Abstract
We profiled microRNAs (miRNAs) in cell-free serum and plasma samples from human volunteers using deep sequencing of barcoded small RNA cDNA libraries. By introducing calibrator synthetic oligonucleotides during library preparation, we were able to calculate the total as well as specific concentrations of circulating miRNA. Studying trios of samples from newborn babies and their parents we detected placental-specific miRNA in both maternal and newborn circulations and quantitated the relative contribution of placental miRNAs to the circulating pool of miRNAs. Furthermore, sequence variation in the placental miRNA profiles could be traced to the specific placenta of origin. These deep sequencing profiles, which may serve as a model for tumor or disease detection, allow us to define the repertoire of miRNA abundance in the circulation and potential uses as biomarkers.
Keywords: pregnancy, cancer
The discovery of novel biomarkers holds the promise of early detection and improved prognosis in many neoplastic and infectious diseases (1–4). A particularly promising class of molecular biomarkers are microRNAs (miRNAs) (5–8). miRNAs are short (22 nt), single-stranded, noncoding RNAs that bind messenger RNA (mRNA) in a sequence-specific manner to regulate gene expression (9, 10). miRNAs play an essential role in fundamental biological processes such as differentiation, proliferation, apoptosis and homeostasis (11). Over 500 miRNAs have been identified in humans (12, 13). Single or multiple miRNA precursors are produced in the nucleus through cleavage of longer transcripts by the endoribonuclease III enzyme, Drosha (14, 15). These products then undergo further processing in the cytoplasm by another RNase III endoribonuclease, Dicer.
Alternate processing versions of the predominant miRNA are termed isomiRs and result in mature miRNAs that can vary from the genomic sequence (16). IsomiRs are products of several processes, including modification of the 3′ end by nucleotide additions (typically adenines or uridines), internal editing of nucleotides, and 5′ end deletion/addition resulting from variability in the precise cleavage site by Drosha and Dicer (17). Changes in the relative distribution of isomiRs have been found in different developmental and disease states. For example, nontemplate additions of adenosines to the 3′ ends of miRNAs are highly abundant early in Drosophila development (18); some isomiRs preferentially occur in benign gastric tissue, whereas other isomiRs preferentially occur in gastric tumors (19).
miRNAs have tissue-specific expression patterns and hence are appealing biomarker candidates. miRNAs, including tissue-specific miRNAs, are shed into circulation where they have been detected by sensitive RT-PCR–based amplification and microarray techniques (20, 21). In the area of infectious diseases, viral-specific miRNAs or miRNAs common to a class of microbes can be identified in the blood (22, 23). In cancer, miRNA expression is dysregulated, with some miRNAs normally expressed during embryogenesis being reactivated and other miRNAs with tumor-suppressor activity being down-regulated (24, 25). Expression patterns may serve as a fingerprint of the tumor source and marker of disease aggressiveness (26). However, recent theoretical predictions have suggested that a given tumor would have to reach a diameter of 1 cm, ∼9 y of growth for a typical ovarian cancer, to be detectable using current techniques for detecting circulating biomarkers (27).
To evaluate the potential use of circulating miRNAs as biomarkers of disease, we characterized miRNAs present in circulation using a deep-sequencing approach (28). RNA sequencing allows quantification of miRNAs, identification of isomiRs, and detection of novel miRNA sequences (29). We chose human placenta as a model to study tissue-specific miRNAs in the blood because it is present exclusively during a well defined period, namely pregnancy, is relatively large (400–500 g) and well perfused, receiving 20% of the total maternal cardiac output thereby being more likely to contribute to circulating miRNAs (30). Thus, the placenta could serve as a model for studying solid tumor contribution of miRNA to circulation.
Several pregnancy-related diseases have been associated with changes in the miRNA profile in placenta. Therefore, in addition to serving as a model for solid tumors, detection of placental miRNAs can be used as a biomarker for pregnancy-related disorders. Preeclampsia is a common and potentially serious complication of pregnancy that affects 5–8% of all pregnancies and is characterized by the development of hypertension and proteinuria (31). Although different hypotheses have been put forth, the underlying etiology of preeclampsia is unknown (32). Analysis of miRNAs from formalin-fixed placental specimens of preeclampsia patients has demonstrated altered miRNA profiles (33–35). Deep sequencing of miRNAs obtained from placental tissues suggests that, in the setting of preeclampsia, the isomiR profile is altered with increased 3′ end addition of an adenosine (36). Although these studies provided biological insight, their clinical utility is limited by the necessity of obtaining placental tissue after delivery. Attempts to detect plasma miRNAs in maternal plasma have thus far been limited to microarray and quantitative RT-PCR approaches (28, 37). Detecting placental miRNAs by deep sequencing could direct specific noninvasive diagnosis of these diseases of pregnancy.
To profile circulating miRNA, we developed a nanogram-scale small RNA deep-sequencing method with sample multiplexing. We applied this method on plasma of men and nonpregnant women as well as pregnant women’s plasma, the umbilical cord blood of their offspring, and their placentas. This approach allowed comprehensive characterization of the plasma miRNA constituents and determination of the tissue-specific contribution from placenta.
Results
Sample Statistics.
