Abstract
Thiamine pyrophosphate (TPP)-sensitive mRNA domains are the most prevalent riboswitches known. Despite intensive investigation, the complex ligand recognition and concomitant folding processes in the TPP riboswitch that culminate in the regulation of gene expression remain elusive. Here, we used single-molecule fluorescence resonance energy transfer imaging to probe the folding landscape of the TPP aptamer domain in the absence and presence of magnesium and TPP. To do so, distinct labeling patterns were used to sense the dynamics of the switch helix (P1) and the two sensor arms (P2/P3 and P4/P5) of the aptamer domain. The latter structural elements make interdomain tertiary contacts (L5/P3) that span a region immediately adjacent to the ligand-binding site. In each instance, conformational dynamics of the TPP riboswitch were influenced by ligand binding. The P1 switch helix, formed by the 5′ and 3′ ends of the aptamer domain, adopts a predominantly folded structure in the presence of Mg2+ alone. However, even at saturating concentrations of Mg2+ and TPP, the P1 helix, as well as distal regions surrounding the TPP-binding site, exhibit an unexpected degree of residual dynamics and disperse kinetic behaviors. Such plasticity results in a persistent exchange of the P3/P5 forearms between open and closed configurations that is likely to facilitate entry and exit of the TPP ligand. Correspondingly, we posit that such features of the TPP aptamer domain contribute directly to the mechanism of riboswitch-mediated translational regulation.
Keywords: RNA, site-specific labeling, allosteric effect, structural preorganization, ergodicity
Riboswitch elements located within noncoding regions of mRNA bind metabolites with high selectivity and specificity to mediate control of transcription, translation, or RNA processing (1, 2). In a manner that is controlled by metabolite concentration, nascent mRNAs containing riboswitch domains can enter one of two mutually exclusive folding pathways to impart regulatory control: the outcomes of these folding pathways correspond to ligand-bound or ligand-free states (3–5). These aptamer folds trigger structural cues into the adjoining expression platform, which, in turn, transduce a signal for “on” or “off” gene expression (6–9).
The thiamine pyrophosphate (TPP)-sensing riboswitch is one of the earliest discovered regulatory elements in mRNA that is prevalent among bacteria, archaea, fungi, and plants (10–12). TPP riboswitches, sometimes present in tandem (13, 14), control genes that are involved in the transport or synthesis of thiamine and its phosphorylated derivatives. The TPP-bound aptamer adopts a uniquely folded structure in which one sensor helix arm (P2/P3) forms an intercalation pocket for the pyrimidine moiety of TPP, and the other sensor helix arm (P4/P5) offers a water-lined binding pocket for the pyrophosphate moiety of TPP that engages bivalent metal ions (Fig. 1) (15–17). Like most riboswitch domains, structural information pertaining to the ligand-free TPP riboswitch is relatively lacking. Consequently, little is presently known about the determinants of alternative riboswitch folding pathways and how ligands regulate these events. A deeper understanding of both bound and unbound forms of the aptamer and expression platforms is required to gain mechanistic insights into the regulatory switch that they induce (9, 18–20). The nature and timing of the folding-recognition process in riboswitches imply potentially complex and rapid dynamic processes within the nascent RNA chain (21). Knowledge of these events is therefore critical to achieving a complete understanding of riboswitch-mediated gene regulation.
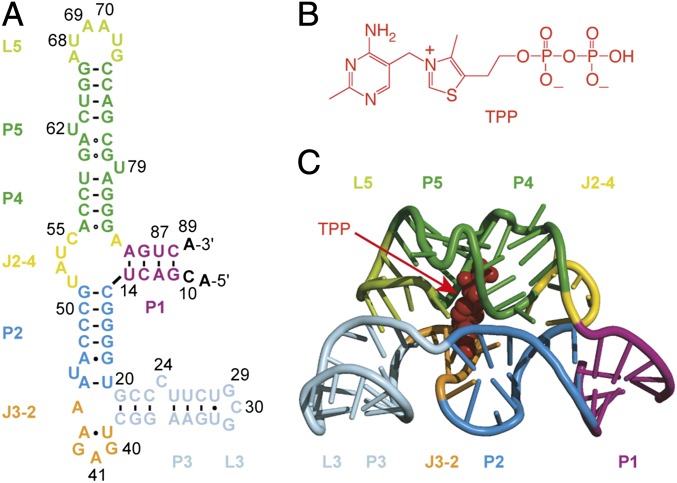
Fig. 1.
E. coli thiM riboswitch aptamer. (A) Secondary structure representation of the native RNA aptamer domain under investigation. (B) Chemical structure of thiamine pyrophosphate (TPP). (C) Cartoon-rendered representation of the RNA aptamer/TPP complex (Protein Data Bank 2GDI) containing the thermodynamically enforced 5-bp stem P1 (15).
