Abstract
Biofilms are antibiotic-resistant, sessile bacterial communities that occupy most moist surfaces on Earth and cause chronic and medical device-associated infections. Despite their importance, basic information about biofilm dynamics in common ecological environments is lacking. Here, we demonstrate that flow through soil-like porous materials, industrial filters, and medical stents dramatically modifies the morphology of Pseudomonas aeruginosa biofilms to form 3D streamers, which, over time, bridge the spaces between obstacles and corners in nonuniform environments. We discovered that accumulation of surface-attached biofilm has little effect on flow through such environments, whereas biofilm streamers cause sudden and rapid clogging. We demonstrate that flow-induced shedding of extracellular matrix from surface-attached biofilms generates a sieve-like network that captures cells and other biomass, which add to the existing network, causing exponentially fast clogging independent of growth. These results suggest that biofilm streamers are ubiquitous in nature and strongly affect flow through porous materials in environmental, industrial, and medical systems.
Keywords: bioclogging, biofouling, porous media
In the laboratory, bacteria are usually grown as planktonic cells in shaken suspensions, which differs dramatically from the natural environments of most microbes. In their natural habitats, bacteria often live in biofilms (1–3), which are tightly packed, surface-associated assemblies of bacteria that are bound together by extracellular polymeric substances (4, 5). Although biofilms are desirable in waste-water treatment (6), biofilms primarily cause undesirable effects such as chronic infections or clogging of industrial flow systems (1–3). Cells in biofilms display many behavioral differences from planktonic cells, such as a 1,000-fold increase in tolerance to antibiotics (7, 8), an altered transcriptome (9–11), and spatially heterogeneous metabolic activity (12, 13). Some of these physiological peculiarities of biofilm-dwelling cells may be due to strong gradients of nutrients and metabolites, which also affect biofilm morphology and composition (14, 15). However, little is known about how physical aspects of the environment affect biofilm dynamics.
The opportunistic pathogen Pseudomonas aeruginosa has become a model organism for biofilm studies largely because it forms biofilms in diverse habitats, including soil, rivers, sewage, and medical devices in humans (1, 2, 16). Two features are common to all of these environments: First, the presence of rough surfaces, which at the microscopic level reduce to surfaces with many corners, and second, a pressure-driven flow. The standard assay for growing biofilms in the laboratory abstracts from these realistic environments by typically using a smooth surface as a substrate, and either no flow or a pump to force nutritious medium across the biofilm at a constant flow rate (17–19). These standard assays have enabled the identification of several genes involved in biofilm development (10, 19–24), yet it is unclear to what extent these results are relevant in natural habitats, as the standard assays neglect the different surface chemistries, interactions with other species, and physical constraints of natural environments.
To investigate biofilm morphologies under more realistic physical conditions, we developed a microfluidic system that combines two shared features of P. aeruginosa habitats, i.e., a sequence of corners (25) and a flow driven by a constant pressure. We discovered that in this system biofilm streamers cause rapid clogging transitions, and we used a combination of experiments and theory to explain the timescales of the clogging dynamics. We further show that biofilm formation under the physical constraints of our model system does not require all of the genes that have been identified as essential in standard biofilm assays. Finally, we demonstrate that biofilm streamers are ubiquitous in soil-like porous materials, feed spacer meshes of water filters, and medical stents.
Results and Discussion
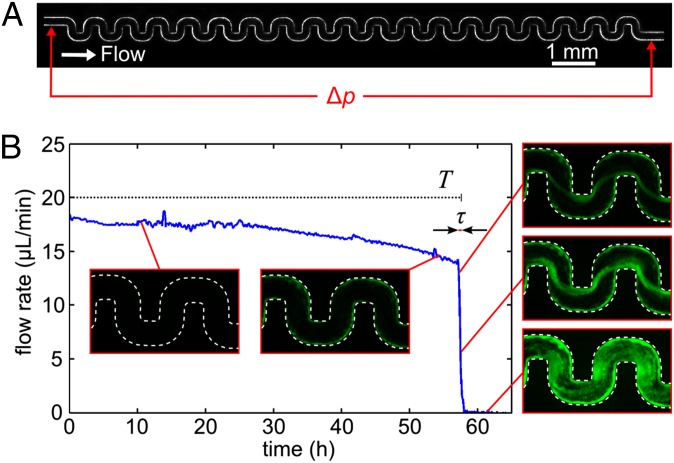
Using our model microfluidic flow system (Fig. 1A, Fig. S1), we discovered that biofilm growth on the walls of the chamber, which has been the focus of much previous work (26, 27), only modestly affects the flow rate through this channel over a period of T ≈ 50 h. By contrast, biofilm streamers that initiate on corners (25, 28, 29) rapidly expand and cause a catastrophic disruption of the flow on timescales as short as τ ≈ 30 min (Fig. 1B) in our model channels, which are 200 μm wide and 90 μm high. A streamer causes a dramatic decrease in flow rate, even in a 3D environment where the flow can pass above and below the streamer, because it consists of immotile biomass, suspended in the center of the channel where the flow speed would be highest in the absence of a streamer. A model calculation (SI Text and Fig. S2) confirms that for flow driven by a constant pressure, a biofilm growing on the walls of the channel has a significantly weaker effect on flow than the same volume of biofilm positioned in the center of the channel. However, such arguments cannot explain why the time until clogging T is long, whereas the duration of the clogging transition τ is short.