A summary of the collected specimens, total RNA, and miRNA content is shown in Table 1. A full description of individual sample characteristics and small RNA annotation data are provided in SI Appendix, Table S1. Median total RNA recovery was 18 ng/mL of maternal plasma [interquartile range (IQR) 14–20], 45 ng/mL of fetal plasma (IQR 34–91), 15 ng/mL of paternal plasma (IQR 13–18), and 23 ng/mL in nonpregnant women’s sera or plasma (IQR 22–28). Two fetal plasma samples had outlying RNA content; 500 and 1,250 ng/mL, possibly explained by cell lysis and release of RNA from the placenta or collection of umbilical cord blood close to the placenta resulting in capture of plasma containing a higher concentration of RNA.
Table 1.
Study sample characteristics
| Samples |
Extraction statistics |
miRNA |
|||||||
| Group | n | Statistic, percentile | RNA content, ng/mL plasma | Total reads | Unique reads | Reads | % of total reads | Plasma concentration, fM | fmol/mcg of RNA |
| Placenta | 8 | 25th | NA | 2,584,217 | 113,079 | 2,271,615 | 94.4 | NA | 28.1 |
| 50th | NA | 3,213,653 | 127,909 | 2,866,397 | 95.3 | NA | 37.2 | ||
| 75th | NA | 4,320,407 | 150,472 | 3,906,444 | 95.5 | NA | 45.6 | ||
| Mothers' plasma | 7 | 25th | 14 | 893,796 | 173,900 | 580,484 | 50.7 | 219 | 4.7 |
| 50th | 18 | 1,461,557 | 242,325 | 1,144,456 | 68.7 | 563 | 9.3 | ||
| 75th | 20 | 3,633,998 | 588,876 | 1,432,651 | 75.4 | 844 | 17.1 | ||
| Fetal plasma | 10 | 25th | 34 | 1,418,002 | 117,158 | 1,113,805 | 82.2 | 1,359 | 14.6 |
| 50th | 45 | 2,998,968 | 144,820 | 1,619,137 | 87.5 | 2,406 | 24.8 | ||
| 75th | 91 | 7,491,092 | 377,490 | 4,081,198 | 92.2 | 8,188 | 35.0 | ||
| Fathers' plasma | 5 | 25th | 13 | 1,490,566 | 280,408 | 981,143 | 52.5 | 250 | 7.3 |
| 50th | 15 | 2,178,003 | 362,306 | 1,122,571 | 56.1 | 250 | 8.9 | ||
| 75th | 18 | 3,074,738 | 514,689 | 1,407,053 | 62.5 | 500 | 11.5 | ||
| Control plasma* | 5 | 25th | 23 | 1,289,020 | 267,413 | 128,434 | 13.1 | 63 | 0.6 |
| 50th | 23 | 1,305,137 | 283,307 | 149,013 | 17.7 | 63 | 0.9 | ||
| 75th | 28 | 2,045,575 | 329,169 | 194,526 | 66.2 | 375 | 7.4 | ||
Tissue source and RNA content of samples. Total RNA concentrations were determined by Qubit fluorometric dye assay (and 260 nm spectroscopy for placental samples), whereas miRNA content was calculated by comparing counts to the known amount of synthetic calibrator oligonucleotides that were sequenced concomitantly. Complete concentration and read count data for individual samples are provided in SI Appendix, Table S1.
*Among nonpregnant control women, two samples were plasma and three samples were sera.
Read count per sample, excluding reads corresponding to the calibrator oligoribonucleotides, was 2.1 million (IQR 1.2–3.7 million). Median percentage of small RNA reads corresponding to miRNA was higher in placental tissue samples compared with maternal plasma (95% vs. 69%, P < 0.0001), paternal plasma (56%, P = 0.002), nonpregnant women’s plasma or sera (18%, P = 0.002) and fetal plasma (87%, P = 0.043) samples. Lower relative miRNA content was accompanied by higher rates of nonmapping RNA sequences in nonplacental samples. Plasma samples had low input of total RNA and, therefore, a higher incidence of adapter–adapter byproducts that are classified as unmappable reads. These byproducts were filtered out from further analysis. The higher amount of input total RNA from placenta tissue compared with blood samples (400-fold) likely resulted in fewer adapter-derived byproducts and lower incidence of unmapped reads.
In terms of absolute abundance, median miRNA recovery (Table 1) was 37 fmol/µg of total RNA in placenta (IQR 28–46), 9 fmol/µg of total RNA in maternal plasma (IQR 5–17), 25 fmol/µg in fetal plasma (IQR 15–35), 9 fmol/µg (IQR 7–12) in paternal plasma and 1 fmol/µg (IQR 1–7) in nonpregnant women’s plasma or sera. Correspondingly, median miRNA concentrations were 563 fM in maternal plasma, 2.4 pM in fetal plasma, 250 fM in paternal plasma, and 63 fM in nonpregnant women’s plasma or sera.
Identification of Plasma miRNA Clusters Specific to Placenta.
