Abstract
Proteins of the Bcl-2 family either enhance or suppress programmed cell death and are centrally involved in cancer development and resistance to chemotherapy. BH3 (Bcl-2 homology 3)-only Bcl-2 proteins promote cell death by docking an α-helix into a hydrophobic groove on the surface of one or more of five pro-survival Bcl-2 receptor proteins. There is high structural homology within the pro-death and pro-survival families, yet a high degree of interaction specificity is nevertheless encoded, posing an interesting and important molecular recognition problem. Understanding protein features that dictate Bcl-2 interaction specificity is critical for designing peptide-based cancer therapeutics and diagnostics. In this study, we present peptide SPOT arrays and deep sequencing data from yeast display screening experiments that significantly expand the BH3 sequence space that has been experimentally tested for interaction with five human anti-apoptotic receptors. These data provide rich information about the determinants of Bcl-2 family specificity. To interpret and use the information, we constructed two simple data-based models that can predict affinity and specificity when evaluated on independent data sets within a limited sequence space. We also constructed a novel structure-based statistical potential, called STATIUM, which is remarkably good at predicting Bcl-2 affinity and specificity, especially considering it is not trained on experimental data. We compare the performance of our three models to each other and to alternative structure-based methods and discuss how such tools can guide prediction and design of new Bcl-2 family complexes.
Keywords: protein–protein interactions, binding specificity, statistical potential, SPOT arrays, yeast-surface display
Introduction
Bcl-2 family proteins play an important role regulating programmed cell death. Overexpression of pro-survival Bcl-2 proteins contributes to evasion of apoptosis in tumor cells and is an important characteristic of cancer.1–3 Bcl-2 homologues are also present in several cancer-associated viruses that include Epstein–Barr and Kaposi sarcoma, suggesting that anti-apoptotic Bcl-2 proteins may contribute to virally induced cancer by subverting the normal host apoptosis mechanism.4,5 For these reasons, the Bcl-2 family has been an important target for the design of cancer therapeutics,6–13 and sequence-structure-binding relationships that can guide the design of therapeutics or diagnostics have been extensively studied.14–23
The Bcl-2 family includes five human pro-survival Bcl-2 receptors (Bcl-xL, Bcl-2, Bcl-w, Mcl-1, and Bfl-1) that interact with ~16 known pro-death BH3 (Bcl-2 homology 3) domains (Bim, Bad, Noxa, etc.).24 Structural and biophysical investigations have shown that helical peptides of ~16–25 residues, corresponding to BH3 motifs, dock in a groove on the surface of pro-survival receptors.19,25,26 The strength of this interaction is specific to the receptor/BH3 pairing, such that some BH3 peptides show promiscuous binding to all receptors, whereas others are highly selective. Examples include the BH3 region of Noxa, which interacts strongly with pro-survival receptor Mcl-1 but weakly or not detectably with other receptors, and pro-apoptotic Bim, which promiscuously interacts with many pro-survival family members.27–31 The selectivity of binding is important for the regulation of apoptosis, because only those BH3 motifs that can bind to anti-apoptotic receptors are capable of antagonizing their activity.32,33
Due to its important role regulating cell life versus death decisions, the Bcl-2 receptor–BH3 interaction is an attractive target for drug design. In mouse models, the small-molecule Bcl-2 family antagonist ABT-737, which binds the same hydrophobic groove as the BH3 motif, can regress or eliminate small cell lung carcinoma, suggesting great promise for therapeutics that antagonize pro-survival Bcl-2 family proteins.34 Work using chemically stabilized α-helices and α/β-peptide foldamers indicates that these, too, may provide a route to therapies.35–38 The receptor-binding specificities of small molecules and peptides determine which types of cancers they may be effective against. For example, ABT-737 and related molecules do not bind to Mcl-1 or Bfl-1 and thus are ineffective in inducing death in cancer cells when these anti-apoptotic proteins are overexpressed.39
In addition to therapeutic applications, designed selective BH3 peptides hold promise as tools for cancer diagnosis. Panels of natural BH3 peptides have been applied by Certo et al. to detect specific mechanisms by which Bcl-2 family members confer resistance to apoptosis in tumor cells.27 This technique, called BH3 profiling, involves determining the pattern of mitochondrial outer membrane permeabilization in cells exposed to different BH3 peptides. Because BH3 peptides have different receptor-binding profiles, they can be used to report on a tumor cell's “addiction” to distinct anti-apoptotic receptors. BH3 profiling could be rendered more sensitive and distinct by discovering or designing new BH3-mimetic peptides with novel receptor-binding specificities.
Protein engineering and design would benefit enormously from accurate computational methods for predicting protein–protein interactions. The development of such methods relies on experimental data to benchmark accuracy. For example, systematic studies using large data sets of coiled-coil, Ras–Ras effector, PDZ–peptide, SH2–peptide, and SH3–peptide interactions have led to computational work exploring different approaches to predicting or recapitulating experimentally observed interactions.40–46 Methods have ranged from structure-based prediction using different types of energy functions to sophisticated machine learning.41,46–50 A critical distinction between different approaches is whether or not they rely on the availability of large amounts of experimental data for model derivation or training. Another distinction is whether the model is intended to be general, that is, applicable to any protein complex, or whether it is specific to a certain protein or protein family. A particularly simple yet often-effective model that is protein specific and based directly on experimental data is a position-specific scoring matrix (PSSM). Such a matrix is constructed using the observed frequencies of individual residues at different sites in known binders. PSSMs can be powerful and have been applied to describing protein–peptide interactions and identifying new SH2 and PDZ binding partners.51,52
Dutta et al. previously identified binding specificity determinants for Bcl-2 receptors Bcl-xL and Mcl-1 using experimental BH3 peptide SPOT arrays and yeast-surface display screening.14 The study focused on two anti-apoptotic receptors and on mutational variants of the native BH3 motif of Bim. A simple PSSM model based on peptide array data showed good performance predicting binding specificity for peptides identified in yeast screening. More general methods may be required to make predictions for a more diverse binding space. It is therefore interesting to consider structure-based models. Such models can be built using only the structures of Bcl-2 family complexes, solved by X-ray crystallography or NMR, which are available for all five human Bcl-2 receptors.17,25,53–55 Large experimental binding data sets are not required, except for model testing. London et al. recently used structures of Bcl-xL and Mcl-1 bound to Bim BH3 to apply a peptide-modeling module implemented in Rosetta, FlexPepBind, to predict Bcl-2 family interactions (N. London et al., submitted for publication). A version of this protocol that was optimized using data from Dutta et al. was capable of discriminating binding to Bcl-xL versus Mcl-1 with impressive accuracy and could accurately rank the specificities of known natural BH3 domains. However, FlexPepBind is computationally expensive and cannot easily be used to evaluate very large data sets or entire proteomes.
In this study, we extend the work of Dutta et al. by presenting additional experimental data that can be used to construct PSSM models for the five human anti-apoptotic receptors Bcl-xL, Mcl-1, Bcl-2, Bcl-w, and Bfl-1. The data include BH3 peptide SPOT arrays and sequences derived from deep sequencing of thousands of BH3 peptides screened for binding to Bcl-2 family proteins. The new data sets provide rich information about determinants of Bcl-2 family specificity and an opportunity to test different computational schemes. We also present and evaluate a novel structure-based protein–protein interaction statistical potential called STATIUM that can score interactions of BH3-like peptides with all five Bcl-2 receptors and is rapid enough to evaluate data sets containing more than 106 sequences in less than 1 s. The very general structure-based STATIUM model shows remarkably good performance compared to the experimentally derived PSSM models. STATIUM demonstrates great potential for evaluating candidate protein–protein interactions and can be used to complement other structure-based modeling techniques such as Rosetta, DFIRE, or MM/PBSA that require accurate construction of all-atom models.56–59
Results
We used a combination of experimental assays and computational models to explore BH3 peptide interactions with Bcl-2 family receptors. Our goal was to elucidate determinants of binding affinity and specificity and to capture these in predictive models that can be used for protein interaction prediction and design. In this section, we describe new experimental binding data before providing an overview of three different models, two of which were derived directly from experiments and one of which is a novel structure-based potential. The bulk of this section reports the performance of different models for predicting different data sets, and in Discussion, we present things we have learned about Bcl-2 family protein interactions and different strategies for modeling them.
SPOT arrays for Bim–Bcl-w, Bim–Bfl-1, Bad–Bcl-xL, and Noxa–Mcl-1
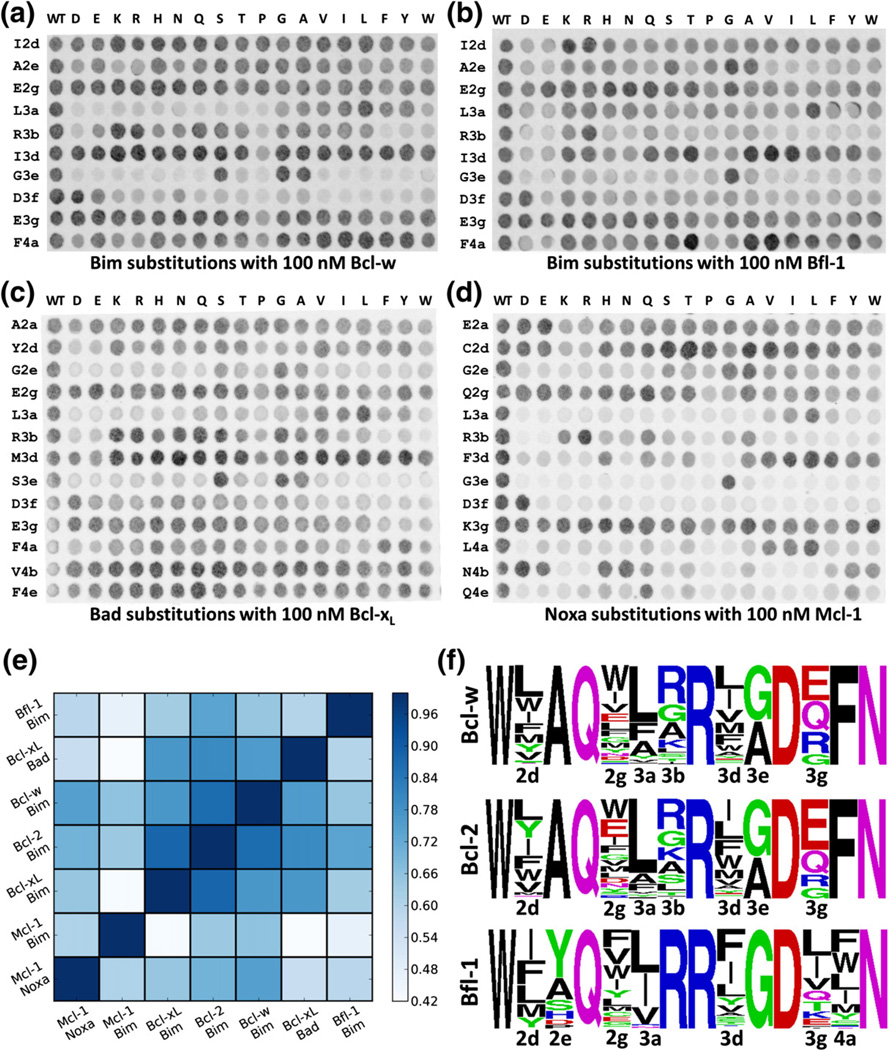
SPOT substitution arrays allow concurrent evaluation of binding for hundreds of peptides, each different by a single substitution, to a given receptor. Membranes displaying synthesized peptides are incubated with Myc-tagged receptor and then probed with anti-Myc antibodies labeled with Cy3. This is not an equilibrium binding assay, but SPOT fluorescence intensities associated with peptides on the arrays are empirically correlated with binding affinities in solution, at least among a series of similar peptides. This can be seen in comparisons of SPOT intensities to binding data from Boersma et al.,15 who reported solution binding of single-residue variant Bim BH3 peptides to Bcl-xL (R = 0.74, Fig. S1a) and Mcl-1 (R = 0.88, Fig. S1b). In this study, we present SPOT substitution arrays that report the binding of single-residue variants of a 26-residue BH3 peptide from Bim to anti-apoptotic receptors Bcl-w and Bfl-1 (Fig. 1a and b). When combined with publication of the substitution arrays for Bcl-xL, Mcl-1, and Bcl-214 (London et al., submitted for publication), these data provide comprehensive data for Bim and 170 of its point mutants binding to five human Bcl-2 receptors.
Fig. 1.
