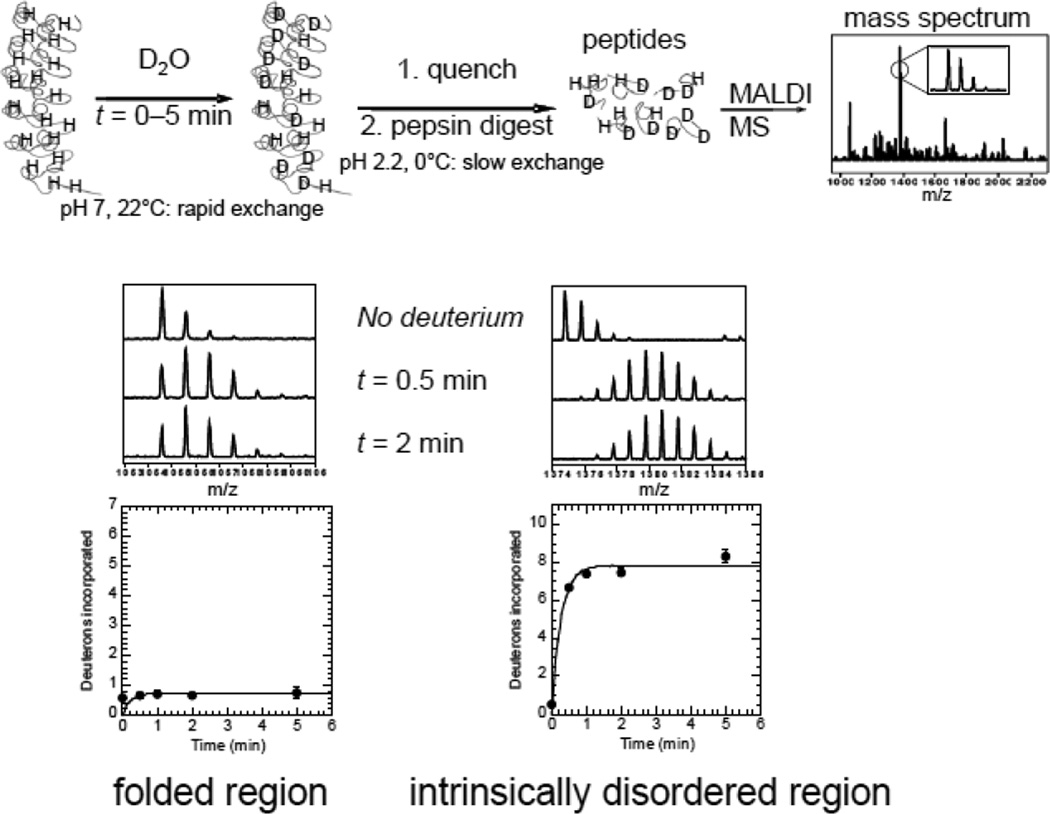
HXMS is being increasingly used to discover problematic disordered regions for construct optimization in structure determination. As outlined schematically in Figure 1, regions of intrinsic disorder exchange within a few minutes, and are readily identified by a protocol in which the protein is incubated for short periods of time in deuterated buffer to exchange the exposed amides, then the reaction is quenched by decreasing the pH to 2.5 and the temperature to 0°C, the sample is digested with pepsin, and finally mass spectrometry is used to determine the deuterium content of the resulting peptides (Figure 1).
Figure 1.
Schematic diagram of the HXMS experiment used to discover intrinsically disordered protein regions. A generic example is given of a repeat protein for which the structure was known only when the protein was in complex with its binding partner. Intrinsic disorder is readily characterized by rapid amide exchange after only a few minutes. There is some residual deuteration at t=0 due to the experimental set-up. The data were fitted to single or double exponential equations. Typically, the rate of the first exponential was set to 30 s−1 and the second exponential as well as the amplitude of each exponential phase were fitted parameters. The amplitude (number of amides exchanging) of the rapidly exchanging group can be taken as approximately the number of amides exchanging at the intrinsic rate.
Pantazatos et al. have outlined an improved HXMS method to allow rapid identification of unstructured regions in proteins and have demonstrated the use of this technique in improving crystallographic success 43. As part of a large scale structural genomics project, they screened 24 proteins from T. maritima by HXMS. The proteins were recombinantly expressed and purified from E. coli and protein fragmentation maps after pepsin digestion and ESI LC MS analysis before and after exchange with D2O for 10 s were analyzed to determine the amides that readily exchanged and identify unstructured and highly dynamic parts of the proteins. The entire analysis was completed rapidly (within 2 weeks) and data were obtained for 21 of the 24 proteins screened. Complete coverage was obtained for 16 of the 21 proteins. Comparison of the HXMS data to the known crystal structure of the T. maritima thy1 protein TM0449 showed that they were able to accurately localize disordered regions within the protein. They identified two regions in the protein that exchanged with D2O rapidly that corresponded to parts in the crystal structure that were disordered. One of the highly exchanging regions corresponded to the substrate binding region in the protein demonstrating how HXMS can provide information on binding sites within proteins 32. They also compared the exchange results for the T. maritima GroES heat shock protein TM0505, which produced poorly diffracting crystals to the homologous M. tuberculosis structure and showed that the highly dynamic regions of the protein mapped mostly to the disordered residues in GroES that are involved in binding to GroEL. From the results of the HXMS analysis the authors were able to design constructs that would have better crystallographic success. Indeed, truncation of highly disordered C-termini detected by HXMS in both TM0160 and TM171 led to improved crystals leading to higher resolution structures of 1.7 Å and 2.3 Å, respectively. Overall the authors were able to successfully crystallize 12 of the 21 proteins for which HXMS data was obtained.