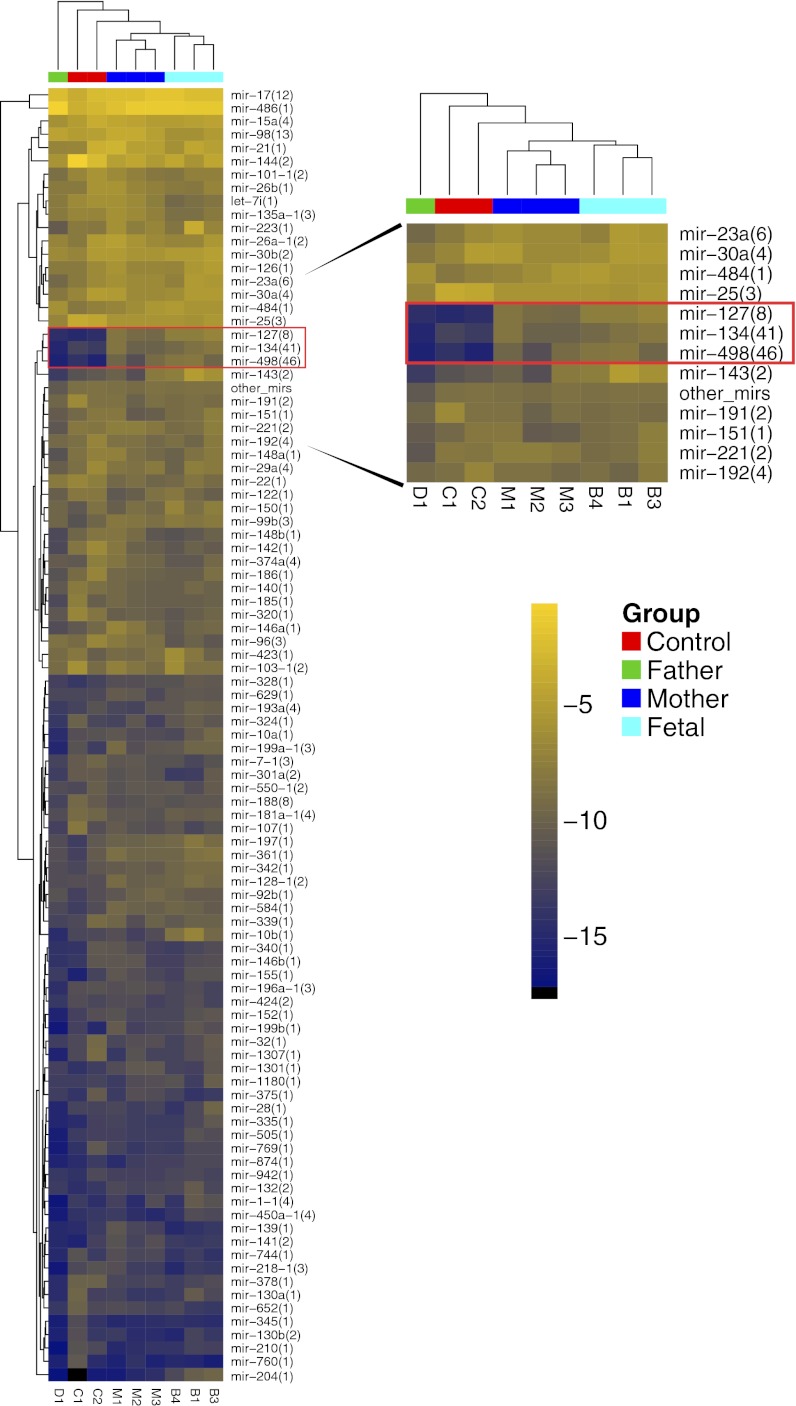
miRNA precursor clusters represent groups of miRNA processed from a single cistron-like pri-miRNA transcript (12). To identify miRNA precursor clusters specific to pregnancy, we performed unsupervised hierarchical clustering of miRNAs isolated from the plasma of three mothers, their umbilical cord plasma samples, as well as one father and two nonpregnant women (Fig. 1, SI Appendix, Fig. S1, and Dataset S1). Relative levels of miRNA cluster mir-498(46) [where “mir-498” denotes the miRNA cistronic cluster and “(46)” the number of cistron members] and, to a lesser extent, mir-127(8) and mir-134(41), were high in placental tissue and in samples from umbilical cord and mothers but were very low in nonpregnant women and fathers (Fig. 1), a finding consistent with what is known about mir-498 representing a cluster of placenta-specific miRNAs in primates (38). Unsupervised sample clustering also revealed a close association within each group of samples. Umbilical cord miRNA profiles were most closely related to the maternal samples. Tissue expression profiling demonstrated that, although mir-127(8) and mir-134(41) were expressed in liver and kidney, mir-498(46) expression was unique to placenta (SI Appendix, Fig. S2).
Fig. 1.
Mir-498, -127, and -134 clusters are pregnancy-associated. Heatmap of hierarchically clustered plasma samples (columns) and miRNA precursor groups (rows). High relative read frequency (log2-transformed) is indicated by bright yellow shades, low frequencies are shown in dark blue. Corresponding numerical data are presented in Dataset S1.
To examine relatedness between placental miRNA profiles and plasma profiles of mothers and fetuses, we compared RNA from plasma collected from mother, father, and umbilical cord trios to RNA from biopsy samples of the corresponding placentas. miRNA abundance was based on normalized read frequency, where the denominator for normalization was the total number of miRNA reads. Distance mapping based on complete miRNA profiles correctly segregated placentas, maternal plasma, paternal plasma, and fetal plasma samples (Fig. 2 and SI Appendix, Fig. S3). Similarity was closest between placenta and fetal plasma, followed by placenta and maternal plasma. Paternal and nonpregnant women’s plasma samples were most distantly situated. In this trio-based set of samples, 36.7% of all miRNA reads obtained from the placentas were members of the mir-498(46) cluster, and this cluster comprised 0.20% and 1.26% of all reads in maternal and fetal plasma, respectively. In contrast, in paternal plasma, only 0.007% of all miRNA reads were members of the mir-498(46) cluster. Thus, compared with paternal plasma, mir-498(46) members are 5,240-fold more abundant in the placenta, 28-fold more abundant in maternal plasma, and 180-fold more abundant in fetal plasma, in accordance with placental-specific expression of this cluster. This finding implies transmission of placental miRNAs into fetal and maternal circulation.
