Abstract
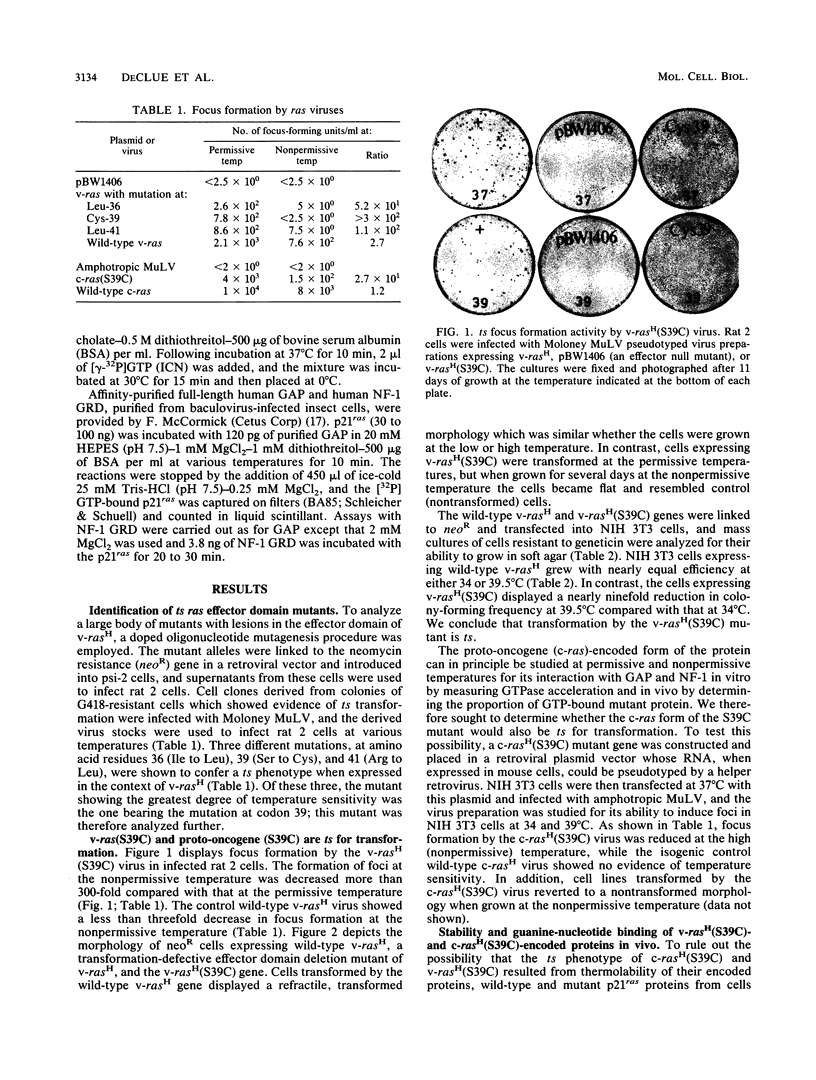
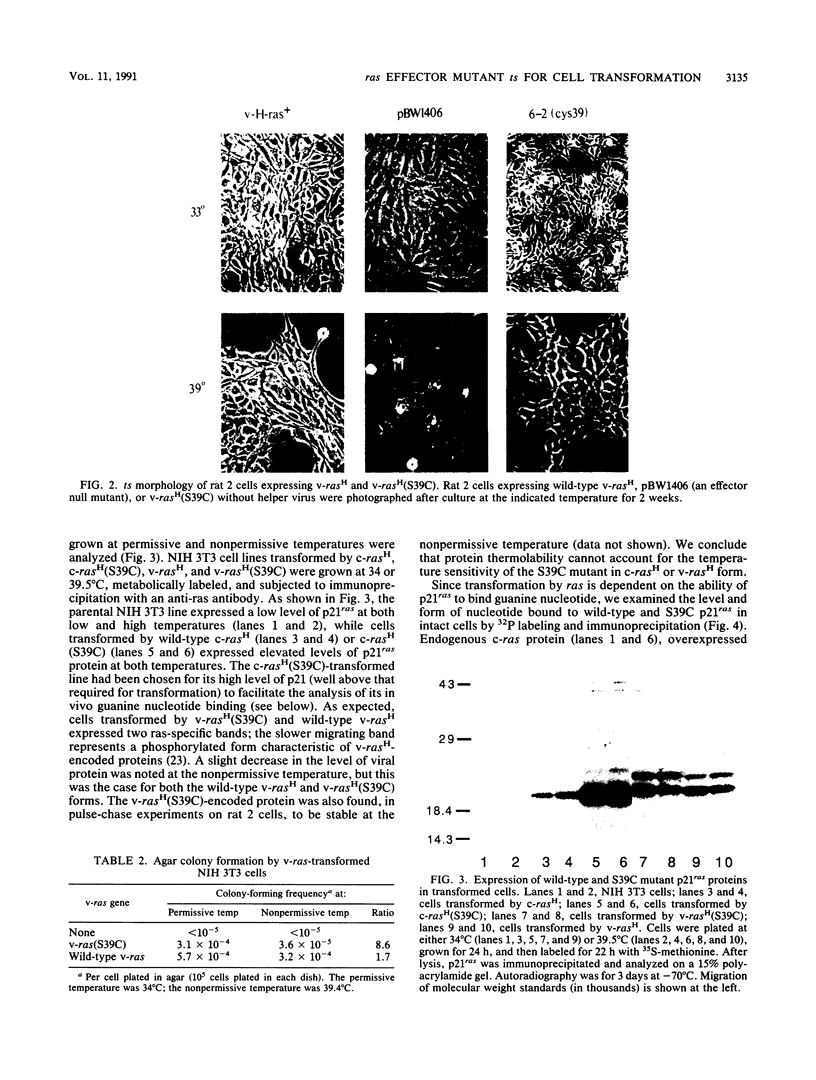
A series of v-rasH effector domain mutants were analyzed for their ability to transform rat 2 cells at either low or high temperatures. Three mutants were found to be significantly temperature sensitive: Ile-36 changed to Leu, Ser-39 changed to Cys (S39C), and Arg-41 changed to Leu. Of these, the codon 39 mutant (S39C) showed the greatest degree of temperature sensitivity. When the same mutation was analyzed in the proto-oncogene form of ras(c-rasH), this gene was also found to be temperature sensitive for transformation. Biochemical analysis of the proteins encoded by v-rasH(S39C) and c-rasH(S39C) demonstrated that the encoded p21ras proteins were stable and bound guanine nucleotides in vivo at permissive and nonpermissive temperatures. On the basis of these findings, it is likely that the temperature-sensitive phenotype results from an inability of the mutant (S39C) p21ras to interact properly with the ras target effector molecule(s) at the nonpermissive temperature. We therefore analyzed the interaction between the c-rasH(S39C) protein and the potential target molecules GTPase-activating protein (GAP) and the GAP-related domain of NF-1, on the basis of stimulation of the mutant p21ras GTPase activity by these molecules in vitro. Assays conducted across a range of temperatures revealed no temperature sensitivity for stimulation of the mutant protein, compared with that of authentic c-rasH protein. We conclude that for this mutant, there is a dissociation between the stimulation of p21ras GTPase activity by GAP and the GAP-related domain NF-1 and their potential target function. Our results are also consistent with the existence of a distinct, as-yet-unidentified effector for mammalian ras proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H., Lowy D. R., Willumsen B. M., Der C. J., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988 Apr 22;240(4851):518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov 16;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Bos J. L. The ras gene family and human carcinogenesis. Mutat Res. 1988 May;195(3):255–271. doi: 10.1016/0165-1110(88)90004-8. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Calés C., Hancock J. F., Marshall C. J., Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988 Apr 7;332(6164):548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Field J., Broek D., Kataoka T., Wigler M. Guanine nucleotide activation of, and competition between, RAS proteins from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2128–2133. doi: 10.1128/mcb.7.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech M., John J., Pizon V., Chardin P., Tavitian A., Clark R., McCormick F., Wittinghofer A. Inhibition of GTPase activating protein stimulation of Ras-p21 GTPase by the Krev-1 gene product. Science. 1990 Jul 13;249(4965):169–171. doi: 10.1126/science.2164710. [DOI] [PubMed] [Google Scholar]
