Summary
Kaposi’s sarcoma-associated herpesvirus (KSHV) is linked to human malignancies. The majority of tumor cells harbor latent virus and a small percentage undergo spontaneous lytic replication. Both latency and lytic replication are important for viral pathogenesis and spread but the cellular players involved in the switch between the two viral lifecycle phases are not clearly understood. We conducted a siRNA screen targeting the cellular kinome and identified Tousled-like kinases (TLKs) as cellular kinases that control KSHV reactivation from latency. Upon treatment of latent KSHV-infected cells with siRNAs targeting TLKs, we saw robust viral reactivation. Knockdown of TLKs in latent KSHV-infected cells induced expression of viral lytic proteins and production of infectious virus. TLKs were also found to play a role in regulating reactivation from latency of another related oncogenic gammaherpesvirus, Epstein-Barr virus (EBV). Our results establish the TLKs as cellular repressors of gammaherpesviral reactivation.
Keywords: KSHV, EBV, TLK1, TLK2, Latency, Reactivation, siRNA screen
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV) is linked to three human malignancies including Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman’s disease (Chang et al., 1994, Songyang et al., 1994, Stürzl et al., 1997, Sodhi et al., 2004, Cesarman et al., 1995, Soulier et al., 1995). The majority of tumor cells harbor latent virus and a small percentage of these cells undergo spontaneous lytic replication. Both latency and lytic replication are important phases for viral pathogenesis and spread but the cellular players involved in the switch between the two phases of the viral lifecycle are not clearly understood.
In the majority of infected cells KSHV remains latent, but a small percentage of cells undergo spontaneous lytic replication at any given time (Zhong et al., 1996, Poyet et al., 2001). This low level of viral reactivation is believed to be important for persistence and pathogenesis (Grundhoff and Ganem, 2004). In cell culture, reactivation of KSHV occurs following treatment with chemical compounds such as the phorbol ester, 12-O-tetradecanoyl-phorbol-13-acetate (TPA/PMA), and the histone deacetylase inhibitor, sodium butyrate. We reported that activation of Toll-like receptors 7 and 8 in PEL cells led to KSHV reactivation (Gregory et al., 2009). In an overexpression system using transient transfection of kinase cDNAs, the Pim family kinases as well as the Ras family kinases have been shown to be involved in KSHV reactivation (Cheng et al., 2009, Yu et al., 2007).
To determine cellular kinases that control KSHV reactivation from latency we performed a siRNA screen of the cellular kinome in the absence of any chemical inducers. This allowed us to assess which kinases are needed for the virus to maintain latency or to induce reactivation. A siRNA library containing siRNAs against 720 different human kinases was used for the screen. We identified the Tousled like kinases (TLKs) as cellular kinases involved in reactivation of KSHV.
Originally described in the plant Arabidopsis thaliana, the Tousled gene encodes a nuclear serine/threonine kinase that is essential for flower and leaf development (Roe et al., 1997, Roe et al., 1993). Two mammalian homologs, termed Tousled-like kinase 1 and 2 (TLK1 and TLK2), show 84% sequence similarity to each other (Takahata et al., 2009). The TLKs are regulated by cell cycle-dependent phosphorylation, their activity is tightly linked to DNA replication with maximal activity during S phase, and they are sensitive to DNA-damaging agents and inhibitors of DNA replication (Takahata et al., 2009). TLKs are also involved in chromatin assembly. TLK1 and TLK2 bind to, and phosphorylate, the human chromatin assembly factors Asf1a and Asf1b (Silljé and Nigg, 2001). The TLKs have been implicated in numerous replicative and transcriptional processes including chromosome condensation and segregation (Sunavala-Dossabhoy et al., 2003, Hashimoto et al., 2008), gene silencing (Wang et al., 2007), and DNA repair (Sunavala-Dossabhoy et al., 2005, Canfield C, 2009).
In this study, we identified TLKs as modulators of KSHV reactivation. Our results show that depletion of TLK2 in KSHV latently infected epithelial cells leads to robust viral reactivation. Knockdown of TLK2 induces ORF50/RTA activation, expression of viral lytic mRNA and proteins, and the production of infectious progeny virions. Depletion of TLK2, and to a lesser extent TLK1, also led to KSHV viral reactivation from latently infected B cells. Moreover, we found that knockdown of TLK1, and to a lesser extent TLK2, results in reactivation of another gammaherpesvirus family member, Epstein-Barr virus (EBV). Our results implicate the TLKs as being key regulators of KSHV and EBV reactivation, whose expression is required for the maintenance of viral latency.