Sharma et al. described the use of HXMS to optimize constructs for NMR analysis of five targets chosen from the Northeast Structural Genomic Project: brain specific protein C32E8.3 from Caenorhabditis elegans; DUF896 family protein YnzC from Bacillus subtilis; protein YjcQ from Bacillus subtilis; cytoplasmic protein Q8ZRJ2 from Salmonella typhimurium, and Escherichia coli lipoprotein YiaD 44. YjcQ and Q8ZRJ2 are highly structured proteins and the HXMS results obtained for them were consistent with the predominantly ordered solution structures of these targets. Removal of disordered tail regions in C32E8.3 and YnzC improved the quality of the HSQC spectra and NMR 3D structural data obtained for these proteins allowing structure determination of the folded regions. YnzC is only 77 residues long and HXMS analysis showed that two regions, one in the middle of the protein and one at the C-terminus were highly disordered. In order to optimize the NMR conditions, five different truncations were made. Remarkably, removal of almost 50% of the sequence (up to 37 amino acids from the C-terminal end of the protein) did not affect the chemical shifts and ensuing structure determination of the remaining small structured region. Determination of the structure of the full-length E. coli lipoprotein YiaD, a desirable structural genomics target, proved impossible. Interestingly, neither disorder predictors nor secondary structure predictors identified disorder within the N-terminal region of YiaD, however, HXMS analysis suggested that it was in fact, disordered. Removal of this sequence led to determination of the structure of this here-to-fore inaccessible structural target.
Recently, Ertekin et al. have applied the HXMS/ NMR technique to optimize constructs for NMR analysis of CDK2AP1 (cyclin-dependent kinase 2-associated protein 1), a tumor suppressor gene that is deleted in oral cancer 45. The CDK2AP1 protein is 115 residues long and had been proposed to form a homodimer with a disulfide bond between the Cys105 residues. HXMS results showed that almost 50% of the N-terminal end of the protein was highly dynamic and unstructured. This information helped researchers to design a construct comprised only of the C-terminal half of the protein. Subsequent structural analysis by NMR showed that this protein is a homodimer with a 4 helix bundle structure and Cys105 is positioned to form a disulfide between the two monomers.
2.1.2 Characterization of receptor cytoplasmic tails and the effects of post-translational modifications
Determination of structures of receptor cytoplasmic tails is difficult as they are usually small, have several disordered regions, and are subject to post translational modifications and conformational changes upon ligand binding 46. HXMS can be extremely useful in determining folded and unfolded regions of cytoplasmic tails of receptors. An interesting example that was studied by HXMS is the cytoplasmic tail of the low density lipoprotein receptor-related protein, LRP1 (LRP1 CT) 47. This short intracellular region of 100 amino acids has two NPXY motifs, NPXY4507 and NPXY4473. The NPXY sequence motif is found in many tyrosine-phosphorylated proteins and is required for the specific binding of the protein interaction domains (PIDs, also known as phosphotyrosine binding domains or PTBs). In LRP1 the membrane proximal NPXY motif binds to Snx17 (sorting nexin 17) when it is not phosphorylated and binds to Shp2 when both of the NPXY motifs are phosphorylated. HXMS analysis of GST tagged LRP1 CT constructs showed that the membrane-distal NPXY4507 motif is unstructured in the protein and easily phosphorylated by v-Src, whereas the membrane-proximal NPXY4473 is not exposed to exchange nor is it readily phosphorylated. However, upon phosphorylation of Y4507, the NPXY4473 motif became more solvent-exposed and also more susceptible to v-Src phosphorylation. The results were further confirmed using a Y4507E mutant of LRP1 CT. In order to directly compare the solvent accessibility of the phosphorylated and unphosphorylated forms in the same reaction, the authors phosphorylated 15N-labeled protein and then mixed it with unlabeled, unphosphorylated protein prior to HXMS. The exposure of Y4473 to become phosphorylated only after Y4507 is phosphorylated has implications with respect to the binding and signaling by Snx17 and Shp2 that are regulated by phosphorylation. HXMS can be very useful in determining the dynamics of such regions as shown in the above example.
