Abstract
Set1 is a conserved histone H3 lysine 4 (H3K4) methyltransferase that exists as a multisubunit complex. Although H3K4 methylation is located on many actively transcribed genes, few studies have established a direct connection showing that loss of Set1 and H3K4 methylation results in a phenotype caused by disruption of gene expression. In this study, we determined that cells lacking Set1 or Set1 complex members that disrupt H3K4 methylation have a growth defect when grown in the presence of the antifungal drug Brefeldin A (BFA), indicating that H3K4 methylation is needed for BFA resistance. To determine the role of Set1 in BFA resistance, we discovered that Set1 is important for the expression of genes in the ergosterol biosynthetic pathway, including the rate-limiting enzyme HMG-CoA reductase. Consequently, deletion of SET1 leads to a reduction in HMG-CoA reductase protein and total cellular ergosterol. In addition, the lack of Set1 results in an increase in the expression of DAN1 and PDR11, two genes involved in ergosterol uptake. The increase in expression of uptake genes in set1Δ cells allows sterols such as cholesterol and ergosterol to be actively taken up under aerobic conditions. Interestingly, when grown in the presence of ergosterol set1Δ cells become resistant to BFA, indicating that proper ergosterol levels are needed for antifungal drug resistance. These data show that H3K4 methylation impacts gene expression and output of a biologically and medically relevant pathway and determines why cells lacking H3K4 methylation have antifungal drug sensitivity.
Keywords: chromatin, gene regulation, histone methylation, epigenetics
Modifications on histones, such as phosphorylation, acetylation, methylation, and ubiquitination, have been implicated in gene expression and silencing (1, 2). One modification on histone H3 that is associated with active gene expression is the methylation of lysine 4 (H3K4) (3, 4). H3K4 methylation is present concurrently in mono-, di-, and trimethyl forms in the cell (1, 2). Primarily, H3K4 methylation is maintained in a pattern that has trimethylation enriched at the 5′ end of ORFs, dimethylation throughout the gene, and monomethylation enriched near the 3′ end (3, 4). In humans, H3K4 methylation is mediated by methyltransferases MLL1–4 and Set1A and -B complexes, which are homologous to the yeast Set1 H3K4 methyltransferase complex (5–12). In addition, Set1 and the human homologs are known to interact with the Ser5-phosphorylated C-terminal tail domain of RNA polymerase II (3). More recently, di- and trimethylated H3K4 have been shown to be docking sites for chromodomain and plant homeodomain (PHD) finger-containing proteins (1, 13, 14). Taken together, this and other studies suggest that the methylation of histone H3K4 plays a role in mediating gene expression by recruiting effector proteins.
In Saccharomyces cerevisiae, the Set1 complex (Set1C or COMPASS) is responsible for the methylation of histone H3K4 and the kinetochore protein Dam1 (5, 15–19). Loss of the catalytic protein Set1 results in the complete loss of global histone H3K4 mono-, di-, and trimethylation and alters expression levels at both euchromatic and subtelomeric genes (4, 5, 15, 17, 20–27). Several studies have been performed to understand the genes that are regulated by Set1 and histone H3K4 methylation (4, 15, 22, 27, 28). In general, genome-wide studies have shown both increases and decreases in expression when H3K4 methylation is abolished (15, 22, 27–29). However, comparing the results from these studies reveals that there is little overlap in what genes are affected by the loss of Set1 and H3K4 methylation and gives no clues to specific signaling or metabolic pathways that may be disrupted by the loss of H3K4 methylation. In addition, to our knowledge, these studies have not made a direct connection between changes in gene expression caused by the loss of Set1-mediated methylation and a biological phenotype. In fact, most of the studies that observed changes in gene expression in an set1Δ strain did not show an associated phenotype, suggesting that the observed changes did not significantly impact the cell (15, 22, 24, 26, 27, 30). In addition, studies that have looked at phenotypes associated with the set1Δ strain have not determined a direct role for Set1 or identified the specific endogenous gene(s) causing the phenotype (17, 20, 23, 25, 31–33).
Therefore, we wanted to connect a direct role of Set1 and H3K4 methylation with changes in gene expression and a biologically significant phenotype. To determine a biological role for Set1, we investigated the role of Set1-mediated H3K4 methylation in Brefeldin A (BFA) sensitivity and ergosterol homeostasis. In this study, we show that the Set1 complex and H3K4 methylation are needed for resistance to the antifungal drug BFA and maintenance of mRNA levels of rate-limiting enzyme HMG-CoA reductase (HMG1) and ERG11, two highly conserved genes needed for the ergosterol and cholesterol biosynthetic pathways. We also show that Set1 directly targets HMG1 and ERG11. More importantly, we determine that the loss of Set1 and/or H3K4 methylation leads to a decrease in HMG1 expression and protein levels as well as total cellular ergosterol levels. We also show that cells lacking Set1-mediated H3K4 methylation also have increased expression of DAN1 and PDR11, two genes involved in ergosterol uptake. Interestingly, the major consequence of increased expression of DAN1 and PDR11 is a gain-of-function phenotype, where set1Δ cells can take up exogenous ergosterol under aerobic conditions. Finally, we determined that set1Δ strains that are sensitive to BFA become resistant to BFA if allowed to take up exogenous ergosterol. Our results strongly indicate that the reduced ergosterol levels in an set1Δ strain result in a BFA-sensitive phenotype, thus making a key connection between BFA resistance and ergosterol homeostasis. Overall, our study provides insight into a biologically and medically relevant pathway that is dependent on Set1 and will provide additional opportunities to better understand the role of Set1-mediated methylation in gene expression.
Results
Histone H3K4 Methylation Is Necessary for Resistance to BFA.
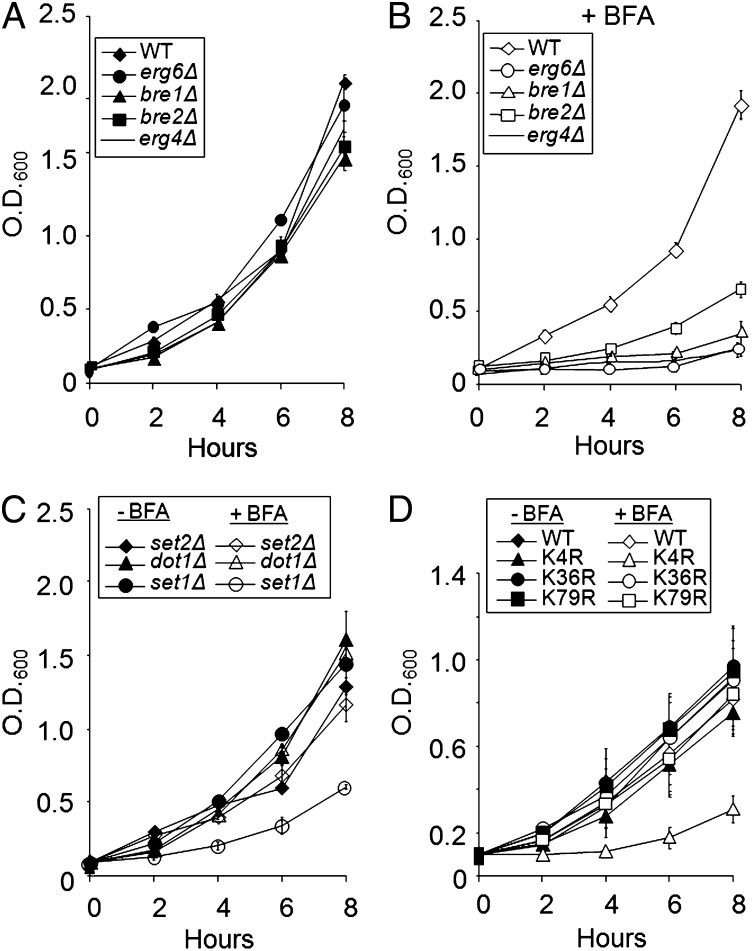
In yeast, BRE1 and BRE2 were named BFA-sensitive proteins 1 and 2, respectively, because it was determined in a BFA drug screen that deletion of either gene resulted in a hypersensitive growth defect (34). To confirm these results, liquid cell growth assays in the presence or absence of BFA were performed on bre1Δ and bre2Δ strains as well as other gene knockouts (KOs) known to be sensitive to BFA the erg4Δ and erg6Δ strains. As previously reported by plate assays, these deletion strains exhibit a growth defect or drug hypersensitivity when grown in the presence of 100 μg/mL (0.3 mM) BFA (Fig. 1 A and B) (34).
Fig. 1.