Previous investigations have used a battery of distinct biophysical methods to explore the nature of the TPP ligand recognition process. Such studies include 2-aminopurine fold analysis (22), small-angle X-ray scattering (SAXS) (23–25), RNase-detected selective 2′-hydroxyl acylation (26, 27), isothermal titration calorimetry (28, 29), as well as single-molecule optical-trapping methods in which force was applied via the 5′ and 3′ ends of the RNA to directly monitor the energy landscape of TPP riboswitch folding and unfolding (30). Investigations of this kind have provided an important framework for understanding global features of the TPP riboswitch aptamer domain, revealing that its structural compaction is enabled by physiological concentrations of Mg2+ ions and enforced by TPP binding. Two additional, generally agreed upon, features of the TPP riboswitch have been derived from these experiments. First, secondary structures of the P2/P3 and P4/P5 ligand sensor arms form in the presence of Mg2+ alone. Second, tertiary interactions between the two sensor arms (e.g., P3/L5) do not form in the absence of TPP binding. However, it is not yet clear how the collapse of the two helical domains and the formation of these tertiary interactions are influenced by ligand binding or whether they are essential to binding pocket formation. Here, we have used single-molecule fluorescence resonance energy transfer (smFRET) (31–33) imaging to track ligand-dependent changes in the TPP riboswitch from multiple structural perspectives to elucidate the relationship between TPP recognition and aptamer folding.
Results and Discussion
We initiated our smFRET investigations with the aim of sensing the dynamics of switch helix P1 within the minimal TPP aptamer construct derived from the Escherichia coli thiM motif (Fig. 1A). Formation of helix P1 in the TPP aptamer domain is essential for ligand binding (22). In thiM, the 3′-terminal nucleotides of the P1 helix (A85 through C88) function as the anti–anti-Shine–Dalgarno sequence (anti-anti-SD) that can alternatively pair with the anti-SD in the expression platform to release the ribosomal binding site (SD) from a large repressor stem (Fig. S1) to initiate translation (10, 22). Correspondingly, “breathing” of stem P1, which is the thermodynamically weakest stem among all stems (P1–P5) in the TPP aptamer (30), contributes directly to the switching and ligand recognition mechanisms of the TPP riboswitch.
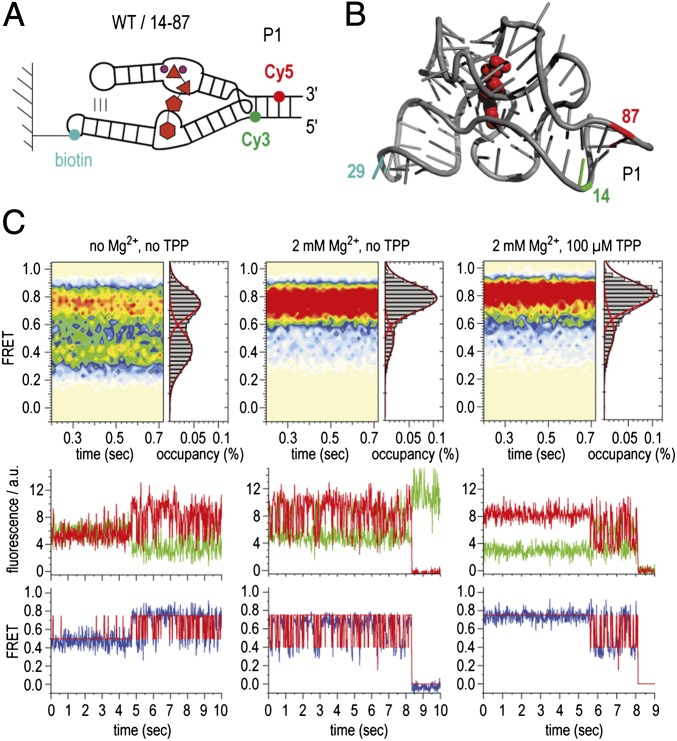
To probe dynamics in this region, donor and acceptor fluorescent probes (Cy3 and Cy5) were covalently linked to positions U14 and U87, respectively, on opposite sides of helix P1 (Fig. 2 A and B). Two RNA molecules were chemically synthesized bearing a 5-aminoallyl uridine modification at each of these positions. Following purification, both RNAs were fluorescently labeled in isolation, followed by enzymatic ligation (34). To enable single-molecule imaging over extended periods using a wide-field total internal reflection fluorescence microscopy setup, biotin was conjugated to position 29 within loop L3, a residue distal to tertiary interactions within the folded aptamer domain (Fig. 2 A and B). Using this approach, dynamics of helix P1 could be tracked over time by recording the emission intensities of both Cy3 and Cy5 fluorophores within hundreds (>500 for each experiment) of individual surface-immobilized molecules simultaneously (35). From these data, fluorescence resonance energy transfer efficiency (FRET) was calculated ratiometrically (ICy5/(ICy5 + ICy3) to reveal estimates of time-dependent changes in distance between the individual fluorophores. Data were obtained at a high signal-to-noise ratio (>5:1) and at an imaging rate of 66 frames per second (15-ms integration time) in both the absence and the presence of ligands under conditions that support extended stabilization of the Cy3 and Cy5 fluorophores (36) (Materials and Methods). Here, τFRET was ∼3.5 s, limited predominantly by Cy5 fluorophore photobleaching. The dynamic behaviors of individual molecules were assessed using hidden Markov modeling procedures (Materials and Methods); ensemble information was obtained by assembling smFRET trajectories into population FRET histograms.