SPOT substitution arrays and Illumina sequencing of yeast library pools. Shown are array images for Bim BH3 peptides binding to Bcl-w (a) and Bfl-1 (b), Bad BH3 peptides binding to Bcl-xL (c), and Noxa BH3 peptides binding to Mcl-1 (d). The leftmost column of each array consists of repeats of wild-type Bim BH3. All other spots along each row are single-residue substitutions or a replicate of the wild-type sequence. Rows indicate the substituted position, labeled as defined in Fig. S2, and columns indicate the substituted amino acid. (e) Pearson correlation of seven SPOT substitution data sets described in the text. (f) Sequence logos for pools of yeast clones binding tightly to Bcl-w (top), Bcl-2 (middle), and Bfl-1 (bottom) were generated using the program WebLogo.60 Positions that were varied in each library are labeled.
To study the effect of sequence variation in peptides not based on Bim, we probed arrays printed with single-residue mutations of Bad and Noxa BH3 peptides with Bcl-xL and Mcl-1, respectively (Fig. 1c and d). Bad BH3 is specific for binding Bcl-xL over Mcl-1, and Noxa BH3 shows the opposite specificity.28 The sequences of Bim, Bad, and Noxa BH3 motifs are considerably different (Fig. S2); hence, these experiments allow us to assess the role of peptide context for numerous point substitutions. The central residues in BH3 peptides can be labeled as 2a–4g, based on three heptad repeats with positions abcdefg that capture the periodicity of an amphipathic α-helix (Fig. S2). In addition to the 10 positions in the core region of the BH3 peptides that were sampled on the Bim BH3 substitution arrays (2d, 2e, 2g, 3a, 3b, 3d, 3e, 3f, 3g, and 4a), the Bad and Noxa BH3 arrays also included variation at an additional N-terminal site (2a) and two additional C-terminal sites (4b and 4e), providing 222 peptide variants. Residues in these positions make contacts with the receptor in many structures.19,25,26,54
With Bim BH3 substitution arrays now available for five human anti-apoptotic Bcl-2 receptors, we compared these data sets. We computed the correlation coefficients for signals resulting from equivalent peptides on each of 21 pairs of SPOT arrays (Fig. 1e). For Bim BH3-based SPOT arrays, the equivalent peptides had identical sequences. For the Bad- and Noxa-based BH3 peptide arrays, we defined equivalence between the same point mutation made in the context of Bim, Bad, or Noxa. Unsurprisingly, receptors with similar binding profiles for known native BH3s, such as Bcl-xL and Bcl-2,28,30 gave interaction profiles on the arrays that were highly correlated (R = 0.88). Receptor pairs with divergent natural binding profiles, such as Bcl-xL and Mcl-1 or Bcl-xL and Bfl-1, gave profiles that were less correlated (R = 0.43 and 0.47). SPOT analysis of substitutions in Bad or Noxa BH3 gave profiles that correlated moderately with Bim BH3 substitution arrays14 probed using the same receptor (R = 0.77 for Bad versus Bim BH3 variants binding to Bcl-xL, and R = 0.61 for Noxa versus Bim BH3 variants binding to Mcl-1). Differences are apparent at individual sites; for example, in the context of Noxa, position 4a appears less permissive for interactions with Mcl-1, compared to the context of Bim. In contrast, position 3d in Noxa is more tolerant of large hydrophobic amino acids in this assay than the same site in Bim.
Dutta et al., and others, have noted the importance of certain positions for determining binding to Bcl-xL and Mcl-1.14,15,17 For example, position 4a is very permissive for Mcl-1 binding, but a much more limited range of amino acids can be accommodated at this site for binding to Bcl-xL.14,15,21 In the SPOT array assay, Bcl-w and Bfl-1 were more similar to Mcl-1 at position 4a and permitted many point substitutions (Fig. 1a and b). At position 3d, many amino acids restrict binding to Mcl-1 but not Bcl-xL,14 and our Bcl-w SPOT data appear very similar to the Bcl-xL profile. At 3d, Bfl-1 was more tolerant than Mcl-1 to substitutions in array peptides, but not as relaxed as the other receptors at this site. Position 3a is strictly conserved as leucine in natural BH3s, and the SPOT arrays similarly showed a strong leucine preference.
The Noxa substitution array showed interesting trends supported by previous structural observations. Position 2d is one of four sites conserved as mostly hydrophobic in native BH3 motifs, although it is occupied by cysteine in human Noxa. On the Noxa SPOT array, position 2d could accommodate negatively charged residues but not positively charged residues for Mcl-1 binding (Fig. 1d). Mcl-1 has a basic patch consisting of Arg233 and Lys234 (human Mcl-1 numbering) at the end of helix 3 in close proximity to position 2d.22,61 Mouse NoxaB has a glutamate at position 2d and binds to mouse Mcl-1 with only ~3-fold weaker affinity than mouse Noxa A, which has a phenylalanine at the same position.18 Noxa B does not show any binding to Bfl-1, which has an acidic patch near this region.18 We also observed disruption of binding to Bfl-1 for I2dD and I2dE in the sequence context of Bim (Fig. 1b).
In the Noxa and Bad substitution arrays, we included three additional residues compared to the Bim BH3 arrays. Surprisingly, the two C-terminal positions 4b and 4e in Noxa could only tolerate a restricted set of residues for binding to Mcl-1, with 4b exhibiting preferences for Asp, Glu, and His in addition to the wild-type residue Asn, while 4e was constrained predominantly to the wild-type residue Gln, and Tyr. In contrast, the N-terminal 2a and C-terminal 4b and 4e positions in Bad could accommodate many residues for binding to Bcl-xL.
Deep sequencing of BH3 peptide libraries screened for Bcl-2 binding
Dutta et al. reported the use of yeast-surface display to screen libraries of yeast cells with >450,000 different surface-displayed Bim BH3 variants for those that bound selectively to Mcl-1 versus Bcl-xL or vice versa.14 In that work, 33 sequences were identified as selective for Mcl-1 and 40 were identified as selective for Bcl-xL, based on testing of individual yeast clones displaying the peptides of interest (labeled as “Specific tight binders” in Table 1). To access a larger set of Bim BH3 peptide variants that bound to these two receptors, we used Illumina technology to sequence many yeast clones from previously isolated pools.14 We sequenced pools that bound to each receptor individually (from affinity screening), as well as pools that bound preferentially to Bcl-xL versus Mcl-1 or vice versa (from specificity screening). Data from independent re-sequencing of the same pools indicated high reproducibility (Fig. S3). We processed the Illumina data using strict filters to identify unique sequences satisfying several quality criteria (see Methods). Acknowledging that peptides in these pools were not individually confirmed to be true binders, we labeled the sequence sets as “Likely binders” and “Likely specific binders,” as indicated in Table 1. These sets may be contaminated by false positives at an unknown rate. However, overall, the pools should be highly enriched in true binders. Supporting this, the frequencies of amino acids from conventional sequencing of true binders and deep sequencing of likely binders were highly correlated (R = 0.97). Further, a scoring function derived from the deep sequencing data showed good performance predicting affinity and specificity for unrelated SPOT array data (see below).
Table 1.
Experimental data sets used in this work and models derived from them
| Data set | Description | Receptor-binding models derived from data |
|---|---|---|
| SPOT substitution arrays | 170 one-residue variants of Bim BH3 (binding to all 5 receptors), 222 one-residue variants of Bad BH3 (binding Bcl-xL) and Noxa BH3 (binding Mcl-1) | Bcl-xL: PSSMSPOTX_Bim, PSSMSPOTX_Bad Mcl-1: PSSMSPOTM_Bim, PSSMSPOTM_Noxa Bcl-2: PSSMSPOT2_Bim, Bcl-w: PSSMSPOTW_Bim Bfl-1: PSSMSPOTF_Bim |
| SPOT library arrays | 359 sequences including 1–5 mutations in Bim BH3 (binding to Mcl-1 and Bcl-xL) | N/A |
| Specific tight binders | Conventional sequencing of individual yeast clones assessed for binding to Bcl-xL and Mcl-1: 40 Bcl-xL and 33 Mcl-1-specific sequences | N/A |
| Likely binders | Illumina deep sequencing of yeast clones screened for high-affinity binding to receptors | Bcl-xL: PSSMDEEPX_AFFIN, Mcl-1: PSSMDEEPM_AFFIN Bcl-2: PSSMDEEP2_AFFIN, Bcl-w: PSSMDEEPW_AFFIN Bfl-1: PSSMDEEPF_AFFIN |
| Likely specific binders | Illumina deep sequencing of yeast clones screened for Mcl-1 versus Bcl-xL specific binding and vice versa | Bcl-xL: PSSMDEEPX_SPEC, Mcl-1: PSSMDEEPM_SPEC |
Mcl-1 and Bcl-xL substitution and library SPOT array data and yeast screening experiments are from Dutta et al.14
N/A - Not applicable.
We now report results from preliminary screens to identify Bim BH3 variants that bind to Bcl-w, Bcl-2, and Bfl-1 (full data from these studies will be reported elsewhere). Two yeast libraries, one designed to favor preferential binding to Bcl-w, Bcl-xL, and Bcl-2 over Mcl-1/Bfl-1, and the second to favor binding to Bfl-1 over each of the other four receptors,62 were screened using FACS (fluorescence-activated cell sorting) at an initial receptor concentration of 1 µM, followed by more stringent screening at lower concentrations (final stringency: 1, 10, or 100 nM). Table S1 describes the pools used to generate data sets for all five Bcl-2 receptors. Sequence logos based on deep sequencing of likely binders to Bcl-w, Bcl-2, and Bfl-1 identified in this way are shown in Fig. 1f.
Models for Bcl-2 interactions
We present three models that can be used to score the interaction between a BH3 peptide and five of the human Bcl-2 receptors. PSSMSPOT models are based on SPOT substitution array data, PSSMDEEP models are based on deep sequencing of yeast screening experiments, and STATIUM is a structure-based statistical potential derived from the structures of Bcl-2 receptor–peptide complexes. The derivation of these models is detailed in Methods, and we briefly summarize the important features of each here and in Table 1 and Fig. 2. To test the models, we used a large amount of experimental data, also summarized in Table 1. In addition to previously published SPOT substitution arrays and new data described here, we used previously published array data for Bcl-xL and Mcl-1 binding that included 359 combinatorial variants of Bim BH3.14 These arrays are referred to as the SPOT library arrays in Table 1 and below, and only five of the library array sequences were identical with those included on the substitution arrays. Library array Bim BH3 variants included sequences with up to five mutations compared to wild-type Bim and were designed to maximize the number of sequences specific for binding either Bcl-xL or Mcl-1.
Fig. 2.
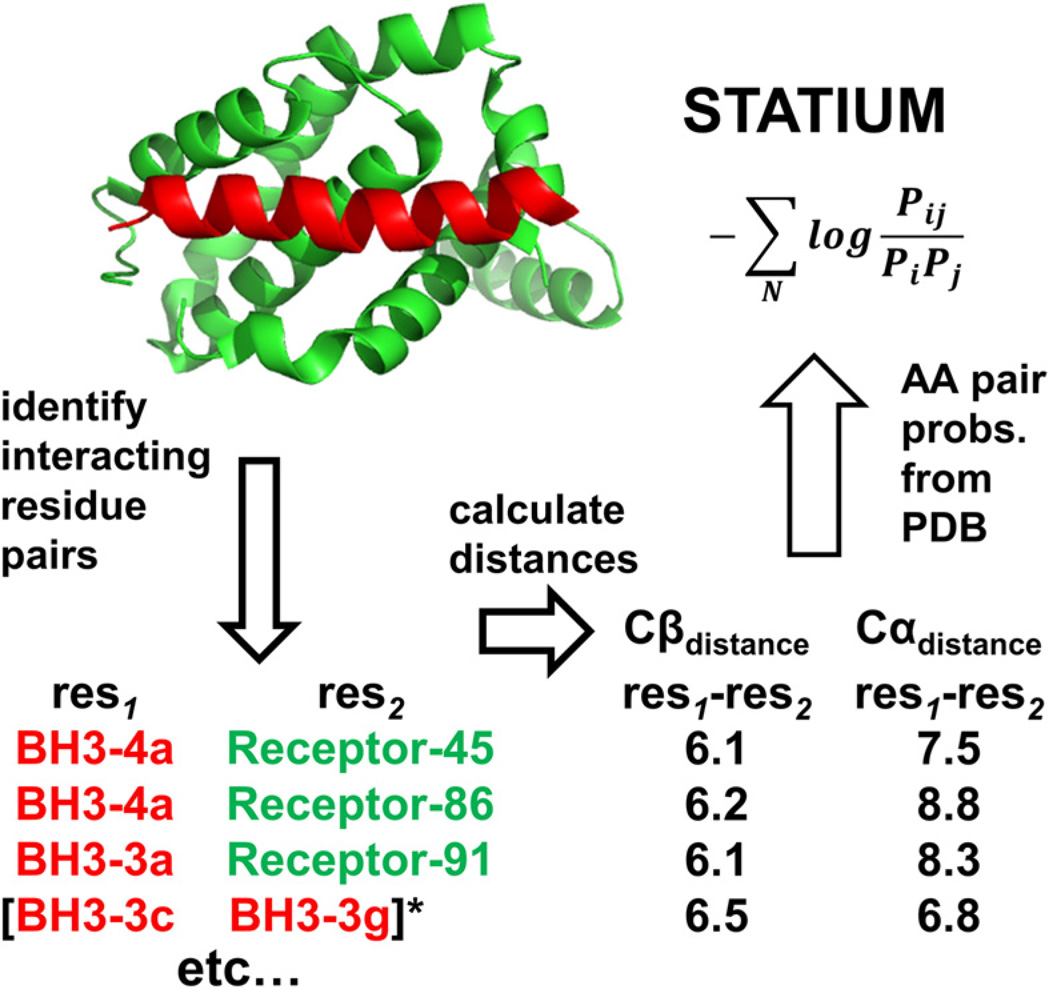
Derivation of the STATIUM potential. The crystal structure of a Bcl-2 receptor bound to a BH3 peptide (here, PDB ID 3IO8) is used to identify interacting residue pairs (see Methods). The Cα–Cα and Cβ–Cβ distances for each interacting pair are measured, and pairs with similar distances are sought in a database of experimentally determined structures. The STATIUM score for amino acid i at position 1 and amino acid j at position 2 is the probability of that amino acid pair occurring in all pairs discovered, divided by the individual probabilities of finding i and j in the PDB. The total STATIUM score for a receptor–peptide interaction is the sum of all residue-pair scores.