Fig. 1.
Biofilm streamers cause rapid and sudden clogging. (A) A constant pressure difference Δp drives a suspension of P. aeruginosa cells through the model microfluidic channel, which is 200 μm wide and 90 μm high. (B) Measurement of flow rate versus time. The flow rate through this channel only changes slowly during biofilm buildup on the walls of the channel for the time period T. Channel walls are indicated by dashed white lines, and cells constitutively express gfp. Biofilm streamers expand rapidly and cause clogging over a short time τ.
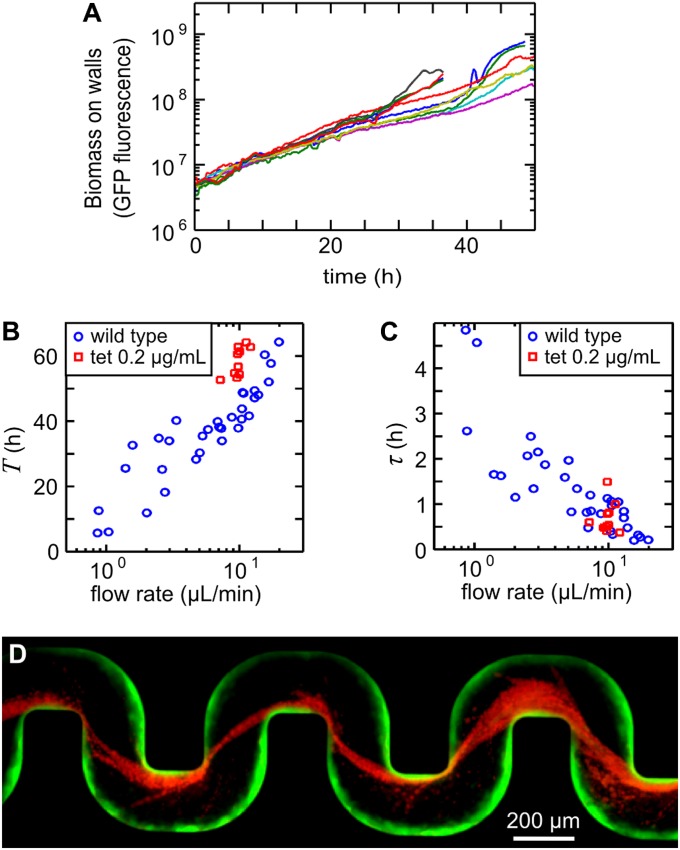
The exponential accumulation of cells on the walls of the channels (Fig. 2A) indicates that the accumulation process is dominated by growth (doubling time 6.5 ± 1 h) rather than attachment of cells that are flowing by, because attachment would result in a subexponential accumulation rate. For example, a constant attachment probability for each cell per unit time would only yield a linear accumulation of wall-attached biomass with time, to first approximation. However, cells must be able to attach to the walls as the channel is initially seeded with sterile medium. We observed that flow shears off streamers from the biofilm positioned at the corners (Fig. S3). These streamers initially consist primarily of extracellular polymeric substances (EPS; refs. 4, 5) and over time, these filaments bridge the distances between corners and capture cells flowing by. Flow therefore affects the biofilm structure not just by providing nutrients (14, 15) but also by actively shaping the biofilm (30, 29, 25).
Fig. 2.
Cell growth sets T, while τ is due to a transport process. (A) Semilogarithmic plot of the accumulation of cells on the walls, measured via GFP fluorescence. Different colors represent data from n = 10 independent experiments. (B) T depends on flow rate, and can be prolonged by slowing growth with a low concentration of the growth-inhibitor tetracycline (tet). (C) Tetracycline has no effect on τ. (D) For the first 43 h, cells expressing gfp are flowed through the channel at a rate 18.1 ± 0.05 μL/min. Subsequently, the in-flowing culture is exchanged to contain only cells producing the red fluorescent protein mCherry. Biofilm streamers are exclusively composed of red cells, whereas very few red cells attach to the resident green biofilm on the wall, indicating that streamers consist of cells that were transported to the eventual clogging site by flow (Movie S1).
Because the wall-attached biofilm is a necessary precondition for streamer formation, slowing growth should delay clogging. Indeed, we found that T is prolonged by the addition of low levels of tetracycline, a bacteriostatic compound (Fig. 2B; see Fig. S4 for the effect on growth rate), which indicates that T is determined by cell growth. As τ is independent of growth (Fig. 2C), some other mechanism must be responsible for the clogging duration. We wondered whether advective transport of cells to the clogging site could be responsible for τ. To test this idea, we loaded the apparatus with cells expressing gfp for the first 43 h, a time that is significantly before the clogging transition is expected. At this time, we exchanged the in-flowing culture to one that exclusively contains cells that express mCherry rather than gfp, but are otherwise isogenic. We discovered (Fig. 2D, Movie S1) that streamers contain only mCherry expressing cells, while only very few mCherry expressing cells attached to the resident (green) biofilm on the walls of the channel. The rapid clogging transition is therefore due to cells that are transported to the clog-forming streamers.
