Abstract
Following the addition of ions to trigger folding, RNA molecules transition from rigid, extended states to a compact ensemble. Determining the time scale for this collapse provides important insights into electrostatic contributions to RNA folding; however it can be challenging to isolate the effects of purely non-specific collapse, e.g. relaxation due to backbone charge compensation, from the concurrent formation of some tertiary contacts. To solve this problem, we decoupled non-specific collapse from tertiary folding using a single point mutation to eliminate tertiary contacts in the small RNA subdomain known as tP5abc. Microfluidic mixing with microsecond time resolution and FRET detection provides insight into the ionic strength dependent transition from extended to compact ensembles. Differences in reaction rates are detected when folding is initiated by monovalent or divalent ions, consistent with equilibrium measurements illustrating the enhanced screening of divalent ions relative to monovalent ions at the same ionic strength. Ion-driven collapse is fast and a comparison of the collapse time of the wild type and mutant tP5abc suggests that site binding of Mg2+ occurs on submillisecond time scales.
RNA plays important biological roles in translation, splicing and enzymatic/catalytic reactions.1 A recent focus on the role of RNA in the control of gene expression indicates that RNA molecules can be exploited for biotechnology applications.2, 3 Growing interest in the use of RNA aptamers and riboswitches as therapeutic and analytic agents4 calls for a process of designing molecules based on insights from RNA folding kinetic mechanisms.5
Structurally, RNA is a collection of short base-paired helices connected by non-base paired regions that include loops, bulges, hinges and junctions.6, 7 Because the RNA backbone carries a high negative charge, strong repulsive electrostatic forces must be overcome for the molecule to fold. RNA folding is induced in vitro by the addition of ions. Crystal structures reveal a small number of site-bound ions in some RNAs;8 however, the majority of counterions form a diffuse cloud around the macromolecule.9 In low salt unfolded states, the helices repel and molecular conformations are extended. Following the addition of counterions to trigger folding, the backbone charge is more locally screened and the molecules relax to compact states. Recent equilibrium studies suggest that this “electrostatic relaxation” is anisotropic; the junctions direct folding by entraining helix motions along certain well-defined pathways.10, 11 Native contacts can then form when the two sides of a tertiary contact come into close proximity. However, studies of short base-paired helices suggest that the negative duplex charge is not fully compensated on intramolecular length scales.12 An outward electrostatic pressure opposes tight compaction in the absence of tertiary contacts, even at moderate to high ionic strength. Thus RNA folding is a balance between weakened, but non-negligible repulsive electrostatic forces and attractive forces, e.g. hydrogen bonding between the two sides of a tertiary contact. The primary goal of this study is to complement the increasing number of RNA folding and kinetic studies13, 14 by focusing on the process of collapse upon addition of charge compensating ions. How does the rapid, initial collapse depend on the valence and concentration of counterions used to trigger it?
Previous small-angle X-ray scattering (SAXS) studies of the Tetrahymena ribozyme and selected mutants reveal a rapid compaction upon the addition of ions.15 Concurrent time-resolved hydroxyl radical footprinting experiments show that the majority (but not all) of tertiary contacts in the molecule remain unformed within the time scale of rapid collapse. However, collapse occurred within the mixing dead times of those kinetic measurements so only an upper limit for the collapse time (milliseconds) was obtained. More extensive time-resolved SAXS studies of the collapse and folding of the Azoarcus ribozyme16 were carried out to focus on this initial rapid collapse. This group I intron displayed heterogeneous folding kinetics when folding was initiated by Mg2+.16 Some subpopulations collapse rapidly with tertiary contacts formed, others undergo non-specific collapse before slower structural rearrangements can occur. Thus, the millisecond time scales reported for this system do not distinguish pure non-specific collapse due to charge compensation from specific collapse. Other attempts to measure pure electrostatic collapse in simplified systems were obscured by the presence of a stiff hinge joining two helical domains that precluded relaxation to a compact ensemble.17
An experiment to measure the time scale of ion-mediated electrostatic collapse in RNA requires a clear delineation between non-specific collapse (purely electrostatic driven) and specific collapse (containing native or non-native tertiary contacts). We accomplish this by choosing a molecule that collapses but is incapable of forming tertiary contacts: the A186U mutant of the tP5abc subdomain of the Tetrahymena ribozyme. In this well-characterized construct, mutation of a specific contact residue for Mg2+ binding18 prevents secondary structure rearrangement, which is a prerequisite for the final steps of tertiary folding.19 Nuclear magnetic resonance (NMR) studies have shown that the A186U mutant has an extended structure similar to that of wild type tP5abc, but does not undergo tertiary folding,20 therefore it is used as a control for unfolded tP5abc or the P4-P6 domain.21-23 A recent publication focusing on the later folding steps of wild type tP5abc speculates about the tight connection between secondary structure rearrangement and tertiary contact formation, suggesting that they occur concomitantly.23 Use of the A186U mutation ensures that we probe only non-specific collapse without the participation of tertiary contacts, allowing us to elucidate the nature of structures within the rapidly formed, compact ensemble.