The PSSMSPOT potentials were constructed using Bim, Bad, and Noxa BH3 SPOT substitution arrays. Similar to Dutta et al., the PSSMSPOT energy of a substitution was calculated by taking the negative logarithm of the ratio of the fluorescence intensity for the corresponding BH3 substitution to the intensity of the wild-type BH3 (averaging over all wild-type spots).14 The PSSMSPOT energy of a BH3 sequence was defined as the sum of the energies of all substitutions. Not all positions were varied in the original SPOT membranes, and substitutions at positions that were not sampled were treated as equivalent for all amino acids. Note that two different types of PSSM models were presented in Dutta et al.14 The model used here corresponds to the first, which was derived directly from SPOT substitution arrays. PSSMSPOT models are named using subscripts that indicate which array and receptor were used; for example, PSSMSPOTX_Bim indicates a model based on data for Bcl-xL binding to Bim variants. In this notation, “X” denotes Bcl-xL, “M” denotes Mcl-1, “2” denotes Bcl-2, “W” denotes Bcl-w, and “F” denotes Bfl-1.
PSSMDEEP potentials were derived from the deep sequencing data described above. We defined models based on yeast library pools that underwent affinity and/or specificity screening. PSSMDEEP_AFFIN models were derived by taking unique sequences from rounds of positive screening for binding to a given receptor. The PSSMDEEP_SPEC models were designed to capture the specificity of a BH3 peptide for a desired receptor in preference to an undesired one. PSSMDEEP_SPEC models were derived using unique sequences resulting from both positive screening for binding to one receptor (e.g., Mcl-1) and negative screening disfavoring binding to another receptor (e.g., Bcl-xL) (Table S1). In PSSMDEEP models, the energy of an amino acid substitution at a BH3 position was computed based on the frequency of that amino acid in the appropriate set of unique sequences (see Methods). Successful screening experiments allowed us to construct affinity models for all five receptors and specificity models for Bcl-xL versus Mcl-1 and vice versa (Table 1). In the same manner as for the PSSMSPOT models, PSSMDEEP models are distinguished with a letter in the subscript to indicate which receptor was used as the target in generating the relevant data; for example, PSSMDEEPX_AFFIN is a model built using affinity pools selected for binding to Bcl-xL.
Our third type of model is STATIUM, which is a statistical potential based on structural modeling that does not incorporate experimental binding data. A STATIUM model can be generated using any structure of a Bcl-2 receptor–peptide complex and then used to score the affinity of other peptides for that receptor. STATIUM aims to reduce structural detail in order to maximize the computational efficiency of scoring, but in a way that preserves accuracy. To that end, the derivation of STATIUM is different from statistical potentials such as DFIRE58 or the statistical components of Rosetta56,63,64 in that it does not define generic energies for structure terms but instead derives energies that are tailored to the residue interaction geometries present in a specific structure.
The derivation of STATIUM is outlined in Fig. 2 and described in full in Methods. STATIUM requires as input a structure of a Bcl-2 receptor in complex with a BH3 peptide; this can be an X-ray or NMR structure, or a homology model. Cα–Cα and Cβ–Cβ distances are used to define the structures of receptor–peptide and intrapeptide interacting residue pairs. A structure database derived from the Protein Data Bank (PDB) is then searched for interacting residue pairs with similar observed distances. The frequencies of observed amino acid pairs that meet the geometry criteria are used to derive the STATIUM score for that pair. The overall STATIUM score for a peptide binding to a given receptor is the sum of all of its residue-pair scores. Based on our observation that increasing the weight of intrapeptide residue pairs relative to receptor–peptide pairs improved affinity prediction but decreased specificity prediction (Fig. S5), different weights were used for affinity and specificity benchmarks presented below, as detailed in Methods and as indicated by the asterisk in Fig. 2.
We constructed numerous STATIUM potentials using different experimental structures of Bcl-2–BH3 peptide complexes. Below, we refer primarily to five models, each one constructed on what we judged to be the best structural template for each receptor. These are referred to as STATIUMM, STATIUMX, STATIUM2, STATIUMW, and STATIUMF using the same single-letter abbreviations defined above.
Prediction of SPOT array binding
The metric we used to assess binding prediction accuracy for the nine SPOT arrays is the area under the curve (AUC) value for a binary classification receiver operating characteristic curve. This approach requires defining two subsets of data, that is, strong binders versus weak binders. The AUC value then represents the probability that a randomly selected strong binder will have a lower energy than a weak binder; a perfect predictor will give AUC = 1.0. After analyzing the distribution of binding signals, we designated the 30% of sequences with the greatest signals as strong binders and the 30% with the weakest binding signals as non-binders (see Methods). The choice of 30% as the cutoff for top and bottom percentiles ensures we use a majority of data points for predictions but limits incorrect class assignments. Results using other cutoffs are included in Table S2.
AUC values for the prediction of SPOT array data with different scoring functions are shown in Tables 2 and 3. We made predictions for both the SPOT library array data (Table 2), which are available for Bcl-xL and Mcl-1 only, and for the SPOT substitution array data, which are available for seven substitution arrays (Table 3). For the library array predictions, the performance of all models is shown. For example, we include the performance of models such as PSSMSPOT2_Bim, a model based on Bcl-2, for predicting BH3 peptide binding to Mcl-1 or Bcl-xL. For the substitution array predictions, we show data only for the most relevant model, that is, the model built for the appropriate receptor used in the experiments. The full analysis of all models tested on all SPOT substitution data is available in Table S3. Every scoring function had at least some predictive capability. The AUC values when using a model built for the appropriate receptor ranged from 0.7 to 1.0. Interestingly, many scoring functions could predict SPOT library array binding for Bcl-xL and Mcl-1, even if the model was constructed using a different receptor. For example, PSSMSPOTX_Bad gave perfect predictions for Bim variants binding to Bcl-xL in this test (AUC = 1.0). This indicates that the determinants of affinity at the extremes of very weak and very strong binding are similar across receptors.
Table 2.
Predictions for Bcl-xL and Mcl-1 binding to SPOT library arrays using different models
| AUC for discriminating strong binders versus non-binders |
||
|---|---|---|
| Model | Mcl-1 binding | Bcl-xL binding |
| PSSMDEEPM_AFFIN | 0.89 (0.86, 0.93) | 0.8 (0.75, 0.85) |
| PSSMDEEPX_AFFIN | 0.84 (0.79, 0.88) | 0.85 (0.8, 0.89) |
| PSSMDEEP2_AFFIN | 0.93 (0.9, 0.96) | 0.93 (0.9, 0.95) |
| PSSMDEEPW_AFFIN | 0.92 (0.89, 0.94) | 0.93 (0.9, 0.96) |
| PSSMDEEPF_AFFIN | 0.57 (0.51, 0.64) | 0.56 (0.49, 0.62) |
| PSSMDEEPM_SPEC | 0.73 (0.67, 0.79) | 0.41 (0.34, 0.47) |
| PSSMDEEPX_SPEC | 0.65 (0.58, 0.71) | 0.91 (0.87, 0.94) |
| PSSMSPOTM_Bim | 0.94 (0.91, 0.96) | 0.78 (0.73, 0.83) |
| PSSMSPOTX_Bim | 0.87 (0.83, 0.91) | 1.0 (0.99, 1.00) |
| PSSMSPOT2_Bim | 0.93 (0.9, 0.96) | 1.0 (1.00, 1.00) |
| PSSMSPOTW_Bim | 0.96 (0.94, 0.98) | 1.0 (1.00, 1.00) |
| PSSMSPOTF_Bim | 0.98 (0.97, 0.99) | 0.96 (0.94, 0.98) |
| PSSMSPOTM_Noxa | 0.99 (0.98, 1.00) | 0.94 (0.92, 0.96) |
| PSSMSPOTX_Bad | 0.91 (0.88, 0.94) | 1.0 (0.99, 1.00) |
| STATIUMM | 0.94 (0.91, 0.96) | 0.81 (0.76, 0.86) |
| STATIUMX | 0.95 (0.93, 0.97) | 0.94 (0.92, 0.96) |
| STATIUM2 | 0.92 (0.89, 0.95) | 0.93 (0.90, 0.96) |
| STATIUMW | 0.9 (0.87, 0.93) | 0.92 (0.89, 0.95) |
| STATIUMF | 0.91 (0.88, 0.94) | 0.85 (0.81, 0.89) |
Library SPOT array sequences contained one to five mutations in Bim BH3.14 Raw fluorescence was converted to −log(SPOT/SPOTWT), where SPOT is the intensity of the variant and SPOTWT is the average intensity of all wild-type Bim BH3 spots on the membrane. The lowest 30% of variants were considered “binders” and the highest 30% were considered non-binders (see Methods). Bold AUC values designate predictions made using a model that was derived for the appropriate receptor. Ninety percent confidence limits are given in parentheses.
Table 3.
Predictions for five receptors binding to SPOT substitution arrays
| SPOT substitution arraya |
AUC for discriminating strong binders versus non-binders |
||
|---|---|---|---|
| Model | Receptor | BH3 | |
| PSSMDEEPM_AFFIN | Mcl-1 | Bim | 0.97 (0.91, 1.0) |
| Noxa | 0.88b (0.76, 0.97) | ||
| PSSMDEEPX_AFFIN | Bcl-xL | Bim | 0.99 (0.96, 1.0) |
| Bad | 0.9b (0.78, 0.98) | ||
| PSSMDEEP2_AFFIN | Bcl-2 | Bim | 0.82 (0.73, 0.9) |
| PSSMDEEPW_AFFIN | Bcl-w | Bim | 0.7 (0.59, 0.81) |
| PSSMDEEPF_AFFIN | Bfl-1 | Bim | 0.74 (0.63, 0.84) |
| PSSMDEEPM_SPEC | Mcl-1 | Bim | 0.97 (0.92, 1.0) |
| Noxa | 0.77b (0.61, 0.91) | ||
| PSSMDEEPX_SPEC | Bcl-xL | Bim | 1.0 (1.0, 1.0) |
| Bad | 0.75b (0.57, 0.89) | ||
| STATIUMM | Mcl-1 | Bim | 0.71 (0.63, 0.8) |
| Noxa | 0.69 (0.61, 0.67) | ||
| STATIUMX | Bcl-xL | Bim | 0.72 (0.63, 0.81) |
| Bad | 0.59 (0.51, 0.68) | ||
| STATIUM2 | Bcl-2 | Bim | 0.79 (0.71, 0.86) |
| STATIUMW | Bcl-w | Bim | 0.82 (0.74, 0.89) |
| STATIUMF | Bfl-1 | Bim | 0.75 (0.67, 0.83) |
Ninety percent confidence limits are given in parentheses.
SPOT substitution arrays contained single-residue mutations of Bim, Bad, or Noxa BH3. Data were processed as for Table 2.
PSSMDEEP models that predict Bcl-xL and Mcl-1 binding to Noxa or Bad BH3 were derived from screens performed on libraries derived from Bim BH3.