- Furth M. E., Davis L. J., Fleurdelys B., Scolnick E. M. Monoclonal antibodies to the p21 products of the transforming gene of Harvey murine sarcoma virus and of the cellular ras gene family. J Virol. 1982 Jul;43(1):294–304. doi: 10.1128/jvi.43.1.294-304.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Marshall M. S., Scolnick E. M., Sigal I. S. Identification of guanine nucleotides bound to ras-encoded proteins in growing yeast cells. J Biol Chem. 1987 Aug 5;262(22):10426–10429. [PubMed] [Google Scholar]
- Hall A. The cellular functions of small GTP-binding proteins. Science. 1990 Aug 10;249(4969):635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- Hart P. A., Marshall C. J. Amino acid 61 is a determinant of sensitivity of rap proteins to the ras GTPase activating protein. Oncogene. 1990 Jul;5(7):1099–1101. [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Nordeen S. K., Vogt K., Edgell M. H. A complete library of point substitution mutations in the glucocorticoid response element of mouse mammary tumor virus. Proc Natl Acad Sci U S A. 1986 Feb;83(3):710–714. doi: 10.1073/pnas.83.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhappan C., Vande Woude G. F., Robins T. S. Transduction of host cellular sequences by a retroviral shuttle vector. J Virol. 1986 Nov;60(2):750–753. doi: 10.1128/jvi.60.2.750-753.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal J. C., Santos E., Notario V., Barbacid M., Yamazaki S., Kung H., Seamans C., McAndrew S., Crowl R. Expression of normal and transforming H-ras genes in Escherichia coli and purification of their encoded p21 proteins. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5305–5309. doi: 10.1073/pnas.81.17.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O'Connell P., Cawthon R. M. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990 Nov 16;63(4):843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Papageorge A., Lowy D., Scolnick E. M. Comparative biochemical properties of p21 ras molecules coded for by viral and cellular ras genes. J Virol. 1982 Nov;44(2):509–519. doi: 10.1128/jvi.44.2.509-519.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Endo M., Nakafuku M., Nakamura S., Kaziro Y. Platelet-derived growth factor stimulates formation of active p21ras.GTP complex in Swiss mouse 3T3 cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5993–5997. doi: 10.1073/pnas.87.15.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Nakamura S., Kaziro Y. Induction of neurite formation in PC12 cells by microinjection of proto-oncogenic Ha-ras protein preincubated with guanosine-5'-O-(3-thiotriphosphate). Mol Cell Biol. 1987 Dec;7(12):4553–4556. doi: 10.1128/mcb.7.12.