Here, we describe the dependence of the earliest events in RNA folding on ion type, valence and concentration, using the A186U mutation in tP5abc RNA to monitor the formation of the collapsed state. Fluorescence correlation spectroscopy (FCS) measurements verify that this construct collapses to a compact state in high salt solutions. To establish the electrostatic contributions of different ions in facilitating collapse, we quantify the strength of electrostatic interactions using second virial coefficients (A2) of short RNA duplexes determined by SAXS. Since theoretical studies suggest that ion-driven non-specific collapse in RNA should occur in submillisecond time scales,24, 25 we use a microfluidic mixer with microsecond time resolution26 combined with Förster resonance energy transfer (FRET) detection in a confocal microscope to measure the kinetics of RNA collapse due to a rapid jump in salt concentration. Between solutions with the same counterion valence, we measure collapse times that decrease as the ionic strength increases, reflecting the increasing entropy of the collapsed state with ionic strength as suggested by equilibrium measurements.27 Near physiological monovalent ionic strengths, the initial non-specific collapse is fast and comparable to the dead time of our rapid mixing microfluidic device (≈ 200 μs). When Mg2+ is used to initiate folding, collapse times are faster in the wild type than in the mutant, suggesting that site binding of Mg2+ occurs within the first millisecond of folding, faster than previously implied using techniques with slower time resolution.28, 29 Our data suggest that specific contact formation accelerates the earliest steps of folding, and ‘directs’ folding along specific pathways.
MATERIALS AND METHODS
Materials
RNA molecules were synthesized, desalted and purified by Dharmacon RNAi Technologies (Lafayette, CO). The sequence of the 25 base pair (bp) double-stranded RNA used for second virial coefficient measurements is GCA UCU GGG CUA UAA AAG GGC GUCG as in previous studies.30 We used the truncated P5abc (tP5abc) construct and its A186U mutant described by Wu and Tinoco19 for fluorescence correlation spectroscopy (FCS) and rapid mixing experiments. We have added a uracil to the 5' end in order to prevent interactions between the donor and the nearby G-C base pair. The 5' end was labeled with the donor fluorophore (fluorescein) while the A171 nucleotide in the P5c stem-loop was labeled with the acceptor fluorophore (Dy547, with spectral characteristics similar to Cy3). The positions of the labels were chosen such that a change in Förster resonance energy transfer (FRET) is measurable between the extended and collapsed states of tP5abc. The tP5abc RNA molecules were stored in 50 mM K+-MOPS buffer, pH 7, with 0.1 mM EDTA. Prior to measurements, they were annealed in 20 mM K+-MOPS buffer, pH 7, with 8 mM EDTA at 50°C for 5 minutes then slowly cooled to room temperature. The molecules were buffer-exchanged to 20 mM K+-MOPS buffer, pH 7 for all experiments and different salt solutions were added to change ion conditions. Monovalent and divalent ion solutions were prepared from Chloride salts unless stated otherwise. All chemicals were purchased from Sigma (St. Louis, MO). Rotational motion of the dye labels were investigated using fluorescence anisotropy measurements. Results are described in the Supporting Information and shown in Figure S1 (Supporting Information).
Second virial coefficient measurements
The strength of intermolecular interactions between 25 bp RNA duplexes was assessed by extracting the second virial coefficient (A2) from SAXS profiles measured as a function of RNA concentration at varying cation valence and salt concentrations. A2 is a measure of the interaction potential between short RNA helices in solution and takes into account the contributions from excluded volume, electrostatic repulsion and interhelical attraction 12, 30-32. The SAXS experiments to measure A2 were carried out at the Cornell High Energy Synchrotron Source (CHESS) and are described extensively in previous publications.12, 30
Equilibrium Fluorescence measurements
FCS was used to measure the changes in diffusion times of tP5abc under different ionic conditions. Molecules in the collapsed state are more compact and diffuse faster than molecules in extended and unfolded configurations. The standard confocal setup described by Chen et al.33 used a 488 nm laser for fluorescein excitation. Data processing and fitting were performed with Origin software (Microcal, Northampton, MA). Equilibrium FRET was calculated from fluorescence spectra taken with a Cary Eclipse Fluorescence Spectrophotometer (Varian Inc., Australia). The effective efficiency of energy transfer (EFRET) was calculated as A/(A+D), where A and D are the peak intensities of the acceptor and donor emission, respectively.
Rapid Mixing to measure RNA collapse
We used a microfluidic mixer to rapidly change the ionic conditions of the tP5abc RNA.26, 34 The mixer uses hydrodynamic focusing35, 36 to facilitate fast diffusion of ions into the RNA sample. The experimental conditions are described in the Supporting Information with a schematic of the device shown in Figure S2. The mixing dead time for K+ and Rb+ ions was 140 μs, and that for Mg2+ and Sr2+ ions was 235 μs. Additional details of the manufacture, characterization and use of the mixing device were published in previous work.26 Devices were fabricated at the Cornell NanoScale Science and Technology Facility (CNF).