H3K4 methylation is required for resistance to BFA. (A) Growth curve of BY4741 WT, bre1Δ, bre2Δ, erg6Δ, and erg4Δ strains over an 8-h time course using synthetic complete media. (B) Growth curve of strains in A treated with 100 μg/mL BFA. (C) Growth curve of histone H3 methyltransferase deletion strains set1Δ, set2Δ, and dot1Δ in the presence and absence of 0.3 mM BFA. (D) Growth curve of WT and histone mutant strains (H3K4R, H3K36R, and H3K79R) in the presence or absence of 0.3 mM BFA. Data combined from three biological repeats. Error bars are calculated SEM.
These results are intriguing, because deletion of either BRE1 or BRE2 disrupts H3K4 methylation (5, 26, 35). Because bre1Δ and bre2Δ strains show hypersensitivity to BFA and disrupt H3K4 methylation, we hypothesized that strains lacking the Set1 H3K4 methyltransferase would have a similar growth phenotype when treated with BFA. As predicted, an set1Δ strain showed similar hypersensitivity to BFA as bre1Δ and bre2Δ strains, whereas set2Δ and dot1Δ strains did not exhibit a growth defect (compare Fig. 1B with Fig. 1C). Furthermore, deletions of other Set1 complex members that negatively affect H3K4 methylation also have slow-growth phenotypes in the presence of BFA (SI Appendix, Fig. S1; doubling times are shown in SI Appendix, Table S5). To determine if hypersensitivity to BFA is specific to the loss H3K4 methylation and not caused by methylation of a nonhistone target by Set1 or the Set1 complex, lysine 4 on histone H3 was mutated to arginine (H3K4R). Similar to an set1Δ strain, the H3K4R strain also showed a slow-growth phenotype in the presence of BFA. However, an increase in BFA sensitivity was not the case when mutations were made at H3K36 or H3K79, known methylation sites for Set2 and Dot1, suggesting that histone H3K4 methylation is specifically required for resistance to BFA (Fig. 1D).
Set1 Methyltransferase Activity Is Needed for WT Expression of Multiple Genes in the Ergosterol Biosynthetic Pathway.
To rule out the possibility that BFA treatment was altering H3K4 methylation levels and thus, affecting gene expression, histone H3K4 methylation levels were determined in WT cells treated with and without BFA. Western blot analysis using methyl-specific H3K4 antibodies showed that WT cells treated with BFA had no apparent changes in the global levels of H3K4 methylation compared with untreated cells (SI Appendix, Fig. S2A). Because previous studies show that Set1 and H3K4 methylation is needed for proper gene expression (4, 15, 21, 22, 24, 26, 27, 30, 36), we hypothesized that yeast strains lacking H3K4 methylation are sensitive to BFA because of decreased gene expression and production of the known protein targets of BFA. Therefore, we examined the expressions of SEC7 and GEA1, which produce two guanine nucleotide exchange factors that are known to be directly inhibited by BFA (reviewed in ref. 37). Unexpectedly, deletion of SET1 showed no significant defect in mRNA levels of either SEC7 or GEA1 (SI Appendix, Fig. S2B). Altogether, these data suggest that Set1-mediated H3K4 methylation is regulating other target gene(s) required for BFA resistance.
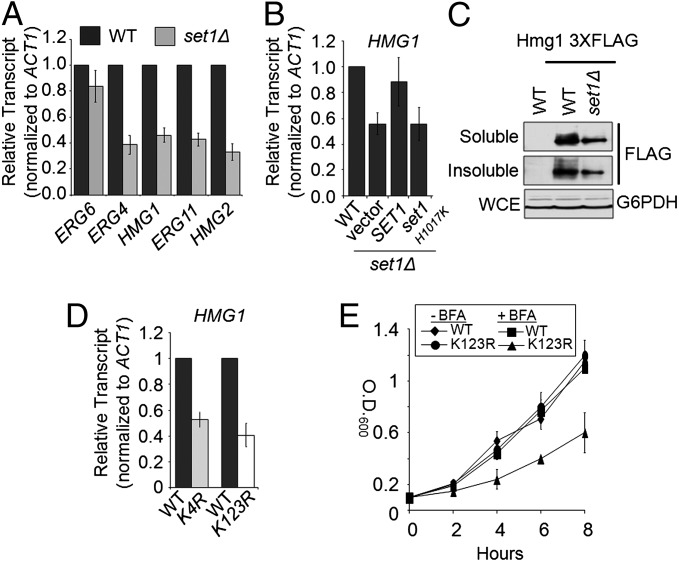
Because deletion of either ERG6 or ERG4 resulted in a slow-growth phenotype in the presence of BFA (Fig. 1B), we decided to examine if there was a decrease in the expression of ERG6 and ERG4 in an set1Δ strain. Quantitative real-time PCR (qRT-PCR) analysis determined that there is no significant change in the transcript levels of ERG6 (Fig. 2A). In contrast, ERG4 expression was 62% lower compared with WT expression (Fig. 2A). After determining that an set1Δ strain showed a decrease in expression of ERG4, other genes in the ergosterol biosynthetic pathway were tested to determine if they had defects in gene expression. The genes that were of particular interest were the genes conserved in both the yeast ergosterol and human cholesterol biosynthetic pathways because of the conserved nature of H3K4 methylation and a possible conserved mechanism for gene regulation. A schematic of the ergosterol and cholesterol pathway and some of the genes examined are shown in SI Appendix, Fig. S2C. Because ergosterol is the yeast equivalent to cholesterol, much of the early part of the ergosterol pathway is conserved from yeast to humans, including the rate-limiting enzyme HMG-CoA reductase (yeast Hmg1 and Hmg2) and the P450 C24 demethylase (Erg11) (38, 39). Interestingly, qRT-PCR analysis determined that, compared with WT, the set1Δ strain was 55% lower in HMG1, 66% lower in HMG2 (the second HMG-CoA reductase isoform in yeast), and 57% lower in ERG11 mRNA levels (Fig. 2A). Actin mRNA levels were used to normalize gene expression analysis.
Fig. 2.
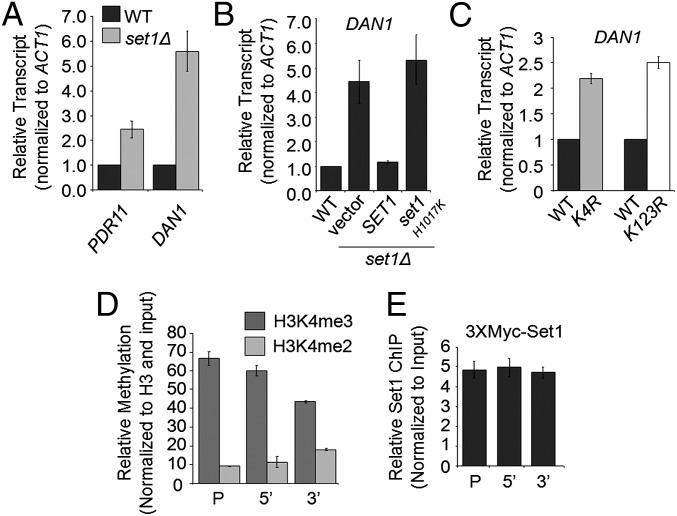
Set1 is necessary for the proper expression of multiple genes in the ergosterol biosynthetic pathway. (A) Expression of ERG6, ERG4, HMG1, HMG2, and ERG11 was determined in the set1Δ strain by qRT-PCR analysis. Statistical analysis determined significant changes in gene expression (P < 0.001) with the exception of ERG6 expression. (B) Expression of HMG1 was determined by qRT-PCR analysis using set1Δ strains transformed with plasmids expressing empty vector, full-length SET1, or catalytically inactive set1 H1017K. Statistical analysis determined significant changes in gene expression (P < 0.001) with the exception of HMG1 expression when full-length Set1 was expressed in the set1Δ strain. (C) Western blot analysis indicating the amount of Hmg1 in WT and set1Δ strains. Glucose 6-phosphate dehydrogenase was used as a loading control. Hmg1 was C-terminally 3xFLAG-tagged at its endogenous locus. (D) Expression of HMG1 was determined by qRT-PCR analysis in the histone H3K4R mutant and H2BK123R mutant strains. All expression analysis was relative to their respective isogenic WT strains using Actin (ACT1) as an internal control to normalize expression levels. Data were analyzed from three biological repeats with three technical repeats. Error bars are SEM. (E) Growth curve of WT and histone H2BK123R mutant strain in the presence or absence of 100 μg/mL BFA.