Fig. 2.
Dynamics of switch helix P1 of the TPP aptamer analyzed by smFRET experiments. (A) Schematics of labeling pattern. (B) Positions of labeling in the 3D structure. (C) (Upper) Population FRET histograms showing the mean FRET values and percentage (%) of occupancies of each state observed for the TPP riboswitch in the absence of Mg2+ and TPP ligand (Left), in the presence of 2 mM Mg2+ ions (Center), and in the presence of 2 mM Mg2+ ions and 100 μM TPP (Right). (Lower) Corresponding fluorescence (green, Cy3; red, Cy5) and FRET (blue) trajectories of individual TPP aptamer molecules under the same conditions, where idealization of the data to a two-state Markov chain is shown in red.
smFRET data obtained from the U14/U87-labeled construct are summarized in Fig. 2C. In the absence of Mg2+, a 4:5 distribution of intermediate- (∼0.4) and high-FRET (∼0.75) states, respectively, was observed (Fig. 2C, Upper Left). Visual inspection of individual smFRET recordings revealed that the P1 switch helix was highly dynamic, rapidly exchanging between intermediate- and high-FRET states on a timescale similar to the imaging-time resolution (∼50–100 s−1). Given the estimated positions of the Cy3 and Cy5 fluorophores within the P1 helix, the high-FRET state was attributed to a conformation in which the P1 helix is formed, whereas the intermediate-FRET state likely corresponds to a configuration in which helix P1 is not base-paired. Transitions to zero-FRET states were absent or rare, suggesting that such dynamics occur within the context of neighboring secondary and tertiary structures. Rarely, dispersed kinetic behaviors were observed in which a single molecule, residing predominantly in an intermediate-FRET state, rapidly converted to a predominantly high-FRET state (Fig. 2C, Lower Left). Such observations suggest that conformational changes elsewhere in the riboswitch influence helix P1 stability. As anticipated from these structural assignments, the high-FRET state became predominant in the presence of physiological concentrations of Mg2+ ions (2 mM) (Fig. 2C, Upper Center). Nonetheless, dynamics persisted in the P1 helix with rapid excursions into, and out of, intermediate-FRET configurations on the millisecond timescale (Fig. 2C, Lower Center). Saturating concentrations of TPP (100 µM) (19, 22) further stabilized the P1 helix. Here, TPP’s predominant impact was to reduce residual dynamics in the system, suggesting a further stabilization of the P1 helix (Fig. 2C, Upper Right). However, dynamic states were observed to persist in the presence of both Mg2+ and TPP, as evidenced by sudden transitions of individual molecules into states that were highly dynamic in nature (Fig. 2C, Lower Right). These data suggest that distal conformational changes (e.g., in the binding pocket as well as the L5/P3 interaction) can occur in the context of bound ligands that regulate helix P1 stability.
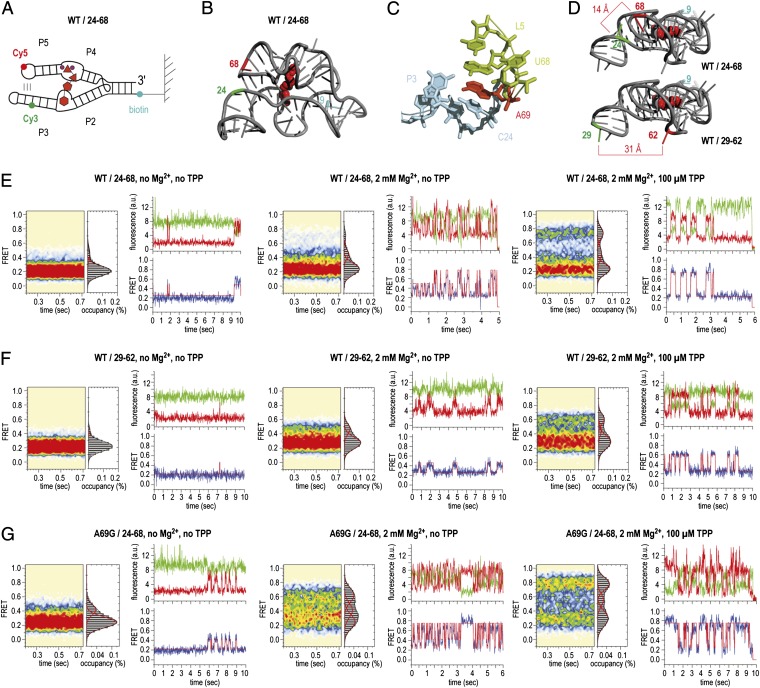
We next set out to investigate aptamer constructs that would report on the dynamics of the forearms of the two ligand-sensor domains, P2/P3 and P4/P5 (Fig. 3A). In the X-ray structure of the TPP-bound aptamer, P2/P3 and P4/P5 surround the TPP ligand, stabilized by a tertiary interaction between P3 and L5 (Fig. 3 B–D). This contact is mediated by nucleotide A69 of L5, which stacks between the neighboring nucleotide A70 and nucleoside C24 of sensor arm P3 (Fig. 3C) (15). We therefore designed a surface immobilization and fluorophore-labeling strategy that would be sensitive to these tertiary contacts. To do so, we functionalized the 2′ hydroxyl group of C24 for Cy3 attachment using a 3-aminopropyl spacer and U68 with a 5-aminoallyl uridine base for Cy5 attachment (SI Materials and Methods). This construct bound TPP with an estimated Kd of about 100 nM (Fig. S2), comparable to the affinity reported for the unmodified aptamer (10, 28, 30).