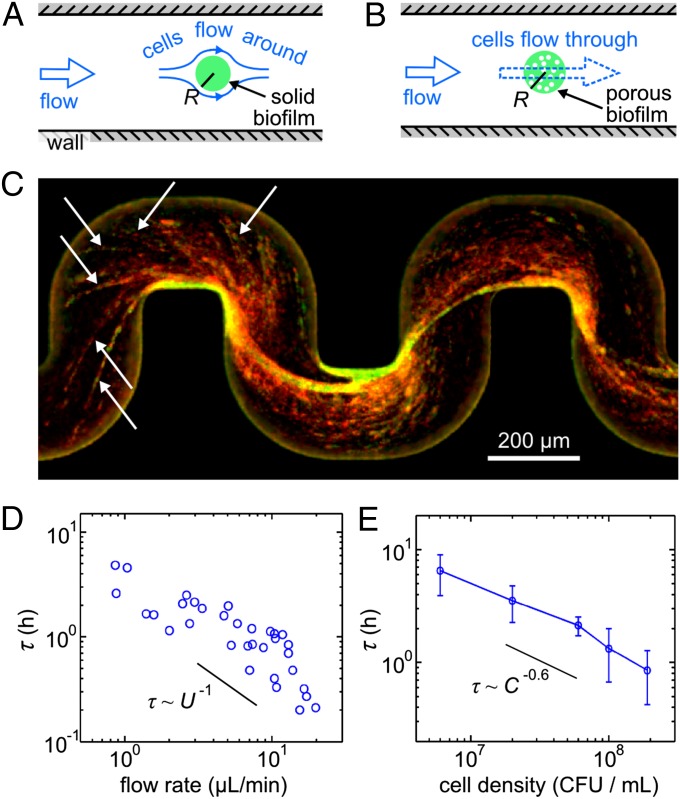
To determine how cells that are transported by flow can cause rapid clogging, we developed quantitative models of streamer growth. Although many theories of biofilm buildup in porous materials have been proposed previously (26, 27, 31–33), they do not apply to our case, as biofilm streamers were not included in these theories, and we have now demonstrated that they are of crucial importance in nonuniform flow systems. Consider first the case of a solid streamer (Fig. 3A). Cells constantly flow past this streamer, and some of them migrate across streamlines and come in contact with the streamer, in which case we assume there is a probability α that the cells get absorbed. For the parameters of our experiments, this advection-diffusion process predicts (SI Text) that the radius R of such a streamer would grow approximately as R ∼ 11 μm (αt)3/4, as a function of time t in hours. Such streamer growth dynamics and the resulting flow rate decrease are slow (Fig. S5) and are therefore unlikely to be the dominant contribution to the experimentally observed rapid clogging. However, if we assume that the biofilm streamer behaves like a permeable, porous material (27, 31), with cells flowing through it (Fig. 3B), the equations for the streamer growth predict (SI Text) that the streamer grows exponentially fast, R ∝ exp(t/τtheory), with a growth timescale τtheory that is of a similar magnitude to the experimentally observed clogging time scales. Fig. S5 shows that the dynamics predicted by the model based on a porous streamer are qualitatively more consistent with the experiments than the results from an advection–diffusion-based model of a solid streamer. High-resolution confocal images of the biofilm structure during streamer growth reveal (Fig. 3C) that the assumption of a porous streamer is indeed justified: the main streamer is a network of smaller biofilm filaments with numerous gaps that create a sieve-like mesh that catches cells, and possibly EPS, flowing through it (Fig. S3 and Movie S2).
Fig. 3.
Biofilm filaments form a sieve-like network that captures cells flowing through. (A) A model based on a nonporous biofilm streamer oriented transverse to the flow direction predicts slow growth rates for the streamer radius R. (B) A model based on a porous streamer, which grows by capturing cells that flow through it, predicts exponential growth for R (SI Text). (C) Image of the biofilm during the clogging transition for an initial flow rate 1.5 ± 0.05 μL/min. P. aeruginosa cells are shown in red, EPS is visualized with green fluorescent dyes conjugated to polysaccharide-binding lectins and a green fluorescent DNA stain. Yellow regions result from the superposition of green and red channels. White arrows point to smaller biofilm streamers that form a network. The thick streamer structures are interspersed with dark regions, indicating that these structures are porous. The porosity is further illustrated by Fig. S3 and Movie S2. (D) Clogging duration τ for different flow rates (which are proportional to U, the average flow speed before streamers emerge) at a fixed concentration of ≈ 2 × 108 cfu/mL, corresponding to midlogarithmic-phase growth. (E) τ for different cell concentrations at a fixed flow rate of 4.8 ± 0.8 μL/min. Error bars: SD of n = 8 independent measurements.
Our model of streamer growth, based on a permeable streamer, can be further tested by noting that this model predicts a functional dependence of the clogging time scale τtheory ∝ U−1C−1, where U is the average flow speed prior to the emergence of streamers, and C is the density of cells in the medium that flows past the streamer (the full expression for τtheory is given in SI Text). Fig. 3D shows that τ ∝ U−0.98, consistent with the prediction for a porous streamer. In addition, τ ∝ C−0.6 (Fig. 3E) at a fixed flow rate of 4.8 ± 0.8 μL/min, which is a weaker functional dependence of τ on C than the model predicts. This discrepancy likely arises because τtheory ∝ C−1 results from the assumption that there is a fixed probability that a cell that flows through the streamer is caught by the streamer, independent of C. This basic physical model does not include biological effects on streamer formation, such as the contributions of particular genes, and their cell-density–dependent regulation (34, 35).