RESULTS
Dependence of charge screening on ion valence for RNA duplexes
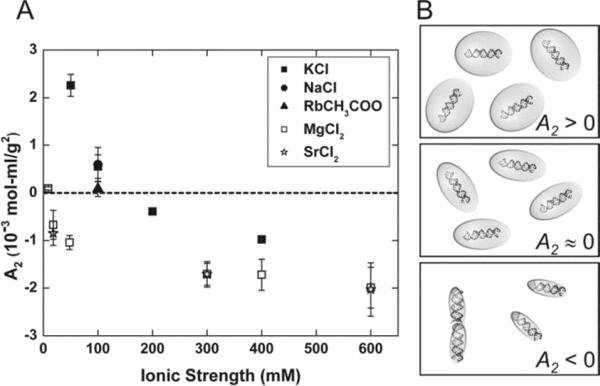
To characterize how ions affect charge screening interactions between RNA helices, we used SAXS to measure A2 between 25 bp RNA duplexes as a function of ion type and ionic strength. End effect contributions to A2 are minimal when duplex lengths exceed ~25 bp,32 allowing us to focus on how ions affect screening lengths measured from the cylindrical axis of the double helix. We find that the charge screening efficiency depends on ion valence but not ion type (Figure 1A). Measured values of A2 in Na+, K+ and Rb+ agree within experimental error while values of A2 in Mg2+ and Sr2+ are indistinguishable. Furthermore, these results are consistent with previous experiments showing that divalent ions provide more efficient charge screening than monovalent ions at the same ionic strengths.9, 27, 37-41 A cartoon illustrating the ionic strength dependence of charge screening efficiency is shown in Figure 1B. Since A2 can be expressed in units of volume,31 we associate an electrostatic excluded volume with each duplex at every ionic strength, denoted by the shaded balloon in the figure. At the lowest ionic strength, where A2 has its largest value and intermolecular repulsion dominates, this excluded volume is large. As ionic strength increases and the duplex negative charge is more efficiently screened, both A2 and the excluded volume decrease, allowing the helices to approach each other more closely. At even higher ionic strengths A2 becomes negative and interhelical attraction consistent with end-to-end stacking is observed.12, 30 In the cartoon representation (Figure 1B) for A2 < 0, the electrostatic excluded volume is small enough to allow other interaction forces to take effect. The A2 measurements shown in Figure 1A enable a quantitative comparison of the screening effectiveness of different ionic solutions. Clearly, ionic strength alone is insufficient to explain the RNA charge screening efficiency of ions with different valences, but comparison between ions of the same valence display similar electrostatic effects.
Figure 1.
We use the second virial coefficient (A2) to quantify the ionic strength dependent intermolecular interaction potential of 25 bp RNA helices. Positive values of A2 denote intermolecular repulsion while negative values of A2 can reflect duplex association via end-to-end stacking.12, 30 (A) A2 measurements of 25 bp RNA duplexes obtained from SAXS data. As ionic strength increases the interhelix repulsion decreases due to greater screening of RNA charges by counterions. A2 in solutions with monovalent cations (KCl (■), NaCl (●), and RbCH3COO (▲)) are indistinguishable, as are those with divalent cations (open symbols: MgCl2 (□) and SrCl2 (*)). However, for a given ionic strength, divalent ions are more effective in screening the charges than monovalent ions. (B) Cartoon illustrating the electrostatic excluded volume at increasing ionic strengths. When A2 > 0, large repulsive forces between helices are represented by large electrostatic excluded volumes. When A2 ≈ 0, the RNA–RNA interactions are negligible. A2 < 0 represents RNA association via end-to-end stacking.30
Equilibrium fluorescence measurements of tP5abc
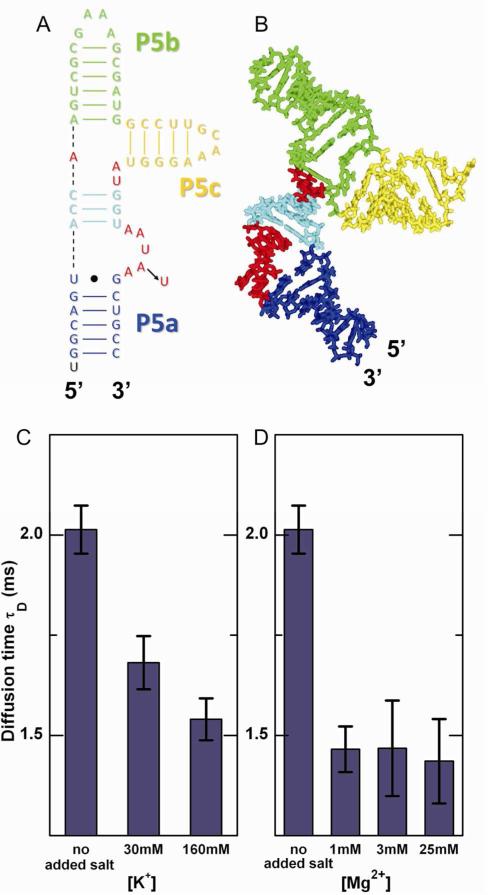
The tP5abc RNA consists of three helices (P5a, P5b and P5c) connected at a junction by single stranded regions as shown in Figure 2A.19 At low salt concentrations the three helices of tP5abc are arranged in an extended state (Figure 2B). Increasing the bulk salt concentration reduces the repulsive forces among the three helices, therefore we expect the RNA to collapse to more compact conformations as salt is added. Since a more extended molecule has a larger hydrodynamic radius and diffuses more slowly, compaction is signaled by a reduction in diffusion time of the RNA.
Figure 2.
tP5abc collapses with increasing ionic strength. (A) The extended state secondary structure of tP5abc.19 The A186U mutation is indicated with an arrow. (B) tP5abc extended state conformation in solution,20 shown with the same color scheme as in (A). (C, D) Diffusion times of A186U mutant obtained from FCS measurements in K+ and Mg2+, respectively. A dramatic decrease in τD is observed upon the addition of salt, demonstrating tP5abc compaction with the increase in bulk counterion concentration.