Fig. 3.
Dynamics of sensor arms P2/P3 and P4/P5 of the TPP aptamer analyzed by smFRET experiments. (A) Schematics of labeling pattern. (B) Positions of labeling in the 3D structure (WT/24–68). (C) Local environment of the interdomain stacking interaction of A69 to C24. (D) Comparison of distances between labeling positions in WT/24–68 and WT/29–62. (E) Population FRET histograms showing the mean FRET values and percentage (%) of occupancies of each state observed for the TPP aptamer and corresponding fluorescence (green, Cy3; red, Cy5) and FRET (blue) trajectories of individual aptamer molecules in the absence of Mg2+ and TPP ligand (Left), in the presence of 2 mM Mg2+ ions (Center), and in the presence of 2-mM Mg2+ ions and 100 μM TPP (Right); labels attached to positions 24 and 68 of the wild-type (WT) TPP aptamer (WT/24-68). (F) Same as E but with Cy3 and Cy5 attached to positions 29 and 62 in forearms P3 and P5 (WT/29–62). (G) Same as E but with an A69G mutant riboswitch (A69G/24–68).
The smFRET data obtained from the C24/U68-labeled RNA are depicted in Fig. 3E. In the absence of Mg2+, a highly populated, low-FRET state (0.2) was observed, flanked by a weakly populated configuration exhibiting intermediate FRET (0.45). From the crystal structure of the closed configuration of the TPP aptamer (15), the distance between the fluorophore attachment sites is estimated to be about 14 Å (Fig. 3D), a distance that should result in a high-FRET signal (>0.8 FRET). Consistent with previous findings (24–28), these data suggest that the sensor arms are largely unable to form tertiary contacts in the absence of ligand.
In the presence of Mg2+, intermediate-FRET configurations also became slightly (∼5%) more pronounced (Fig. 3E, Center). In addition, a high-FRET state (0.75) emerged that was transient in nature (Fig. 3E, Center). Consistent with a structure in which the ligand-sensor arms are compacted toward each other as in the crystal structure (15), the population of molecules occupying the high-FRET state increased substantially (∼25%) upon addition of saturating concentrations of TPP (100 µM). However, in contrast to the SAM-II riboswitch, where pseudoknot collapse was observed to be ∼80% complete in the presence of ligands (37), the fully folded, high-FRET state of the TPP riboswitch plateaued at ∼30% occupancy. Raising the Mg2+ ion concentration further (10 mM) only modestly increased the population of molecules in this configuration.
Here, visual inspection of the smFRET data again revealed evidence of dispersive kinetic behaviors. Two predominant populations were observed: one in which a relatively stable high-FRET state was occupied, whose lifetime was on the order of photobleaching, and one in which a low-FRET state predominated with transient excursions to higher-FRET states. As expected for an ergodic system, individual molecules were observed to display both kinetic signatures before photobleaching, albeit infrequently (Fig. 3E, Right). Together with the observations made while monitoring helix P1 dynamics, these data suggest that conformational changes within the TPP aptamer core and/or sensor arm domains, which occur on a relatively slow timescale, trigger alternate conformations in the TPP aptamer that either promote or disrupt P3/L5 interactions. Residual V1 nuclease cleavage of the L5 backbone under similar conditions provides an independent line of evidence for dynamics in this region (15). In this context, it is noteworthy that nucleotides at the P3/L5 interface are poorly conserved (15).
To further substantiate this finding, a second labeling strategy was designed to probe the relative positions of the two sensor arms of the TPP aptamer and the P3/L5 tertiary interaction. Here, the Cy3 and Cy5 fluorophores were again located within each forearm sequence (Cy3 at position 29 of P5 and Cy5 at position 62 of P3) but spatially distal from the P3/L5 interaction (Fig. 3D). In good agreement with the distances evidenced in the ligand-bound state of folded TPP aptamer (∼30 Å between the dye-attachment points) (15), the absolute values of the FRET configurations observed in this construct (0.2 low FRET; 0.6 high FRET) were altered (Fig. 3F). Importantly, population and kinetic signatures similar to the C24/U68-labeled construct were observed across experimental conditions (Fig. 3F). These data further corroborate the conclusion that residual dynamics between the ligand sensor arms persist in the bound state of the TPP aptamer.