2.1.3 Characterization/quality control of biopharmaceuticals
Mass spectrometry has gained importance in the characterization of biopharmaceuticals in the recent years. Kaltashov et al. have reviewed several different uses of biological mass spectrometry applied to biopharmaceutical characterization 48. Biopharmaceuticals, which are mainly protein-based therapies that vary from 5 to greater than 100 kDa, are becoming more prevalent. Proper folding of these biopharmaceuticals is critical for their effectiveness and safety, and quality control requires thorough characterization of their primary, secondary and tertiary structures as well as post-translational modifications. HXMS is useful in ensuring proper folding and is extremely useful in determining if small parts of the protein vary in solvent exposure. HXMS was used to characterize the recombinant form of acid-β-glucocerebrosidase (GCase) which is used as a treatment for Gaucher's disease by enzyme replacement therapy 49. If the protein is extensively oxidized during the manufacturing process it can be detected using a combination of native ESI-MS and HXMS. The protein’s charge distribution is changed due to a less compact conformation in ESI MS analysis and HXMS revealed further insight into the partial unfolded state of the recombinant GCase by comparing the spectra for intact and oxidized forms of the protein 50. A detailed analysis of the results suggests that upon oxidation the activity of GCase is highly diminished. The ability of HXMS to detect changes in conformation of protein drugs resulting from different aspects of design and processing has led to the use of this technique in biopharmaceutical characterization 51; 52.
2.2 COUPLED FOLDING AND BINDING OF IDPs ANALYZED BY HXMS
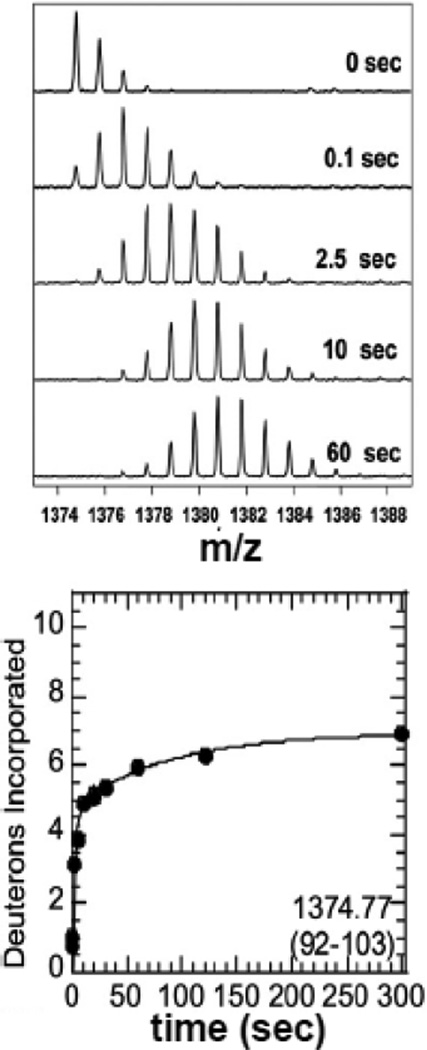
Although some IDPs appear to always exist in an unfolded state, many more are only unfolded until they find their binding partner, and fold upon binding. Since weakly-folded or unfolded proteins exchange rapidly whereas folded proteins exchange orders of magnitude more slowly, HXMS provides a sensitive indicator of coupled folding and binding. One of the first examples of coupled folding and binding detected by HXMS was that of the inhibitor, IκBα, which binds to various members of the NFκB transcription factor family via its ankyrin repeat domain (ARD) 53. The IκBα ARD contains six ankyrin repeats (ARs), and HXMS of IκBα on its own revealed that only the central three ARs were protected from exchange 54. In fact, many of the amides in ARs 1, 5, and 6 exchanged in a few seconds as revealed by flow quench amide exchange coupled to MALDI-TOF mass spectrometry 55 (Figure 2). The flow quench approach, pioneered by Dharmasiri and Smith 56, involves rapid mixing of the protein with D2O so that mixing and quench steps can be accomplished within tens of milliseconds. Flow quench followed by either MALDI-TOF or ESI MS allows measurement of the rates of exchange of even the very rapidly exchanging amides.
Figure 2.
Example of HXMS data from a region of IκBα collected using a quench-flow apparatus that allows rapid mixing and measurement of exchange after 10 msec. The mass spectra show incorporation of deuterons in times under 1 sec can be readily determined allowing the determination of how many amides are exchanging at the intrinsic rate indicating that they are in completely unstructured regions.