PSSMSPOT and PSSMDEEP models performed very well. This is expected considering that both models were derived from experimental data that covered much of the sequence space of the test set. Nonetheless, it is encouraging that PSSMDEEP performed well on the SPOT tests, given that the model and test data came from imperfect and very different experiments. On the other hand, the STATIUM models, which used no system-specific binding data, performed as well as the experimental models in a few cases and were competitive with PSSMDEEP for the SPOT library data set. STATIUM also predicted the Bfl-1 SPOT substitution data as well as the PSSMDEEP models. This is striking, given that much less prior knowledge was needed to derive this model.
Table S2 shows the AUC values from Table 2, but with cutoffs of 40% and 50% used to define binders and non-binders. In general, the AUC values slightly decreased as the cutoff increased. This could be due to the test set becoming more challenging for the prediction models, as well as to the blurring of the experimental class assignments of strong binders and non-binders. Table S4 shows the total number of assigned binders/non-binders for each test.
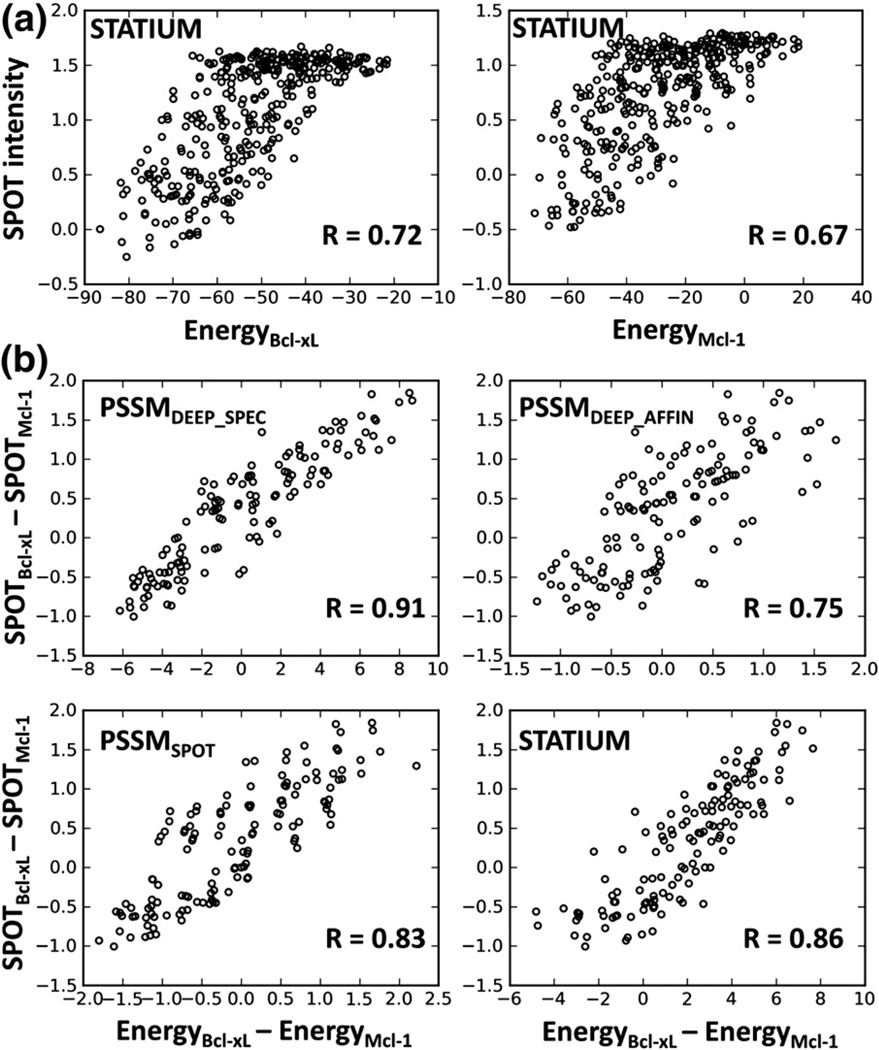
For cases where the STATIUM AUC values were high for all models, we examined the correlation between the predicted energy and the experimental SPOT array signal. Note that the agreement of the SPOT data with solution binding constants was R = 0.74–0.88 (see above), providing an approximate upper limit on the meaningful correlation that can be achieved. Figure 3a shows the correlation of the SPOT library array data with STATIUM (R = 0.72 and 0.67 for Bcl-xL and Mcl-1, respectively). This is exceptionally high for a structure-based approach and compares well with a version of Rosetta optimized for Bcl-2 binding (unoptimized R = 0.54, 0.46; optimized R = 0.8, 0.67; London et al., submitted for publication). As discussed below, it should be noted that STATIUM is much faster due to the simplicity of its derivation.
Fig. 3.
Correlation of SPOT library signals with predicted values. (a) Intensities expressed as −log(SPOT/SPOTWT) for library variants binding to Bcl-xL or Mcl-1 are correlated with the STATIUM energy scores for Bcl-xL or Mcl-1 binding. (b) Differences in SPOT signals for Bcl-xL and Mcl-1 versus differences in the Bcl-xL and Mcl-1 scores from various models, for strong binders on the library arrays. The model tested is indicated in each panel. Bootstrapping analysis gave 90% confidence intervals as follows: PSSMDEEP_SPEC, 0.89–0.94; PSSMDEEP_AFFIN, 0.69–0.8; PSSMSPOT, 0.80–0.87; and STATIUM, 0.82–0.89.
We also used SPOT data to predict binding specificity, that is, peptide binding preferences for Bcl-xL versus Mcl-1. The SPOT library arrays were designed to sample a space rich in sequences with specificity for either Bcl-xL or Mcl-1.14 Thus, we used these data to define a test set of 134 sequences specific for one receptor versus the other (see Methods). We considered the experimental specificity to be the difference between −log(SPOT/SPOTWT) for binding to Bcl-xL versus Mcl-1. The predicted specificity for a sequence was defined as the difference between the energy from the Bcl-xL scoring model and the Mcl-1 scoring model for that sequence.
Figure 3b shows the experimental versus predicted specificity for each sequence using PSSMDEEP_SPEC, PSSMDEEP_AFFIN, PSSMSPOT, and STATIUM. Among the four models, the strongest correlation was found for PSSMDEEP_SPEC (R = 0.91), and the weakest was found for PSSMDEEP_AFFIN (R = 0.75), showing that a model built from the sequences of specific binders did significantly better for specificity prediction. PSSMDEEP_SPEC outperformed PSSMSPOT (R = 0.83), even though the latter scoring function was derived from substitution SPOT array data that contained every individual residue mutation made in the library SPOT array. The performance of the structure-based model STATIUM (R = 0.86) was similar to that of PSSMSPOT, again remarkable for a model that does not incorporate any experimental binding data. Confidence intervals from bootstrapping analysis supported the statistical significance of these comparisons (see Methods and Fig. 3).
Prediction of yeast-display screening results
Data from yeast-surface display screening provided another opportunity to test different binding models (Table 1 and Table S1). Deep sequencing resulted in sequences designated as likely binders for each of the five receptors. Using each model, we evaluated whether these likely binders were ranked highly compared to the full theoretical library of sequences input to the screening experiment. This is related to the extent to which a scoring model can enrich a library in real binders by design, which is an application of increasing interest.65–69 The metric we used to quantify computational library enrichment was the fraction of likely binders identified in the top-scoring 20% of the full theoretical library. For example, a 472,392-member library was screened for interaction with Bcl-xL by Dutta et al.,14 leading to the identification of 5367 unique likely binders (Table S1). Using each model, we defined an energy cutoff such that 20% of the 472,392 sequences scored below this. The fraction of the 5367 likely binder sequences that scored below this cutoff for each model was defined as the enrichment. Values greater than 20% indicate that a model was able to assign high ranks to experimental binders.
In Table 4, we present the library enrichment results for all of the models on appropriate data sets. Predicting library enrichment with the PSSMDEEP model derived from the same experiment is circular; such values are labeled with an asterisk. In all cases except one, the logical choice of receptor scoring model for a given data set provided significant library enrichment, with PSSMSPOT enrichments ranging from 54% to 87% and STATIUM values ranging from 55% to 90%. PSSMSPOTF_Bim prediction of Bfl-1 library enrichment was poor, and discrepancies between the SPOT array data and the sequence logo for likely binders are apparent in Fig. 1. A strong preference for Tyr at position 2e in sequences identified by yeast screening was not recapitulated on the SPOT arrays, and the SPOT arrays indicated a preference for polar residues at 2g, whereas hydrophobic residues were selected in the yeast screening. At this time, we do not have experimental data to resolve these discrepancies. However, three peptides chosen from this pool (one with Tyr at position 2e), which showed significant enrichment over successive screening rounds, competed effectively with wild-type Bim BH3 for binding to Bfl-1, supporting specific binding to the same site (data not shown). Notably, STATIUM showed enrichment of 90% for the Bfl-1 data set.
Table 4.
Predicted enrichment of experimental binders in top 20% of library sequences
| Model | Receptor | Enrichment |
|---|---|---|
| PSSMDEEPM_AFFIN | Mcl-1 | 0.91* (0.9, 0.91) |
| PSSMDEEPX_AFFIN | Bcl-xL | 0.96* (0.95, 0.96) |
| PSSMDEEP2_AFFIN | Bcl-2 | 0.97* (0.96, 0.98) |
| PSSMDEEPW_AFFIN | Bcl-w | 0.98* (0.97, 0.98) |
| PSSMDEEPF_AFFIN | Bfl-1 | 0.93* (0.93, 0.93) |
| PSSMDEEPM_SPEC | Mcl-1 | 0.86 (0.85, 0.87) |
| PSSMDEEPX_SPEC | Bcl-xL | 0.85 (0.84, 0.86) |
| PSSMSPOTM_Bim | Mcl-1 | 0.82 (0.81, 0.82) |
| PSSMSPOTX_Bim | Bcl-xL | 0.87 (0.86, 0.87) |
| PSSMSPOT2_Bim | Bcl-2 | 0.54 (0.52, 0.56) |
| PSSMSPOTW_Bim | Bcl-w | 0.54 (0.52, 0.55) |
| PSSMSPOTF_Bim | Bfl-1 | 0.21 (0.21, 0.22) |
| PSSMSPOTM_Noxa | Mcl-1 | 0.81 (0.8, 0.81) |
| PSSMSPOTX_Bad | Bcl-xL | 0.62 (0.61, 0.63) |
| STATIUMM | Mcl-1 | 0.58 (0.57, 0.59) |
| STATIUMX | Bcl-xL | 0.55 (0.54, 0.56) |
| STATIUM2 | Bcl-2 | 0.63 (0.61, 0.65) |
| STATIUMW | Bcl-w | 0.75 (0.74, 0.77) |
| STATIUMF | Bfl-1 | 0.9 (0.9, 0.91) |
Likely binders were derived from yeast pools screened for BH3 peptides that bound tightly to a given receptor and were deep-sequenced using Illumina technology (see Methods and Table S1). The value in the table is the fraction of binders below the energy threshold that covers 20% of library sequences. Bold values designate enrichments derived from models built for the appropriate receptor. The values labeled with an asterisk designate circular cases where the test data set was used to derive the scoring function. Ninety percent confidence limits are given in parentheses.
We also used yeast-display data to assess predictions of Bcl-xL versus Mcl-1 binding specificity. First, we examined how well different models could distinguish 33 Mcl-1-specific versus 40 Bcl-xL-specific peptides reported by Dutta et al.14 (specific tight binders in Table 1). Results are shown in Fig. 4a. The difference between energies predicted using the BclxL and Mcl-1 models can separate the two groups of sequences, with the best performance coming from the PSSMSPOTX/PSSMSPOTM and PSSMDEEPX_SPEC/PSSMDEEPM_SPEC models, as expected. STATIUMX/STATIUMM also showed good resolution in this test, with a very low rate of incorrect classifications. Corresponding data for likely specific binders (a larger but lower confidence data set) are given in Fig. S6.