Results
Cellular siRNA Kinome Screen
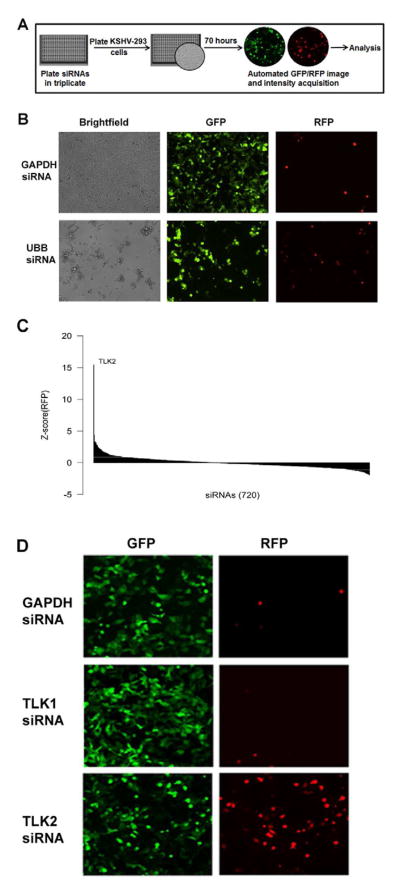
To determine which cellular kinases are important for KSHV reactivation we performed a siRNA screen targeting the human cellular kinome. Over 720 siRNAs against all human protein and lipid kinases were included in the screen. Each well contained a pool of 4 siRNAs with different target sequences to a single cellular kinase. For the screen, we used KSHV-infected 293 (KSHV-293) cells that harbor latent KSHV and constitutively express GFP, while RFP is under the control of a lytic promoter and thus is only expressed upon viral reactivation (Vieira and O’Hearn, 2004). A variety of confounding factors often leads to high false discovery rates among siRNA screens, an inevitable association of high throughput investigations. To lessen the false discovery rate we performed our primary screen in triplicate and used siRNAs that have been previously validated. Cells were reverse transfected with the siRNA pools and seventy hours post-siRNA transfection, GFP and RFP images and fluorescence intensities were acquired using a Cellomics ArrayScan VTI HCS Reader (Fig. 1A). To ensure efficient siRNA transfection, an siRNA against Ubiquitin B (UBB) was included as a control siRNA in the screen as UBB knockdown is known to lead to cell death (Tiedemann et al., 2010) (Fig. 1B). The siRNA screen data was analyzed using the R statistical program environment (Team., 2008). Statistically significant changes in viral reactivation (i.e. RFP intensity) were determined using both the median and mean RFP values for all the wells of the siRNA screen. Figure 1C shows a Waterfall plot of the Z-scores of the median RFP value for each of the siRNAs. A Z-score of 2 or higher was considered significant (Fig. 1C). Additionally, Table 1 lists the cellular kinases that when depleted showed a ≥ 2 standard deviation increase from the overall mean RFP intensity. One isoform of Tousled like kinase (TLK) named TLK2 stood out in both these analyses (Fig. 1C and Table 1) because knockdown of TLK2 showed a Z-score of 15 based on median RFP value, and TLK2 knockdown led to a level of RFP expression that was 13 standard deviations above the mean RFP value for the screen. The next most significant kinases were only 3 standard deviations above the mean. These encompassed 7/720 (1%) of the siRNA targets and attests to the specificity and stringency of our screen. It is possible that some of the siRNAs in our screen do not sufficiently deplete their target protein and thus, potentially provide a false negative result. Figure 1D shows representative GFP and RFP images from the kinome screen. As can be seen in the RFP panels, knockdown of TLK1 shows no change in RFP expression compared to the GAPDH siRNA control in KSHV-293 cells, while TLK2 knockdown results in a large increase in RFP-positive cells indicative of viral reactivation. To rule out the possibility that viral reactivation following TLK2 knockdown is due to off-target effects of one or more of the TLK2 siRNAs, we transfected KSHV-293 cells with each of the 4 individual TLK2 siRNAs that target different regions of the TLK2 transcript. Cells were examined by microscopy (Supp. Fig. 1A) and cell lysates were subjected to Western blots (Supp. Fig. 1B). Three of the four siRNAs showed significant knockdown of TLK2 and robust viral reactivation.