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaber M. D., Garsky V. M., Boylan D., Hill W. S., Scolnick E. M., Marshall M. S., Sigal I. S., Gibbs J. B. Ras interaction with the GTPase-activating protein (GAP). Proteins. 1989;6(3):306–315. doi: 10.1002/prot.340060313. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Stokes P. E., Smythers G. W., Dhar R., Oroszlan S. Characterization of the phosphorylation sites and the surrounding amino acid sequences of the p21 transforming proteins coded for by the Harvey and Kirsten strains of murine sarcoma viruses. J Biol Chem. 1982 Oct 10;257(19):11767–11773. [PubMed] [Google Scholar]
- Shih T. Y., Weeks M. O., Young H. A., Scolnick E. M. p21 of Kirsten murine sarcoma virus is thermolabile in a viral mutant temperature sensitive for the maintenance of transformation. J Virol. 1979 Aug;31(2):546–546. doi: 10.1128/jvi.31.2.546-546.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Scolnick E. M. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stone J. C., Vass W. C., Willumsen B. M., Lowy D. R. p21-ras effector domain mutants constructed by "cassette" mutagenesis. Mol Cell Biol. 1988 Aug;8(8):3565–3569. doi: 10.1128/mcb.8.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp W. C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981 Aug;113(1):408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Velu T. J., Vass W. C., Lowy D. R., Tambourin P. E. Harvey murine sarcoma virus: influences of coding and noncoding sequences on cell transformation in vitro and oncogenicity in vivo. J Virol. 1989 Mar;63(3):1384–1392. doi: 10.1128/jvi.63.3.1384-1392.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U. S., Dixon R. A., Schaber M. D., Diehl R. E., Marshall M. S., Scolnick E. M., Sigal I. S., Gibbs J. B. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988 Sep 1;335(6185):90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Papageorge A. G., Kung H. F., Bekesi E., Robins T., Johnsen M., Vass W. C., Lowy D. R. Mutational analysis of a ras catalytic domain. Mol Cell Biol. 1986 Jul;6(7):2646–2654. doi: 10.1128/mcb.6.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Yatani A., Okabe K., Polakis P., Halenbeck R., McCormick F., Brown A. M. ras p21 and GAP inhibit coupling of muscarinic receptors to atrial K+ channels. Cell. 1990 Jun 1;61(5):769–776. doi: 10.1016/0092-8674(90)90187-j. [DOI] [PubMed] [Google Scholar]
- Zhang K., DeClue J. E., Vass W. C., Papageorge A. G., McCormick F., Lowy D. R. Suppression of c-ras transformation by GTPase-activating protein. Nature. 1990 Aug 23;346(6286):754–756. doi: 10.1038/346754a0. [DOI] [PubMed] [Google Scholar]
- Zhang K., Noda M., Vass W. C., Papageorge A. G., Lowy D. R. Identification of small clusters of divergent amino acids that mediate the opposing effects of ras and Krev-1. Science. 1990 Jul 13;249(4965):162–165. doi: 10.1126/science.2115210. [DOI] [PubMed] [Google Scholar]