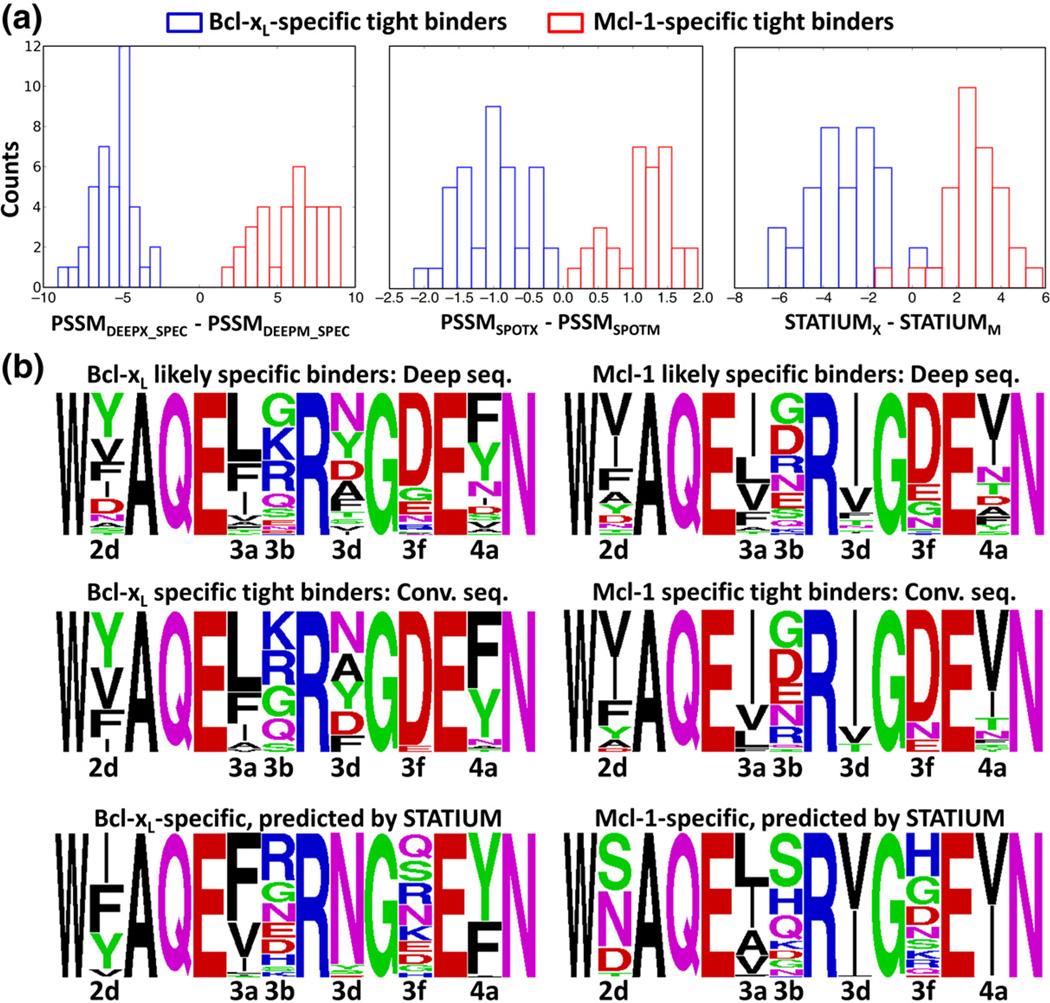
Fig. 4.
Prediction of binding specificity for Mcl-1 and Bcl-xL specific peptides isolated in library screening. (a) Three scoring models can distinguish 40 Bcl-xL and 33 Mcl-1-specific tight binders from Dutta et al.14 (b) STATIUM prediction of the most specific sequences in the full theoretical library that was screened for binding to Bcl-xL or Mcl-1. A sequence logo built from these sequences (bottom row) is compared to logos built from sequences identified by experimental screening and sequencing (top rows). Logos were generated with WebLogo.60
Finally, we used STATIUM to predict which sequences in the full theoretical library input to yeast screening would be specific for either Bcl-xL or Mcl-1. This corresponds to running a computational version of the screening experiment. To do this, we identified all sequences with STATIUM energies in the lowest 10% for the targeted receptor and in the highest 65% for the alternative receptor. We generated a sequence logo for this list and compared it to logos derived experimentally (Fig. 4b). The sequence logo for the entire list of 90,949 sequences that were successfully displayed on the surface of yeast is also provided for comparison (Fig. S4). At most positions in the Bcl-xL-specific and Mcl-1-specific logos, the large variability observed in sequences that were successfully expressed was reduced to four or fewer dominant amino acids by STATIUM. Among those predicted amino acids, many were the dominant residues observed for the experimentally validated sequences. For example, at position 3d, the Bcl-xL predicted logo exclusively consists of asparagine, which was the top amino acid at that position among the likely specific binders. Likewise, at position 3d in the Mcl-1 predicted logo, the two dominant amino acids are isoleucine and valine, which are identical with what was preferred experimentally. A significant failure of STATIUM is at position 2d for the Mcl-1-specific binders, where the model predicts mostly polar residues rather than the apolar residues present in the Mcl-1 likely specific binders. STATIUM also predicts greater variability at site 3f than was observed at this site experimentally or in native BH3 motifs. This same observation was made previously using atomic-level structural modeling.70
Prediction of specificity in natural BH3 peptides
Natural BH3 domains share little sequence similarity beyond a few conserved positions (Fig. S2). Some of these BH3 peptides interact selectively with specific receptors; thus, we used them to test the performance of our interaction models in cases where the sequence diversity is much higher than in our experimental data sets, which consist mostly of Bim BH3 variants. There are four natural BH3 domains with unambiguous specificity profiles: Bad BH3 interacts preferentially with Bcl-xL/Bcl-w/Bcl-2 over Mcl-1/Bfl-1, and Mule/Noxa/Bok BH3 domains interact preferentially with Mcl-1 over all other receptors.27–31,71 In Table 5, we show the top predicted receptor for each specific peptide. PSSMDEEP exhibited mixed results for this test, ranking the top receptor for Bad and Mule correctly, but ranking Mcl-1 as only the third best Noxa binder and the fourth best Bok binder. PSSMSPOT performed significantly better, predicting the top binders for Bad, Mule, and Bok, while assigning Mcl-1 as the second best Noxa-binder. The PSSMSPOT assigned Bfl-1 as the top Noxa binder, although experimentally binding to Mcl-1 is preferred. STATIUM ranked the tightest binding receptor as the top receptor in every case.
Table 5.
Prediction of binding specificity for natural BH3 peptides
| Top-ranked receptor for highly specific BH3 peptidesa | |||
|---|---|---|---|
| BH3 (specific receptor) | PSSMDEEP_SPEC | PSSMSPOT | STATIUM |
| Bad (Bcl-w/Bcl-2/Bcl-xL) | Bcl-w | Bcl-w | Bcl-xL |
| Mule (Mcl-1) | Mcl-1 | Mcl-1 | Mcl-1 |
| Noxa (Mcl-1) | Bfl-1 (3) | Bfl-1 (2) | Mcl-1 |
| Bok (Mcl-1) | Bfl-1 (4) | Mcl-1 | Mcl-1 |
| STATIUM-predicted ranking of natural BH3 peptidesb | |||
| IC50 <100 nM | IC50 <10 µM | IC50 >100 µM | |
| Mcl-1 | Puma, Bim, Noxa | Bid, Bik, Bmf, Hrk | Bad |
| Predicted rank | 3, 4, 5 | 1, 2, 6, 7 | 8 |
| Bcl-xL | Bid, Puma, Bad, Bim, Hrk, Bik, Bmf | N/A | Noxa |
| Predicted rank | 1, 2, 3, 4, 5, 6, 7 | N/A | 8 |
| Bcl-2 | Bim, Puma, Bmf, Bad | Bid, Bik, Hrk | Noxa |
| Predicted rank | 1, 4, 5, 6 | 2, 3, 7 | 8 |
| Bcl-w | Bid, Bim, Bik, Puma, Bmf, Bad, Hrk | N/A | Noxa |
| Predicted rank | 1, 2, 3, 4, 5, 6, 7 | N/A | 8 |
| Bfl-1 | Bid, Bik, Bim, Puma, Hrk | Bad, Bmf, Noxa | N/A |
| Predicted rank | 1, 2, 3, 5, 6 | 4, 7, 8 | N/A |
N/A - Not applicable.
Peptide binding specificities from the literature.27–29,71,72 Correct predictions are italicized. For incorrect predictions, the rank of the correct receptor is shown in parentheses. To compare different receptors binding to a given BH3, we referenced the PSSMSPOT and PSSMDEEP energies to the energy of Puma, which binds all receptors tightly (see the bottom half of the table).
IC50 values are for competition with Bim BH3.28
As an additional comparison, we considered peptides from a comprehensive study of BH3 interaction specificity involving all five Bcl-2 receptors.28 In Table 5, we report the STATIUM rankings for BH3 peptides experimentally assigned as very tight binders (IC50 <100 nM), moderate binders (IC50 <10 µM), and non-binders (IC50 >100 µM). This test is identical with one used by London et al. (submitted for publication) in an assessment of Rosetta prediction performance, which allows us to compare the performance of STATIUM with that model. Noxa BH3, which is specific for Mcl-1, is appropriately ranked last for all receptors other than Mcl-1. Bad BH3, which does not bind Mcl-1, is ranked last for that receptor. These results are slightly better than those achieved by Rosetta, which erroneously ranks Noxa as a better Bcl-xL binder than tight binding Bik BH3. In the same analysis, PSSMSPOT was less accurate (Table S5). For example, Noxa BH3 was ranked first for Bcl-w according to the PSSMSPOT, but in experiments, it does not bind that receptor. We attribute this poor performance to under-sampling of the native BH3 sequence space by PSSMSPOT. PSSMDEEP was unsuitable for use on this test because it sampled a small number of positions (Table S1) and a limited number of amino acids at each position. For that reason, some natural BH3 domains could be evaluated at all sampled positions and others only at a few, making it impossible to fairly assess their relative ranks.
Performance of the STATIUM statistical potential
An important consideration for any structure-based method is how sensitive the method is to the specific structure models used. We compared the prediction accuracy of STATIUM when alternative structural templates were used for Bcl-xL (3IO8 versus 3FDL) and Mcl-1 (3PK1 versus 2PQK). Overall, the prediction capabilities were similar. For example, the yeast specificity prediction AUC decreased from 0.98 to 0.92 and the affinity prediction AUC values for Bcl-xL and Mcl-1 changed from 0.94 and 0.94 to 0.93 and 0.93 using 3FDL and 2PQK.
Because Bcl-2 receptors are all structurally similar,22 we tested how STATIUM performed in a homology modeling mode, where the structure of one receptor was used to model others. Note that the modeling problem is a simple alignment problem, because only Cα and Cβ coordinates are used in STATIUM. We aligned the sequences of Bcl-xL and Mcl-1 and substituted the amino acid identities at each aligned position. For example, the sequence of Bcl-xL was modeled using Mcl-1 structure 3PK1. We evaluated several affinity and specificity prediction metrics in this mode. The results are shown in Table 6. Specificity prediction was more sensitive than affinity prediction to the choice of model. In particular, using Mcl-1 structure 3PK1 to model both Mcl-1 and Bcl-xL had modest effects on affinity prediction performance but dramatically decreased the AUC value (AUC=0.90 to 0.52) and correlation coefficient (R = 0.86 to 0.21) for SPOT specificity prediction. Thus, STATIUM appears to capture important information about specificity that is encoded in the structures of different receptors, and homology modeling reduces accuracy because appropriate structural detail is lost. For Bcl-w, we also tested a homology model based on the X-ray structure of Bcl-2 bound to Bax BH3, rather than an NMR-based model of Bcl-w in complex with Bid BH3. This decreased performance somewhat, reducing the SPOT AUC from 0.82 to 0.78 and the library enrichment from 0.75 to 0.70.
Table 6.
Sensitivity of STATIUM affinity and specificity predictions to the template structures used
| High-affinity prediction | Specificity prediction | ||||||
|---|---|---|---|---|---|---|---|
| Modelsa | Library SPOT AUC |
Substitution SPOT AUCb |
Modelsa | Library SPOTc AUC (R) |
Yeast screeningd AUC |
||
| Bim | Bad/Noxa | ||||||
| STATIUMX | 0.94 | 0.72 | 0.66 | STATIUMX and STATIUMM | 0.9 (0.86) | 0.99 | |
| STATIUMX_Mstruc | 0.82 | 0.67 | 0.64 | STATIUMX and STATIUMM_Xstruc | 0.81 (0.78) | 0.9 | |
| STATIUMM | 0.94 | 0.71 | 0.69 | STATIUMX_Mstruc and STATIUMM | 0.52 (0.21) | 0.37 | |
| STATIUMM_Xstruc | 0.95 | 0.65 | 0.71 | ||||
Two STATIUM models were tested: Bcl-xL sequence with an Mcl-1 structure model (STATIUMX_Mstruc) and Mcl-1 sequence with a Bcl-xL structure model (STATIUMM_Xstruc). Specificity prediction (right column) requires two models, because specificity is predicted using a difference in scores.
The AUC values in the left column are for SPOT data from Bcl-xL or Mcl-1 binding Bim BH3 variants, and the AUC values in the right column are for SPOT data from Bcl-xL binding Bad BH3 variants and Mcl-1 binding Noxa BH3 variants.
The values in parentheses are the Pearson correlation coefficients for the same data set.