Figure 1. Design of siRNA Screen and Analysis of Data.
A) Schematic of cellular kinome siRNA screen. siRNAs from the Dharmacon SMARTpool kinase siRNA library were loaded into 384-well plates in triplicate. KSHV-293 cells were added to each well containing siRNA and incubated for 70 hours at 37°C. Both RFP and GFP images were then taken for 5 fields per well. B) Control siRNAs demonstrate efficient siRNA transfection in our screen. GAPDH and Ubiquitin B (UBB) siRNAs were reverse transfected at 25nM each into 2500 KSHV-293 cells/well in a 384-well plate. At 70 hours post-transfection brightfield, GFP, and RFP pictures were taken on a fluorescent microscope. C) Statistical analysis of primary hits. A Waterfall plot was used to analyze the data acquired from the screen. It shows the Z-score of the median RFP intensity of each siRNA. D) Reactivation by TLK2 knockdown in KSHV-293 cells. GFP and RFP images of a representative field taken during the screen are shown for the wells containing siRNA targeting GAPDH, TLK1, and TLK2. See also Fig. S1.
Table 1.
List of transfected siRNAs that yield increased RFP intensities at least 2 standard deviations from the mean when transfected into KSHV-293 cells
| RFP intensity | 2 Std. Dev | 3 Std. Dev | 13 Std. Dev | |
|---|---|---|---|---|
| TLK2 | Increased | ++ | ++ | ++ |
| ETNK1 | Increased | ++ | ++ | |
| STK17B | Increased | ++ | ++ | |
| FASTK | Increased | ++ | ++ | |
| MAP4K5 | Increased | ++ | ++ | |
| ATR | Increased | ++ | ++ | |
| EPHA4 | Increased | ++ | ++ | |
| LCK | Increased | ++ | ||
| CSNK1G1 | Increased | ++ | ||
| MAP3K7 | Increased | ++ | ||
| TNK2 | Increased | ++ | ||
| FN3K | Increased | ++ | ||
| PDK3 | Increased | ++ | ||
| DGKQ | Increased | ++ | ||
| PTK6 | Increased | ++ | ||
| GK | Increased | ++ | ||
| ABL1 | Increased | ++ | ||
| MAP3K7IP1 | Increased | ++ | ||
| KIAA1765 | Increased | ++ | ||
| DUSP21 | Increased | ++ |
TLK2 Knockdown Leads to KSHV Reactivation in KSHV-293 Cells
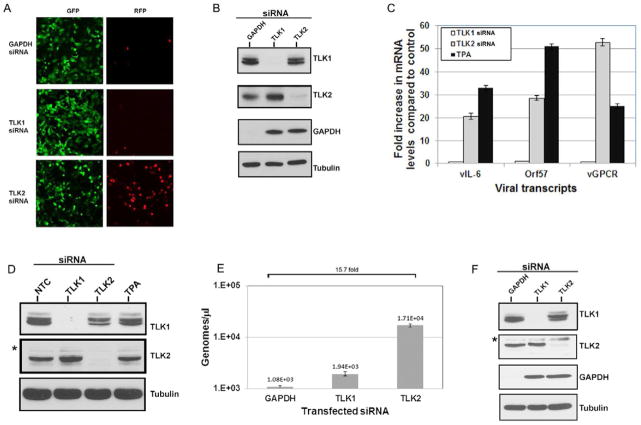
To validate our screen results, we reverse transfected KSHV-293 cells with equivalent amounts of GAPDH siRNA, or a pool of 4 siRNAs against TLK1 and TLK2. Cells were imaged at 70 hours post-transfection for GFP and RFP expression; again knockdown of TLK2 robustly induced RFP expression (Fig. 2A). Cell lysates were also harvested and Western blots were performed to look at kinase expression levels. As seen in Fig. 2B, siRNA transfection led to dramatic decreases in the expression of the target protein.
Figure 2. TLK2 Plays a Role in KSHV Reactivation.