We used FCS to monitor the A186U mutant, which has the same extended-state secondary structure as the wild type, but does not form tertiary contacts and thus does not fold to the native state.18, 19 A single diffusion constant (τD) and two exponential time constants (τA, τB) were needed to adequately fit the FCS data. The presence of only one diffusion component indicates the absence of conformational states with substantially different diffusion times and thus different hydrodynamic radii. In addition to the characteristic diffusion time, two fast characteristic fluctuation time components are required to accurately fit the FCS data. We tentatively attribute these time scales to a combination of internal molecular motions leading to fluctuations in FRET, and dye photophysics. At increased ionic strengths, FCS data report a dramatic decrease in τD (Figure 2C and D). Because τD is proportional to the hydrodynamic radius of the molecule, the different values measured in the initial (low salt concentration, 9 mM K+) buffer and the final solution (containing an additional 30 or 160 mM K+ (Figure 2C) or 1, 3, or 25 mM Mg2+ (Figure 2D)) indicate collapse to a compact state. FCS measurements of wild type tP5abc show the same trends and comparable diffusion times as the mutant (Figure S3, Supporting Information); however, because of its lack of sensitivity FCS cannot further distinguish the folded (wild type) from the collapsed (mutant) states. Figure S4 (Supporting Information) shows the diffusion times of the mutant in solutions containing different ionic species. Similar to second virial measurements, we see no significant dependence of diffusion time on ion type.
Equilibrium FRET measurements were also performed at different ionic conditions. In our case, FRET cannot be used as a quantitative probe of the size of the molecule because fluorescence anisotropy measurements show that the internally labeled acceptor dye was not freely rotating in solution (see Supporting Information for details). Figure S5 (Supporting Information) shows the effective equilibrium EFRET of the tP5abc wild type and mutant in different ionic conditions. We see no significant dependence of EFRET on monovalent ion type but there is a slight difference between Mg2+ and Sr2+ ions. Since FCS and A2 measurements cannot detect this difference, it is possibly due to a change in the local dye environment during divalent ion aided collapse. Nonetheless, a normalized change in FRET efficiency still signals a conformational change upon addition of ions and can be used to compare time scales of collapse for each ion type. Additional equilibrium fluorescence control experiments are discussed in the Supporting Information and shown in Figure S6.
Measurement of collapse times at different ionic conditions
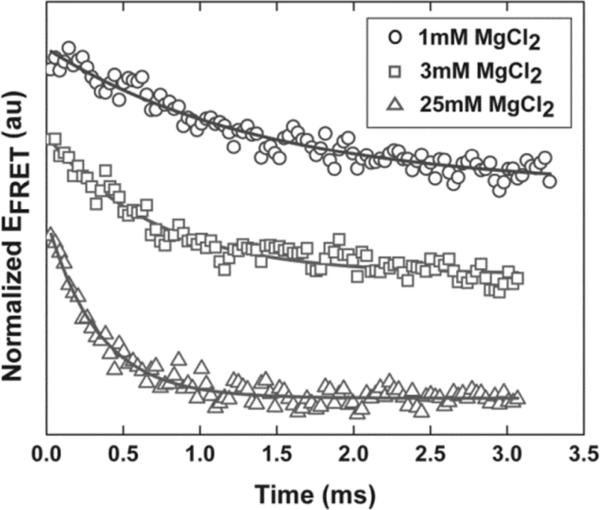
The equilibrium measurements discussed above suggest that tP5abc and its mutant, held in low salt concentration solution, become compact following the addition of ions. To gain insight into the energy landscape of collapse, we measured the collapse time (τc) of the A186U non-folding mutant under several different experiment conditions using microfluidic mixing.26 Mixing of ions is facilitated by diffusion after hydrodynamic focusing35, 36 as described in the Supporting Information. Initially, RNA is equilibrated in a low salt buffer (9 mM K+), where FCS indicates that it populates extended states. To focus on the collapse transition, we measured how the effective EFRET changed in time when the A186U mutant was rapidly mixed with buffers at different ionic conditions. The signal was normalized by the measured EFRET from a control experiment in which the RNA was mixed with the same low salt buffer as in its initial state. The resulting normalized EFRET vs. time was fit to a single exponential decay with a constant offset to extract τc. Figure 3 shows representative, normalized kinetic traces at different concentrations of added divalent ions.
Figure 3.
Typical normalized kinetic traces of the tP5abc A186U mutant after mixing with different Mg2+ concentrations. Solid lines represent single exponential fits to the data. Offsets have been added to the traces to separate them.
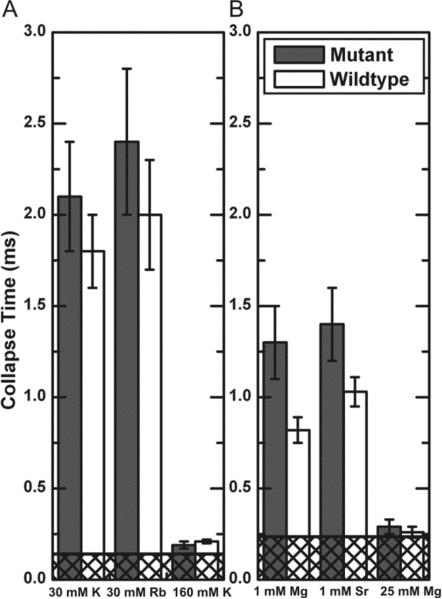
The time scale for collapse (Figure 4) displays a marked decrease with increasing ionic strength. At the highest ionic strengths measured, the collapse time is comparable to the dead times of the mixing instrument (indicated by cross hatched regions in Figure 4). To test its importance on ion type, we measured the collapse timescale in solutions containing 30 mM K+ or 30 mM Rb+ and 1 mM of Mg2+ or 1 mM Sr2+. These lower ionic strengths enhanced our sensitivity to differences by increasing the separation between mixing and collapse time scales.