To confirm that the changes in gene expression were caused by Set1 methyltransferase activity, mRNA levels of ERG11, ERG4, and HMG1 were compared in WT, set1Δ, and an set1Δ strain transformed with WT SET1 or a catalytically inactive set1 H1017K mutant. Again, the loss of SET1 resulted in a reduction of mRNA levels for all three genes tested (Fig. 2B and SI Appendix, Fig. S3 A and B). Importantly, full-length SET1 was able to restore mRNA levels back to near WT levels, whereas the catalytically inactive H1017K mutant of Set1 was unable to rescue the defect in mRNA levels (Fig. 2B and SI Appendix, Fig. S3 A and B). Altogether, these results suggest that the methyltransferase activity of Set1 is necessary to maintain WT levels of mRNA for ERG11, ERG4, and HMG1. To determine if the decrease in mRNA levels correlated with a decrease in protein levels, Hmg1 was 3xFLAG-tagged at its endogenous locus in a WT and set1Δ strain. Western blot analysis of Hmg1–3xFLAG-tagged strains showed a reduction in Hmg1 protein levels of about 57% in an set1Δ strain compared with a WT strain in both soluble fraction and insoluble pellet, which likely contains membrane-bound Hmg1 (Fig. 2C). Protein levels were normalized by a Bradford assay, and glucose 6-phosphate dehydrogenase was used as a loading control (Fig. 2C). Therefore, the decrease in HMG1 mRNA expression strongly correlates with a decrease in Hmg1 protein expression.
To determine if the changes in gene expression observed in the set1Δ strain were directly caused by the loss of H3K4 methylation and not another target of Set1, qRT-PCR analysis of HMG1 and ERG11 was performed in a histone H3K4R mutant strain. Consistent with the set1Δ strain, the H3K4R mutant strain exhibited decreases in HMG1 and ERG11 expression, although the overall expression of ERG genes was higher in the H3K4R strain and the isogenic WT strain (Fig. 2D and SI Appendix, Fig. S3 C and D). Furthermore, to verify that the changes in gene expression were directly caused by the loss of H3K4 methylation, the H2BK123R mutation, which abolishes H2BK123 ubiquitination (H2BK123ub) and causes loss of H3K4 methylation, was used and resulted in BFA hypersensitivity and changes in gene expression of HMG1 and ERG11 similar to the set1Δ and H3K4R mutant strains (Fig. 2 D and E and SI Appendix, Fig. S3C).
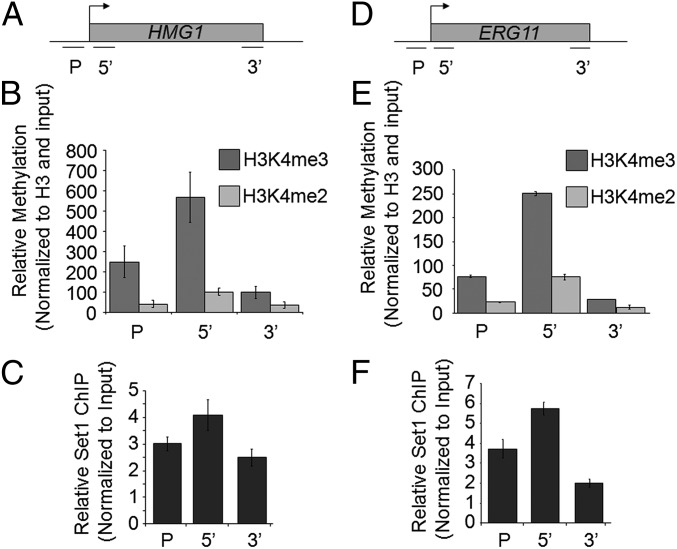
Although several studies indicate that the loss of Set1 can disrupt gene expression and abolish gene-specific histone methylation, few studies show that Set1 binding and histone methylation directly affect gene expression. To determine if Set1 is directly binding to and methylating the HMG1 and ERG11 loci, ChIP analysis was performed. Using an set1Δ strain as a background negative control, the levels of di- and trimethylation at HMG1 and ERG11 loci were analyzed by ChIP and qRT-PCR using gene-specific fluorescent probes (IDT) targeted to the promoter and 5′- (ORF) and 3′-ORF (Fig. 3 A and D). As expected, H3K4 trimethylation peaks at the 5′-ORF of both HMG1 and ERG11 and was reduced at the promoter and 3′-ORF similar to the pattern of methylation observed at other constitutively active genes (Fig. 3 B and E). ChIP analysis also showed that 3xMYC-Set1 locates at the promoter and 5′- and 3′-ORF of HMG1 and ERG11 compared with a nontagged WT control, where Set1 displays a similar pattern of enrichment to H3K4 methylation (Fig. 3 C and F). Altogether, these data indicate that Set1 and H3K4 methylation are directly associated with and needed for the proper expression of these biologically important genes.
Fig. 3.
Set1 and H3K4 methylation localizes to the HMG1 locus. (A and D) Schematics of HMG1 and ERG11 loci with indicated positions of probes for ChIP analysis. Probes were targeted to the promoter (P) and 5′- (ORF) and 3′-ORF regions. (B, C, E, and F) ChIP analyses from the indicated strains were performed using antibodies specific to a 3XMYC tag for detection of Set1, histone H3K4 di- and trimethylation, and histone H3. ChIP analysis for H3K4 di- and trimethylation was normalized to input and histone H3 and relative to a ChIP from an set1Δ strain. ChIP analysis for Set1 3XMYC was normalized to input and relative to WT untagged strain. All ChIP analyses wer performed using three biological repeats with three technical replicates; error bars represent SEM.
Set1 Is Necessary for Maintaining WT Levels of Cellular Ergosterol.
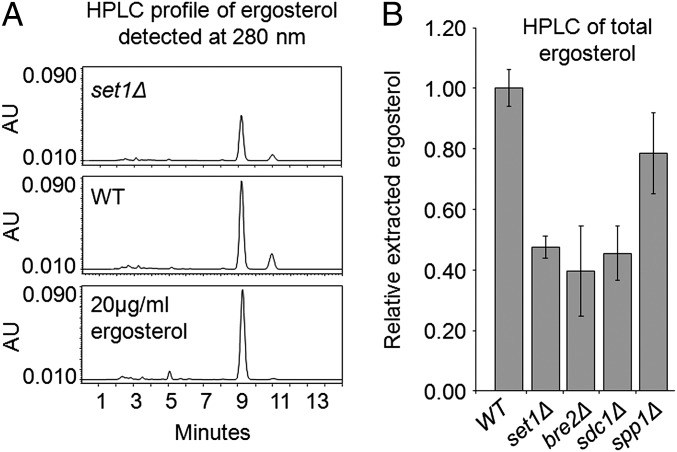
Based on our findings that cells lacking H3K4 methylation are sensitive to BFA, set1Δ cells have reduced expression of multiple genes in the ergosterol biosynthetic pathway, and deletion of ERG genes also shows an increase in the sensitivity of yeast to BFA, the set1Δ strain was tested to determine if there are reduced cellular levels of ergosterol. To determine if there are changes in ergosterol levels in an set1Δ strain, nonpolar lipids were extracted from yeast cultures, and total ergosterol levels were determined using HPLC. HPLC analysis determined that ergosterol peak detection (absorbance = 280 nm) was ∼9 min when using purified ergosterol as a standard. A smaller second peak eluted at 11 min and was present in our WT, set1Δ, and purified ergosterol samples. It is currently unclear what this peak is, but it may be a derivative form of ergosterol. However, comparing the extracted ergosterol level (the area under the 9-min peaks) from a WT with the set1Δ strain, HPLC analysis showed consistently less ergosterol extracted from an set1Δ strain (Fig. 4A). The relative amount of extracted ergosterol was determined by using known concentrations of purified ergosterol to generate a standard curve. In addition, purified cholesterol was added to each sample before extraction as an internal control. This analysis determined that strains lacking Set1 complex subunits SET1, BRE2, and SDC1 exhibit an approximately 50% decrease in cellular ergosterol levels, whereas an spp1Δ strain has only a 20% decrease in ergosterol levels (Fig. 4B). The observed 20% decrease in ergosterol levels in the spp1Δ strain coincides with an intermediate growth defect in the presence of BFA suggesting that the specific level of H3K4 methylation may have an impact on gene expression and ergosterol levels (Fig. 4B and SI Appendix, Fig. S1) Overall, these data not only suggest that H3K4 methylation is needed for proper HMG1 and ERG gene expression but that it also impacts the production of ergosterol and provides a reasonable explanation of why an set1Δ strain or any strain with defects in histone H3K4 methylation has a hypersensitivity phenotype when treated with BFA.
Fig. 4.