A) Knockdown of TLK2 reactivates KSHV. KSHV-293 cells were reverse transfected with a final siRNA concentration of 25nM. A single GAPDH siRNA or a pool of 4 siRNAs against TLK1 or TLK2 were used. At 70 hours post-transfection, images were taken on a fluorescent microscope. B) siRNAs efficiently knock down target. Cellular lysates from the samples imaged in Fig. 2A were harvested and Western blots were performed for TLK1, TLK2, GAPDH, and the loading control tubulin. C) Viral lytic mRNAs are expressed. KSHV-293 cells were either mock transfected or reverse transfected with 50nM of the non-targeting control siRNA or the pooled TLK1 or TLK2 siRNAs. The mock transfected sample was treated with 25 ng/mL of TPA at the time of transfection. Levels of viral lytic transcripts, vIL-6, Orf57, and vGPCR, were measured by qPCR at 54 hours post-transfection. Values are normalized to the control siRNA. D) Western blots were performed for the indicated proteins for the experiment described in 2C. E) KSHV-293 cells were reverse transfected as described above in 2C. At 96 hours post-transfection, DNA was harvested and viral load was determined by qPCR. Primers for Orf57 were used as the indicator of viral genome copies. F) Western blots were performed for the indicated proteins to confirm knockdown in the experiment described in 2E. Non-specific bands are indicated by an asterisk “*”. Error bars represent standard deviation from the mean. Data were analyzed using a two-tailed type II Student’s t test for significance. See also Fig. S2.
To validate this result we also used a TLK2 siRNA from a different source and measured viral reactivation. We saw the same reactivation phenotype as described above (Supp. Fig. 2A and B). In order to determine if TLK2 levels were increased in KSHV-293 latently infected cells compared to uninfected cells, we performed Western blots probing for TLK2 and found that expression levels of TLK2 were similar between both infected and uninfected cells (Supp. Fig. 2C). We also tested whether knockdown of TLK2 affected cell viability of HEK-293 cells in the absence of KSHV. 293 cells were transfected with a non-targeting control (NTC) siRNA or siRNAs against TLK2 or UBB, and cells were examined by microscopy at 72 hours to gauge viability (Supp. Fig. 2D). TLK2 knockdown had only a minor effect on cell viability, and cell death in the TLK2 knockdown cells was similar to the NTC siRNA transfected control cells, and was much less pronounced than the positive control cells transfected with UBB siRNA. We also examined the effect of TLK2 knockdown on HEK-293 cell proliferation using a MTS assay and saw that TLK2 knockdown did not cause an overall loss in proliferation as was seen following UBB knockdown over the time period examined (Supp. Fig. 2E).
Depletion of TLK2 Leads to Expression of KSHV Lytic Genes
To determine if TLK2 knockdown resulted in increased viral lytic gene expression, we transfected KSHV-293 cells with a non-targeting control siRNA or siRNAs against TLK1 or TLK2. We also treated mock transfected KSHV-293 cells with 12-O-Tetradecanoyl-phorbol-13-acetate (TPA), a potent chemical inducer of KSHV lytic reactivation, as a positive control. RNA was harvested 54 hours post-transfection and quantitative real-time PCR (qPCR) was performed for three key viral lytic mRNAs: vIL-6, ORF57, and vGPCR (Fig. 2C). TLK2 knockdown cells induced expression of all three viral lytic mRNAs, and to levels fairly similar to the TPA treated sample. The corresponding Western blots showing protein levels of the targeted genes are also depicted (Fig. 2D). To investigate whether depletion of TLK2 led to an increase in viral genomes, KSHV-293 cells were transfected with siRNAs against GAPDH, TLK1, or TLK2, and total DNA was extracted 94 hours post-transfection. Viral genomes were quantitated by qPCR (Fig. 2E). TLK2 siRNA transfected cells displayed an approximate 16-fold increase in viral genome copy number compared to the GAPDH depleted cells. Knockdown of TLK1, TLK2, and GAPDH was confirmed by Western blot analysis (Fig. 2F). This demonstrates that knockdown of TLK2 results in increased viral lytic gene expression and increased viral genome replication.