Fig. 2.
Placental, fetal, and maternal miRNA profiles are closely related to each other but distantly related to nonpregnant women and paternal samples. Distance mapping of miRNA profiles using Euclidean distance metric of placental biopsy (P) and control plasma (C), paternal plasma (D), maternal plasma (M), and fetal plasma (B) samples is shown. Red color represents a close distance, whereas yellow indicates greater distance. Numeric distance values are shown in SI Appendix, Fig. S3.
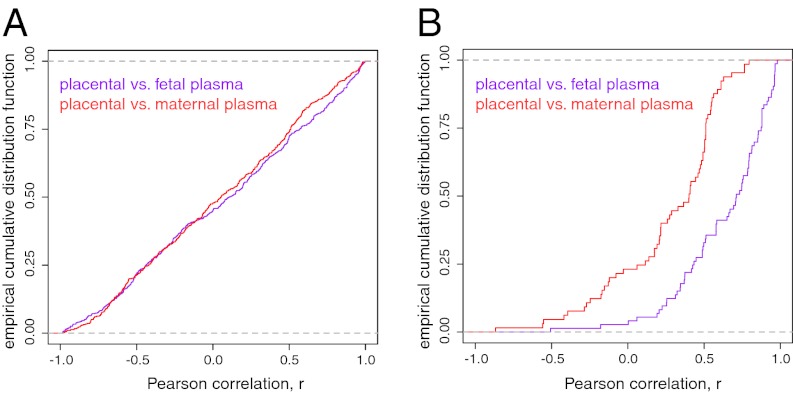
Individual examination of all mature and star miRNA, and how their corresponding read numbers correlate between placental and plasma samples, revealed uniformly distributed correlation coefficients (Fig. 3A), suggesting that among the totality of miRNAs there was no linkage between placentas and corresponding plasma miRNAs. In contrast, limiting the analysis to mir-498(46) cluster members exposed a predominance of positive correlations between maternal plasma or fetal plasma and placental miRNA counts (Fig. 3B). Therefore, the abundance of reads annotating to members of the mir-498(46) cluster in plasma samples reflects the specific placenta from which they originate.
Fig. 3.
Mir-498 cluster reads expose association between maternal and fetal plasma miRNA and the specific placenta from which it originated. Pearson correlation of all mature miRNAs (A) and mir-498 miRNA(46) members (B) between placental and maternal (red) or fetal (purple) plasma samples.
Distribution of Placental-Specific miRNA IsomiRs.
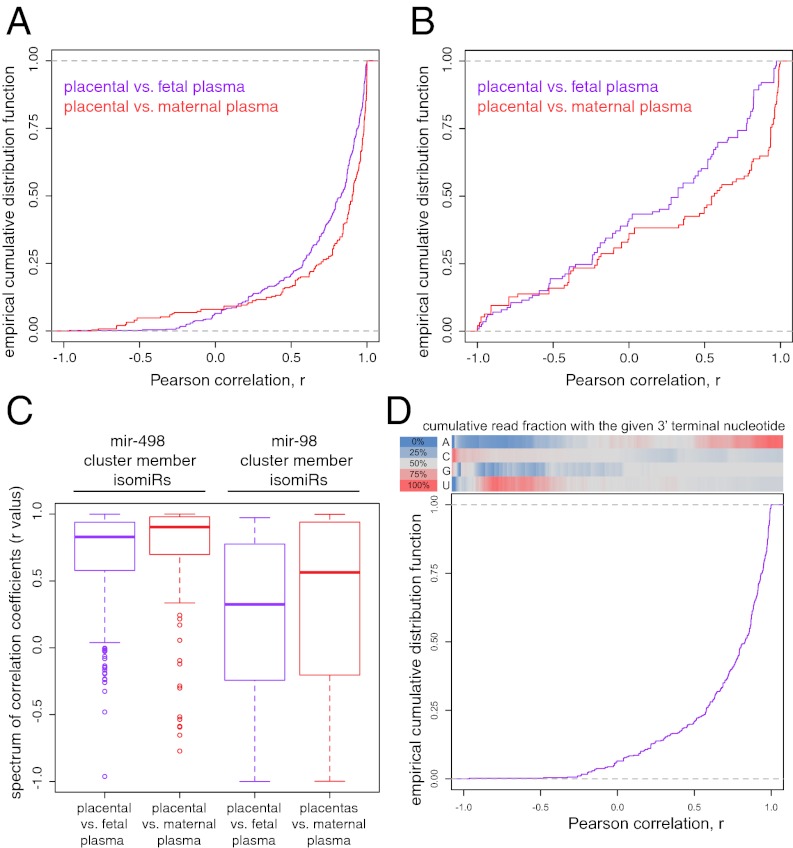
The miRNA cluster mir-498 codes for 46 distinct miRNAs. Individual miRNAs within the mir-498(46) cluster can be differentially excised and processed at different efficiencies to produce different levels of the constituent miRNAs. Posttranscriptional processing also results in isomiRs of variable length and sequence. We examined the count correlation of the most abundant isomiRs of mir-498 members in placenta (∼800 unique sequences) and fetal and maternal plasma samples (Fig. 4A). Correlations between placenta and matching plasma were strong, with over 80% of comparisons having r-values >0.6 for both mothers and fetuses. This distribution revealed that the isomiR fingerprint within plasma reflected the placenta from which it originated. For perspective, we examined the variant distribution of the miR-98(13) cluster, a collection of multicopy and polycistronic miRNA clusters (miR-125∼let-7∼miR-99/100). In comparing placentas to fetuses, placentas to mothers, and mothers to fetuses, both the range and median values of the spectrum of correlation coefficients (r) were poor in the mir-98 cluster compared with the mir-498 cluster, reflecting the shared origin of the mir-498 cluster member isomiRs (Fig. 4 B and C). Further investigation of the basis of higher correlation among mir-498 cluster variants revealed that a different degree of 3′ end adenylation of miRNA in placentas, previously suggested to link with pregnancy complications, was captured in respective plasma samples (Fig. 4D).