HPLC analysis of total cellular ergosterol. (A) Chromatograms of ergosterol extracted from WT and set1Δ cells compared with commercially purified ergosterol. Ergosterol was detected at 280 nm, where peak elution was ∼9 min. (B) Relative amount of extracted ergosterol from the indicated strains compared with WT. Amounts of extracted ergosterol were determined by comparing extracted ergosterol to a standard curve of known concentrations of purified ergosterol. Purified cholesterol was added to each sample before extraction as an internal control. Analysis was performed using three biological replicates; error bars represent SEM.
Set1 Is Important for the Repression of Genes Involved in Ergosterol Uptake.
Many studies indicate that the regulation of ergosterol in yeast and cholesterol levels in metazoans is a balance between synthesis and uptake (40). For example, previous work by others shows that ergosterol uptake in yeast only occurs when ergosterol is depleted by using mutant strains, shifting yeast to anaerobic conditions because ergosterol biosynthesis requires oxygen or depleting ergosterol levels using antifungal drugs (41–53). Therefore, because set1Δ cells have reduced cellular ergosterol levels, exogenous ergosterol uptake by the set1Δ strain could occur by increasing expression of genes involved in ergosterol uptake, even when grown under aerobic conditions. Genes known to be involved in ergosterol uptake include two ABC transporters (AUS1 and PDR11), the transcription factor SUT1, and the cell wall protein DAN1 (44, 54). In contrast to the role of Set1 in mediating proper expression of HMG1 and ergosterol biosynthesis genes, qRT-PCR analysis revealed that set1Δ cells exhibited a significant increase in mRNA levels for PDR11 and DAN1, which are 2.5- and 5.6-fold increased, respectively (Fig. 5A). A small increase in the expression of AUS1 (1.6-fold) was observed, whereas no significant change was observed in the expression of SUT1 (SI Appendix, Fig. S3E). The altered levels of PDR11 and DAN1 expression in an set1Δ strain are restored to WT levels by expressing SET1 in an set1Δ strain. In contrast, catalytically inactive set1 H1017K maintained expression levels of PDR11 and DAN11 similar to an set1Δ strain (Fig. 5B and SI Appendix, Fig. S3F). These data suggest that the catalytic activity of Set1 is needed for repressing PDR11 and DAN1 expression under aerobic conditions. To test if there is a functional relationship between the level of DAN1 mRNA and the mRNA levels of ergosterol biosynthetic genes, the expressions of HMG1 and ERG11 in the dan1Δ and the set1Δdan1Δ strains were tested. Deletion of DAN1 resulted in no significant change in the expression of ERG4, ERG11, or HMG1 in the dan1Δ strain compared with WT or the set1Δdan1Δ compared with the set1Δ strain (compare Fig. 2A with SI Appendix, Fig. S4A).
Fig. 5.
Set1 methyltransferase activity is required for repression of sterol uptake genes. (A–C) Relative transcript levels of DAN1 and PDR11 were determined in the indicated strains by qRT-PCR analysis. All expression analysis was relative to WT cells using ACT1 as an internal control to normalize expression levels. Data were analyzed from three biological repeats with three technical repeats. Each error bar represents SEM. Statistical analysis determined significant changes in gene expression (P < 0.001) with the exception of DAN1 expression when full-length Set1 was expressed in the set1Δ strain. (D and E) ChIP analysis from the indicated strains was performed using antibodies specific to a 3XMYC tag for detection of Set1, histone H3K4 di- and trimethylation, and histone H3. ChIP analysis for H3K4 di- and trimethylation was normalized to input and histone H3 and relative to a ChIP from an set1Δ strain. ChIP analysis for Set1 was normalized to input and relative to WT untagged strain. The ChIP analysis was performed using three biological repeats; error bars represent SEM.
Because Set1 and H3K4 methylation are thought to play an indirect role in repressing genes near the telomere, it was necessary to determine if Set1 and H3K4 methylation played a direct role in repressing the DAN1 locus. Unexpectedly, H3K4 trimethylation was detected by ChIP analysis in WT cells at the promoter and 5′- and 3′-ORF of DAN1 (Fig. 5D). This finding was intriguing considering that DAN1 is mostly repressed under aerobic conditions (compare DAN1 transcript in a WT with dan1Δ) (SI Appendix, Fig. S4A) (41, 54–61). Consistent with the H3K4 methylation ChIP analysis, Set1 protein was also detected at the DAN1 locus at similar levels across the promoter and 5′- and 3′-ORF, suggesting that Set1 plays a direct role in the repression of mRNA expression at the DAN1 locus (Fig. 5E). Using the set1Δ strain as a no methylation control, the levels of H3K4 tri- and dimethylation detected at DAN1 were tested to determine if they were more enriched at the DAN1 locus compared with an intergenic nontranscribed region of the genome. Interestingly, ChIP analysis at an intergenic locus on the right arm of chromosome six revealed that the DAN1 locus has significant enrichment of H3K4 di- and trimethylation and 3xMyc-Set1 compared with this untranscribed region (SI Appendix, Fig. S4 B and C).
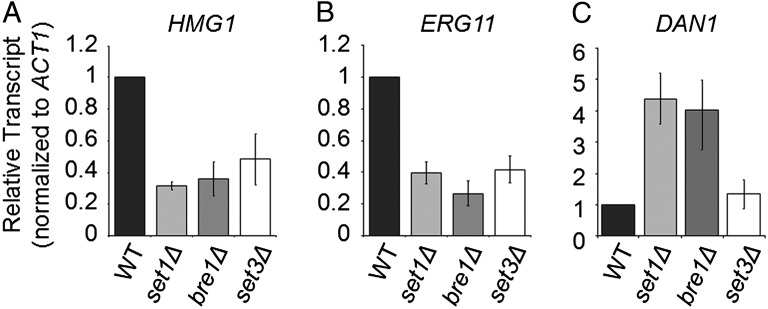
Based on these studies, both increases and decreases in gene expression were observed when SET1 is deleted or H3K4 is mutated to arginine. One way in which Set1 and H3K4 methylation may be involved in the regulation of gene expression is through recruitment of the Set3 histone deacetlyase complex. Interestingly, in the same screen that identified Bre1 and Bre2 as important for resistance to BFA, the Set3 complex member Hos2 was also shown to be necessary for normal growth in the presence of BFA (34). The Set3 complex is known to be involved in transcriptional regulation, and the PHD finger of Set3 has been shown to interact with H3K4 dimethylation (14, 62). Therefore, Set3 may also be necessary for the regulation of genes involved in ergosterol production and/or genes needed for ergosterol uptake. To test if the Set3 complex plays a role in the expression of ergosterol biosynthetic genes HMG1 and ERG11 as well as the sterol uptake gene DAN1, qRT-PCR analysis was performed in an set3Δ strain and as an additional control, the bre1Δ strain, which affects H2BK123ub. Intriguingly, the bre1Δ strain mimicked the set1Δ strain with reductions in HMG1 and ERG11 expression (WT was 2.8- and 3.8-fold higher in expression, respectively) and an increase in DAN1 expression (bre1Δ was 4-fold higher compared with WT) (Fig. 6). Interestingly, the set3Δ strain exhibited a decrease in expression of HMG1 and ERG11, but no significant change in DAN1 expression was observed, suggesting that the role of H3K4 methylation in gene expression acts through multiple mechanisms to either promote or repress expression (Fig. 6).
Fig. 6.
Loss of Set3 leads to lower expression of ergosterol biosynthetic genes. (A–C) Relative transcript levels of DAN1, ERG11, and HMG1 were determined in the indicated strains by qRT-PCR analysis. All expression analysis was relative to WT cells using ACT1 as an internal control to normalize expression levels. Data were analyzed from three biological repeats with three technical repeats. Each error bar represents SEM. Statistical analysis determined significant changes in gene expression (P < 0.001) with the exception of DAN1 expression in the set3Δ strain.
Because Set3 does not seem to play a role in the repression of DAN1 through Set1-mediated H3K4 methylation, the possible role of Set1 being involved in regulating the expression of known transcriptional repressors of DAN1 was examined. qRT-PCR analysis of MOT3 and ROX1, two repressors known to localize to the DAN1 promoter, showed no significant change in expression in the set1Δ strain, suggesting that the derepression of DAN1 observed is because of another mechanism (SI Appendix, Fig. S4D). Another possible role for H3K4 methylation to act as a repressive modification for DAN1 is through the expression of a noncoding RNA. A genome-wide screen for noncoding RNA revealed a putative antisense transcript located in the 5′ region of DAN1 (63). To test if an antisense transcript was involved in the repression of DAN1, gene expression analysis was performed using probes specific to the promoter and 5′ and 3′ regions of DAN1. If a noncoding RNA was transcribed in the 5′ region of DAN1 to repress the sense transcript of DAN1, more detectable 5′ transcripts would be observed compared with the 3′ end. In addition, transcript in the promoter region of DAN1 would likely be detected. qRT-PCR analysis did not show a difference between the 5′ and 3′ regions of DAN1 in a WT strain, and in the set1Δ strain, it showed similar increases in expression at both the 5′ and 3′ regions (SI Appendix, Fig. S4E). The promoter region of DAN1 also had no observable detection of RNA transcript, suggesting that antisense noncoding RNA is not involved in the repression of DAN1 (SI Appendix, Fig. S4E).