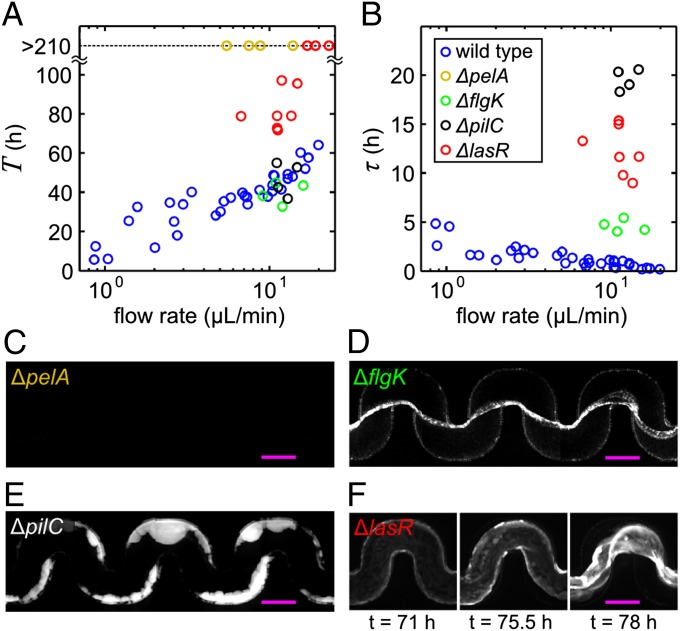
A general question applicable to all microbiological studies of gene regulation is whether the genes that are crucial for a particular phenotype under laboratory conditions are also crucial in natural environments, where organisms often encounter different nutrient sources, surface chemistries, and physical environments. Our system, which is designed to mimic physical aspects of realistic habitats, allows us to compare the biofilm morphology (Fig. 4 C–F) and clogging dynamics (Fig. 4 A and B) of wild type and mutant strains harboring defects in genes required for biofilm formation on smooth surfaces. In standard assays with glucose-based medium, flagellar-mediated swimming motility leads to increased residence times of cells near surfaces (36), and flagella also enhance attachment to surfaces (21, 22, 37). However, we find that a nonmotile flagellar mutant (ΔflgK) forms streamers with a similar T to wild type, even though it has a significantly larger τ (Fig. 4) and it displays a delay in biomass accumulation (Fig. S6). This result suggests that swimming is not required for transport under our experimental conditions because flow provides the necessary transport of cells to the clog-forming biofilm. The delay in biomass buildup on the walls of the channel and the prolonged τ of the ΔflgK strain relative to the wild type support the hypothesis that flagella are important, but not essential, for attaching to surfaces (21, 22, 38), including biofilm streamers. Whether the longer τ is due to a lack of motility, the flagellum, or the decreased effective size of a ΔflgK cell remains unknown.
Fig. 4.
Mutants that affect biofilm formation in nonuniform environments. (A, B) Comparison of the time until clogging T and the duration of the clogging transition τ for different mutants: ΔpelA lacks the major component of the EPS, ΔflgK is nonmotile due to an incomplete flagellum (21), ΔpilC has no type IV pili, and ΔlasR lacks the quorum sensing master regulator. (C) ΔpelA produces no significant biofilm during 210 h of observation. (D) ΔflgK produces biofilm streamers similar to the wild type. (E) ΔpilC forms no streamers, but does form thick biofilms on the walls of the channel. (F) ΔlasR forms biofilms on the walls of the channel, which detach, slowly deform, and reattach to clog the channel. The image lookup table is the same for C–F. Scale bars: 200 μm.
Other previously identified genes that are important for biofilm formation (4, 20–24) do have large effects in our model system. EPS production is required, as a mutant that is deficient in EPS production (ΔpelA) is unable to initiate biofilms and clog the channel (Fig. 4 A and C). Cells lacking type IV pili (ΔpilC) can form biofilms in the presence of flow (38, 39) but do not form streamers, and instead clog channels by forming thick biofilms on the walls (Fig. 4E). If, however, EPS or type IV pili mutants are flowed through a channel that contains a biofilm of wild-type cells on the walls, these mutants can participate in streamer formation (Fig. S7). This observation further supports the idea that the resident biofilm on the walls is required to generate the sieve of EPS that catches cells flowing by. Previous work showing that type IV pili are required for dynamic rearrangement of cells in biofilms (38, 39) may explain why wall-attached biofilms of the ΔpilC mutant cannot form streamers. The lasI–lasR quorum-sensing system activates the pelA gene encoding an enzyme required to make the EPS matrix (40). Thus, mutants lacking lasR are strongly impaired in biofilm development, and they produce frail biofilms consisting of cells that are less tightly bound to one another than wild-type cells (23, 40). In our system, mutants defective for quorum sensing (ΔlasR) display severely delayed clogging or no clogging, and have a dramatically different biofilm morphology than the wild type. Biofilms of the ΔlasR mutant grow on the walls of the channel, and undergo several cycles of wall detachment and deformation before complete clogging. Over time, the quorum-sensing mutant can clog the channel; however, this occurs without the formation of streamers (Fig. 4 A, B, and F), perhaps due to the reduced cell–cell cohesiveness (23, 40).