Fig. 4.
IsomiRs of mir-498 members, but not mir-98 (miR-125-let-7-miR-99/100) members, uniquely fingerprint the placenta of origin. Pearson correlation of mir-498 member isomiR counts (A) and mir-98 member isomiR counts (B) between placentas and maternal (red) or fetal (purple) plasma samples. (C) Box plots summarizing the distribution of Pearson correlations of miR-498 and miR-98 counts. (D) The distribution of Pearson correlations between placental and fetal miR-498 member isomiR reads with reference to 3′ terminal nucleotide (banner) showing that correlation is dependent on the identity of the 3′ terminal nucleotide. The color heatmaps in the banner depict the cumulative read fraction with each 3′ terminal nucleotide along the spectrum of Pearson correlations. Terminal As and Us represented predominantly nontemplated additions. Thus, the highly correlating reads are characterized by a terminal A, whereas the less well correlating reads terminate with G, C, or U.
Cluster Member Distribution in Tissue Compared with Plasma.
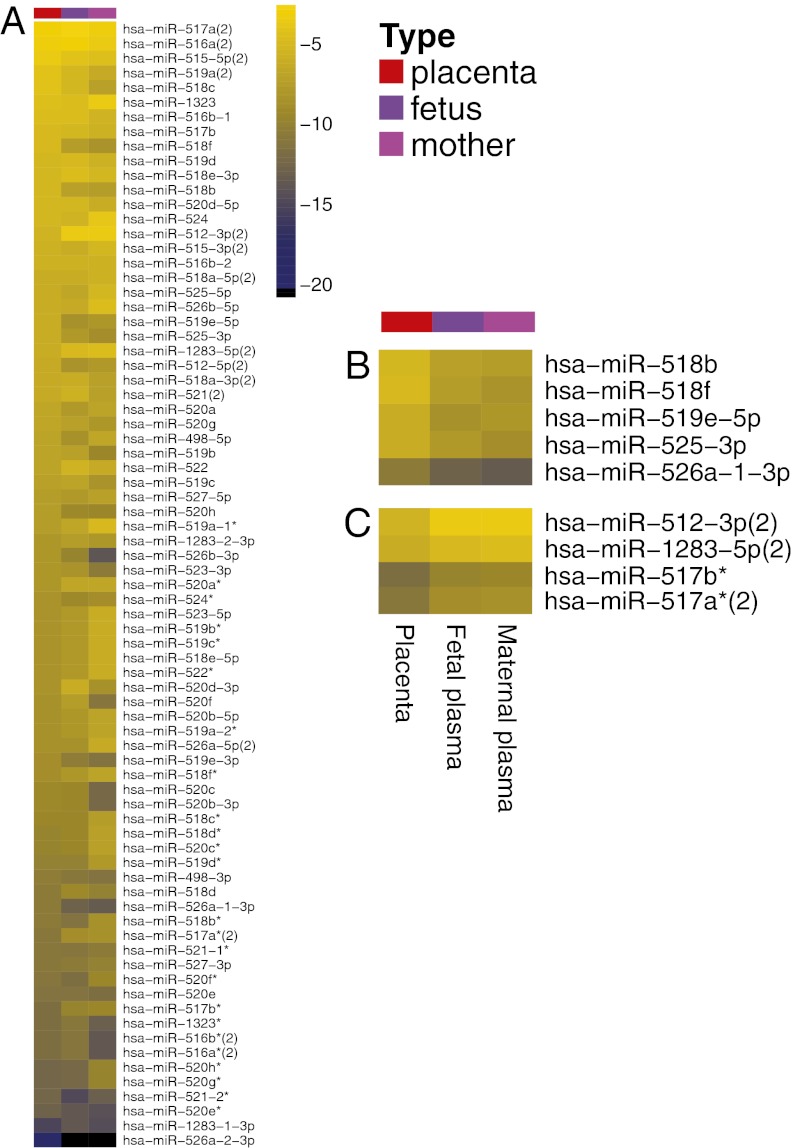
To determine whether miRNA release into plasma, stability in plasma, and clearance from plasma is a sequence-biased or unbiased process, we compared the relative abundance of members of cluster mir-498 in placenta to that of plasma. Fig. 5 shows the read counts for all mir-498 members in the different study groups, normalized to the total number of reads mapping to mir-498(46) (Fig. 5A). Differences were seen in the relative levels of mir-498 cluster members between tissue and plasma samples, and between plasma samples obtained from fetal blood and maternal blood. Highlighted are several mir-498 cluster members that are preferentially abundant in tissue (Fig. 5B) and plasma (Fig. 5C). This finding suggests that factors other than biogenesis rates affect the levels of circulating miRNA (e.g., selective secretion and stability), although we cannot exclude the possibility that placental miRNAs in circulation are reflective of the placental tissue closest to the vasculature. Irrespective of the mechanism, these findings should be taken into account when attempting to capture changes in tissue miRNA by studying their circulating counterparts.