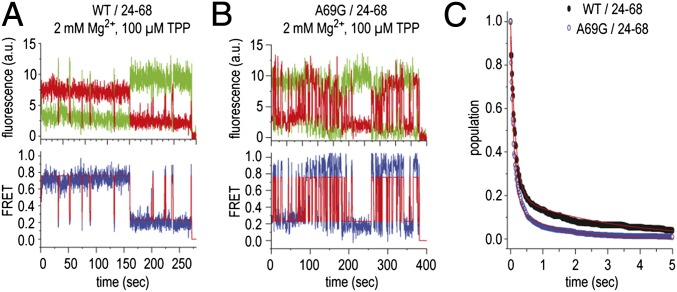
To evaluate the contribution of tertiary interactions between P3 and L5 to the stability of the high-FRET state, we prepared a riboswitch construct in which an A69G mutation was introduced. This mutation was anticipated to reduce ligand-induced stabilization of the fully folded TPP aptamer by about 30% and thereby increase the off rate of the TPP ligand (30). Remarkably, this mutant TPP aptamer displayed substantially increased dynamics in both the absence and presence of Mg2+ and TPP ligands (Fig. 3G). Here, the rates entering high-FRET configurations increased more than approximately threefold. This finding suggests that the A69G mutation globally alters the energies associated with the aptamer folding landscape. Counter to expectation, in the presence of TPP, the A69G construct consequently displayed modestly increased occupancy of the high-FRET state (0.75) (Fig. 3G, Right). To more accurately assess the relative stabilities of the high-FRET state and the contribution of the P3/L5 interaction to this configuration in both the WT and A69G mutant, smFRET data were obtained on both constructs at a lower time resolution (150 ms) to enable the relative stabilities of high-FRET states to be assessed (Fig. 4). Consistent with the P3/L5 tertiary interaction contributing to the stability of the high-FRET state, the A69G mutation reduced the lifetime of the high-FRET state by ∼2.5-fold. However, as the A69G mutation also influenced the folding rates of distal regions of the TPP aptamer (e.g., the rate exiting the low-FRET state), we cannot presently distinguish here whether such effects are direct (e.g., through altered stacking interactions) or allosteric in nature.
Fig. 4.
Comparison of typical smFRET traces for A69G and WT TPP aptamer at low time resolution (150 ms). (A) Fluorescence (green, Cy3; red, Cy5) and FRET (blue) trajectories of an individual WT TPP aptamer molecule, where idealization of the data to a three-state Markov chain is shown in red, observed in the presence of 2 mM Mg2+ ions and 100 μM TPP. (B) Same as A but for the A69G construct. (C) Survival plots showing the bimodal nature of the high-FRET state. Lifetimes for wild-type and A69G constructs were estimated by fitting each distribution to a double exponential decay function and determining the population weighted average of short- (∼100 ms) and long-lived (approximate number of seconds) high-FRET states (WT ∼700 ms; A69G ∼275 ms). For both constructs, the long-lived high-FRET state constituted ∼20% of the dwells observed. The lifetimes of both short- and long-lived dwells were similarly reduced in the A69G context.
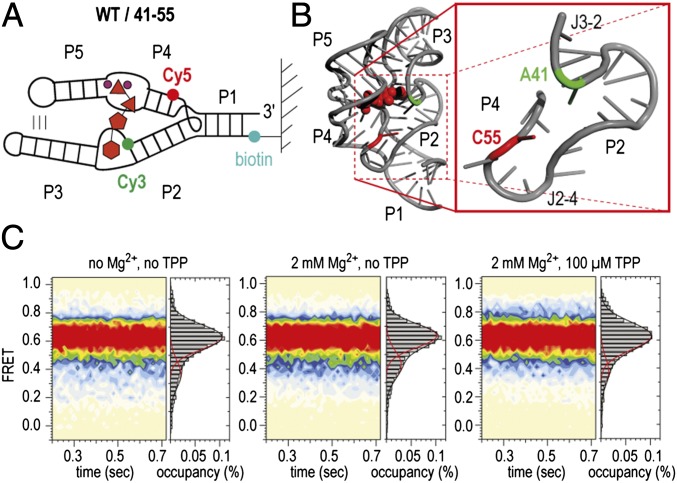
To further explore and understand the complex dynamics of the TPP aptamer, we investigated another labeling pattern that was selected to report on the upper arm (P2/P4) orientation (WT/41–55, Fig. 5A). Strikingly, this construct occupied a stable high-FRET configuration in both the ligand-free and the TPP-bound states. Rare transitions to lower-FRET states were consistent with the persistent dynamics observed in the other constructs examined. Such behaviors can be explained if the three-way junctional region (J3-2, P2, J2-4, P4) is prefolded in the absence of ligand. We consider this observation important for the overall TPP folding model because it implies that a preorganized P1/P2/P4 region serves as a platform for initial TPP recognition, where the P2/P4 elements are significantly less dynamic than the relatively dynamic P3/P5 forearms.
Fig. 5.
Prefolding of P2 and P4 of the TPP aptamer. (A) Schematics of labeling pattern. (B) Positions of labeling in the 3D structure (WT/41–55). (C) Population FRET histograms. Conditions as indicated.