AUC value for prediction of the specific sequences used in Fig. 4a, when one class is Bcl-xL specific (40 sequences) and the other is Mcl-1 specific (33 sequences) and the difference in energies is used to predict the state.
We compared our STATIUM results to Rosetta FlexPepBind, which was used by London et al. to predict binding data reported by Dutta et al.14 The affinity prediction performance of STATIUM on the SPOT library data is summarized in Table 2 and Fig. 3. STATIUM achieved a Pearson correlation of 0.72 for the Bcl-xL data and 0.67 for the Mcl-1 data. The performance of Rosetta “out of the box” was not very good on this test but improved markedly when small adjustments to the electrostatic part of the energy function and the structure building protocol were made to improve fit to the Bcl-2 data (unoptimized R = 0.54, 0.46; optimized R = 0.8, 0.67, discussed in London et al., submitted for publication). For the specific tight binders (Table 1), STATIUM correctly classified the specificity of virtually all sequences (AUC = 0.98), but the optimized version of Rosetta could distinguish all (AUC=1.0). Keeping in mind the advantages of a generally applicable protein–protein interaction model, we did not extensively optimize this version of STATIUM for the Bcl-2 system. We did compare two different approaches for geometrical description of residue–residue interactions, and we adjusted the weighting of inter- and intracomplex pairs differently for affinity versus specificity prediction, as described in Methods. Overall, compared to the all-atom Rosetta method, the reduction in structural detail that makes STATIUM so fast does not significantly reduce its performance on the types of tests presented here.
Discussion
To better understand features of BH3 motifs that govern specific binding to Bcl-2 family receptors, we collected large amounts of experimental data that report the effects of single and multiple amino acid substitutions in BH3 peptides. Although SPOT arrays do not directly measure equilibrium binding constants, they do provide one-at-a-time binding information for large numbers of peptides without laborious synthesis and purification and therefore can be applied in high throughput.14 Similar approaches have provided important insights for other protein domains.73 We found that SPOT array intensities can correlate well with solution measurements of BH3 affinity and specificity (Fig. S1). Furthermore, the SPOT and yeast-screening methods gave consistent views of BH3 interaction preferences, as illustrated by the good performance of SPOT-derived models on yeast-screening data and vice versa.
Large data sets such as those we present here are difficult to interpret or use without a model. Of the several modeling strategies we applied, two involved converting data from the experiments directly into position-specific scoring matrices and testing the ability of these models to predict data not used in their derivation. This mimics a possible long-term approach focused on measuring as much of the BH3 sequence space as necessary to describe the entire BH3 sequence universe. A third model was derived directly from protein structure data, and it utilized experimental binding data only for testing. This approach has the potential to be much more general and also has the advantage of not requiring extensive experimental measurements. Applying different models to different data sets provided insights into both the data and the models, which we discuss below.
Modeling affinity
An interesting finding was that our models behaved differently when modeling affinity versus specificity. In particular, our tests based on discriminating strong binders from non-binders were quite insensitive to which model was used. Table 2 reports predictions for Bcl-xL and Mcl-1 binding to SPOT library array peptides, and even models that were not developed to describe Bcl-xL or Mcl-1 binding gave good performance. For example, models PSSMDEEP2_AFFIN, PSSMSPOTW_Bim, and even PSSMSPOTM_Noxa could describe either Bcl-xL or Mcl-1 binding to Bim BH3 variants, and STATIUM performance was not strongly dependent on which Bcl-2 family structure was used as a template. Similar trends were observed for SPOT substitution array predictions (Table S3) and library affinity enrichment (data not shown). These observations are not entirely surprising, given that the Bcl-2 receptors overlap in the types of sequences they bind. In the SPOT substitution arrays, where 18 mutations were made at each site, many of these stabilized or destabilized binding to many receptors similarly. Thus, any receptor model good at recognizing such globally stabilizing or destabilizing influences on binding performed well. The many high correlations among SPOT substitution arrays, shown in Fig. 1e, support this.
The experiment-based PSSM models provided insights into which mutations were broadly stabilizing or destabilizing. In general, Bim BH3 variants that bound well to many receptors on SPOT arrays were quite similar to wild type (e.g., Bim I2dV or F4aL), and peptides that had weakened binding to all of the receptors included polar substitutions at buried sites (e.g., L3aD), charge incompatibility on the surface (e.g., R3bD), large residues at positions normally occupied by small ones (e.g., A2eF), or a proline in the central region of the BH3 helix. STATIUM correctly predicted many of these types of effects. For example, position 3a is a conserved buried leucine in known BH3 sequences and is the best residue at this position according to the SPOT arrays. STATIUM predicted leucine to be preferred by a large energy gap to all polar/charged residues but predicted a much smaller gap to some hydrophobic alternatives. A preference for small residues Ala, Gly, or Ser at position 2e was captured well, and STATIUM also recognized that negative charge is disfavored at position 3b. Position 3f is conserved as an aspartic acid in known BH3 domains, and the SPOT arrays confirmed that all other substitutions decrease binding for all receptors. STATIUM, however, predicted no preference for aspartic acid over most other hydrophilic residues at that site (Fig. 4b). This could be due to the fact that STATIUM does not model long-range electrostatic interactions, which may be critical for that position. An alternative explanation may be that the conservation of aspartic acid at position 3f is caused by a multi-residue structural context that is not captured in the pairwise derivation of STATIUM.
For predicting library enrichment in tight binders, models had to do more than recognize strongly destabilizing residues. This is because the libraries input into yeast-surface screening were already depleted in such residues by design (see Methods). This test placed a greater emphasis on identifying the most stabilizing substitutions. Even so, performance in library affinity enrichment was good when many different models were used (Table 4).
Modeling specificity
For predicting interaction specificity, sequence features that affect all receptors similarly are unimportant; models must distinguish possibly subtle differences between proteins. In contrast to affinity tests, in specificity tests requiring discrimination of Mcl-1 versus Bcl-xL binding, the computational model constructed for the appropriate receptor performed the best (data not shown). Similarly, when using STATIUM, homology modeling of the template gave inferior specificity prediction results to using an appropriate experimental structure (Table 6). Another difference is that intrapeptide interactions are expected largely to cancel when comparing binding of one peptide to different receptors, whereas these make important contributions to determining binding affinity. Following testing that confirmed this expectation (Fig. S5), we developed slightly different versions of STATIUM for predicting affinity versus specificity (see Methods).
An interesting result from the specificity tests in Fig. 3, which use SPOT library array binding specificity data, is that the PSSMDEEP_SPEC (R = 0.91) outperformed PSSMSPOT (R = 0.83), even though the latter scoring function was derived from SPOT substitution array data that included every individual residue mutation made in the SPOT library array. PSSMDEEP_SPEC was derived from sequences selected to be specific for binding one receptor versus the other and thus directly uncovered specificity determinants. For the PSSMSPOT models, specificity was computed using differences in scores from affinity models (PSSMSPOTX−PSSMSPOTM or PSSMSPOTM−PSSMSPOTX). The greater effectiveness of PSSMDEEP_SPEC highlights the advantages of a specialized model. Figure 3 demonstrates that specificity in at least some cases can be better resolved than affinity, possibly due to cancellation of errors in the models for different receptors.
Dutta et al. elucidated several features that make BH3 peptides specific for Bcl-xL versus Mcl-1, and vice versa.14 PSSMSPOT, PSSMDEEP, and STATIUM often agreed on such sequence determinants of this specificity. For example, a valine substitution at position 4a in Bim was universally predicted to favor binding to Mcl-1 in preference to Bcl-xL. With models now available for five receptors, we reexamined previously isolated Mcl-1-specific BH3 peptides to identify candidate specificity determinants. Several Mcl-1-specific peptides exhibited specificity over Bfl-1, Bcl-2, Bcl-w, and Bcl-xL, even though negative selection was only applied against Bcl-xL in screening.14 Position 3b may be an important specificity determinant in this regard, as most of the Mcl-1-specific sequences from yeast selection included Gly, Asn, Asp, or Glu at this site. Both STATIUM and the PSSMSPOT models predicted that three of these four substitutions (Asp, Asn, and Gly) favor Mcl-1 binding over all the other receptors and predicted Glu at 3b to favor Mcl-1 binding over Bcl-xL, Bcl-2, and Bfl-1 but not Bcl-w. Interestingly, Asp and Glu at position 3b are mutations of the type labeled “class 2” for Mcl-1 versus Bcl-xL by Dutta et al.14 Class 2 substitutions destabilize binding to all receptors, but unequally. Asp at 3b destabilizes interactions with all receptors, but the effect on Mcl-1 binding is smaller, making this a specificity determinant favoring Mcl-1 binding. Additionally, an Ile at position 3a, found in a high percentage of Mcl-1-specific peptides,14 scores favorably for binding Mcl-1 over any of the other four receptors with all three models. These features appear likely to explain the observed specificities of some Mcl-1-specific peptides.14
Data-based versus structure-based models
In many cases, the models derived from the experimental data outperformed STATIUM. This is expected because the experimental models were built using highly relevant measurements for the prediction tasks at hand and because STATIUM makes numerous approximations in extracting a binding score from a single structural model. However, there are tasks for which STATIUM was equally good or occasionally better than the PSSM models. These included predicting strong binders versus non-binders on the SPOT library arrays (Table 2) and ranking the native BH3 motif binding preferences of different receptors (Table 5). The library enrichment performance of STATIUM was also good for some receptors (Table 4).
Detailed analyses of the test data did uncover some important deficiencies in STATIUM. Indeed, the main strength of STATIUM, which is generalizing structure information to model interactions, can become a weakness when making predictions for idiosyncratic structure contexts. An example is the substitution of arginine for leucine at buried position 3a, which is not tolerated for binding to any receptor except Bfl-1 on the SPOT arrays (Fig. 1b). Bfl-1 contains a buried glutamic acid that may interact favorably with an arginine at position 3a,54,74 allowing this unusual substitution.54 STATIUM recognizes charge compatibility between Arg and Glu at these sites, but because that specific charge–charge pairing is one out of many pairs involving position 3a, it is outweighed by other interactions that prefer apolar residues at buried sites. Other groups have approached such problems by optimizing their methods to suit the specific system that is the subject of prediction. For example, the Kortemme group has modified the parameters of the side-chain hydrogen-bonding potential in Rosetta to improve modeling of PDZ–peptide interactions41 and the Furman group has likewise optimized the parameters of Rosetta for Bcl-2 interactions (London et al., submitted for publication). We could potentially improve STATIUM in a similar manner, by up-weighting interactions involving buried charges. For now, we choose not to take this approach because of the negative effect it may have on the general applicability of STATIUM.
STATIUM offers two important advantages over data-derived methods that cannot be easily overcome by increasing the complexity of those models. First and most obvious, it does not require large amounts of data for model derivation. Large data sets can be costly and time-consuming to obtain; thus, this is a significant benefit. Second, structure-based methods such as STATIUM have potentially greater generality. Experimentally derived models can only describe interactions for the sequence space that is sampled. In our libraries, technical limitations constrained sampling to ~107 sequences distributed over ~10 sites of Bim BH3. In contrast, STATIUM can be used to model interactions at any site, although performance at some sites may be better than at others. The good performance of STATIUM and Rosetta at predicting the natural specificity profiles of BH3 peptides, shown in Table 5 for STATIUM, highlights this ability to score highly diverse sequences and has important implications for the use of these predictive models in the design of novel BH3 peptides and in genomic searches for undiscovered BH3 domains. An important caveat is that structure-based models assume a highly conserved binding mode. For cases where there are significant changes in binding geometry, more structural templates would be required to make effective predictions.
Implications for Bcl-2 interaction prediction and design
STATIUM provides a practical combination of speed and accuracy that allows a user to efficiently survey large numbers of sequences. This is important for applications such as scanning the proteome for new BH3 motifs, and protein design. The results described in this study demonstrate that STATIUM can evaluate Bcl-2 interactions at a rate of ~106 sequences per second. This is similar to the computational efficiency of the simple PSSM models and orders of magnitude faster than methods that require full flexible side-chain and/or backbone modeling. A legitimate question is how much accuracy is lost when structural detail is reduced by STATIUM. Our comparisons with the work of London et al. suggest that similar accuracy can in fact be achieved with much higher speed, although with less insight into exact structural details that may be responsible. What our results suggest is that STATIUM and atomic-resolution modeling can serve complementary roles, with STATIUM narrowing down vast regions of sequence space to a smaller number of sequences that Rosetta, or other methods, can handle. Structure-based models have great potential for designing novel BH3 domains with nonnative sequences and desired specificities,75 and combining STATIUM with higher-resolution methods and using it to design combinatorial libraries for screening are promising directions that we are pursuing.