Fig. 5.
miRNA cluster members have a different distribution in plasma compared with tissue. (A) Heatmap of mir-498 cluster member read counts in pooled study samples. Read counts were pooled from placenta biopsies (red), fetal plasma (purple), and maternal plasma (magenta), and the relative contribution of each cluster member to the total reads in the cluster was calculated. Subsets of miRNA showing relative enrichment (B) or depletion (C) among cluster members in placenta vs. plasma are depicted separately. Corresponding numerical data are presented in Dataset S1.
Discussion
In this study, we profiled circulating miRNA from blood samples of human volunteers and compared expression levels to that of tissue. An overview inspection of the circulating miRNA profiles revealed that the vast majority of circulating miRNA molecules originate from blood components and endothelial cells. However, tissue-specific miRNAs, for example, from liver and gut, were represented as well, indicating a broad source of tissue contribution to total circulating miRNAs and confirming prior observations (39, 40). During a state of pregnancy, placental miRNAs were also among the more highly detected miRNAs. In contrast, skeletal muscle-, heart-, and most brain-specific miRNA are present at very low levels in healthy plasma, affording a wide dynamic range for potential biomarker applications (SI Appendix, Fig. S4).
The 10 most abundant miRNAs in plasma were miR-451-DICER1, miR-486, miR-92a(2), miR-16(2), let-7b, miR-21, miR-19b(2), miR-25, miR-22, and miR-144. miR-451, a Dicer-independent miRNA, is the most abundant miRNA within red blood cells (RBC), comprising ∼50% of the miRNA content in these cells (SI Appendix, Fig. S5). Its presence in plasma thus likely represents leakage from RBC or their progenitors, in which it regulates erythroid maturation (41). The second most abundant miRNA in plasma is miR-486, accounting for 13% (IQR 7–30%) of all miRNA (42, 43). It is transcribed from an intron in the ankyrin 1 gene (ANK1) expressed in endothelial cells, erythrocyte precursors, and skeletal muscle. It is therefore expected to be detected in plasma, but its plasma levels are higher than other miRNAs that are more abundant in erythrocytes and endothelial cells (e.g., miR-16, miR-144, let-7f), suggesting selective secretion to plasma or increased stability in plasma. Further investigation will be required to elucidate the precise explanation for the vast abundance of this miRNA and the difference in relative abundance seen with this and other miRNA clusters.
The full-term placenta weighs ∼450 g (30). Based on relative read frequencies of our spike-in calibrators, we estimate that the full-term placenta contains 17 nmol of total miRNA and 6 nmol of mir-498(46) members collectively, whereas the circulation of a woman at full term (assuming pregnancy plasma volume of 3.7 L) contains 2.1 pmol total miRNAs and 4.2 fmol of mir-498(46). Thus, in a woman carrying a full-term pregnancy, there is 1.4 × 106-fold more placental-specific miRNA in her placenta than in her circulation.
We can view the placenta as a model for a solid tumor and members of cluster mir-486 as a model for a tumor-specific miRNA. In our experiments, 1 mg of placenta contains ∼2 × 108 molecules of an individual member of the mir-486 cluster and results in 0.03 molecules of an individual member of mir-486 cluster per mL of plasma. At the technical limits of current day approaches, highly optimized RT-PCR has been able to detect miRNA at a concentration of ∼10 copies per ml. Assuming that a tumor sheds miRNA at similar rate as placenta and that the abundance of the index miRNA is similar to that of a miR-498 member in placenta, a tumor would have to reach a mass of ∼0.3 g to produce sufficient quantities of a tumor-specific miRNA to be theoretically capable of detection by processing 1 mL of plasma and performing RT-PCR. This estimate is similar to the results of mathematical modeling by Hori et al. predicting that an ovarian tumor would have to grow for ∼8.8 y and reach a size of 610 m3 (0.6 g) to reach the threshold for detection (27). Both our experimental estimates and the mathematical modeling of Hori et al. assume that the biomarker is both unique to the pathology in question and abundant in the source tissue. Techniques allowing capture of more miRNAs from circulation would improve the sensitivity and feasibility of using miRNAs as screening tools. Circulating miRNAs may also prove useful in nononcological setting in which the abundance is higher, such as in Duchenne muscular dystrophy patients (44).