Loss of Set1 Activity Allows for Sterol Uptake and Rescues BFA Sensitivity in the Presence of Ergosterol.
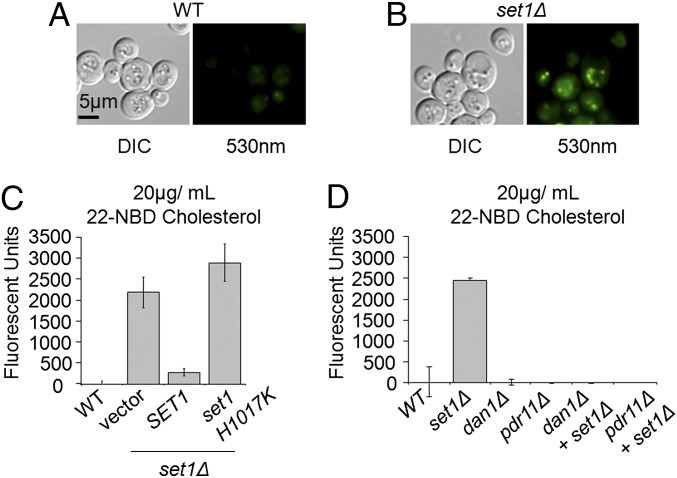
To determine if the increase in mRNA levels of PDR11 and DAN1 is functionally important, uptake of sterols from culture media was determined. In budding yeast, uptake of sterols does not normally occur under aerobic conditions but is observed in some mutants (64, 65). To determine if an set1Δ strain can import sterols from the media, WT and the set1Δ strain were grown in synthetic complete media (SC) supplemented with 20 μg/mL 22-NBD cholesterol (a fluorescent sterol analog). Previous work has shown that 22-NBD cholesterol can be imported into yeast (66, 67). Using fluorescent microscopy to visualize sterol uptake, set1Δ cells showed an increased in fluorescence compared with WT (Fig. 7 A and B). To determine the relative amount of imported cholesterol, the amount of 22-NBD cholesterol uptake was quantified using a Bio-Tek Synergy 4 multimode plate reader. An increase in fluorescence was detected from set1Δ cells relative to WT cells (Fig. 7 C and D). Consistent with the qRT-PCR data, expression of WT SET1 in an set1Δ strain significantly reduced the level of cholesterol uptake, whereas catalytically inactive Set1 H1017K was able to maintain cholesterol uptake, suggesting that the catalytic activity of Set1 is important for limiting sterol uptake (Fig. 7C). To determine if the increase in expressions of DAN1 and PDR11 are actively contributing to sterol uptake in an set1Δ strain, both genes were deleted in an set1Δ background. Using the quantitative fluorescent 22-NBD cholesterol uptake assay, deletion of DAN1 or PDR11 resulted in a failure to import cholesterol from the media in WT and set1Δ strains (Fig. 7D). These data show that the increased mRNA levels of DAN1 and PRD11 in an set1Δ strain result in the functional import of ergosterol, indicating the biological significance of Set1 in mediating ergosterol homeostasis.
Fig. 7.
Loss of Set1 leads to aerobic ergosterol uptake. (A and B) DIC and fluorescent images of WT and set1Δ strains grown in the presence of 20 μg/mL 22-NBD cholesterol. Fluorescent images were observed using a filter for FITC (530 nm), and the corresponding DIC image was visualized at 100× oil. (C and D) Quantification of fluorescent units in the indicated strains. Fluorescence was detected from cells at an excitation wavelength of 485 nm and emission wavelength of 528 nm. Cells were normalized to OD600, and WT background was subtracted from total fluorescent units; error bars represent SEM of three biological replicates.
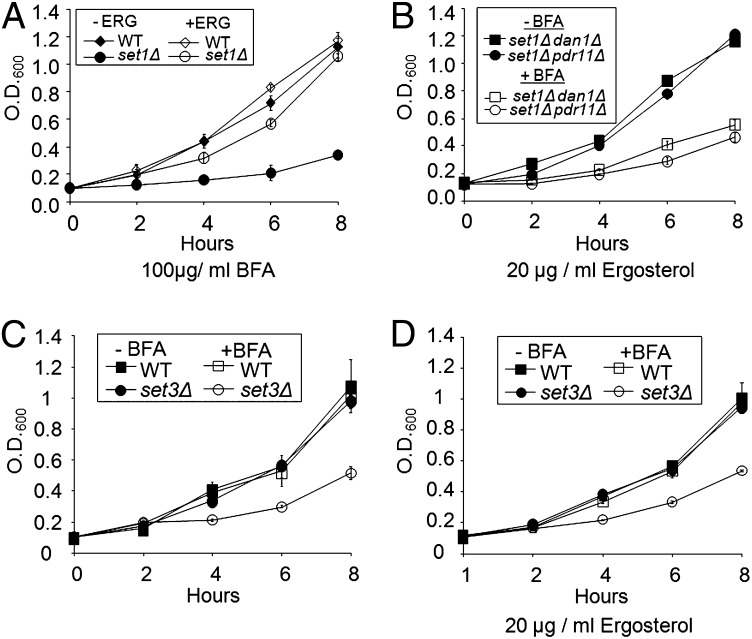
To determine if reduced ergosterol levels were the cause for an set1Δ strain to be sensitive to BFA, growth assays were performed in the presence or absence of BFA and supplemented with 20 μg/mL ergosterol. Interestingly, the set1Δ strain was no longer sensitive to BFA and grew at a similar rate as WT strains in the presence and absence of BFA (Fig. 8A). In addition, deletion of DAN1 or PDR11 in an set1Δ strain that failed to uptake ergosterol remained sensitive to BFA treatment even in the presence of ergosterol, indicating that ergosterol must be imported into the cell for BFA resistance and ergosterol in the media does not have an indirect effect on BFA uptake (Fig. 8B). We also determined that deletion of DAN1 had no effect on growth in the presence of BFA without ergosterol (SI Appendix, Fig. S5A). Deletion of PDR11 exhibits a mild slow-growth phenotype in the presence of BFA without ergosterol, consistent with the role of other ABC transporters in drug resistance (SI Appendix, Fig. S5B). In line with the observed changes in HMG1 and ERG11 expression, the set3Δ strain displayed a hypersensitivity to BFA in our growth assays (Fig. 8C). In addition, when the set3Δ strain was grown in the presence of BFA and ergosterol, there was no observed rescue of growth, which is expected because of both Set1 and Set3 playing a role in expression of ergosterol biosynthesis genes, but the loss of Set3 does not significantly contribute to the derepression of DAN1, which is important for sterol uptake (Fig. 8D).
Fig. 8.
Aerobic uptake of ergosterol can rescue resistance to BFA. (A–D) Growth curve of the indicated strains grown with or without 20 μg/mL ergosterol in the presence or absence of 0.3 mM (100 μg/mL) BFA. Error bars indicated are SEM of three biological replicates.
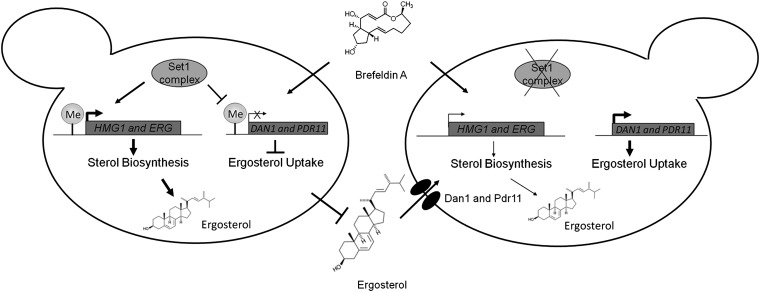
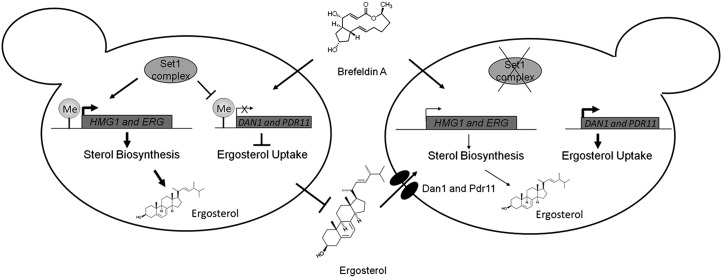
Overall, these data show that Set1 and H3K4 methylation is important for the regulation of ergosterol homeostasis of the cell and BFA resistance. The loss of H3K4 methylation in an set1Δ strain led to lower expression of several of the genes needed for the production of ergosterol and increased expression of genes important for ergosterol uptake through two different regulatory mechanisms. These alterations in gene expression in an set1Δ strain are functionally and biologically important, because the lower levels of ergosterol in these cells make them more susceptible to BFA, causing a slow-growth phenotype. Because of increased expression of ergosterol transport genes, the BFA-induced slow-growth phenotype can be rescued by supplementing the cells with exogenous ergosterol (Fig. 9).