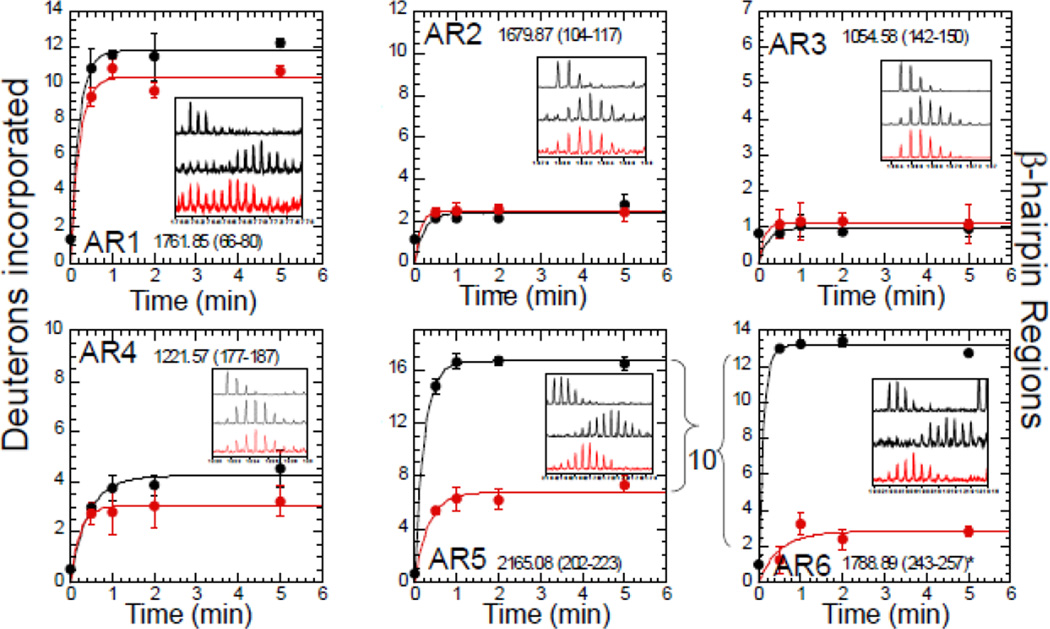
In studying coupled folding and binding by HXMS, some issues arise regarding interpretation of the data. Most HXMS data represents an ensemble of rates since deuterium exchange is quantified at the peptide rather than residue level. Thus, individual rates of exchange cannot be discerned from the data, however it is possible to “bin” the data into the number of amides exchanging at the intrinsic rate, at an intermediate rate, and at a slow rate 34. Using such an analysis, it was possible to demonstrate that a relatively large number of amides in ARs of IκBα exchange at the intrinsic rate reported for amides in completely unstructured peptides 36. Later, it was shown that although amides in the β-hairpins of AR5-AR6 exchange rapidly in the free state, they exchange much more slowly when bound to NFκB (Figure 3). The marked decrease in the number of amides that exchanged by 10 min (over 10 in the beta hairpins of AR5 and AR6) could only be explained if these repeats folded upon binding to NFκB 57.
Figure 3.
Example of how HXMS data can reveal a coupled folding and binding event. Each panel shows the HXMS data from one of the six β-hairpins of IκBα in the free state (black symbols) compared to the NFκB-bound state (red symbols). It can be seen that the folded regions (from repeats 2, 3, and 4) do not change much upon binding whereas the intrinsically disordered regions (AR5–6) exchange completely in the free state, but in the bound state their exchange resembles that of the folded ARs 2, 3, and 4. The data are from Truhlar et al., (2006) 57.
The intrinsically disordered proteins CREB-binding protein (CBP) and activator of thyroid and retinoid receptors (ACTR) both fold upon binding to one another and these proteins and their binding have been well-studied by NMR 58. Weis’s group used HXMS to measure amide exchange of CBP, ACTR, and the complex between 5 sec and 12 h and then fitted the results with stretched exponentials. After defining a segment averaged protection factor, they were able to define regions of each protein that contained amides that exchange at the intrinsic rate 59. In this case, nearly all of the CBP exchanges more slowly in the bound form as compared to the free. The region defined by the peptic fragment spanning residues 33–47 was completely exchanged by the first measurement, and others, such as 48–59 were nearly completely exchanged (cf. Figure 2 in Keppel et al).
Both AR5–AR6 in IκBα and CBP are only marginally stable. CBP appears to undergo two-state unfolding with a ΔGu of 6.1 ± 0.4 kJ/mol 60, and therefore approximately 8% of the CBP population can be expected to be unfolded at any given time at 300 K. Oas and colleagues have analyzed in full kinetic detail the ramifications of small amounts of unfolded populations based on NMR-determined protection factors (which are derived accurately from the exchange rates of individual amides) 61. Essentially, since the unfolded portion exchanges so much more quickly than the folded portion, molten globules will appear to be more unfolded than they really are. Only if the exchange between states is extremely slow, or if one population of states is much more highly exchanging than another, will two separable mass envelopes be observed in the HXMS spectra, a phenomenon that is referred to as EX1 behavior. In most cases, molten globules are a population of related states that interconvert on relatively fast timescales and then as the time of deuteration is increased, a single mass envelope is observed that gradually increases in mass, a phenomenon that is referred to as EX2. Because the molten globule is spending some time in a largely unfolded state, the amount of exchange will appear more rapid than expected due to the increased exchange from sparsely populated unfolded states.