Ever since the initial discovery of circulating miRNAs, there has been a question of whether they may exert a hormone-like effect on target tissue (45). Our data argue against this view. The plasma concentration of total miRNA was found to be in the 100 fM range, and any single miRNA was present as a fraction of that concentration. In contrast, steroid hormones such as testosterone, progesterone, and estrogen are present at nM concentrations, whereas even the most trace peptide hormones such as parathyroid hormone and adrenocorticotropic hormone are present in the pM range. Moreover, although typical hormones bind receptors that amplify their signals, miRNAs act on a 1:1 basis with their mRNA targets and are thought to require intracellular levels of greater than ∼1,000 copies per cell to exert measureable activity (38, 46). Thus, in the absence of a known and highly sensitive miRNA receptor, it appears unlikely that miRNAs can function as hormones.
With our methods, placental miRNA fingerprints are sufficiently unique to enable matching of a plasma sample from either the fetus or mother to the specific placenta from which they originated. Moreover, deep sequencing of the plasma miRNAs allows detection of modifications to the miRNA that, in terms of relative frequency, are also specific to the placenta from which they originated. Applying barcoded adapters allows us to multiplex 20 samples in a single sequencing reaction, thereby dramatically lowering the cost (∼75 USD per sample for reagents and sequencing at 5 million reads per sample plus ∼75 USD per sample for labor), limiting batch effects, and improving the prospects for clinical research utility.
Within our pool of samples from healthy volunteers, variability was observed between samples in the relative levels of miRNA and their isomiRs. We were able to use this variability to match the relative miRNA levels and isomiR fingerprints in plasma to the source placenta, suggesting that clinically meaningful changes, such as those observed in preeclampsia, can be detectable in plasma. Therefore, miRNA biomarkers may not only be assessed for the relative abundance of specific miRNAs, but differences in the relative isomiR abundance (or even the presence of a unique isomiR) may also be clinically useful.
Collectively, our results suggest that deep sequencing of barcoded small RNA cDNA libraries from maternal plasma is a robust and relatively inexpensive assay to detect and distinguish tissue- and condition-unique miRNA fingerprints. Further studies will be required using larger sample sizes to determine whether the miRNA profiles from placentas of women with pregnancy complications match the profile seen in maternal plasma and the degree to which a miRNA profile “fingerprint” exists for specific diseases of the placenta. Placental miRNAs in maternal plasma may, in the latter case, be used for noninvasively detecting changes indicative of placental disease in a timely manner so as to allow for closer monitoring or intervention. It will also be of interest to determine whether the altered miRNAs are merely a marker for the placental disease or may participate in its pathogenesis.
Materials and Methods
Patient Samples.
EDTA-preserved peripheral blood samples (3 mL) were collected (BD Vacutainer, K2 EDTA, Ref 367856) from pregnant women presenting to the labor floor either for scheduled cesarean section or in labor. Blood samples were concomitantly collected from their respective male partners and, following delivery, from umbilical cord blood (fetal blood). In some cases, a 2-cm section of placenta was collected as well. RNA was isolated from 7 venous blood samples of mothers, 5 venous blood samples from fathers, 10 umbilical cord blood samples (“fetus”), 8 placental biopsy samples, and 5 venous blood samples (3 without anticoagulant, i.e., sera) from nonpregnant women (controls). Blood and placental samples were immediately placed at 4 °C and were processed within 3 h of collection. Blood samples from fathers were selected as controls because processing times and sample conditions could be matched to the maternal and fetal samples. Informed consent was obtained before collecting samples. All aspects of this study were reviewed and approved by the institutional review boards at the Weill-Cornell Medical Center and The Rockefeller University Hospital.
RNA Isolation.
Total RNA isolation from placenta biopsy samples was performed with the miRNeasy mini kit (Qiagen) following lysis and homogenization in TRIzol (Invitrogen). Processing of 50 mg of placental tissue with 1 mL of TRIzol yielded ∼56 µg of RNA, as determined by 260-nm spectrometry. For isolation of total RNA from peripheral blood, samples were centrifuged at 500 × g (Sorvall Legend RT Rotor 75006441K) for 5 min. Next, 500 µL of plasma (or serum) supernatants were removed, placed in 1.5 mL Eppendorf tubes and centrifuged again at 16,060 × g (Sorvall Biofuge fresco) for 5 min to pellet residual cells and debris. A total of 400 µL of supernatant was then transferred to 2-mL Eppendorf tubes. TRIzol LS (1.2 mL) was added and vortexed in each sample. Chloroform (320 µL) was added and mixed. Total RNA was purified using the miRNeasy kit by applying the aqueous phase following 15 min centrifugation at 12,000 × g (Sorvall Biofuge fresco) at 4 °C. Total RNA was eluted with 50 µL of water into siliconized (low retention) microcentrifuge tubes (G-Tube Siliconized, Bio Plastic), dried using speed-vacuum, and resuspended in 11 µL of water. One or 2 µL was used to quantify total RNA using Qubit fluorometric dye assay (Invitrogen), which correlated well with quantities reported by picogram-scale capillary electrophoresis (Agilent Bioanalyzer).
cDNA Library Preparation, Sequencing, and Data Analyses.