The Major Viral Lytic Switch Protein, ORF50/RTA, is Activated Upon TLK2 Knockdown
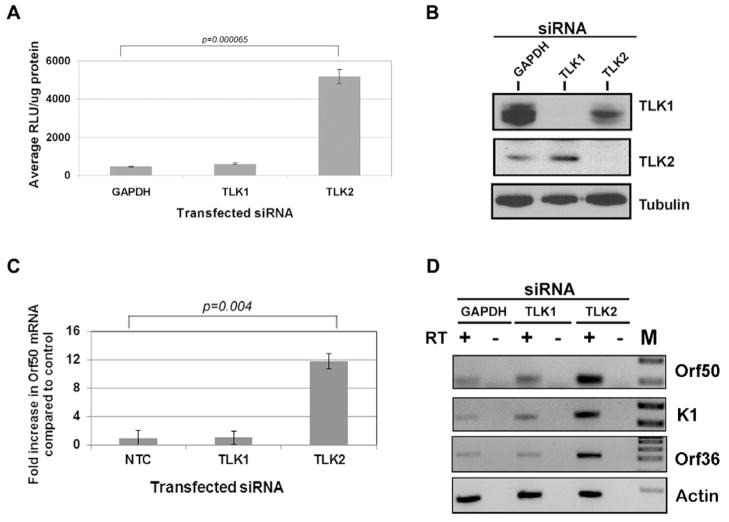
The KSHV replication and transcription activator protein (RTA), encoded by ORF50, plays an essential role in the initiation of viral lytic gene expression. Since knockdown of TLK2 led to the expression of viral lytic genes that were regulated by KSHV ORF50, we wanted to determine if ORF50 levels were also increased. To determine if ORF50 promoter activity was increased following TLK2 knockdown, we performed a reporter gene assay using a luciferase construct under the control of the ORF50 promoter. 293 cells were transfected with siRNA against GAPDH, TLK1, or TLK2 and then 24 hours later the cells were transfected with an ORF50-promoter luciferase construct (Damania et al., 2004). Luciferase expression was measured 48 hours post-transfection (Fig. 3A). Depletion of TLK2 resulted in an approximate 10-fold increase in ORF50 promoter activity compared to GAPDH and TLK1 depleted cells. Knockdown of the targeted proteins was confirmed (Fig. 3B). This indicates that TLK2 knockdown leads to the activation of the ORF50 promoter, even in the absence of other viral proteins. Next, we examined ORF50 transcript levels. KSHV-293 cells were transfected with a non-targeting control (NTC) siRNA or siRNAs against TLK1 or TLK2. Total RNA was harvested 54 hours post-transfection and qPCR for ORF50 mRNA was performed. As can be seen in Figure 3C, there was a 12-fold increase in ORF50 mRNA levels when TLK2 siRNA was transfected into KSHV-293 cells compared to the control siRNA. Additionally, lytic gene expression of two other viral genes, K1 and ORF36, were also increased in TLK2 siRNA transfected KSHV-293 cells (Fig. 3D).
Figure 3. The Major Lytic Switch Protein, KSHV ORF50/RTA, is Activated Following TLK2 Knockdown.
A) The KSHV ORF50 promoter is activated by TLK2 knockdown. HEK-293 cells were transfected with GAPDH siRNA or a pool of TLK1 or TLK2 siRNAs (50nM) for 24 hours followed by transfection with an ORF50 luciferase reporter construct. Lysate was harvested 48 hours after ORF50-luciferase transfection and a luciferase assay was performed. Values are normalized to protein levels determined by Bradford assay. B) Western blots were performed for TLK1, TLK2, and tubulin to confirm knockdown. C) Increase in ORF50 mRNA. Levels of ORF50 viral transcript were measured by qPCR as described in 2C. D) Induction of other viral lytic genes. KSHV-293 cells were reverse transfected with GAPDH siRNA or a pool of TLK1 or TLK2 siRNAs (50nM) for 96 hours. Total RNA was harvested and reverse transcriptase PCR was performed on the RNA (+/− reverse transcriptase (RT)) and PCR products were run out on agarose gels. M, Marker lane. Error bars represent standard deviation from the mean. Data were analyzed using a two-tailed type II Student’s t test for significance.