In this context, we note that our results are in accordance with the basic three-state model that was deduced from SAXS measurements, namely compaction in the presence of Mg2+ and further compaction upon TPP binding (23, 25). However, models that were built for the free TPP riboswitch by superposition on the corresponding SAXS density maps favored more elongated conformations between P2 and P4. We speculate that the unexpected dynamics of the TPP system may give rise to SAXS density maps that are difficult to interpret (23).
Collectively, these observations reveal that both the apo- and ligand-bound TPP aptamer exhibit dynamics across a dispersed range of timescales. We interpret this finding in the context of a growing number of studies that report that RNA polymers can display structural transitions and hysteretic behaviors (38). For the group I intron and the hairpin ribozyme, dynamics of this kind have been attributed to conformational “memory” (39) or what has been more recently described as a “persistent ruggedness” of the RNA folding landscape (40). Such complexities imply “hidden” conformational changes in the molecule that have a direct impact on the observed signal. In the case of the TPP riboswitch, switch-like behaviors in dynamics were observed for the constructs designed to monitor formation of helix P1 (Fig. 2) as well as of P3/L5 tertiary contacts (Fig. 3). We hypothesize that such behaviors arise from relatively slow conformational changes [also observed in other riboswitch systems (19, 33)] within the core domain of the TPP aptamer within the J2-4 junction and related distortions in the RNA backbone at the union of the P4 and P5 helices (Fig. 1). In the TPP aptamer crystal structure (15), this site is at the base of the TPP-binding cleft, and alterations in this region are anticipated to propagate in the direction of both the P1 helix and the P3/L5 tertiary contact. This model stipulates that the TPP aptamer follows a multistep folding pathway, a conclusion that is directly supported by evidence to this effect obtained through optical trapping studies of the TPP aptamer domain under force (30). It also suggests that the global stability of the aptamer fold is controlled by both TPP binding as well as hidden and intrinsic conformational events in the system.
Such dynamics allow the aptamer sensor arms to exchange between a relatively open, “Y-shaped” configuration that has yet to be structurally defined and a configuration in line with the TPP aptamer domain crystal structure (15), where the sensor arms fully collapse around the TPP ligand and P3/L5 interactions are formed. Opening of the sensor arms is likely to provide solvent access to the binding pocket and thus a plausible route for TPP entry and exit. This model implies that hinge-like movements of helix P5 relative to P4 (or P3 relative to P2), and the conformational events underpinning this exchange process, directly contribute to the global stability of the aptamer domain. Data obtained on the A69G mutant construct (Fig. 3G) suggest that such processes are influenced at a distance by the L5 loop sequence. Notably, structural processes of this kind may also provide a plausible explanation for the dynamic instability observed in helix P1 in the presence of saturating ligand concentrations (Fig. 2). Conversely, the intrinsic instability of the native helix P1 sequence contained within the TPP aptamer domain investigated here (4 bp) may also contribute allosterically to the dynamics of the P4/P5 junction. For many previous folding and structural investigations of the TPP aptamer domain, helix P1 was either extended or altered (15, 22, 24–28) to promote aptamer stability and compaction. In this view, the inherent instability of helix P1 may contribute directly to the mechanism of TPP riboswitch-mediated translation.
To test the hypothesis that remote parts of the molecule (P1 and L5/P3) are dynamically coupled, we investigated the smFRET behavior of an additional construct with a thermodynamically stabilized (6 bp) stem P1 (WTP1stab/24–68, Fig. 6). Consistent with our proposed model, this RNA exhibited a 15% increased population of the fully folded, high-FRET state compared with the WT counterpart with a native 4-bp stem.
Fig. 6.
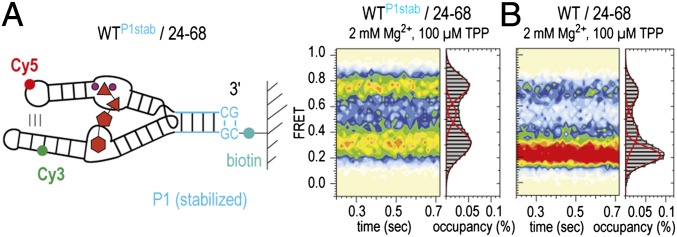
Dynamic coupling of P1 and P3/L5 of the TPP aptamer. (A) Labeling pattern and population FRET histogram of the TPP aptamer with an extended stem P1 (WTP1stab/24–68). (B) Histogram of WT/24–68 for comparison. Conditions as indicated.