2.3 CHARACTERIZATION OF OLIGOMERS AND AGGREGATES OF IDPS
HXMS is a powerful tool for probing the solvent accessibility of regions of IDPs when they form oligomers or aggregates. Typically, only a portion of the protein will be buried in the oligomer, and this portion will have decreased amide exchange. Studies of IκBα were some of the first to demonstrate this phenomenon. IκBα is a marginally stable protein, and its thermal denaturation is irreversible. When monomeric IκBα is incubated for as little as 10 min at 37°C, a soluble aggregate forms that elutes from a size exclusion column in the void volume 54. To characterize this soluble aggregate, we performed HXMS experiments at room temperature and after the protein had been incubated at 37°C for 10 min. Because the amide exchange experiment was carried out at 37°C, regions that remained in the same folded state as at 25°C would be expected to exchange more rapidly because amide exchange is a temperature-dependent reaction. Regions of ARs 2 and 3 showed this expected increase in HDX rate for the higher temperature (37 vs. 25°C) (Figure 4 peptides shown in green). On the other hand, regions of the protein that became buried in the core of the soluble aggregate that forms at 37°C would be expected to exchange LESS than they exchanged at 25°C, when the protein was in the monomeric state. Regions of ARs 1, 5, and 6 showed this behavior as they became much less solvent accessible at the higher temperature (Figure 4 peptides shown in red). These results made a lot of sense because the regions of ARs 2 and 3 (green peptides) are the regions of IκBα that are most well-folded, and so were not expected to unfold and form an aggregate. Conversely, regions of ARs 1, 5, and 6 (red peptides) are the regions of IκBα that deviate from the consensus for stable ankyrin repeats, and might be expected to unfold at 37°C and become buried in the aggregate. Amide exchange is a powerful method that can sample the protein backbone as it engages in aggregate formation and provide a “read-out” of which regions of the protein actually participate in aggregate formation and which retain their folded state and go along for the ride.
Figure 4.
An example of how HXMS data can be used to identify the regions of a protein that are buried in an aggregate vs. those that are not. Incubation of IκBα at 37°C rapidly promotes aggregate formation. When HDX is carried out at 37°C, the exchange rate is expected to be faster than at 25°C. The β-hairpins from ARs 2 and 3 show this behavior (green lines) however segments from ARs 1, 5, and 6 exchange more slowly at 37 °C than at 25 °C indicating they have become buried in the aggregate that forms at the higher temperature. The data are from Hughes et al., (2004) 54. The regions of IκBα marked with green bars had an increase in the number of amides exchanged in 5 min from an average of 1.4 at 25°C to an average of 5.4 at 37°C, consistent with the expected increase due to the increased temperature of the exchange reaction. The regions of IκBα marked with red bars had a decrease in the number of amides exchanged in 5 min from 5.6 to 4.6 in the N-terminus, from 12 to 10 in AR4 and from 18 to 15 in the PEST.
Jorgensen’s group characterized a proteolytic fragment of β2macroglobulin (ΔK58-β2m), which is known to cause dialysis-related amyloidosis 62. Operating under the well-established assumption that propensity to unfold is linked to propensity to aggregate into amyloid fibrils, these researchers used HXMS to characterize the proteolyzed variant, ΔK58-β2m. Their results showed that a significant proportion of this variant which was much less stable, was also unfolded at 37°C. Since the ΔK58-β2m variant had a different molecular weight, they were able to directly compare it to the full-length protein in the same reaction. HXMS of the ΔK58-β2m variant showed the evolution of a bimodal mass distribution with 18 of about 29 previously protected amide hydrogens exchanging in a correlated manner forming a separate mass envelope from the more highly exchanging protein population (ie. EX1 behavior). The full-length protein also showed bimodal mass envelopes indicative of such EX1 exchange, but on a much longer time scale. Thus, it was possible to surmise that the region of the protein that unfolded in a correlated manner, as indicated by HXMS, would be the region that would initially participate in amyloid formation. In addition, thioflavin-T dye binding, a classic indicator of amyloid formation, showed that the ΔK58-β2m variant formed amyloid-like aggregates orders of magnitude faster than the full-length protein.
PrP is a monomeric, α-helical protein that is called a prion because it can transition to a misfolded, aggregated form termed PrPSc, resulting in transmissible spongiform encephalopathy (TSE), or prion disease 63; 64. HDX approaches are becoming widely used to discover which regions of IDPs are buried in various oligomeric states of prion proteins. Wemmer’s group used both HXMS and exchange-quenched NMR to probe the regions of the prion protein, PrP (89–143, P101L) that participate in fibril formation 65. They found two regions, residues 102–109 and 117–136 that were highly protected (with a half-life of exchange longer than one week!) in the fibril form. In addition, the NMR experiments revealed that the intervening segment, residues 110–116, had multi-exponential exchange behavior indicating that it was sampling different chemical environments.
Smirnovas et al., recently reported the characterization of the transformation of the cellular form of PrP, PrPC to the amyloid form, PrPSc, in protein isolated from infected mouse brains 66. The authors took advantage of a mouse model that does not have the capability of attaching glycosyl-phosphatidylinositol (GPI) linkers into the PrPC. The PrPC that does not have the GPI linker attached was better proteolyzed, and therefore the coverage of the C-terminal part of the protein in the HXMS experiments was greatly improved. The prions isolated from the brains of these mice are highly infective suggesting that the lack of post-translational modifications does not affect the structure of the disease-causing protein oligomer. HXMS results allowed the authors to discriminate between different theoretical models of the oligomerization process, providing strong evidence for refolding of the entire C-terminal part of the protein. These researchers also provided evidence that the structure (based on HXMS evidence) of the native oligomer resembled the aggregates that could be generated in vitro confirming earlier HXMS experiments 67as well as the model by Dima and Thirumalai 68. The results highlight the fact that HXMS is the ONLY label-free approach that can be used to study disease-causing IDPs isolated from natural sources.