Methods
Expression of recombinant pro-survival Bcl-2 proteins
Pro-survival Bcl-2 proteins with a c-Myc tag at the amino terminus were used for all studies, with the exception of yeast-display screening experiments involving Bcl-xL, where an amino-terminal His-tagged protein was used. The Bcl-2 protein constructs used by Dutta et al. were expressed in Escherichia coli BL21(DE3) strains as described previously.14 The oligomerization state of each protein after purification was analyzed using a Superdex S75 column (GE Healthcare) in 20mMTris, 300mMNaCl, and 10% glycerol, pH 8. All proteins were predominantly monomeric, the monomeric fraction ranging from ~85% to 100%.
Construction of combinatorial libraries for screening
The BH3 peptide libraries used to generate the affinity and specificity pools for Bcl-xL and Mcl-1 were described previously.14 The peptide libraries used for screening against Bfl-1, Bcl-w, and Bcl-2 were designed computationally based on the SPOT substitution array data.62 The library screened for binding to Bfl-1 was based on the Bim BH3 sequence, with the following residues encoded by degenerate codons at seven randomized positions: F, I, K, L, M, N, and Y at position 2d; A, D, H, P, S, and Y at position 2e; C, D, E, F, G, I, K, L, M, N, R, S, V, W, and Y at position 2g; D, H, I, L, N, and V at position 3a; A, C, D, F, G, H, I, L, N, P, R, S, T, V, and Y at position 3d; A, E, I, K, L, P, Q, T, and V at position 3g; C, F, I, K, L, M, N, R, S, W, and Y at position 4a. A separate BH3 peptide library based on the Bim BH3 sequence was used to screen for binders to Bcl-2 and Bcl-w with the following composition: C, D, E, F, G, H, I, K, L, M, N, Q, R, S, V, W, and Y at 2d; C, D, E, F, G, I, K, L, M, N, R, S, V, W, and Y at position 2g; A, D, F, H, L, P, S, V, and Y at position 3a; A, E, G, I, K, L, R, S, T, and V at position 3b; A, C, D, E, F, G, I, K, L, M, N, R, S, T, V, W, and Y at position 3d; G and A at position 3e; E, G, Q, and R at position 3g. All combinatorial BH3 libraries for screening were constructed using homologous recombination in yeast as described previously and were expressed on the yeast cell surface as Aga2p fusion proteins, bearing a C-terminal FLAG tag.14
Flow cytometric analysis and screening
Yeast cells for flow cytometric analysis and screening were labeled as described previously.14 Briefly, yeast cells were incubated with Myc-tagged anti-apoptotic proteins (starting at 1 µM and using reduced concentration in successive rounds, as shown in Table S1) for 1–2 h and labeled with primary antibodies (anti-FLAG rabbit and anti-c-myc mouse) (Sigma), and then after washing, they were incubated with secondary antibodies fluorescein-isothiocyanate-conjugated goat anti-rabbit antibody and R-phycoerythrin (PE)-conjugated goat anti-mouse IgG (Sigma). For the first round of FACS screening for Bcl-xL, Bcl-w, and Bfl-1, ~108 cells (10 times the theoretical library size) were screened based on their fluorescein isothiocyanate and PE intensities, to quantify BH3 surface expression and receptor binding, respectively. Typically, expression-positive cells with the top 5% of PE intensities were recovered in the first round. For each successive round, the number of cells screened was maintained at least 10× greater than the number collected in the previous round, and the percentage of recovered cells was reduced to those exhibiting the top 1–2% PE intensity. For library screening at low concentrations of Bcl-2 and Bcl-w, 6 × 106 cells were incubated with 5 ml of 1 nM receptor for ~2 h to maintain excess molar concentration of receptor before staining with antibodies. Recovered cell populations were validated to bind specifically to the desired anti-apoptotic protein by confirming the absence of nonspecific binding to antibodies or by confirming that competition with unlabeled anti-apoptotic proteins reduced binding by at least 90%. All screening was performed on a BD FACSAria or Cytomation MoFlo using 488 nm and/or 561 nm excitation.
SPOT arrays
SPOT arrays were synthesized and processed following the procedures described previously.14
Library preparation for Illumina sequencing
Yeast pellets from all library pools screened for binding to Bcl-2, Bcl-w, Bcl-xL, or Mcl-1 were lysed by heating at 95 °C. DNA from library pools for binding to Bfl-1 was extracted using the Zymoprep Yeast Plasmid Miniprep I kit (Zymoresearch). Subsequently, all samples were PCR amplified, employing Platinum Taq DNA Polymerase (Invitrogen) for Bcl-xL and Mcl-1 pools and High Fidelity Platinum Taq DNA Polymerase (Invitrogen) for Bcl-2, Bcl-w, and Bfl-1 pools. Errors will be introduced in PCR amplification. We conservatively estimated the error rate per BH3 sequence of Taq polymerase, considering varied sequence regions only, as 3.6 × 10−3 to 4.2 × 10−3.76 Thus, the fraction of erroneous sequences after 31 PCR cycles ranges between 0.05 and 0.06. For HiFi Taq polymerase, using an error per base of 3.3 × 10−5 gives a fraction of erroneous sequences after 31 PCR cycles of 0.009–0.011.
The forward primer had the sequence 5′AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACGGCCGTCCGGAAATTTGG3′ (containing flow cell annealing site and the 5′ sequencing primer binding site) and the reverse primer had the sequence 5′CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGGCGACGCGCATAATACGCATT3′, where NNNNNN represents a six-base-pair barcode region (containing a flow cell annealing site and the 3′ sequencing primer binding site). The product was purified using the Qiagen PCR Purification Kit (Qiagen). The quality of all DNA samples was checked using the Agilent 2100 BioAnalyzer at the MIT BioMicro Center.
Illumina sequencing
Pools for Bcl-2, Bcl-w, Bcl-xL, and Mcl-1 were sequenced on an Illumina Genome Analyzer II. Pools for Bfl-1 were sequenced on the Illumina HiSeq system. We multiplexed DNA samples using 12 barcodes per lane. Each barcode was different in at least three positions from any other barcode. Data used for model building met three conditions. First, we only considered reads with perfect barcodes. Second, bases had to be called in all positions of a sequence and any non-mutated nucleotide had to be read without error. Third, we applied stringent thresholds for the Illumina quality scores amounting to a confidence probability of 0.995 that the variable nucleotides of a read collectively were correct. Further criteria were imposed on sequences used for model building and testing, as described for PSSMDEEP below.
PSSMSPOT
The PSSMSPOT potentials were constructed using fluorescence intensities resulting from the binding of receptors to single amino acid variants of Bim, Bad, and Noxa BH3 peptides printed on SPOT membranes (substitution arrays). Similar to Dutta et al., the PSSMSPOT energy of a substitution was calculated by taking the negative logarithm of the ratio of the fluorescence intensity for the corresponding BH3 substitution to the intensity of wild-type BH3 (averaging over all wild-type spots).14 The PSSMSPOT energy of a BH3 sequence was defined as the sum over all substitutions. Not all positions were varied on the SPOT membranes, and substitutions at unsampled positions were equivalent for all amino acids.
PSSMDEEP
PSSMDEEP potentials were derived from Illumina sequencing of previously published yeast display cell screening experiments (for Bcl-xL and Mcl-1)14 and screening experiments that we present in this article (for Bcl-2, Bcl-w, and Bfl-1). Sequenced pools included those selected for binding to specific receptors (positive screening) and those further selected for not binding to a specific receptor (negative screening).
The PSSMDEEP_AFFIN models were developed to describe affinity towards a receptor. For these models, we used unique sequences that were consistently present, in at least one copy, in all available pools from positive screening rounds (we used only positive rounds that were not separated by rounds of negative screening; see Table S1). At least two such pools were used in all cases. The PSSMDEEP_AFFIN energy of an amino acid substitution at a BH3 position was computed as the frequency of occurrence of that amino acid in the list of unique sequences. The total PSSMDEEP_AFFIN score was obtained by summing over all positions that were varied in each screening experiment:
| (1) |
The PSSMDEEP_SPEC models were developed to predict binding affinity for targets Bcl-xL and Mcl-1 and also to capture information about the specificity towards each receptor relative to the other. The PSSMDEEP_SPEC models were derived using unique sequences from rounds of yeast screening that included both positive screening for binding to one receptor and negative screening disfavoring binding to the other receptor. Table S1 shows the successive screening requirements imposed on each sequenced pool used in the benchmark. Again, sequences used for model building appeared at least once in all pools analyzed.
STATIUM potential
Defining interacting residue pairs in a template complex
The STATIUM potential for a protein–protein interaction consists of the sum of contributions from all interacting residue pairs in a structure of the protein complex (Fig. 2). A residue pair is considered interacting if the distance between the Cβ atoms of the two residues is less than 10 Å, and the angle between the Cα–Cβ vectors is less than 120° or the distance between the Cβ atoms of the two residues is less than the distance between the two Cα atoms of the two residues.
For a residue pair considered interacting according to these criteria, the Cα atom distance (Cα1Cα2) and the Cβ atom distance (Cβ1Cβ2) are used to describe the 3D structure of the pair. Because receptor residues never changed in this study, we did not consider interactions between residues within the receptor subunit of the complex. Thus, all interacting pairs were interface contacts between the receptor and peptide or within the peptide.
Generating a STATIUM potential for a complex
The STATIUM scoring function was generated by searching through all chains in an experimental structure database to identify residue–residue interactions geometrically similar to interactions in the template complex. The database used for this work consisted of X-ray crystal structure models with resolution less than 2.5 Å and sequence similarity less than 90%. It was generated using the program PISCES.77 The protein chains of multimeric complexes were separated; hence, no Bcl-2 receptor interactions were present. There were 19,309 chains in the database.
An interacting pair in a unique chain was considered similar to an interacting pair in the template complex if the Cα1Cα2 and Cβ1Cβ2 distances of the PDB chain pair were each within ±0.1 Å of the Cα1Cα2 and Cβ1Cβ2 distances of the template pair. Every time a structurally similar pair was found in the database, the amino acid identities i and j of the pair were tabulated in a 20 × 20 matrix of amino acid counts for the appropriate residue pair in the template complex. The probability of an amino acid pair, Pij, for a given template structure pair was computed as the counts for that amino acid pair divided by the sum of all amino acid pair counts for that template structure pair. Also tabulated were the frequencies of each of the 20 amino acids in the database, and the resulting individual probabilities were Pi and Pj. The total STATIUM score for a complex is the sum of the score of all pairs in the complex, and is represented by Eq. (2). We found that affinity prediction was improved by increased weighting of the intrapeptide pair terms, but specificity prediction was not (Fig. S5), so for affinity prediction we increased the weighting of those terms by a factor of 5.
| (2) |
The time it takes to derive a STATIUM potential for a given protein–protein interaction depends on the total number of interacting residue pairs in the template. For Bcl-2 receptors with BH3 ligands, the total number of residue–residue interactions ranged from 215 to 227, and it took ~10 h to generate a potential by searching through all of the interacting residues in the 19,309 PDB chains in the database on a single processor. This process is highly parallelizable. After the potential was generated, the STATIUM energy of the interaction between a Bcl-2 receptor and a BH3 sequence could be determined at a rate of ~2 × 106 sequences per second on a single processor.
Template structures
The PDB accession codes for the human receptor–BH3 template structure models were as follows: 2XA055 for Bcl-2 (Bax BH3 ligand), 1ZY325 for Bcl-w (Bid BH3 ligand), 3IO817 for Bcl-xL (Bim BH3 ligand), 3PK153 for Mcl-1 (Bax BH3 ligand), and 3MQP for Bfl-1 (Noxa BH3 ligand). 2XA0 is the only crystal structure of a BH3-bound Bcl-2 and 3MQP is the only Bfl-1 crystal structure with the full range of BH3 residues under consideration; thus, we only used this structure for the Bfl-1 STATIUM model. There is no crystal structure available for a Bcl-w complex; hence, we used the NMR-based model of the Bcl-w complex with Bid (1ZY3). 3IO8 for Bcl-xL and 3PK1 for Mcl-1 were chosen because they included the full-length BH3 peptide and a preliminary analysis showed that they performed slightly better than alternative structures.
Affinity prediction for SPOT array data
For the binary prediction of affinity or specificity classes, we used SPOT array data from Dutta et al.14 and from this work. The fluorescence intensity (SPOT) of Bim, Bad, and Noxa BH3 variants on SPOT arrays were referenced to the average wild-type fluorescence intensity:
| (3) |
The signal distributions did not have strong features that could be used to separate binders from non-binders, and absolute intensities varied from array to array. Therefore, we defined peptides with SSPOT values in the lowest 30% of scores as binders and peptides with SSPOT values in the highest 30% as non-binders. Extracting upper and lower percentiles ensures equal populations of binders versus non-binders, which is useful for interpreting receiver operating characteristic curves.