Depletion of TLK2 Leads to Complete Viral Reactivation and Production of Infectious Virus
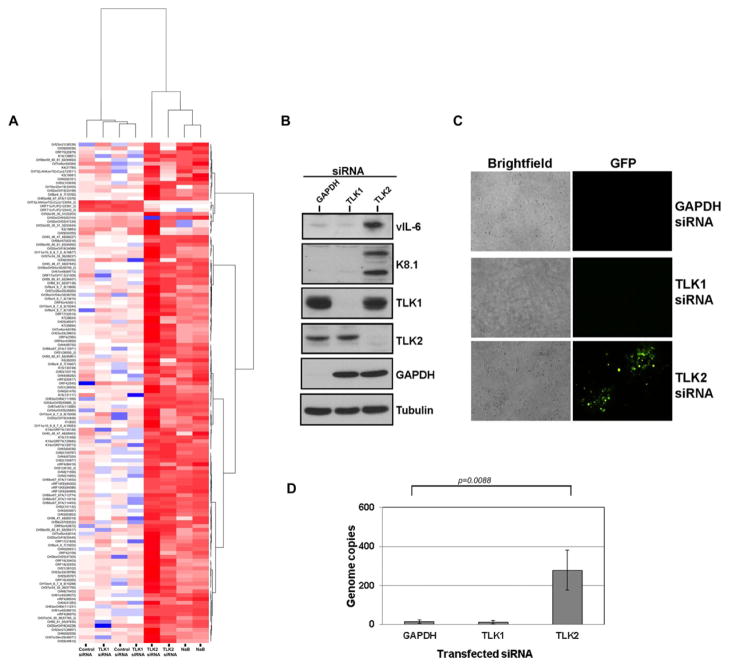
To test the effect of TLK2 knockdown on overall viral gene expression, we performed genome-wide viral profiling using our KSHV qPCR array (Dittmer, 2003). KSHV-293 cells were transfected with the non-targeting control siRNA or a siRNA targeting TLK1 or TLK2. As a positive control for viral reactivation, we treated untransfected cells with 0.1mM sodium butyrate, a known inducer of KSHV reactivation. Cells were incubated for 96 hours and then KSHV genome-wide transcription was measured. When TLK2 was depleted from the KSHV-293 cells there was upregulation of nearly all viral genes, similar to the sodium butyrate-treated cells, indicative of complete viral reactivation (Fig. 4A; also see Supp. Fig. S3). This was in contrast to the control siRNA and TLK1 siRNA treated cells which showed minimal levels of upregulated viral transcripts in KSHV-293 cells.
Figure 4. Genome-wide Upregulation of Viral Transcripts, Induction of Lytic Proteins, and Production of Infectious Progeny Virions Following TLK2 Knockdown.
A) TLK2 knockdown induces KSHV reactivation. KSHV-293 cells were either treated with 0.1mM sodium butyrate or transfected with 50nM of a single control siRNA or siRNA targeting TLK1 or TLK2 for 96 hours. RNA was isolated and a KSHV viral array was performed to determine viral transcript levels. This array has multiple qPCR primer pairs for each annotated KSHV ORF. A heat map for the viral array is shown. Higher transcript levels are indicated by red, lower levels by blue. Each primer was scaled independently, such that the median across all experiments for this primer is indicated by white color. Genes on the vertical axis and samples on the horizontal axis were clustered by similarity of their transcription profile using a Euclidian based distance matrix and Ward’s algorithm. The corresponding dendrograms (tree) are shown outside the heatmap. The branch length corresponds to relative similarity of samples and genes, e.g. induced (NaB) and TLK2 siRNA treated samples form the right branch and cluster together. Control siRNA and TLK1 siRNA treated samples form the left main branch and cluster together. B) Viral lytic proteins are expressed. KSHV-293 cells were reverse transfected with GAPDH siRNA or a pool of TLK1 or TLK2 siRNAs (50nM). Cell lysates were harvested 96 hours post-transfection and Western blots were performed for the lytic proteins vIL-6 and K8.1A, cellular TLK1, TLK2, and GAPDH, and the loading control, tubulin. C) Infectious virus is produced. Cell supernatants were harvested from the same samples as in 4B and used to infect 80,000 Vero cells/well. Images were taken 96 hours post-infection. D) Viral load assay. Cell-associated DNA was harvested from the Vero cells described in 4C above at 96 hours post-infection and intracellular viral genome copy number was determined for the whole cell population. Error bars represent standard deviation from the mean. Data were analyzed using a Student’s t test for significance. See also Supp. Fig. S3.