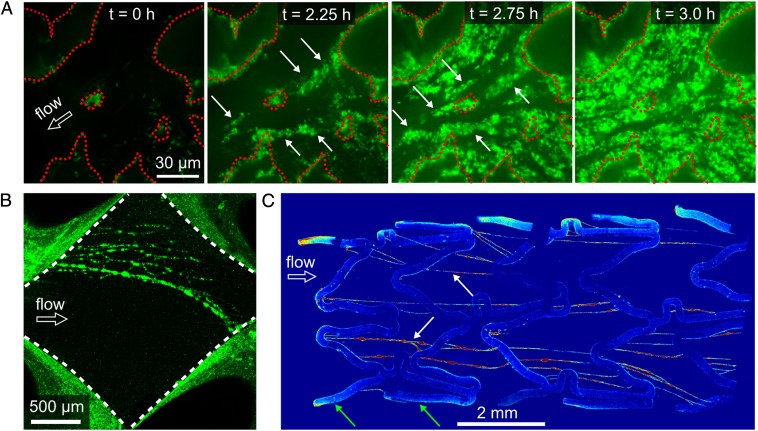
Our results suggest that biofilm streamer formation could be critical in natural settings as streamers can cause rapid and sudden clogging. To examine whether these processes are general phenomena that are independent of our model system, we investigated biofilm morphologies in flow systems in which biofilm-induced clogging is well-documented but a mechanistic understanding is lacking. Soil is a major habitat for bacteria, including P. aeruginosa (16, 41). Clogging of soil-like porous materials is a primary concern in waste-water treatment reactors (6) and has been studied extensively (26, 27, 31, 32, 42–46). Direct observations of biofilm dynamics inside soil have not been possible because these environments are generally opaque beyond the first layer of granules. To overcome this limitation, we used a recently developed method to generate a transparent porous material with geometric features similar to multiple layers of fine sand (47), and observed that biofilm streamers are ubiquitous (Fig. 5A), consistent with observations in the first layer of granules in packed-bead experiments (43). We further found that in soil-like environments, biofilm streamers precede rapid clogging events of the 3D pores (Fig. 5A). Whether streamers are involved in the clogging of smaller pores in this artificial soil could not be resolved.
Fig. 5.
Biofilm streamers form in diverse environments. (A) Time series of biofilm buildup in a 3D soil-like porous material made from transparent Nafion granules (outlined by red dashed lines). Green indicates P. aeruginosa cells constitutively expressing gfp. Arrows point toward streamers, which are heterogeneous in thickness at this magnification. (B) Networks of biofilm streamers form in a feed spacer mesh, which is a component of spiral-wound reverse osmosis water filters. The image is a maximum-intensity projection of a confocal z-stack, which visualizes biofilms on the surface of the mesh, located outside the white dashed lines. (C) Biofilm streamers form in bare-metal stents. White arrows point to streamers; green arrows point to wire mesh of the stent. The image is stitched together from the maximum intensity projections of 83 z-stacks. A false-color scheme is used to illustrate that different color intensity scales were used for visualizing the stent surface and the streamers because the fluorescence from the stent surface was significantly brighter due to the large amount of biomass on the surface. The resulting two images of the stent surface and the streamers were overlaid, giving the displayed image. Green indicates P. aeruginosa cells constitutively expressing gfp. Arrows point toward streamers, which are heterogeneous in thickness and biomass at this magnification.
Other systems that suffer from biofilm-induced clogging are spiral-wound reverse osmosis filters (48, 49), which are used, e.g., to purify drinking water. Feed spacer mesh, the element within these filtration devices that is prone to biofilm formation, also displays biofilm streamers that form a network (Fig. 5B). Finally, prosthetic devices can host biofilms that cause chronic infections and other symptoms (1, 2). In particular, biliary stents regularly clog due to slime made up of cholesterol and multispecies biofilms that include P. aeruginosa (50). We found that biofilm streamers develop on bare-metal stents within 12 h after inoculation, and span the gaps in the wire mesh (Fig. 5C).
Conclusions
These examples illustrate that biofilm streamers are likely a major feature of biofilms in natural, industrial, and medical environments, and we demonstrated that they can cause rapid clogging without warning (Fig. 1B). We also showed that streamers cause a much stronger disruption of flow than wall-attached biofilms. Using a microfluidic model system, we demonstrated that the timing when clogging occurs depends on growth, while the duration of clogging is determined by transport and trapping of cells in the streamer network positioned at the clog site. Only a subset of the genes required for biofilm formation in standard laboratory assays is essential in our model system, which is designed to mimic physical constraints of natural habitats. For example, flagellar motility, a prerequisite to biofilm formation in the standard biofilm assays, is dispensable in our system. These findings underscore the need to investigate bacterial behavior in realistic environments as biofilm morphology and regulation may change in industrial and clinical settings, where biofilm prevention is critical.
Materials and Methods
A detailed description of the mathematical models is provided in SI Text. The experiments are summarized below.
Bacterial Strains and Culture Conditions.