Figure 4.
Collapse times of tP5abc when rapidly mixed with (A) monovalent and (B) divalent ions. Collapse of the mutant does not depend on ion type (30 mM K+ and Rb+ or 1 mM Mg2+ and Sr2+). The wild type collapses faster than the mutant in 1 mM Mg2+, suggesting early binding of Mg2+ to the specific binding site. Cross-hatched regions represent the mixer dead times (140 μs in (A) and 235 μs in (B)).
Results for the non-folding A186U mutant show no significant dependence of collapse time on ion type. Thus for both monovalent and divalent ions, non-specific collapse depends on the valence and concentration, but not the identity of the ion. Similarly, as we expect, there are no differences between the collapse times of the wild type and the mutant in monovalent salt solutions (Figure 4A).
Previous studies using hydroxyl radical footprinting have found folding times for the isolated P5abc domain to be slow, on the time scale of tens to hundreds of milliseconds.17, 42 Other studies used stopped-flow fluorescence to measure a folding time of 240 ms for wild type tP5abc in 1 mM Mg2+ and show that secondary structure rearrangement is rate-limiting for the tertiary folding of wild type tP5abc.19, 20, 23 Since our rapid mixing results gives us faster time resolution than the previous measurements cited, we are observing a transition of the extended wild type molecule to a compact set of states in which tertiary contacts and secondary structure rearrangement have not yet occurred.
To discern the importance of specific Mg2+ binding to rapid compaction, we compare the collapse time of the mutant with that of the wild type tP5abc. At 1 mM Mg2+, the collapse time of the wild type is 0.82 ± 0.07 ms compared with 1.3 ± 0.2 ms for the mutant (Figure 4B). The small but significant difference between collapse times of the wild type and the non-folding mutant suggests that something other than electrostatic screening stabilizes the collapsed state of the wild type. We also investigated the effect of ion type in collapse of wild type tP5abc and measured 1.03 ± 0.08 ms in 1 mM Sr2+, which is slower than the collapse in 1 mM Mg2+ (0.82 ± 0.07 ms). The difference in collapse time of the wild type due to different divalent ions is small and just outside the error bars. While it is only in the wild type where we can confidently distinguish between collapse times due to ions of different types, we note the same general trend for both mutant and wildtype.
DISCUSSION
Impact on collapse times of ion valence and type
Measurements of second virial coefficients of model systems detect minimal differences in intermolecular interactions based on ion type. At a given ionic strength, K+, Rb+ and Na+ have identical second virial coefficients, as do Mg2+ and Sr2+ (Figure 1A). We also tested the effectiveness of various ions in inducing the collapse transition in the A186U mutant. No ion type effects were observed in non-specific collapse: τc does not distinguish 30 mM K+ from 30 mM Rb+, or 1 mM Mg2+ from 1 mM Sr2+. However, these effects do not scale across solutions of different valence ions. The FCS, A2 and previous persistence length measurements38 confirm that charge screening of the RNA phosphate backbone is more efficient in divalent than monovalent ions. This is, in fact, what we observe in the kinetic measurements (see Figure 4).
From Debye-Hückel (DH) arguments alone, we expect non-specific electrostatic effects to be determined solely by ionic strength. However, the anomalous behavior of divalent relative to monovalent ions is a frequent theme in studies of nucleic acid electrostatics.22, 27, 37, 38, 43 Consistent with literature reports, in the kinetic experiments reported here mixing with divalent ions leads to a shorter collapse time than mixing with monovalent ions at a given ionic strength. Although some studies may suggest that spatially correlated counterions can lead to enhanced screening of polyelectrolytes, these effects seem to be more evident when the counterion valence is greater than two 44, 45. For the case of divalent ions, some insight can be gleaned from recent MD simulations of ions around RNA.46 Notably, simulation results agree with DH models at large distances from the RNA: screening is a function of ionic strength at distances larger than 16 Å from the central axis of the molecule. However, to achieve this agreement requires adjustment of the surface charge density near the RNA surface. Close to the surface, both nucleic acid topologies as well as the distinctive features of counterions (and co-ions) are important. Mean field models are inapplicable because they cannot account for atomic detail ab initio. Our measurements suggest that the surface potential is smaller in the presence of divalent ions, due to their tighter localization and the larger degree of charge compensation. As a result, there is simply less charge to screen at larger distances where the mean field theory can be applied. In terms of the model shown in Figure 1B, the electrostatic excluded volume is smaller for divalent than monovalent ions at a given ionic strength, because the ‘near charge’ on the duplex is more completely neutralized. Thus differences result from the ions that are in closest proximity to the RNA surface.