Fig. 9.
Model indicating the role of Set1 in ergosterol homeostasis and BFA sensitivity. Set1, the catalytic subunit of the Set1 complex, localizes to and methylates histone H3K4 of genes involved in ergosterol biosynthesis and genes involved in sterol uptake. Set1-mediated methylation of HMG1 and other ERG genes promote proper expression and production of ergosterol. Histone H3K4 methylation at DAN1 and PDR11 by Set1 limits the expression and prevents aerobic sterol uptake, which allows cells to maintain resistance to BFA. Deletion of SET1 leads to complete loss of histone H3K4 methylation, reduction in expression of HMG1 and other ERG genes (leading to lower ergosterol levels), and increase in expression of ergosterol transport genes DAN1 and PDR11. A reduction of ergosterol causes set1Δ cells to become BFA-sensitive. In contrast, set1Δ cells treated with exogenous ergosterol for uptake under aerobic conditions cause set1Δ cells to become resistant to BFA.
Discussion
In this study, we determined that Set1-mediated H3K4 methylation plays an important role in maintaining ergosterol homeostasis and drug resistance to BFA. Bre1, an E3 ubiquitin ligase that is needed for H2BK123ub, and Bre2, an Set1 complex subunit, were originally identified in a drug screen, where yeast strains lacking either BRE1 or BRE2 grew slower on plates containing BFA compared with a WT strain. However, until this study, the reason why bre1Δ and bre2Δ strains were sensitive to the drug BFA remained undetermined. We identified that loss of H3K4 methylation mediated by Set1 causes a BFA-sensitive phenotype, and we have shown a direct connection between Set1 and genes involved in ergosterol production and uptake. We also show that the level of ergosterol in an set1Δ strain determines resistance to BFA and likely, other antifungal drugs. Altogether, our study has identified target genes of Set1 and established that loss of SET1 can alter a biologically and medically relevant pathway. Therefore, Set1-mediated H3K4 methylation may play a vital role in yeast developing resistance to antifungal drugs. In addition, the conservation between cholesterol and ergosterol pathways suggests that the human H3K4 methyltransferases may also impact cholesterol homoeostasis.
Our combined data also suggest that the regulation of ergosterol homeostasis through Set1-mediated H3K4 methylation is through different mechanisms. The observation that Set3 is important for expression of ergosterol biosynthetic genes HMG1 and ERG11 but not the repression of DAN1 may indicate why we observe a different pattern of methylation at HMG1 and ERG11 compared with DAN1. Active genes that have 5′ enrichment of H3K4 trimethylation may be regulated through recruitment of Set3, and genes with alternate patterns of H3K4 methylation may be regulated by a different mechanism. More recently, it has been discovered that Set1 is involved in the repression of gene expression through the promotion of noncoding RNA expression (29, 68, 69). One interesting aspect of the role of Set1 in noncoding RNA expression is a 3′ enrichment of H3K4 trimethylation of the gene in which the noncoding RNA represses (29, 68, 69). Our data would indicate the DAN1 locus does not have a noncoding RNA or 3′ enrichment of histone H3K4 trimethylation. Intriguingly, another possible mechanism for Set1-mediated repression of DAN1 could be the decrease in transcript of transcriptional repressors. Our data indicate that Set1 does not play a role in the proper expression of DAN1 repressors Rox1 or Mot3, but these data could suggest that Set1 and H3K4 methylation may play a role in recruitment of repressors to DAN1. Alternatively, Set1-mediated H3K4 methylation may be playing a role in how yeast sense oxygen or heme levels, which in turn, could affect the expression of genes in the ergosterol biosynthesis and uptake pathways. Additional studies will be needed to determine the different mechanisms involved in Set1-mediated gene expression and repression.
More recently, a genome-wide drug screen was performed on homozygous and heterozygous yeast deletion collections. However, this genome-wide screen failed to identify a connection between Set1-mediated H3K4 methylation and BFA sensitivity, because the BFA drug screen was limited only to a heterozygous deletion library (70). In contrast, the homozygous deletion screen using other drug compounds did reveal that strains lacking Set1 complex components can lead to growth defects when treated with multiple toxic compounds, suggesting that Set1-mediated H3K4 methylation may also be needed for multidrug resistance. These data are consistent with our results showing that strains lacking SET1 can increase the expression of the ergosterol transport gene PDR11, a gene that is classified as a member of the ABC family of transporters (71). Additional analysis would be needed to determine if expression of other ABC transporters is controlled by Set1 and/or if Set1 plays a role in antifungal drug resistance. Because ABC transporters are important in multidrug resistance, the human H3K4 methyltransferases may play a clinically important role in multidrug resistance and cancer treatment by controlling the levels of ABC transporters (70).
Our study not only shows that Set1 is needed for the proper expression of PDR11 but also, that it is needed for full expression of ERG11, a conserved gene needed for ergosterol and cholesterol biosynthesis. Interestingly, both ABC transporters and Erg11 are important factors in the study of antifungal drug resistance; ABC transporters are involved in the active pumping of antifungal drugs out of yeast cells, and Erg11 is the main target for the antifungal azole drugs such as Ketoconazole and Fluconazole (72–74). In addition, the expressions of ABC transporters and ERG11 are increased in cells that are resistant to antifungal drugs. Because of these observations, our findings suggest that Set1-mediated H3K4 methylation could play an important but unexplored role in the mechanism and development of antifungal drug resistance. More importantly, our results showing that the sensitivity to BFA in an set1Δ strain can be overcome when supplemented with exogenous ergosterol would strongly indicate that ergosterol levels are directly connected to resistance to BFA and other antifungals. Although the increased sensitivity to BFA in ERG gene KOs has been suggested to be caused by the altered sterol composition of the cell, to our knowledge, the relationship between ergosterol levels and BFA sensitivity has never been directly tested (34, 75, 76). Therefore, ergosterol levels of drug-resistant yeast need to be further explored, and the use of Statin HMG-CoA inhibitors combined with antifungal drugs may need to be evaluated as an efficacious treatment of drug-resistant yeast.
Regulation of the ergosterol and cholesterol pathways is controlled transcriptionally and posttranscriptionally (40, 72, 77). Much of the work done to elucidate the cholesterol biosynthetic pathway has come from studying ergosterol in yeast (40, 66, 72). Understanding ergosterol biosynthesis is also of medical importance, because the ergosterol pathway is the target of many antifungal drugs. In both yeast and humans, there is a transcriptional response to changes in sterol levels (66, 72, 78, 79). Sterol regulatory element (SRE) binding proteins in mammals bind to DNA SREs in response to lower levels of cellular cholesterol (72). In yeast, there are no direct homologs to SRE binding proteins, but the transcription factors Upc2 and Ecm22 are both regulators of the ergosterol pathway and bind to SRE sites (78, 79). In addition to Upc2 and Ecm22, the heme-dependent transcription factor Hap1 is also needed in the regulation of ergosterol genes (78–80). Until now, little was known about the role of H3K4 methylation in the regulation of genes important for ergosterol or cholesterol biosynthesis. Our data, along with a study that shows RNAi knockdown of human H3K4 methyltransferase component WDR5, result in a defect in the expression of HMG-CoA reductase, suggesting that H3K4 methylation-mediated regulation of sterol biosynthesis is conserved from yeast to humans (81). Additional studies will be needed to determine the impact of H3K4 methylation on cholesterol homoeostasis in humans.