All strains are derivatives of Pseudomonas aeruginosa PA14. Overnight cultures were grown in tryptone broth (1% tryptone in H2O, refs. 25, 28) at 37 °C with shaking. The mutant strains ΔflgK (sad-36, ref. 21), ΔpilC (sad-29, ref. 21), and ΔpelA (24) were characterized previously. A ΔlasR::aacC1 mutant (AFS20) was a gift from A. Siryaporn (Princeton University). The PA14 strain harboring PA1/04/03::gfp inserted in the intergenic region between the glmS and PA14_73160 genes is designated PA14-gfp (25) and was a gift from the Kolter Laboratory, Harvard University, Cambridge, MA. The lac-derived promoter PA1/04/03 results in constitutive expression of gfp (51, 52). Strains carrying the PA1/04/03::mCherry construct at the identical chromosomal site were engineered by mating a pUC-mini-Tn7 (53) derived plasmid, pAS08.2E (gift from A. Siryaporn), into the chromosome. This method resulted in strains PA14-mCherry (AFS27E, gift from A. Siryaporn), ΔflgK mCherry, ΔpilC mCherry, ΔpelA mCherry, and ΔlasR mCherry.
Microfluidic Pressure-Driven Flow Assay and Microscopy.
A schematic diagram of the apparatus used to investigate biofilm dynamics in a microfluidic model system is shown in Fig. S1. Overnight P. aeruginosa cultures were back-diluted 1:100 in tryptone broth and grown to midlogarithmic phase (OD600 = 0.5). This culture (100 mL) was used to fill a reservoir connected via Tygon tubing (inner diameter 2.4 mm) to a microfluidic channel made from polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning). The channels (Fig. 1A) are 90 μm high, 200 μm wide, and constitute the narrowest part of the experimental flow system. Analogous to the inlet, the outlet of the microfluidic channel is also connected to Tygon tubing, and the effluent culture is collected in a dish on an analytical balance. By changing the elevation of the culture reservoir above the effluent collection dish, the applied pressure difference Δp is altered, which alters the flow rate through the channel. Operating this system for 24 h with H2O at a flow rate of 15.7 ± 0.05 μL/min reduced the liquid level in the reservoir to decrease the pressure drop across the channel by 2.1%. For experiments in which the cell concentration that is flowed through the channel is varied (Fig. 3E), we maintained the flow rate approximately constant (4.8 ± 0.8 μL/min; ± SD between independent channels), and changed the cell concentration in the reservoir by diluting or concentrating (via centrifugation) midlogarithmic phase cells. Images of the biofilms were acquired using epifluorescence microscopy on a Nikon Ti-Eclipse microscope and a CCD camera (iXon, Andor), or a confocal laser scanning microscope (TCS SP5, Leica). Although the cfu and OD600 in the reservoir are approximately constant for more than 72 h at 22 °C (Fig. S8A), the state of the inflowing culture may change over time. Exchanging the culture in the reservoir every 24 h does not significantly change T or τ, compared with experiments in which the culture was not exchanged (Fig. S8 B and C). We thus exchanged the reservoir culture approximately every 24 h in long experiments, but used all data points shown in Fig. S8 B and C. In some experiments, the in-flowing culture was exchanged to contain a different strain (Fig. 2D and Fig. S7). These experiments were performed at a flow rate of ≈18 μL/min. When the in-flowing strain was exchanged, the entire reservoir container was exchanged.
Analysis of Flow-Rate Time Series.
The weight of the effluent culture was measured every 4 s on an analytical balance (GD503, Sartorius), controlled with LabVIEW. To obtain the flow rate time series Q(t) from the effluent weight time series w(t), we computed
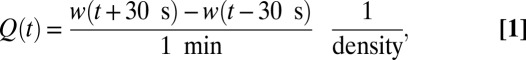 |
where the density was assumed to be the density of water, 1 kg/L. T and τ were calculated by fitting the function 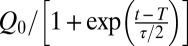 to the measured flow rate Q(t), where Q0 is the flow rate before the clogging transition. The time τ is therefore defined as the time period in which the fitted flow rate decreases from 76% to 27% of Q0, and T is the time at which the fitted flow rate is Q0/2.
to the measured flow rate Q(t), where Q0 is the flow rate before the clogging transition. The time τ is therefore defined as the time period in which the fitted flow rate decreases from 76% to 27% of Q0, and T is the time at which the fitted flow rate is Q0/2.
Measuring the Accumulation of Wall-Attached Biofilm.
Using a flow rate of 18 ± 1 μL/min (± SD of n = 10 independent experiments) and the PA14-gfp strain, we acquired GFP fluorescence images of the complete channel until streamers emerged. The pixel intensities at the walls were summed up to measure the wall-attached biomass.
Staining of EPS In Situ.
To visualize the different components of a streamer while it expands, we fluorescently stained the EPS produced by P. aeruginosa PA14-mCherry. For this assay, we used a microfluidic channel containing only 5 bends instead of the 36 bends that make up the standard channel. In addition, this channel had three inlet ports, which could be opened and closed using multilayer microfluidic gates (54). One of these inlet ports was connected to the P. aeruginosa PA14-mCherry culture, as in the previous assays. The second port was connected to a PBS solution, and the third port was connected to a mixture of several EPS stains dissolved in PBS. These EPS stains included 5 μM of the green fluorescent nucleic acid stain SYTO 9 (Molecular Probes) (55, 56), 20 μg/mL fluorescein isothiocyanate (FITC)-conjugated Concanavalin A (ConA), and 20 μg/mL FITC-conjugated wheat germ agglutinin (WGA). The lectin ConA (Sigma) binds to α-D-mannose, α-D-glucose, and likely alginate (56), whereas the lectin WGA (Sigma) binds to N-acetyl-D-glucosamine and N-acetylneuraminic acid (56). The nucleic acid stain was used to visualize extracellular DNA (57). High-magnification (100×) images of P. aeruginosa biofilms stained with SYTO 9 showed that the stain predominantly binds to extracellular, rather than intracellular, nucleic acids. To stain the EPS of the streamer before complete clogging occurred, we flowed P. aeruginosa PA14-mCherry cells through the channel until the streamer was rapidly expanding. Supply of the cell culture was then terminated by closing the relevant gate, and PBS was flowed through the channel to clear unattached cells. Subsequently, the PBS supply was stopped by closing the appropriate gate and the staining solution was flowed through the channel for approximately 5 min, after which the unbound stain was washed out by once again flushing the channel with PBS.