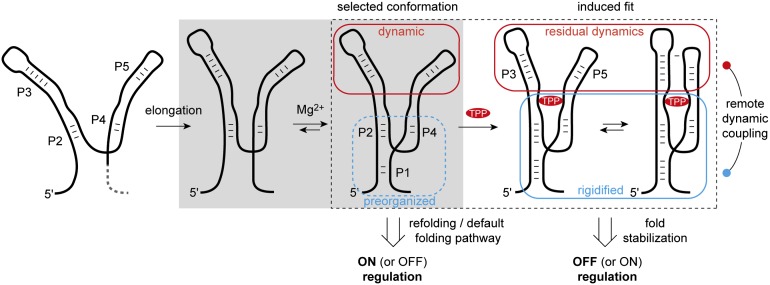
Previous investigations have shown that the process of TPP recognition is contingent on the formation of helix P1 (15, 22). Formation of helix P1 orients stems P2 and P4 into the “Y-shaped” P2/P1/P4 junction, which is relatively rigid at physiological Mg2+ concentrations (Fig. 7). This platform provides an initial interaction module for TPP that is composed of the junction J2-3 and the interface of P4 and P5. TPP binding favors a parallel alignment of the P3 and P5 sensor forearms and the formation of P3/L5 tertiary interactions. These contacts result in further stabilization of the P2/P1/P4 junction, sequestering the anti-anti-SD sequence encoded in the 3′ terminal sequence of the aptamer fold (Fig. S1). In so doing, TPP binding diminishes the probability that the 3′ sequence element of the P1 helix is released to form competing interactions with the anti-SD sequence within the expression platform. As a consequence, TPP binding prevents translation initiation (off-regulation). Conversely, residual dynamics within the TPP aptamer enables TPP to exit its binding site temporally downstream of initial binding and TPP aptamer domain folding. In this context, it is important to note that RNA polymerase has been shown to pause immediately after synthesis of the E. coli thiM TPP aptamer within the expression platform at the sequence surrounding the ribosome-binding site (20). As pausing of this nature would provide time for the equilibration of alternative folds and TPP sensing, the thiM riboswitch, like other riboswitch domains (9, 19, 20, 41), is likely to exhibit aspects of both kinetic and thermodynamic control (9, 42). The observed instability of the TPP aptamer fold and helix P1—in both the absence and the presence of saturating concentrations of the TPP ligand—also suggests that the thiM-mediated regulatory outcome during translation initiation may also be strongly influenced by sequence modifications and environmental variables (temperature and ionic conditions) and the extent to which conformational selection and adaptive—or induced-fit—processes (43–46) contribute to ligand recognition.
Fig. 7.
Proposed model for TPP riboswitch folding and ligand recognition. Gray area: Minimal aptamer motif that preorganizes into a Y-shaped platform (P1/P2/P4) in the presence of Mg2+. Dotted rectangle: Platform representing the conformation that is selected by the ligand TPP and thereby becomes further conformationally adapted. A high population with open forearms (P3, P5) and residual dynamics in the bound state are characteristic of this riboswitch.
Finally, the dynamic behavior of the TPP riboswitch is surprisingly distinct from other junctional riboswitches, such as purine and c-di-GMP riboswitches previously investigated by smFRET methods (32, 33, 47). In those systems, long-range tertiary interactions formed between remote parts of the molecule distant from the actual ligand-binding sites were shown to be stably formed to efficiently interact with the ligand and, in turn, to allow for a mutually exclusive gene response.
Materials and Methods
For chemical solid-phase RNA synthesis, Cy3/Cy5 labeling, enzymatic ligations, and Kd measurements, see SI Materials and Methods.
smFRET data were acquired using a prism-based total internal reflection microscope, where the biotinylated TPP riboswitch was surface-immobilized within PEG-passivated, strepatividin-coated quartz microfluidic devices (35). The Cy3 fluorophore was directly illuminated under 1.5-kW⋅cm−2 intensity at 532 nm (Laser Quantum). Photons emitted from both Cy3 and Cy5 were collected using a 1.2 N.A. 60× Plan-APO water-immersion objective (Nikon), where optical treatments were used to spatially separate Cy3 and Cy5 frequencies onto two synchronized EMCCD devices (Evolve 512; Photometrics). Fluorescence data were acquired using MetaMorph acquisition software (Universal Imaging) at a rate of 66.7 frames per second (15-ms integration). Fluorescence trajectories were selected from the movie files for analysis using automated image analysis software coded in Matlab (MathWorks). Fluorescence trajectories were selected on the basis of the following criteria: a single catastrophic photobleaching event, at least 6:1 signal-to-background noise ratio calculated from the total fluorescence intensity, and a FRET lifetime of at least 30 frames (450 ms) in any FRET state ≥0.15. smFRET trajectories were calculated from the acquired fluorescence data using the formula FRET = ICy5/(ICy3 + ICy5), where ICy3 and ICy5 represent the Cy3 and Cy5 fluorescence intensities, respectively. Equilibrium smFRET experiments were performed in 50 mM potassium 3-(N-morpholino)propanesulfonate (KMOPS), 100 mM KCl, pH 7.5 buffer in the presence of an optimized triplet state quenching mixture and an oxygen-scavenging environment (1 unit protocatchuate-3,4-dioxygenase, 2 mM protocatechuic acid; 1 mM Trolox, 1 mM cyclooctatetraene, 1 mM nitrobenzyl-alcohol) (36). Concentrations of MgCl2 and TPP are as specified in the individual figure captions. FRET state occupancies and transition rates were estimated by idealization to a two- or three-state Markov chain model according to the FRET values obtained for each system by fitting to Gaussian distributions using the segmental k-means algorithm implemented in QuB (48).