Ionic strength dependence of collapse reflects increasing conformational entropy in the collapsed ensemble
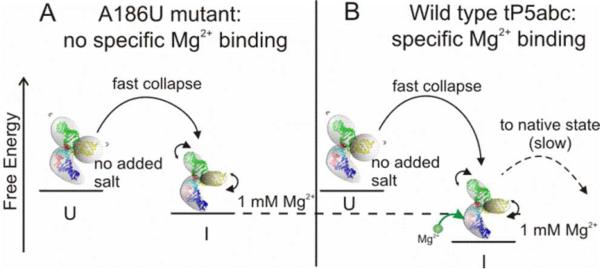
Our results can be placed into the context of recent reports suggesting that tP5abc folding is a sequential process with at least two barriers.23 While the previous work focused on understanding the transition from the collapsed intermediate to the folded state,23 our use of a submillisecond mixing device and a non-folding mutant allowed us to focus on the formation of the collapsed state from an extended conformation. For a given valence of ion, we find that collapse times decrease with increasing salt concentration (Figure 3 and 4). To explain our results, we propose a simple model based on the concept of electrostatic excluded volume, derived from the second virial coefficients (see Results and Figure 1B). Within this model, the ionic strength dependence of τc is readily explained. With no added salt, the A186U mutant exists in an extended (or unfolded) state (U) due to the large electrostatic excluded volume of its three helices. With the addition of charge compensating ions, the electrostatic excluded volume decreases as illustrated in Figure 5A and more conformations are available to the molecule. The more flexible, collapsed collection of states (I) have increased conformational entropy relative to U, as suggested by Bai et al.27 In the absence of tertiary contacts, there are minimal changes to the enthalpy, so the free energy decreases with respect to the extended state. Therefore population of the collapsed state(s) is more favorable at increased ionic strength and the kinetic transition time τc decreases. It is interesting to note a recent report discussing other favorable entropic effects of tertiary folding as a function of increasing [Mg2+].47 Clearly, the role of thermodynamic contributions in ion-dependent RNA folding is still an open question and should be subject to further investigations. Future directions should involve temperature dependent kinetic experiments with microsecond time resolution to examine Arrhenius-type dependence of ion concentration.
Figure 5.
Possible schematic of the early stages of tP5abc collapse. (A) With no added salt, the A186U mutant exists in an extended (or unfolded) state (U) due to the large electrostatic excluded volume of its three helices. When rapidly mixed with ions, the molecule collapses to a compact intermediate state (I). The increase in ion concentration reduces the electrostatic excluded volume allowing the molecule to become more flexible. This change increases its conformational entropy and reduces its free energy, which all lead to faster collapse times. (B) The same arguments as in (A) apply to the wild type tP5abc, however, specific binding of Mg2+ ions reduces the enthalpy of the collapsed state, which can manifest as a faster collapse time as observed in the mixing experiments. Once specific binding has occurred, the wild type can proceed to fold to its native state, although this step is slow due to the large free energy barrier of secondary structure rearrangement.
Site bound Mg2+ ions
To determine the effect of specific interaction on the initial stages of RNA collapse, we compared the collapse time of mutant and wild type tP5abc in 1 mM Mg2+ and 1 mM Sr2+. The crystal structure of the Tetrahymena thermophila ribozyme's full P4-P6 domain shows 5 specific binding sites for Mg2+ ions within P5abc.18 The A186U mutation disrupts one of these sites and prevents secondary structure rearrangement.19 It is possible that the altered secondary structure is necessary for full coordination of the remaining bound Mg2+ ions, thus this single mutation may disrupt all site binding. We speculate that the shorter collapse time in 1 mM Mg2+ reflects a more stable compact intermediate state of the wild type relative to the mutant (Figure 5B). We attribute the reduced free energy of the collapsed state of the wild type to the lower enthalpy contributed by binding of at least one Mg2+ ion. Thus we suggest that Mg2+ binding occurs prior to either tertiary contact formation or secondary structure rearrangement, since these are known to occur more slowly.17, 23, 42 Recent results by Koculi et al. showed that the slow step of wild type tP5abc folding does not require binding of additional Mg2+ ions to the RNA.23 This also suggests that Mg2+ binds early in the folding process. The submillisecond time scale measured here is much faster than previously suggested for Mg2+ binding,28, 29 however those studies only had millisecond time resolution.
Our measurements indicate that the wild type collapses faster in 1 mM Mg2+ than in 1 mM Sr2+. Evidence that Sr2+ cannot occupy the Mg2+ binding sites of P5abc48 supports our proposal that the faster collapse of the wild type in Mg2+ is due to bound ions. We note that while the difference in the collapse times of the wild type RNA due to 1 mM Mg2+ and Sr2+ ions is just outside the error bars, the same trend (albeit statistically insignificant) can also be discerned in the mutant. An alternate explanation for this observation may be found in previous theoretical work which suggests that the presence of smaller ions (like Mg2+) in the ion atmosphere increase the stability of the collapsed ensemble relative to larger ions with the same valence (Sr2+).49 However, the size of diffuse ions alone is not sufficient to account for the small difference in collapse timescales observed between the wild type and the mutant in the presence of 1 mM Sr2+, where no specific binding occurs. Experimental techniques that specifically probe binding of Mg2+ and Sr2+ ions are needed to fully understand the contribution of Sr2+ ions in tertiary folding.