To compute the correlation between predicted and experimental binding, we used all data points available in a given experiment. For the calculation of the correlation between predicted and experimental specificity for binding to receptor 1 versus receptor 2 for the SPOT data sets, we required that SSPOT1 or SSPOT2 be less than 0.5. This approach excluded sequences that bound neither receptor strongly but, due to the difference in dynamic range of each experiment, had a large difference in signal intensity.
Ranking natural BH3 domains
To compare the results from PSSM models derived for different receptors, it was necessary to define a reference state. For the data in the top section of Table 5, we referenced PSSMSPOT and PSSMDEEP scores to the scores of these models for Puma, a universal tight binder. STATIUM scores were not normalized. Normalization was not necessary for the bottom section of Table 5 because scores were compared for the same receptor in those tests.
Bootstrapping analysis to provide confidence intervals
To compute confidence intervals for the metrics that we report (R, AUC, and library enrichment), we used bootstrapping. For each data set, we resampled the data 2000 times to generate the bootstrap distribution.78 In the tables and figure legends, we report the limits of the 90% confidence interval resulting from this procedure.
Supplementary Material
Acknowledgements
We gratefully acknowledge use of the Biopolymers and Flow Cytometry core facilities of the David H. Koch Institute Swanson Biotechnology Center and the BioMicro Center in the MIT Biology Department. We thank the staff of these facilities, especially S. Levine and R. Cook for assistance with Illumina sequencing and SPOT arrays, respectively. We thank S. Gullá for processing SPOT arrays and members of the Keating laboratory, especially T.-C. S. Chen, for thoughtful discussions. This study was funded by National Institutes of Health award GM084181 to A.E.K. Computational resources to support this work were provided by the National Science Foundation under Grant No. 0821391.
Abbreviations used
- BH3
Bcl-2 homology 3
- PSSM
position-specific scoring matrix
- PDB
Protein Data Bank
- AUC
area under the curve
- PE
R-phycoerythrin
Footnotes
Supplementary Data
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2012.05.022
References
- 1.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 2.Greenhough A, Wallam CA, Hicks DJ, Moorghen M, Williams AC, Paraskeva C. The proapoptotic BH3-only protein Bim is downregulated in a subset of colorectal cancers and is repressed by antiapoptotic COX-2/PGE(2) signalling in colorectal adenoma cells. Oncogene. 2010;29:3398–3410. doi: 10.1038/onc.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letai A, Sorcinelli MD, Beard C, Korsmeyer SJ. Antiapoptotic BCL-2 is required for maintenance of a model leukemia. Cancer Cell. 2004;6:241–249. doi: 10.1016/j.ccr.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of the BHRF1 protein from Epstein–Barr virus, a homolog of human Bcl-2. J. Mol. Biol. 2003;332:1123–1130. doi: 10.1016/j.jmb.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc. Natl Acad. Sci. USA. 2002;99:3428–3433. doi: 10.1073/pnas.062525799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat. Rev. Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 7.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin. Cancer Res. 2009;15:1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leber B, Geng F, Kale J, Andrews DW. Drugs targeting Bcl-2 family members as an emerging strategy in cancer. Expert Rev. Mol. Med. 2010;12:e28. doi: 10.1017/S1462399410001572. [DOI] [PubMed] [Google Scholar]
- 9.Esposti MD. Bcl-2 antagonists and cancer: from the clinic, back to the bench. Cell Death Dis. 2010;1:e37. doi: 10.1038/cddis.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrington EM, Vikstrom IB, Light A, Sutherland RM, Londrigan SL, Mason KD, et al. BH3 mimetics antagonizing restricted prosurvival Bcl-2 proteins represent another class of selective immune modulatory drugs. Proc. Natl Acad. Sci. USA. 2010;107:10967–10971. doi: 10.1073/pnas.1005256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldsmith KC, Liu X, Dam V, Morgan BT, Shabbout M, Cnaan A, et al. BH3 peptidomimetics potently activate apoptosis and demonstrate single agent efficacy in neuroblastoma. Oncogene. 2006;25:4525–4533. doi: 10.1038/sj.onc.1209489. [DOI] [PubMed] [Google Scholar]
- 12.High LM, Szymanska B, Wilczynska-Kalak U, Barber N, O'Brien R, Khaw SL, et al. The Bcl-2 homology domain 3 mimetic ABT-737 targets the apoptotic machinery in acute lymphoblastic leukemia resulting in synergistic in vitro and in vivo interactions with established drugs. Mol. Pharmacol. 2010;77:483–494. doi: 10.1124/mol.109.060780. [DOI] [PubMed] [Google Scholar]
- 13.Letai A. BH3 domains as BCL-2 inhibitors: prototype cancer therapeutics. Expert Opin. Biol. Ther. 2003;3:293–304. doi: 10.1517/14712598.3.2.293. [DOI] [PubMed] [Google Scholar]
- 14.Dutta S, Gulla S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J. Mol. Biol. 2010;398:747–762. doi: 10.1016/j.jmb.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boersma MD, Sadowsky JD, Tomita YA, Gellman SH. Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein–protein recognition: application to the Bim BH3 domain. Protein Sci. 2008;17:1232–1240. doi: 10.1110/ps.032896.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee EF, Fedorova A, Zobel K, Boyle MJ, Yang H, Perugini MA, et al. Novel Bcl-2 homology-3 domain-like sequences identified from screening randomized peptide libraries for inhibitors of the pro-survival Bcl-2 proteins. J. Biol. Chem. 2009;284:31315–31326. doi: 10.1074/jbc.M109.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee EF, Czabotar PE, Yang H, Sleebs BE, Lessene G, Colman PM, et al. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J. Biol. Chem. 2009;284:30508–30517. doi: 10.1074/jbc.M109.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day CL, Smits C, Fan FC, Lee EF, Fairlie WD, Hinds MG. Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J. Mol. Biol. 2008;380:958–971. doi: 10.1016/j.jmb.2008.05.071. [DOI] [PubMed] [Google Scholar]
- 19.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of Bcl-xL–Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 20.Petros AM, Nettesheim DG, Wang Y, Olejniczak ET, Meadows RP, Mack J, et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee EF, Czabotar PE, Smith BJ, Deshayes K, Zobel K, Colman PM, Fairlie WD. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 2007;14:1711–1713. doi: 10.1038/sj.cdd.4402178. [DOI] [PubMed] [Google Scholar]
- 22.Fire E, Gulla SV, Grant RA, Keating AE. Mcl-1–Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci. 2010;19:507–519. doi: 10.1002/pro.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 25.Denisov AY, Chen G, Sprules T, Moldoveanu T, Beauparlant P, Gehring K. Structural model of the BCL-w–BID peptide complex and its interactions with phospholipid micelles. Biochemistry. 2006;45:2250–2256. doi: 10.1021/bi052332s. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 27.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 29.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 31.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vo TT, Letai A. BH3-only proteins and their effects on cancer. Adv. Exp. Med. Biol. 2010;687:49–63. doi: 10.1007/978-1-4419-6706-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 35.Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol. Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 37.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boersma MD, Haase HS, Peterson-Kaufman KJ, Lee EF, Clarke OB, Colman PM, et al. Evaluation of diverse alpha/beta-backbone patterns for functional alpha-helix mimicry: analogues of the Bim BH3 domain. J. Am. Chem. Soc. 2012;134:315–323. doi: 10.1021/ja207148m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yecies D, Carlson NE, Deng J, Letai A. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115:3304–3313. doi: 10.1182/blood-2009-07-233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. J. Mol. Biol. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 41.Smith CA, Kortemme T. Structure-based prediction of the peptide sequence space recognized by natural and synthetic PDZ domains. J. Mol. Biol. 2010;402:460–474. doi: 10.1016/j.jmb.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 42.Hou T, Xu Z, Zhang W, McLaughlin WA, Case DA, Xu Y, Wang W. Characterization of domain-peptide interaction interface: a generic structure-based model to decipher the binding specificity of SH3 domains. Mol. Cell. Proteomics. 2009;8:639–649. doi: 10.1074/mcp.M800450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 44.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 45.Wohlgemuth S, Kiel C, Kramer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J. Mol. Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 46.Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, Serrano L. Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. J. Mol. Biol. 2005;348:759–775. doi: 10.1016/j.jmb.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 47.Beuming T, Farid R, Sherman W. High-energy water sites determine peptide binding affinity and specificity of PDZ domains. Protein Sci. 2009;18:1609–1619. doi: 10.1002/pro.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen JR, Chang BH, Allen JE, Stiffler MA, MacBeath G. Predicting PDZ domain–peptide interactions from primary sequences. Nat. Biotechnol. 2008;26:1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez IE, Beltrao P, Stricher F, Schymkowitz J, Ferkinghoff-Borg J, Rousseau F, Serrano L. Genome-wide prediction of SH2 domain targets using structural information and the FoldX algorithm. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000052. e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 52.Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Czabotar PE, Lee EF, Thompson GV, Wardak AZ, Fairlie WD, Colman PM. Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis. J. Biol. Chem. 2011;286:7123–7131. doi: 10.1074/jbc.M110.161281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herman MD, Nyman T, Welin M, Lehtio L, Flodin S, Tresaugues L, et al. Completing the family portrait of the anti-apoptotic Bcl-2 proteins: crystal structure of human Bfl-1 in complex with Bim. FEBS Lett. 2008;582:3590–3594. doi: 10.1016/j.febslet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 55.Ku B, Liang C, Jung JU, Oh BH. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21:627–641. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raveh B, London N, Zimmerman L, Schueler-Furman O. Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One. 2011;6:e18934. doi: 10.1371/journal.pone.0018934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.King CA, Bradley P. Structure-based prediction of protein–peptide specificity in Rosetta. Proteins. 2010;78:3437–3449. doi: 10.1002/prot.22851. [DOI] [PubMed] [Google Scholar]
- 58.Zhang C, Liu S, Zhou H, Zhou Y. An accurate, residue-level, pair potential of mean force for folding and binding based on the distance-scaled, ideal-gas reference state. Protein Sci. 2004;13:400–411. doi: 10.1110/ps.03348304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 60.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DC, et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl Acad. Sci. USA. 2007;104:6217–6222. doi: 10.1073/pnas.0701297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen TS. MIT Department of Biology Doctoral Dissertation. 2012. Design of Protein–Protein Interaction Specificity Using Computational Methods and Experimental Library Screening. [Google Scholar]
- 63.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 64.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 65.Guntas G, Purbeck C, Kuhlman B. Engineering a protein–protein interface using a computationally designed library. Proc. Natl Acad. Sci. USA. 2010;107:19296–19301. doi: 10.1073/pnas.1006528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allen BD, Nisthal A, Mayo SL. Experimental library screening demonstrates the successful application of computational protein design to large structural ensembles. Proc. Natl Acad. Sci. USA. 2010;107:19838–19843. doi: 10.1073/pnas.1012985107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treynor TP, Vizcarra CL, Nedelcu D, Mayo SL. Computationally designed libraries of fluorescent proteins evaluated by preservation and diversity of function. Proc. Natl Acad. Sci. USA. 2007;104:48–53. doi: 10.1073/pnas.0609647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hayes RJ, Bentzien J, Ary ML, Hwang MY, Jacinto JM, Vielmetter J, et al. Combining computational and experimental screening for rapid optimization of protein properties. Proc. Natl Acad. Sci. USA. 2002;99:15926–15931. doi: 10.1073/pnas.212627499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fazelinia H, Cirino PC, Maranas CD. Extending Iterative Protein Redesign and Optimization (IPRO) in protein library design for ligand specificity. Biophys. J. 2007;92:2120–2130. doi: 10.1529/biophysj.106.096016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu X, Apgar JR, Keating AE. Modeling backbone flexibility to achieve sequence diversity: the design of novel alpha-helical ligands for Bcl-xL. J. Mol. Biol. 2007;371:1099–1117. doi: 10.1016/j.jmb.2007.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu SY, Kaipia A, McGee E, Lomeli M, Hsueh AJ. Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc. Natl Acad. Sci. USA. 1997;94:12401–12406. doi: 10.1073/pnas.94.23.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 73.Wiedemann U, Boisguerin P, Leben R, Leitner D, Krause G, Moelling K, et al. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J. Mol. Biol. 2004;343:703–718. doi: 10.1016/j.jmb.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 74.Smits C, Czabotar PE, Hinds MG, Day CL. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure. 2008;16:818–829. doi: 10.1016/j.str.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 75.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eckert KA, Kunkel TA. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang G, Dunbrack RL., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 78.Efron B. Nonparametric estimates of standard error: the jackknife, the bootstrap and other methods. Biometrika. 1981;68:589–599. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.