Supplementary Material
Acknowledgments
This work was funded by the Austrian Science Foundation (Fonds zur Förderung der wissenschaftlichen Forschung) Projects I1040 and P21641 (R.M.) and M1449 (M.F.S.), the National Science Foundation (Project 1223732; S.C.B.), and the Irma T. Hirschl/Monique Weill Caulier Trust (S.C.B. and R.B.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218062110/-/DCSupplemental.
References
- 1.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43(6):867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garst AD, Edwards AL, Batey RT (2011) Riboswitches: Structures and mechanisms. Cold Spring Harb Perspect Biol 3(6):pii: a003533, 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed]
- 3.Deigan KE, Ferré-D’Amaré AR. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44(12):1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serganov A, Patel DJ. Molecular recognition and function of riboswitches. Curr Opin Struct Biol. 2012;22(3):279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blouin S, Mulhbacher J, Penedo JC, Lafontaine DA. Riboswitches: Ancient and promising genetic regulators. ChemBioChem. 2009;10(3):400–416. doi: 10.1002/cbic.200800593. [DOI] [PubMed] [Google Scholar]
- 6.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29(1):11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA switches: Insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl. 2007;46(8):1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 8.Serganov A. Determination of riboswitch structures: Light at the end of the tunnel? RNA Biol. 2010;7(1):98–103. doi: 10.4161/rna.7.1.10756. [DOI] [PubMed] [Google Scholar]
- 9.Haller A, Soulière MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res. 2011;44(12):1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 10.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419(6910):952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 11.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9(6):644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447(7143):497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 13.Sudarsan N, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314(5797):300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 14.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13(4):573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441(7097):1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312(5777):1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 17.Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14(9):1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state. Wiley Interdiscip Rev RNA. 2012;3(3):369–384. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perdrizet GA, II, Artsimovitch I, Furman R, Sosnick TR, Pan T (2012) Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci USA 109(9):3323–3328. [DOI] [PMC free article] [PubMed]
- 20.Wong TN, Pan T. RNA folding during transcription: Protocols and studies. Methods Enzymol. 2009;468:167–193. doi: 10.1016/S0076-6879(09)68009-5. [DOI] [PubMed] [Google Scholar]
- 21.Al-Hashimi HM, Walter NG. RNA dynamics: It is about time. Curr Opin Struct Biol. 2008;18(3):321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35(16):5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali M, Lipfert J, Seifert S, Herschlag D, Doniach SJ. The ligand-free state of the TPP riboswitch: A partially folded RNA structure. J Mol Biol. 2010;396(1):153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baird NJ, Kulshina N, Ferré-D’Amaré AR. Riboswitch function: Flipping the switch or tuning the dimmer? RNA Biol. 2010;7(3):328–332. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird NJ, Ferré-D’Amaré AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16(3):598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steen K-A, Malhotra A, Weeks KM. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease. J Am Chem Soc. 2010;132(29):9940–9943. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steen K-A, Rice GM, Weeks KM. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J Am Chem Soc. 2012;134(32):13160–13163. doi: 10.1021/ja304027m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulshina N, Edwards TE, Ferré-D’Amaré AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16(1):186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burnouf D, et al. kinITC: A new method for obtaining joint thermodynamic and kinetic data by isothermal titration calorimetry. J Am Chem Soc. 2012;134(1):559–565. doi: 10.1021/ja209057d. [DOI] [PubMed] [Google Scholar]
- 30.Anthony PC, Perez CF, García-García C, Block SM. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci USA. 2012;109(5):1485–1489. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13(8):857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Wood S, Ferré-D’Amaré AR, Rueda D. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem Biol. 2012;7(5):920–927. doi: 10.1021/cb300014u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang K, Micura R. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat Protoc. 2008;3(9):1457–1466. doi: 10.1038/nprot.2008.135. [DOI] [PubMed] [Google Scholar]
- 35.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25(4):505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96(6):2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7(6):393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 38.Frieden C. Actin and tubulin polymerization: The use of kinetic methods to determine mechanism. Annu Rev Biophys Biophys Chem. 1985;14:189–210. doi: 10.1146/annurev.bb.14.060185.001201. [DOI] [PubMed] [Google Scholar]
- 39.Zhuang X, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288(5473):2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 40.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463(7281):681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18(1):49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 42.Garst AD, Batey RT. A switch in time: Detailing the life of a riboswitch. Biochim Biophys Acta. 2009;1789(9–10):584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leulliot N, Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40(27):7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 45.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287(5454):820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 46.Duchardt-Ferner E, et al. Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch. Angew Chem Int Ed Engl. 2010;49(35):6216–6219. doi: 10.1002/anie.201001339. [DOI] [PubMed] [Google Scholar]
- 47.Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49(8):1596–1605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin F, Li L. Model-based fitting of single-channel dwell-time distributions. Biophys J. 2004;87(3):1657–1671. doi: 10.1529/biophysj.103.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.