A model of tP5abc collapse and folding
Taken together, and in the context of other folding measurements on the same molecule, our data present a unified picture for how this small domain acquires tertiary structure. In the initial, low salt state, the molecule possesses the secondary structure that minimizes its free energy. Following the addition of ions to reduce the electrostatic repulsion between helices, the molecule favors more compact conformations. Mixing experiments indicate that formation of this collapsed ensemble occurs on the submillecond time scales predicted by theoretical studies of non-specific collapse of RNA.24, 25 In the wild type molecule, our results suggest that Mg2+ ion interactions with specific nucleotides in the A-rich bulge are concurrent with the rapid collapse. It is interesting to speculate that the early binding of Mg2+ may be the cause of the energetically expensive shift in secondary structure that distinguishes non-specifically collapsed from folded states. The bound Mg2+ ion coordinated to A186 also interacts specifically with nucleotides in the nearby strands of the P5c stem loop (Figure 2A). This interaction, coupled with dynamic fluctuations of the non-Watson-Crick base pairs near the three helix junction, may assist with the secondary structure shift measured in fully folded tP5abc. The A186U mutation disrupts the binding of this Mg2+ ion, and the barrier for rearrangement can never be overcome. The very large barrier associated with the folding transition may account for the wide separation in time of the collapse and folding transitions, and enables their separate study. Interestingly, the proposed Mg2+ binding site in the VS ribozyme is also near the sequence where a secondary structure shift occurs,50 hinting that the binding of Mg2+ may facilitate energetically expensive shifts in RNA secondary structure.
CONCLUSIONS
We used a number of different experimental techniques (equilibrium SAXS and FCS, rapid mixing with FRET detection) to relate the electrostatic properties of isolated RNA elements (double-stranded and non-base paired regions) to the kinetic folding behavior of a small, RNA domain. We find that non-specific electrostatic interactions are important in determining the conformational space accessed by a compact molecule that lacks tertiary contacts, which in turn is related to the time scale for collapse. Divalent ions are very effective in electrostatic screening and generate comparable rapid collapse rates to monovalent ions at much higher ionic strength. We propose that divalent ions are more effective than monovalent ions in charge screening due to their tighter localization around the RNA helix. Comparison of the collapse times of the wild type and A186U mutant of tP5abc RNA in divalent ions show that folding RNA sequences collapse faster than their non-folding mutants, supporting the concept that specific interactions lead to increased efficiency of RNA collapse. We suggest that specific binding of Mg2+ ions can occur even during the initial collapse and bias the reaction. Our results highlight the differing roles of counterions in RNA folding kinetics and may aid the design and use of synthetic RNA for biotechnology applications.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge useful discussions with Steve Meisburger, Ron Elber and Serdal Kirmizialtin, and experimental assistance from Ken Finkelstein, Arthur Woll, Watt Webb, Warren Zipfel, Avtar Singh, Christopher Jones, Hye Yoon Park, David Rigie and Natalie Paquette. We thank Ignacio Tinoco for sharing extended state structures.
FUNDING SOURCES
This work was supported by National Institutes of Health grant number [R01-GM085062]. CHESS is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences NSF & NIH/NIGMS via NSF award DMR-0936384. CNF, a member of the National Nanotechnology Infrastructure Network, is supported by the National Science Foundation [ECS-0335765].
ABBREVIATIONS
- FRET
Förster resonance energy transfer
- FCS
fluorescence correlation spectroscopy
- SAXS
small-angle X-ray scattering
- A2
second virial coefficient
- bp
base pair
Footnotes
SUPPORTING INFORMATION AVAILABLE
Discussion of fluorescence anisotropy measurements; discussion of equilibrium FRET; experimental details of the microfluidic mixer; diffusion coefficients of salt solutions used in mixing experiments (Table S1); plot of fluorescence anisotropy results (Figure S1); schematic of mixing device (Figure S2); additional FCS results (Figure S3-S4); equilibrium FRET results (Figure S5). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 2.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 3.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 4.Link KH, Breaker RR. Engineering ligand-responsive gene-control elements: lessons learned from natural riboswitches. Gene Ther. 2009;16:1189–1201. doi: 10.1038/gt.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold. Des. 1996;1:R78–88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 6.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 7.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Bowman JC, Lenz TK, Hud NV, Williams LD. Cations in charge: magnesium ions in RNA folding and catalysis. Curr. Opin. Struct. Biol. 2012;22:262–272. doi: 10.1016/j.sbi.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu VB, Lipfert J, Bai Y, Pande VS, Doniach S, Herschlag D. Do conformational biases of simple helical junctions influence RNA folding stability and specificity? RNA. 2009;15:2195–2205. doi: 10.1261/rna.1747509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Pabit SA, Lamb JS, Park HY, Pollack L. Closing the lid on DNA end-to-end stacking interactions. Appl. Phys. Lett. 2008;92:223901–2239013. doi: 10.1063/1.2937402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodson SA. Compact intermediates in RNA folding. Annu. Rev. Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, Smith H, Brenowitz M, Pollack L. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J. Mol. Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- 16.Roh JH, Guo L, Kilburn JD, Briber RM, Irving T, Woodson SA. Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle X-ray scattering. J. Am. Chem. Soc. 2010;132:10148–10154. doi: 10.1021/ja103867p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlatterer JC, Kwok LW, Lamb JS, Park HY, Andresen K, Brenowitz M, Pollack L. Hinge stiffness is a barrier to RNA folding. J. Mol. Biol. 2008;379:859–870. doi: 10.1016/j.jmb.