One of the pioneers of using HXMS to study amyloids was Carol Robinson. By carefully devising a pulse-labeling approach, she was able to discern the kinetics of formation and recycling of oligomers during the formation of Aβ fibrils. This work highlighted for the first time the dynamic nature of the fibrils, which many researchers had mistakenly imagined were static once fibrils had formed 69.
2.1 FUTURE OUTLOOK
HXMS is finding many useful applications within the realm of discovery of intrinsically disordered regions in proteins. The advent of automated instrumentation will allow high throughput analysis to become routine 70, and in the next few years it is likely that quality control of all protein biopharmaceuticals will require HXMS characterization since it reveals site-specific defects in expressed proteins whereas the more commonly used analyses such as HPLC retention time or CD only report defects in the globally averaged structure. It is also likely that crystallographers and structural biologists in general will routinely screen their constructs by HXMS to discover problematic disordered regions. High throughput instrumentation will also enable the use of HXMS for fragment screening in therapeutic discovery because often drug binding results in decreased exchange of dynamic/disordered loop regions. The study of coupled folding and binding, that still relies mostly on NMR approaches will benefit greatly from advances in ionization/fragmentation that allow single amide resolution in HXMS studies 71; 72; 73. For these detailed biophysical characterizations, the ability to measure rates of exchange of individual amides in the free and bound states will provide a wealth of kinetic and thermodynamic information that remains elusive. Such developments will likely also be critical to the discovery of the actual disease-causing oligomers of amyloidogenic IDPs. Here, the ability of HXMS to characterize proteins isolated from natural sources promises to be essential for understanding the behavior of these disease-causing proteins in vivo.
Highlights for Balasubramaniam and Komives.
HXMS is useful for discovery of intrinisically disordered (ID) regions in proteins.
ID discovery and removal facilitates structure determination.
ID discovery facilitates biopharmaceutical quality control.
HXMS reveals coupled folding and binding of IDPs.
HXMS reveals the regions of IDPs involved in aggregation.
HXMS reveals structures of early oligomers, likely causative agents in amyloid diseases.
ACKNOWLEDGEMENTS
This work was supported in part by P01- GM071862.
Abbreviations
- HXMS
hydrogen/deuterium exchange coupled to mass spectrometry
- IDP
intrinsically disordered protein
- LRP1 CT
the cytoplasmic tail of the low density lipoprotein receptor-related protein, LRP1
- AR
ankyrin repeat.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Romero P, Obradovic Z, Dunker AK. Folding minimal sequences: the lower bound for sequence complexity of globular proteins. FEBS Letters. 1999;462:363–367. doi: 10.1016/s0014-5793(99)01557-4. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Information Series Workshop on Genome Information. 2000;11:161–171. [PubMed] [Google Scholar]
- 3.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state : conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, s28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 5.Daughdrill GW, Hanely LJ, Dahlquist FW. The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry. 1998;37:1076–1082. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- 6.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator: coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 7.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structurefunction paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 8.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nuc Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weathers EA, Paulaitis ME, Woolf TB, Hoh JH. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Letters. 2004;576:348–352. doi: 10.1016/j.febslet.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 11.Ward JJ, Sodhi JS, Mcguffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 13.Prilusky J, Felder CE, Zeev-Ben-Mordehei T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 14.Sickmeier M, Hamilton JA, LegallL T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the database of disordered proteins. Nuc Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue B, Oldfield CJ, Dunker AK, Uversky VN. CDF it all : consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Letters. 2009;583:1469–1474. doi: 10.1016/j.febslet.2009.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 17.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Unfoldomics of human genetic diseases: illustrative examples of ordered and intrinsically disordered members of the human diseasome. Prot Pept Lett. 2009;16:1533–1547. doi: 10.2174/092986609789839377. [DOI] [PubMed] [Google Scholar]