Artificial Soil, Feed Spacer Mesh, and Stents.
To manufacture a 3D porous material that is geometrically similar to soil, we used granules made from the fluoropolymer Nafion, which is transparent in water and thus allows microscopic imaging deep inside the porous material (47). Nafion granules were generated from dissolved Nafion (Ion Power). The solvent was evaporated on glass slides at room temperature and the dried Nafion was cut repeatedly to generate heterogeneous granules. Before packing the Nafion granules into a straight microfluidic channel (300 μm high, 1 mm wide), the channel was air-plasma treated. The PDMS channel was subsequently bonded to glass by a heat treatment at 95 °C for 1 min. Welded polypropylene feed spacer mesh (gap size 1.8 × 1.8 mm; Industrial Netting), similar to that used in industrial reverse osmosis filters (48), was placed in a rectangular PDMS channel (900 μm high, 10 mm wide), which was subsequently bonded to a glass microscope slide. A bare-metal stent (diameter 2.5 mm, length 28 mm) was placed into a nearly circular PDMS channel, which was molded onto tubing that was later withdrawn. The porous material, feed spacer, and stent were all exposed to P. aeruginosa PA14-gfp culture, which was flowed through the channels for approximately 12 h.
Supplementary Material
Acknowledgments
We are grateful to A. Siryaporn for providing P. aeruginosa strains and advice and C. Nadell, C. O’Loughlin, Y. Shao, N.S. Wingreen, and members of the B.L.B. and H.A.S. laboratories for discussions and advice. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health Grant 5R01GM065859, National Science Foundation (NSF) Grant MCB-0343821 (to B.L.B.), NSF Grant MCB-1119232 (to H.A.S. and B.L.B.), and the Human Frontier Science Program (K.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300321110/-/DCSupplemental.
References
- 1.Donlan RM, Costerton JW. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, et al. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol Med Microbiol. 2012;65(2):127–145. doi: 10.1111/j.1574-695X.2012.00968.x. [DOI] [PubMed] [Google Scholar]
- 3.Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441(7091):300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 4.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 6.Escudié R, Cresson R, Delgenès JP, Bernet N. Control of start-up and operation of anaerobic biofilm reactors:A overview of 15 years of research. Water Res. 2011;45(1):1–10. doi: 10.1016/j.watres.2010.07.081. [DOI] [PubMed] [Google Scholar]
- 7.Mah TF, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426(6964):306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334(6058):982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteley M, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413(6858):860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 10.Folsom JP, et al. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 2010;10:294. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dötsch A, et al. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS ONE. 2012;7(2):e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teal TK, Lies DP, Wold BJ, Newman DK. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl Environ Microbiol. 2006;72(11):7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rani SA, et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189(11):4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLOS Comput Biol. 2010;6(3):e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picioreanu C, van Loosdrecht MCM, Heijnen JJ. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol Bioeng. 1998;58(1):101–116. doi: 10.1002/(sici)1097-0290(19980405)58:1<101::aid-bit11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 16.Botzenhardt K, Doring G. Ecology and epidemiology of Pseudomonas aeruginosa. In: Campa M, Bendinelli M, Friedman H, editors. Pseudomonas Aeruginosa as an Opportunistic Pathogen. New York: Springer; 1993. pp. 1–7. [Google Scholar]
- 17.Sternberg C, Tolker-Nielsen T. Growing and analyzing biofilms in flow cells. Curr Protocols Molec Biol. 2006 doi: 10.1002/9780471729259.mc01b02s00. 10.1002/9780471729259. [DOI] [PubMed] [Google Scholar]
- 18.Busscher HJ, van der Mei HC. Microbial adhesion in flow displacement systems. Clin Microbiol Rev. 2006;19(1):127–141. doi: 10.1128/CMR.19.1.127-141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsek MR, Tolker-Nielsen T. Pattern formation in Pseudomonas aeruginosa biofilms. Curr Opin Microbiol. 2008;11(6):560–566. doi: 10.1016/j.mib.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Harmsen M, Yang LA, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol. 2010;59(3):253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 21.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 22.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184(4):1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies DG, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 24.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51(3):675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 25.Rusconi R, Lecuyer S, Guglielmini L, Stone HA. Laminar flow around corners triggers the formation of biofilm streamers. J R Soc Interface. 2010;7(50):1293–1299. doi: 10.1098/rsif.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thullner M. Comparison of bioclogging effects in saturated porous media within one- and two-dimensional flow systems. Ecol Eng. 2010;36(2):176–196. [Google Scholar]