A surprising but important result from our study was that adding ergosterol to culture media rescued the BFA sensitivity phenotype of an set1Δ strain. These data clearly showed that the set1Δ strain was able to uptake sterols from the media under aerobic conditions (Fig. 8). The observed aerobic uptake of sterols is highly unusual, because yeast do not normally take up sterols from media unless under hypoxic or anaerobic growth environments. Interestingly, the sterol uptake that we observe is dependent on DAN1 expression, which is increased 5.6-fold in the set1Δ strain as opposed to the more significant increase in transcript observed under anaerobic conditions, suggesting that a small increase in DAN1 transcript is sufficient for sterol uptake to occur (55, 56). The sterol uptake that we observed is similar to the uptake of serum cholesterol that occurs in human cells when HMG-CoA reductase is inhibited by Statin drugs (40). Because of the conserved nature of many genes involved in sterol biosynthesis and the fact that the human histone methyltransferase component WDR5 is important for HMG-CoA reductase expression, it is likely that inhibiting H3K4 methylation could potentially reduce serum cholesterol levels in humans. In addition, based on our genetic data, treatment with Statin drugs and H3K4 methyltransferase inhibitors could be an effective drug treatment. Previous work by others suggests that a combinatorial treatment with Statins and additional chemothereputic agents, such as Doxorubicin and Cisplatin, is efficacious (82–84). Additionally, it is quite possible that treatments for pathogenic fungal infections, such as Candida albicans, could have an increased efficacy using a combination of Statin and a histone H3K4 methyltransferase inhibitor. Understanding how Set1-mediated H3K4 methylation plays a role in cholesterol homeostasis in humans and antifungal drug resistance can address issues in heart disease, diabetes, cancer development, and neurological disorders, such as Parkinson and Alzheimer’s diseases, as well as develop novel strategies for combating fungal infections and antifungal drug resistance.
Materials and Methods
Plasmids and Yeast Strains.
All yeast strains and plasmids used in this study are described in SI Appendix, Tables S1 and S2. Yeast plasmids were constructed as previously described (26, 85). Transformations and deletion strains were generated as previously described (26, 85). Verification of plasmids and strains was performed by the Purdue University Center for Cancer Research DNA sequencing facility.
BFA Sensitivity Assay.
Yeast strains were grown in SC supplemented with either 0.3 mM (100 μg/mL) BFA dissolved in ethanol or an equal volume of ethanol as a control. Log-phase yeast cultures were back-diluted in 100 μL media to an OD of 0.1 (OD600 = 0.1), and absorbance readings were taken every 2 h using a Bio-Tek Synergy 4 multimode plate reader (www.biotek.com) for a total of 8 h. Each experiment was done in triplicate. For the rescue of sensitivity assay, the experiment was repeated as stated above, but the SC was also supplemented with 20 μg/mL ergosterol (stock #B23840; Alfa Aesar) dissolved in a 1:1 ratio of ethanol and Tween 80 (P1754-500; Sigma). Doubling times from growth assays were calculated and shown in SI Appendix, Table S5. Light microscopy was used to determine that changes in OD were not caused by changes in cell size or flocculation of the yeast when grown in the presence of ergosterol, ethanol, or BFA.
Gene Expression Analysis.
qRT-PCR was performed as previously described (26, 36). Three biological repeats, each including three technical repeats, were performed for all samples. Data were analyzed using the ΔΔCt method, in which actin was used as the endogenous control, and the relative quantity of ERG6, ERG4, ERG11, HMG1, HMG2, DAN1, AUS1, SUT1, PDR11, SEC7, and GEA1 transcript was determined for each deletion strain compared with transcript in a WT strain. Primer sequences used for PCR amplification are described in SI Appendix, Table S3. Relative fold changes in mRNA levels and statistical significance values are described in SI Appendix, Tables S6–S11.
HPLC Analysis of Nonpolar Sterols.
Five-milliliter cultures were grown overnight in SC, spun down, and washed two times with water. Sterols were extracted from yeast using 4 M potassium hydroxide in 70% (vol/vol) ethanol at 80 °C for 1 h. After extraction, nonpolar lipids were separated using N-hexane two times. Nonpolar sterols were crystallized after evaporation of the N-hexane and redissolved in 100% methanol. Samples were then analyzed using a Waters HPLC system (www.waters.com) and separated using a C-18 column with the flow rate set at 1 mL/min 100% methanol. Ergosterol was detected at 280 nm. Cholesterol was used as an injected internal control and detected at 210 nm. A standard curve was determined using known concentrations of purified ergosterol.
Fluorescent Microscopy and Sterol Uptake Analysis.
For visualization of sterol uptake, 5-mL cultures were grown overnight (16 h) in SC supplemented with 20 μg/mL 22-NBD cholesterol dissolved in a 1:1 ratio of ethanol and Tween 80. Samples were washed two times in SC media and one time in PBS. Samples were visualized for differential interference contrast (DIC) using an Olympus microscope at 1,000× oil magnification and fluorescent images using FITC filters. Quantification of cholesterol uptake was performed using the Bio-Tek Synergy 4 multimode plate reader. Cells were grown as stated above and washed. OD was determined for each sample before determining fluorescent intensity to normalize each well. Each strain was analyzed with three technical repeats for each of three biological replicates. Fluorescent readings were done using a filter for 476-nm excitation and 528-nm emission wavelengths.
ChIP.
ChIP was performed as previously described with additional modifications (26, 36). Briefly, 50-mL cultures were grown to midlog phase in selective media. Cells were harvested and cross-linked with 1% formaldehyde for 15 min. ChIP was performed using MYC (MYC 9E10; Sigma), H3K4 tri- or dimethyl-specific antibodies (07–030 and 07–473; Millipore), or an antibody specific to H3 (Ab1791; Abcam) and DYNAL magnetic Protein G beads (Invitrogen). Cross-linked immunoprecipitated DNA bound to DYNAL beads was amplified by qRT-PCR using a Step-one plus PCR machine (Life Technologies) and dual-labeled fluorescent probe sets to the promoter 5′ and 3′ regions of HMG1, ERG11, DAN1, and ERG6 (IDT). Probe sets with the indicated fluorescent dye (FAM) and quenchers (ZEN/Iowa Black FQ) used in qRT-PCR are described in SI Appendix, Table S4.
Supplementary Material
Acknowledgments
We thank Dr. Beth Tran for review of the manuscript. This work was supported in whole or part by Bilsland Strategic Initiatives Fellowship (to P.F.S.), National Institutes of Health Grant GM74183 (to S.D.B.), and a Purdue University Center for Cancer Research fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 4174 (volume 110, number 11).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215768110/-/DCSupplemental.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Sci Aging Knowledge Environ. 2003;2003(14):RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 3.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11(3):709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 4.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 5.Roguev A, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20(24):7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dou YL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 7.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13(9):852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10(5):1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 9.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280(50):41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282(18):13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24(13):5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143(3):470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137(2):259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boa S, Coert C, Patterton HG. Saccharomyces cerevisiae Set1p is a methyltransferase specific for lysine 4 of histone H3 and is required for efficient gene expression. Yeast. 2003;20(9):827–835. doi: 10.1002/yea.995. [DOI] [PubMed] [Google Scholar]
- 16.Zhang K, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122(5):723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briggs SD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15(24):3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA. 2002;99(1):90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latham JA, Chosed RJ, Wang S, Dent SY. Chromatin signaling to kinetochores: Transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146(5):709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryk M, et al. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol. 2002;12(2):165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- 21.Dietvorst J, Brandt A. Flocculation in Saccharomyces cerevisiae is repressed by the COMPASS methylation complex during high-gravity fermentation. Yeast. 2008;25(12):891–901. doi: 10.1002/yea.1643. [DOI] [PubMed] [Google Scholar]
- 22.Guillemette B, et al. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011;7(3):e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogan NJ, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277(13):10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 24.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J Biol Chem. 2012;287(4):2652–2665. doi: 10.1074/jbc.M111.280867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8(12):2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.South PF, Fingerman IM, Mersman DP, Du HN, Briggs SD. A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J Biol Chem. 2010;285(1):595–607. doi: 10.1074/jbc.M109.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci USA. 2007;104(42):16609–16614. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiner A, et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012;10(7):e1001369. doi: 10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margaritis T, et al. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 2012;8(9):e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SS, Zhou BO, Zhou JQ. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol Cell Biol. 2011;31(15):3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6(8):e1001082. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung A, et al. Histone H2B ubiquitylation and H3 lysine 4 methylation prevent ectopic silencing of euchromatic loci important for the cellular response to heat. Mol Biol Cell. 2011;22(15):2741–2753. doi: 10.1091/mbc.E11-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morohashi N, Mitchell AP, Shimizu M. Effect of histone methyltransferase gene mutations on sporulation in S. cerevisiae. Nucleic Acids Symp Ser (Oxf) 2005;2005(49):325–326. doi: 10.1093/nass/49.1.325. [DOI] [PubMed] [Google Scholar]