- 18.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Frontiers in Bioscience. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 19.Uversky VN, Eliezer D. Biophysics of Parkinson’s disease: structure and aggregation of asynuclein. Curr Prot Pept Sci. 2009;10:483–499. doi: 10.2174/138920309789351921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Iakoucheva LM, Mooney SD, Radivojac P. Loss of post-translational modification sites in disease. Pacific Symposium on Biocomputing. 2010;15:337–347. doi: 10.1142/9789814295291_0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Ann Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 22.Gog GK, Dunker AK, Uversky VN. Protein intrinsic disorder and influenza virulence: the 1918 H1N1 and H5N1 viruses. Virology J. 2009;6:1–12. doi: 10.1186/1743-422X-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue B, Williams RW, Oldfield CJ, Goh GK, Dunker AK, Uversky VN. Viral disorder or disordered viruses : do viral proteins possess unique features? Prot Pept Lett. 2010;17:932–951. doi: 10.2174/092986610791498984. [DOI] [PubMed] [Google Scholar]
- 24.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyson HJ, Wright PE. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv Protein Chem. 2002;62:311–340. doi: 10.1016/s0065-3233(02)62012-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandell JG, Falick AM, Komives EA. Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Analytical Chemistry. 1998;70:3987–3995. doi: 10.1021/ac980553g. [DOI] [PubMed] [Google Scholar]
- 28.Eyles SJ, Dresch T, Gierasch LM, Kaltashov IA. Unfolding dynamics of a beta-sheet protein studied by mass spectrometry. J Mass Spectrom. 1999;34:1289–1295. doi: 10.1002/(SICI)1096-9888(199912)34:12<1289::AID-JMS882>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Powell KD, Fitzgerald MC. Measurements of protein stability by H/D exchange and matrix-assisted laser desorption/ionization mass spectrometry using picomoles of material. Anal Chem. 2001;73:3300–3304. doi: 10.1021/ac0100805. [DOI] [PubMed] [Google Scholar]
- 30.Rosa JJ, Richards FM. An experimental procedure for increasing the structural resolution of chemical hydrogen-exchange measurements on proteins: application to ribonuclease S peptide. J Mol Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- 31.Englander S, Mayne L, Bai Y, Sosnick T. Hydrogen exchange: the modern legacy of Linderstrøm-Lang. Protein Science. 1997;6:1101–1109. doi: 10.1002/pro.5560060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandell JG, Falick AM, Komives EA. Identification of protein-protein interfaces by decreased amide proton solvent accessibility. Proc. Nat. Acad. Sci. U. S. A. 1998;95:14705–14710. doi: 10.1073/pnas.95.25.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Scapin G, Blanchard JS, Angeletti RH. Substrate binding and conformational changes of Clostridium glutamicum diaminopimelate dehydrogenase revealed by hydrogen/deuterium exchange and electrospray mass spectrometry. Protein Sci. 1998;7:293–299. doi: 10.1002/pro.5560070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandell JG, Baerga-Ortiz A, Akashi S, Takio K, Komives EA. Solvent accessibility of the thrombin-thrombomodulin interface. J Mol Biol. 2001;306:575–589. doi: 10.1006/jmbi.2000.4416. [DOI] [PubMed] [Google Scholar]
- 35.Baerga-Ortiz A, Hughes CA, Mandell JG, Komives EA. Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein. Protein Sci. 2002;11:1300–1308. doi: 10.1110/ps.4670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Smith DL. Probing Protein Structure and Dynamics by Hydrogen-Exchange-Mass Spectrometry. Current Protocols in Protein Science. 2002:17.6.1–17.6.18. doi: 10.1002/0471140864.ps1706s28. [DOI] [PubMed] [Google Scholar]
- 38.Rand KD, Zehl M, Jensen ON, Jørgensen TJ. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 39.Rand KD, Bache N, Nedertoft MM, Jørgensen TJ. Spatially resolved protein hydrogen exchange measured by matrix-assisted laser desorption ionization in-source decay. Anal Chem. 2011;83:8859–8862. doi: 10.1021/ac202468v. [DOI] [PubMed] [Google Scholar]
- 40.Vendruscolo M, Paci E, Dobson CM, Karplus M. Rare fluctuations of native proteins sampled by equilibrium hydrogen exchange. J Am Chem Soc. 2003;125:15686–15687. doi: 10.1021/ja036523z. [DOI] [PubMed] [Google Scholar]
- 41.Liu T, Pantazatos D, Li S, Hamuro Y, Hilser VJ, Woods VL. Quantitative assessment of protein structural models by comparison of H/D exchange MS data with exchange behavior accurately predicted by DXCOREX. J Am Soc Mass Spectrom. 2012;23:43–56. doi: 10.1007/s13361-011-0267-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Craig PO, Lätzer J, Weinkam P, Hoffman RM, Ferreiro DU, Komives EA, Wolynes PG. Prediction of native-state hydrogen exchange from perfectly funneled energy landscapes. J Am Chem Soc. 2011;133:17463–17472. doi: 10.1021/ja207506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pantazatos D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL. Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci U S A. 2004;101:751. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, Zheng H, Huang YJ, Ertekin A, Hamuro Y, Rossi P, Tejero R, Acton TB, Xiao R, Jiang M, Zhao L, Ma LC, Swapna GV, Aramini JM, Montelione GT. Construct optimization for protein NMR structure analysis using amide hydrogen/deuterium exchange mass spectrometry. Proteins. 2009;76:882–894. doi: 10.1002/prot.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ertekin A, Aramini JM, Rossi P, Leonard PG, Janjua H, Xiao R, Maglaqui M, Lee HW, Prestegard JH, Montelione GT. Human Cyclin-dependent Kinase 2-associated Protein 1 (CDK2AP1) Is Dimeric in Its Disulfide-reduced State, with Natively Disordered N-terminal Region. J Biol Chem. 2012;287:16541–16549. doi: 10.1074/jbc.M112.343863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stavropoulos I, Khaldi N, Davey NE, O’Brien K, Martin F, Shields DC. Protein disorder and short conserved motifs in disordered regions are enriched near the cytoplasmic side of single-pass transmembrane proteins. Plos One. 2012;7:e44389, 1–8. doi: 10.1371/journal.pone.0044389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Betts GN, Van Der Geer P, Komives EA. Structural and functional consequences of tyrosine phosphorylation in the LRP1 cytoplasmic domain. J Biol Chem. 2008;283:15656–15664. doi: 10.1074/jbc.M709514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaltashov IA, Bobst CE, Abzalimov RR, Wang G, Baykal B, Wang S. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotech Adv. 2011 doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.GA G. Phenotype, diagnosis, and treatment of Gaucher's disease. Lancet. 2008;372:1263–1271. doi: 10.1016/S0140-6736(08)61522-6. [DOI] [PubMed] [Google Scholar]