- 27.Pintelon TRR, Picioreanu C, Loosdrecht MC, Johns ML. The effect of biofilm permeability on bio-clogging of porous media. Biotechnol Bioeng. 2012;109(4):1031–1042. doi: 10.1002/bit.24381. [DOI] [PubMed] [Google Scholar]
- 28.Rusconi R, Lecuyer S, Autrusson N, Guglielmini L, Stone HA. Secondary flow as a mechanism for the formation of biofilm streamers. Biophys J. 2011;100(6):1392–1399. doi: 10.1016/j.bpj.2011.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoodley P, Lewandowski Z, Boyle JD, Lappin-Scott HM. Oscillation characteristics of biofilm streamers in turbulent flowing water as related to drag and pressure drop. Biotechnol Bioeng. 1998;57(5):536–544. doi: 10.1002/(sici)1097-0290(19980305)57:5<536::aid-bit5>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 30.Stewart PS. Mini-review: Convection around biofilms. Biofouling. 2012;28(2):187–198. doi: 10.1080/08927014.2012.662641. [DOI] [PubMed] [Google Scholar]
- 31.Thullner M, Baveye P. Computational pore network modeling of the influence of biofilm permeability on bioclogging in porous media. Biotechnol Bioeng. 2008;99(6):1337–1351. doi: 10.1002/bit.21708. [DOI] [PubMed] [Google Scholar]
- 32.Dupin HJ, McCarty PL. Pore-scale modeling of biological clogging due to aggregate expansion: A material mechanics approach. Water Resour Res. 2001;37(12):2965–2979. [Google Scholar]
- 33.Stewart TL, Kim DS. Modeling of biomass-plug development and propagation in porous media. Biochem Eng J. 2004;17(2):107–119. [Google Scholar]
- 34.Williams P, Cámara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12(2):182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drescher K, Dunkel J, Cisneros LH, Ganguly S, Goldstein RE. Fluid dynamics and noise in bacterial cell-cell and cell-surface scattering. Proc Natl Acad Sci USA. 2011;108(27):10940–10945. doi: 10.1073/pnas.1019079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toutain CM, Caizza NC, Zegans ME, O’Toole GA. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res Microbiol. 2007;158(5):471–477. doi: 10.1016/j.resmic.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Klausen M, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003;50(1):61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 39.Barken KB, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10(9):2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 40.Sakuragi Y, Kolter R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol. 2007;189(14):5383–5386. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88(6):1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 42.Brown DG, Stencel JR, Jaffé PR. Effects of porous media preparation on bacteria transport through laboratory columns. Water Res. 2002;36(1):105–114. doi: 10.1016/s0043-1354(01)00208-1. [DOI] [PubMed] [Google Scholar]
- 43.Stoodley P, Dodds I, De Beer D, Scott HL, Boyle JD. Flowing biofilms as a transport mechanism for biomass through porous media under laminar and turbulent conditions in a laboratory reactor system. Biofouling. 2005;21(3-4):161–168. doi: 10.1080/08927010500375524. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C, et al. Effects of pore-scale heterogeneity and transverse mixing on bacterial growth in porous media. Environ Sci Technol. 2010;44(8):3085–3092. doi: 10.1021/es903396h. [DOI] [PubMed] [Google Scholar]
- 45.Kim JW, Choi H, Pachepsky YA. Biofilm morphology as related to the porous media clogging. Water Res. 2010;44(4):1193–1201. doi: 10.1016/j.watres.2009.05.049. [DOI] [PubMed] [Google Scholar]
- 46.Kapellos GE, Alexiou TS, Payatakes AC. Hierarchical simulator of biofilm growth and dynamics in granular porous materials. Adv Water Resour. 2007;30(6–7):1648–1667. [Google Scholar]
- 47.Leis AP, Schlicher S, Franke H, Strathmann M. Optically transparent porous medium for nondestructive studies of microbial biofilm architecture and transport dynamics. Appl Environ Microbiol. 2005;71(8):4801–4808. doi: 10.1128/AEM.71.8.4801-4808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vrouwenvelder JS, Graf von der Schulenburg DA, Kruithof JC, Johns ML, van Loosdrecht MCM. Biofouling of spiral-wound nanofiltration and reverse osmosis membranes: A feed spacer problem. Water Res. 2009;43(3):583–594. doi: 10.1016/j.watres.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Marty A, Roques C, Causserand C, Bacchin P. Formation of bacterial streamers during filtration in microfluidic systems. Biofouling. 2012;28(6):551–562. doi: 10.1080/08927014.2012.695351. [DOI] [PubMed] [Google Scholar]
- 50.Guaglianone E, et al. Microbial biofilms associated with biliary stent clogging. FEMS Immunol Med Microbiol. 2010;59(3):410–420. doi: 10.1111/j.1574-695X.2010.00686.x. [DOI] [PubMed] [Google Scholar]
- 51.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6(7):726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 53.Choi KH, et al. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2(6):443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 54.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288(5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 55.Chen MY, Lee DJ, Tay JH. Distribution of extracellular polymeric substances in aerobic granules. Appl Microbiol Biotechnol. 2007;73(6):1463–1469. doi: 10.1007/s00253-006-0617-x. [DOI] [PubMed] [Google Scholar]
- 56.Strathmann M, Wingender J, Flemming HC. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J Microbiol Methods. 2002;50(3):237–248. doi: 10.1016/s0167-7012(02)00032-5. [DOI] [PubMed] [Google Scholar]
- 57.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487–1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.