A detailed description of the methods can be found in SI Appendix (47). Briefly, a barcoded small RNA cDNA library was prepared from placenta-extracted RNA as described, with 2 µg of total RNA per sample (29). Conversely, 5 ng of total RNA from each plasma or serum sample were used for small RNA cDNA library preparation. When less than 5 ng were obtained, we used the available amount, taking record for subsequent calculation of quantities. Barcoded 3′-adapters were ligated to RNA in each sample. The 3′-adapter ligation reaction was based on our published method (48), with modifications that adapt to low input RNA quantities: the final concentrations of 3′ adenylated adapters (5 µM) and mutated Rnl2 ligase (0.05 µg/µL) were unchanged; radiolabeled size markers were omitted from the reaction (size identification on subsequent gel purification steps relied on an independently ligated sample of size markers); and a synthetic calibrator mixture consisting of 10 22-nt oligoribonucleotides was added, with sequences noncognate to the human miRNA transcriptome, at a final amount of 0.25 fmol for each of the calibrator oligonucleotides per 1 µg total RNA from the biological sample (0.0125 fmol pooled amount per 3′ ligation reaction on 5 ng total RNA). This mixture allowed absolute quantification of miRNA content in samples within the previously validated molar range (48). Subsequent pooled library preparation steps were performed as described (48). Amplified cDNA libraries underwent 50 cycles of single-end sequencing by synthesis (Illumina GA II or HiSEq. 2000). We used an in-house computer pipeline to extract barcodes, align to the genome (human genome assembly Hg19) and assign small RNA annotations to the reads. miRNA abundance was determined as the sum of all reads with up to two annotation mismatches (49). In-house curated definitions were used for miRNA annotation as well as sequence family and precursor cluster grouping (50). Summary data are expressed as median and IQR unless otherwise stated. Numerical data for data presented in heatmaps is provided in Dataset S1.
Supplementary Material
Acknowledgments
We thank members of the T.T. laboratory for helpful discussions and advice. This work was supported by the Howard Hughes Medical Institute (T.T.), National Institutes of Health Grant HD068546 and the Center for Reproductive Medicine and Infertility (to Z.W.), and also in part by National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health Grant 8UL1TR000043 (I.Z.B.-D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214046110/-/DCSupplemental.
References
- 1.Etzioni R, et al. The case for early detection. Nat Rev Cancer. 2003;3(4):243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone A, Krapcho M, Neyman N. SEER Cancer Statistics Review, 1975–2008. Bethesda: National Cancer Institute; 2011. [Google Scholar]
- 3.Homsi J, Kashani-Sabet M, Messina JL, Daud A. Cutaneous melanoma: Prognostic factors. Cancer Contr. 2005;12(4):223–229. doi: 10.1177/107327480501200403. [DOI] [PubMed] [Google Scholar]
- 4.van de Beek D, de Gans J, Tunkel AR, Wijdicks EFM. Community-acquired bacterial meningitis in adults. N Engl J Med. 2006;354(1):44–53. doi: 10.1056/NEJMra052116. [DOI] [PubMed] [Google Scholar]
- 5.Gilad S, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 7.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3(11):e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10):2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Farazi TA, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 16.Morin RD, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. RNA. 2010;16(10):1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S-C, et al. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics. 2012;13(Suppl 1):S13. doi: 10.1186/1471-2164-13-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat Med. 2005;11(7):712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- 22.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pegtel DM, van de Garde MDB, Middeldorp JM. Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim Biophys Acta. 2011;1809(11-12):715–721. doi: 10.1016/j.bbagrm.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 25.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 26.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11(9):644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 27.Hori SS, Gambhir SS. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med. 2011;3:109–116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chim SSC, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54(3):482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 29.Hafner M, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44(1):3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aherne W, Dunnill MS. Morphometry of the human placenta. Br Med Bull. 1966;22(1):5–8. doi: 10.1093/oxfordjournals.bmb.a070437. [DOI] [PubMed] [Google Scholar]
- 31.ACOG Committee on Practice Bulletins--Obstetrics ACOG practice bulletin. Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. Obstet Gynecol. 2002;99(1):159–167. doi: 10.1016/s0029-7844(01)01747-1. [DOI] [PubMed] [Google Scholar]
- 32.Cunningham F, et al. Williams Obstetrics. 23rd Ed. New York: McGraw-Hill Professional; 2009. [Google Scholar]
- 33.Enquobahrie DA, et al. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204(2):178.e12–178.e21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noack F, et al. miRNA expression profiling in formalin-fixed and paraffin-embedded placental tissue samples from pregnancies with severe preeclampsia. J Perinat Med. 2011;39(3):267–271. doi: 10.1515/jpm.2011.012. [DOI] [PubMed] [Google Scholar]
- 35.Wu L, et al. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction. 2012;143(3):389–397. doi: 10.1530/REP-11-0304. [DOI] [PubMed] [Google Scholar]
- 36.Guo L, et al. A comprehensive survey of miRNA repertoire and 3′ addition events in the placentas of patients with pre-eclampsia from high-throughput sequencing. PLoS ONE. 2011;6(6):e21072. doi: 10.1371/journal.pone.0021072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miura K, et al. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem. 2010;56(11):1767–1771. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 38.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS ONE. 2011;6(6):e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard CC, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5(3):492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen KD, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207(7):1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrie CH, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts TC, Blomberg K, McClorey G. Molecular Therapy — Nucleic Acids - Abstract of article: Expression Analysis in Multiple Muscle Groups and Serum Reveals Complexity in the MicroRNA Transcriptome of the mdx Mouse with Implications for Therapy. Mol Ther. 2012 doi: 10.1038/mtna.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cortez MA, et al. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25(12):1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 47.Hafner M, et al. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods. 2012;58(2):164–170. doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hafner M, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17(9):1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2008;44(1):13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Farazi TA, et al. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods. 2012;58(2):171–187. doi: 10.1016/j.ymeth.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.