- 50.Bobst CE, Thomas JJ, Salinas PA, Savickas P, Kaltashov IA. Impact of oxidation on protein therapeutics: conformational dynamics of intact and oxidized acid-β-glucocerebrosidase at near-physiological pH. Protein Sci. 2010;19:2366–2378. doi: 10.1002/pro.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaltashov IA, Bobst CE, Abzalimov RR, Berkowitz SA, Houde D. Conformation and dynamics of biopharmaceuticals: transition of mass spectrometry-based tools from academe to industry. J Am Soc Mass Spectrom. 2010;21:323–337. doi: 10.1016/j.jasms.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 54.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Truhlar SME, Croy CH, Torpey JW, Koeppe JR, Komives EA. Solvent accessibility of protein surfaces by amide H/2H exchange MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1490–1497. doi: 10.1016/j.jasms.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 56.Dharmasiri K, Smith DL. Mass spectrometric determination of isotopic exchange rates of amide hydrogens located on the surfaces of proteins. Analytical Chemistry. 1996;68:2340–2344. doi: 10.1021/ac9601526. [DOI] [PubMed] [Google Scholar]
- 57.Truhlar SM, Torpey JW, Komives EA. Regions of IkappaBalpha that are critical for its inhibition of NF-kappaB.DNA interaction fold upon binding to NF-kappaB. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 59.Keppel TR, Howard BA, Weis DD. Mapping unstructured regions and synergistic folding in intrinsically disordered proteins with amide H/D exchange mass spectrometry. Biochemistry. 2011;50:8722–8732. doi: 10.1021/bi200875p. [DOI] [PubMed] [Google Scholar]
- 60.Kjaergaard M, Teilum K, Poulsen FM. Conformational selection in the molten globule state of the nuclear coactivator binding domain of CBP. Proc Natl Acad Sci U S A. 2010;107:12535–12540. doi: 10.1073/pnas.1001693107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henkels CH, Oas TG. Ligation-state hydrogen exchange: coupled binding and folding equilibria in ribonuclease P protein. J Am Chem Soc. 2006;128:7772–7781. doi: 10.1021/ja057279+. [DOI] [PubMed] [Google Scholar]
- 62.Heegaard NH, Jørgensen TJ, Rozlosnik N, Corlin DB, Pedersen JS, Tempesta AG, Roepstorff P, Bauer R, Nissen MH. Unfolding, aggregation, and seeded amyloid formation of lysine-58-cleaved beta 2-microglobulin. Biochemistry. 2005;44:4397–4407. doi: 10.1021/bi047594t. [DOI] [PubMed] [Google Scholar]
- 63.Prusiner SB, Groth DF, Bolton DC, Kent SB, Hood LE. Purification and structural studies of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 64.Sim VL, Caughey B. Ultrastructures and strain comparison of under-glycosylated scrapie prion fibrils. Neurobiol Aging. 2009;30:2031–2042. doi: 10.1016/j.neurobiolaging.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 65.Damo SM, Phillips AH, Young AL, Li S, Woods VL, Wemmer DE. Probing the conformation of a prion protein fibril with hydrogen dxchange. J Biol Chem. 2010;285:32303–32311. doi: 10.1074/jbc.M110.114504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2011;18:504–506. doi: 10.1038/nsmb.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu X, Wintrode PL, Surewicz WK. Beta-Sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci U S A. 2007;104:1510–1515. doi: 10.1073/pnas.0608447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dima RI, Thirumalai D. Probing the instabilities in the dynamics of helical fragments from mouse PrPC. Proc Natl Acad Sci U S A. 2004;101:15335–15340. doi: 10.1073/pnas.0404235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carulla N, Zhou M, Giralt E, Robinson CV, Dobson CM. Structure and intermolecular dynamics of aggregates populated during amyloid fibril formation studied by hydrogen/deuterium exchange. Acc Chem Res. 2010;43:1072–1079. doi: 10.1021/ar9002784. [DOI] [PubMed] [Google Scholar]
- 70.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-Speed and High-Resolution UPLC Separation at Zero Degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rand KD, Zehl M, Jensen ON, Jørgensen TJ. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 72.Rand KD, Pringle SD, Morris M, Engen JR, Brown JM. ETD in a traveling wave ion guide at tuned Z-spray ion source conditions allows for site-specific hydrogen/deuterium exchange measurements. J Am Soc Mass Spectrom. 2011;22:1784–1793. doi: 10.1007/s13361-011-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amon S, Trelle MB, Jensen ON, Jørgensen TJ. Spatially resolved protein hydrogen exchange measured by subzero-cooled chip-based nanoelectrospray ionization tandem mass spectrometry. Anal Chem. 2012;84:4467–4473. doi: 10.1021/ac300268r. [DOI] [PubMed] [Google Scholar]