- 34.Murén E, Oyen M, Barmark G, Ronne H. Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast. 2001;18(2):163–172. doi: 10.1002/1097-0061(20010130)18:2<163::AID-YEA659>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 35.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278(37):34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 36.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009;23(8):951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chardin P, McCormick F. Brefeldin A: The advantage of being uncompetitive. Cell. 1999;97(2):153–155. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- 38.Basson ME, Thorsness M, Finer-Moore J, Stroud RM, Rine J. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol. 1988;8(9):3797–3808. doi: 10.1128/mcb.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturley SL. Conservation of eukaryotic sterol homeostasis: New insights from studies in budding yeast. Biochim Biophys Acta. 2000;1529(1–3):155–163. doi: 10.1016/s1388-1981(00)00145-1. [DOI] [PubMed] [Google Scholar]
- 40.Mouritsen OG, Zuckermann MJ. What’s so special about cholesterol? Lipids. 2004;39(11):1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 41.Sertil O, Vemula A, Salmon SL, Morse RH, Lowry CV. Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast. Mol Cell Biol. 2007;27(6):2037–2047. doi: 10.1128/MCB.02297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal AK, et al. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J Biol Chem. 2003;278(37):34998–35015. doi: 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- 43.Bourot S, Karst F. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene. 1995;165(1):97–102. doi: 10.1016/0378-1119(95)00478-o. [DOI] [PubMed] [Google Scholar]
- 44.Kohut P, et al. The role of ABC proteins Aus1p and Pdr11p in the uptake of external sterols in yeast: Dehydroergosterol fluorescence study. Biochem Biophys Res Commun. 2011;404(1):233–238. doi: 10.1016/j.bbrc.2010.11.099. [DOI] [PubMed] [Google Scholar]
- 45.Lorenz RT, Parks LW. Regulation of ergosterol biosynthesis and sterol uptake in a sterol-auxotrophic yeast. J Bacteriol. 1987;169(8):3707–3711. doi: 10.1128/jb.169.8.3707-3711.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenz RT, Parks LW. Physiological effects of fenpropimorph on wild-type Saccharomyces cerevisiae and fenpropimorph-resistant mutants. Antimicrob Agents Chemother. 1991;35(8):1532–1537. doi: 10.1128/aac.35.8.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loubbardi A, Marcireau C, Karst F, Guilloton M. Sterol uptake induced by an impairment of pyridoxal phosphate synthesis in Saccharomyces cerevisiae: Cloning and sequencing of the PDX3 gene encoding pyridoxine (pyridoxamine) phosphate oxidase. J Bacteriol. 1995;177(7):1817–1823. doi: 10.1128/jb.177.7.1817-1823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ness F, et al. Sterol uptake in Saccharomyces cerevisiae heme auxotrophic mutants is affected by ergosterol and oleate but not by palmitoleate or by sterol esterification. J Bacteriol. 1998;180(7):1913–1919. doi: 10.1128/jb.180.7.1913-1919.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otero JM, et al. Whole genome sequencing of Saccharomyces cerevisiae: From genotype to phenotype for improved metabolic engineering applications. BMC Genomics. 2010;11:723–2753. doi: 10.1186/1471-2164-11-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinabarger DL, Keesler GA, Parks LW. Regulation by heme of sterol uptake in Saccharomyces cerevisiae. Steroids. 1989;53(3–5):607–623. doi: 10.1016/0039-128x(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 51.Soustre I, Girard P, Karst F. Biosynthesis and transport of sterols in the yeast Saccharomyces cerevisiae. C R Seances Soc Biol Fil. 1998;192(5):977–990. [PubMed] [Google Scholar]
- 52.van den Hazel HB, et al. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J Biol Chem. 1999;274(4):1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- 53.Youings A, Rose AH. Sterol uptake by anaerobically grown Saccharomyces cerevisiae. Yeast. 1989;5(6):S459–S463. [PubMed] [Google Scholar]
- 54.Régnacq M, Alimardani P, El Moudni B, Bergès T. SUT1p interaction with Cyc8p(Ssn6p) relieves hypoxic genes from Cyc8p-Tup1p repression in Saccharomyces cerevisiae. Mol Microbiol. 2001;40(5):1085–1096. doi: 10.1046/j.1365-2958.2001.02450.x. [DOI] [PubMed] [Google Scholar]
- 55.Abramova N, Sertil O, Mehta S, Lowry CV. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J Bacteriol. 2001;183(9):2881–2887. doi: 10.1128/JB.183.9.2881-2887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abramova NE, et al. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics. 2001;157(3):1169–1177. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen BD, Sertil O, Abramova NE, Davies KJ, Lowry CV. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 2001;29(3):799–808. doi: 10.1093/nar/29.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mrsa V, et al. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J Bacteriol. 1999;181(10):3076–3086. doi: 10.1128/jb.181.10.3076-3086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sertil O, Cohen BD, Davies KJ, Lowry CV. The DAN1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene. 1997;192(2):199–205. doi: 10.1016/s0378-1119(97)00028-0. [DOI] [PubMed] [Google Scholar]
- 60.Sertil O, Kapoor R, Cohen BD, Abramova N, Lowry CV. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31(20):5831–5837. doi: 10.1093/nar/gkg792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilcox LJ, et al. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem. 2002;277(36):32466–32472. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- 62.Pijnappel WW, et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15(22):2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yassour M, et al. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11(8):R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shianna KV, Dotson WD, Tove S, Parks LW. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J Bacteriol. 2001;183(3):830–834. doi: 10.1128/JB.183.3.830-834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crowley JH, Leak FW, Jr, Shianna KV, Tove S, Parks LW. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J Bacteriol. 1998;180(16):4177–4183. doi: 10.1128/jb.180.16.4177-4183.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiner S, Micolod D, Schneiter R. Saccharomyces cerevisiae, a model to study sterol uptake and transport in eukaryotes. Biochem Soc Trans. 2005;33(Pt 5):1186–1188. doi: 10.1042/BST20051186. [DOI] [PubMed] [Google Scholar]
- 67.Reiner S, Micolod D, Zellnig G, Schneiter R. A genomewide screen reveals a role of mitochondria in anaerobic uptake of sterols in yeast. Mol Biol Cell. 2006;17(1):90–103. doi: 10.1091/mbc.E05-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Biol. 2011;31(17):3569–3583. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Dijk EL, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475(7354):114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 70.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rogers B, et al. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J Mol Microbiol Biotechnol. 2001;3(2):207–214. [PubMed] [Google Scholar]
- 72.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 73.Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev. 2011;75(2):213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshida Y. Cytochrome P450 of fungi: Primary target for azole antifungal agents. Curr Top Med Mycol. 1988;2:388–418. doi: 10.1007/978-1-4612-3730-3_11. [DOI] [PubMed] [Google Scholar]
- 75.Shah N, Klausner RD. Brefeldin A reversibly inhibits secretion in Saccharomyces cerevisiae. J Biol Chem. 1993;268(8):5345–5348. [PubMed] [Google Scholar]
- 76.Zweytick D, Hrastnik C, Kohlwein SD, Daum G. Biochemical characterization and subcellular localization of the sterol C-24(28) reductase, erg4p, from the yeast saccharomyces cerevisiae. FEBS Lett. 2000;470(1):83–87. doi: 10.1016/s0014-5793(00)01290-4. [DOI] [PubMed] [Google Scholar]
- 77.Bennett MK, Osborne TF. Nutrient regulation of gene expression by the sterol regulatory element binding proteins: Increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc Natl Acad Sci USA. 2000;97(12):6340–6344. doi: 10.1073/pnas.97.12.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies BS, Wang HS, Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: Similar activation/regulatory domains but different response mechanisms. Mol Cell Biol. 2005;25(16):7375–7385. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vik A, Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(19):6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hickman MJ, Winston F. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol Cell Biol. 2007;27(21):7414–7424. doi: 10.1128/MCB.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 82.Nilsson S, Huelsenbeck J, Fritz G. Mevalonate pathway inhibitors affect anticancer drug-induced cell death and DNA damage response of human sarcoma cells. Cancer Lett. 2011;304(1):60–69. doi: 10.1016/j.canlet.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 83.Martirosyan A, Clendening JW, Goard CA, Penn LZ. Lovastatin induces apoptosis of ovarian cancer cells and synergizes with doxorubicin: Potential therapeutic relevance. BMC Cancer. 2010;10:103. doi: 10.1186/1471-2407-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feleszko W, Jakóbisiak M. Lovastatin augments apoptosis induced by chemotherapeutic agents in colon cancer cells. Clin Cancer Res. 2000;6(3):1198–1199. [PubMed] [Google Scholar]
- 85.Fingerman IM, Wu CL, Wilson BD, Briggs SD. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem. 2005;280(31):28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]