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat. Struct. Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 19.Wu M, Tinoco I., Jr. RNA folding causes secondary structure rearrangement. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng M, Wu M, Tinoco I., Jr Formation of a GNRA tetraloop in P5abc can disrupt an interdomain interaction in the Tetrahymena group I ribozyme. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3695–3700. doi: 10.1073/pnas.051608598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silverman SK, Zheng M, Wu M, Tinoco I, Jr, Cech TR. Quantifying the energetic interplay of RNA tertiary and secondary structure interactions. RNA. 1999;5:1665–1674. doi: 10.1017/s1355838299991823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenfeld M, Solomatin SV, Herschlag D. Removal of covalent heterogeneity reveals simple folding behavior for P4-P6 RNA. J. Biol. Chem. 2011;286:19872–19879. doi: 10.1074/jbc.M111.235465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koculi E, Cho SS, Desai R, Thirumalai D, Woodson SA. Folding path of P5abc RNA involves direct coupling of secondary and tertiary structures. Nucleic Acids Res. 2012;40:8011–8020. doi: 10.1093/nar/gks468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee N, Thirumalai D. Dynamics of collapse of flexible polyampholytes. J. Chem. Phys. 2000;113:5126–5129. [Google Scholar]
- 25.Thirumalai D, Lee N, Woodson S, Klimov D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 26.Park HY, Qiu X, Rhoades E, Korlach J, Kwok LW, Zipfel WR, Webb WW, Pollack L. Achieving uniform mixing in a microfluidic device: hydrodynamic focusing prior to mixing. Analytical Chemistry. 2006;78:4465–4473. doi: 10.1021/ac060572n. [DOI] [PubMed] [Google Scholar]
- 27.Bai Y, Chu VB, Lipfert J, Pande VS, Herschlag D, Doniach S. Critical assessment of nucleic acid electrostatics via experimental and computational investigation of an unfolded state ensemble. J. Am. Chem. Soc. 2008;130:12334–12341. doi: 10.1021/ja800854u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman SK, Cech TR. An early transition state for folding of the P4-P6 RNA domain. RNA. 2001;7:161–166. doi: 10.1017/s1355838201001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pabit SA, Qiu X, Lamb JS, Li L, Meisburger SP, Pollack L. Both helix topology and counterion distribution contribute to the more effective charge screening in dsRNA compared with dsDNA. Nucleic Acids Res. 2009;37:3887–3896. doi: 10.1093/nar/gkp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonnete F, Vivares D. Interest of the normalized second virial coefficient and interaction potentials for crystallizing large macromolecules. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1571–1575. doi: 10.1107/s090744490201418x. [DOI] [PubMed] [Google Scholar]
- 32.Nicolai T, Mandel M. The ionic strength dependence of the second virial coefficient of low molar mass DNA fragments in aqueous solutions. Macromolecules. 1989;22:438–444. [Google Scholar]
- 33.Chen H, Rhoades E, Butler JS, Loh SN, Webb WW. Dynamics of equilibrium structural fluctuations of apomyoglobin measured by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10459–10464. doi: 10.1073/pnas.0704073104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park HY, Kim SA, Korlach J, Rhoades E, Kwok LW, Zipfel WR, Waxham MN, Webb WW, Pollack L. Conformational changes of calmodulin upon Ca2+ binding studied with a microfluidic mixer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:542–547. doi: 10.1073/pnas.0710810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight JB, Vishwanath A, Brody JP, Austin RH. Hydrodynamic Focusing on a Silicon Chip: Mixing Nanoliters in Microseconds. Physical Review Letters. 1998;80:3863–3866. [Google Scholar]
- 36.Pollack L, Tate MW, Darnton NC, Knight JB, Gruner SM, Eaton WA, Austin RH. Compactness of the denatured state of a fast-folding protein measured by submillisecond small-angle x-ray scattering. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10115–10117. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu X, Andresen K, Kwok LW, Lamb JS, Park HY, Pollack L. Inter-DNA attraction mediated by divalent counterions. Phys. Rev. Lett. 2007;99:038104. doi: 10.1103/PhysRevLett.99.038104. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Meisburger SP, Pabit SA, Sutton JL, Webb WW, Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Chu VB, Bai Y, Lipfert J, Herschlag D, Doniach S. A repulsive field: advances in the electrostatics of the ion atmosphere. Curr. Opin. Chem. Biol. 2008;12:619–625. doi: 10.1016/j.cbpa.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipfert J, Sim AY, Herschlag D, Doniach S. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA. 2010;16:708–719. doi: 10.1261/rna.1985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. Folding mechanism of the Tetrahymena ribozyme P4-P6 domain. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 43.McIntosh DB, Saleh OA. Salt Species-Dependent Electrostatic Effects on ssDNA Elasticity. Macromolecules. 2011;44:2328–2333. [Google Scholar]
- 44.Ha B, Thirumalai D. Bending Rigidity of Stiff Polyelectrolyte Chains: A Single Chain and a Bundle of Multichains. Macromolecules. 2003;36:9658–9666. [Google Scholar]
- 45.Andresen K, Qiu X, Pabit SA, Lamb JS, Park HY, Kwok LW, Pollack L. Mono- and trivalent ions around DNA: a small-angle scattering study of competition and interactions. Biophys. J. 2008;95:287–295. doi: 10.1529/biophysj.107.123174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirmizialtin S, Elber R. Computational exploration of mobile ion distributions around RNA duplex. J Phys Chem B. 2010;114:8207–8220. doi: 10.1021/jp911992t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiore JL, Holmstrom ED, Nesbitt DJ. Entropic origin of Mg2+-facilitated RNA folding. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2902–2907. doi: 10.1073/pnas.1114859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Travers KJ, Boyd N, Herschlag D. Low specificity of metal ion binding in the metal ion core of a folded RNA. RNA. 2007;13:1205–1213. doi: 10.1261/rna.566007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge density of divalent metal cations determines RNA stability. J. Am. Chem. Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersen AA, Collins RA. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.