Complete KSHV replication results in virion production and the release of infectious virions from the cell which can subsequently infect naïve cells. In order to determine if TLK2 knockdown led to the production of infectious virions, we transfected KSHV-293 cells with siRNAs targeting GAPDH, TLK1, or TLK2. Both the cell lysates and supernatants were collected 96 hours post-transfection. The lysates were used to perform Western blots examining expression of two viral lytic proteins, an early lytic protein (vIL-6) and a late lytic protein (K8.1A), which were highly expressed only in the TLK2 siRNA transfected cells (Fig. 4B), and not in the TLK1 or GAPDH siRNA transfected cells.
The supernatants collected from the KSHV-293 transfected cells were clarified to remove cellular debris and then used to infect naïve Vero cells to determine if infectious virus was present. The Vero cells were monitored by fluorescence microscopy for GFP-positive cells, indicative of viral infection. Pockets of GFP-positive cells can clearly be seen in the Vero cells treated with the supernatants from the TLK2 knockdown KSHV-293 cells (Fig. 4C) indicating that infectious virus was produced. These newly infected Vero cells were harvested and total intracellular DNA was extracted in order to perform a viral load assay. In agreement with our other data, naive Vero cells incubated with the supernatants from the TLK2 knockdown KSHV-293 cells displayed much higher levels of viral genomes than the GAPDH knockdown cells (Fig. 4D).
TLK Depletion Can Also Reactivate KSHV From PEL
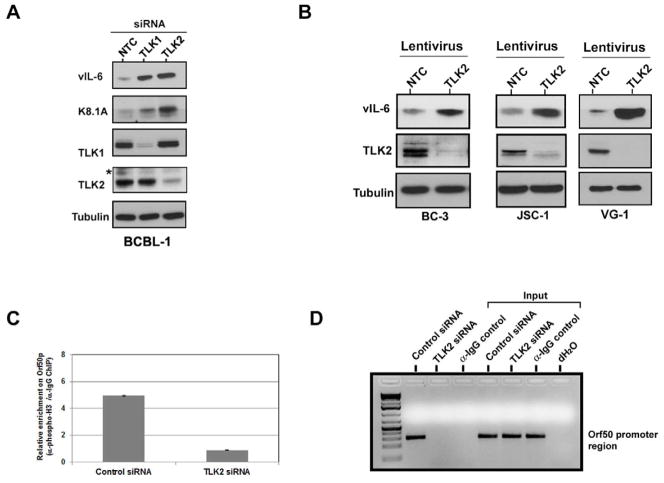
To examine the effect of TLK knockdown in KSHV-infected B cells, BCBL-1 PEL cells were transfected with a non-targeting control siRNA or siRNAs against TLK1 or TLK2. At 120 hours post-transfection, the cells were harvested and protein lysate was extracted and subjected to Western blot analysis against the viral lytic proteins, vIL-6 and K8.1A. As can be seen in Fig. 5A, BCBL-1 cells transfected with the TLK2 siRNA showed viral reactivation as indicated by the increased expression of vIL-6 and K8.1A lytic proteins compared to the control siRNA-transfected cells. Cells transfected with TLK1 siRNA also showed a certain degree of viral reactivation in PEL but not to the same extent as TLK2 depletion, suggesting that depending on the particular cell type, one of the two TLK genes may be predominantly involved in modulating viral latency and suppressing reactivation. To investigate the effect of TLK2 depletion on a variety of PEL cell lines, we infected a panel of PEL cells (BC-3, JSC-1, VG-1) with either a control lentivirus or one targeting TLK2 for knockdown. Infections proceeded for 96 hours and then Western blots for vIL-6 were performed on the harvested lysates (Fig. 5B). Viral reactivation was seen in each of the examined PEL cell lines. This data shows that depletion of TLKs in natural KSHV-infected PEL cells, leads to KSHV reactivation.
Figure 5. Knockdown of TLKs Leads to KSHV Reactivation in PEL and Reduction of Phospho-histone H3 Associated with the KSHV ORF50 Promoter.
A) Depletion of TLKs reactivates KSHV in PEL cells. BCBL-1 cells were transfected with single siRNAs (150mM) against TLK1, TLK2, or a non-targeting control (NTC) and cell lysates were harvested 120 hours post-transfection. Western blots were performed for the viral lytic proteins, vIL-6 and K8.1A, as well as TLK1, TLK2, and tubulin. B) Reactivation in a panel of PEL lines. BC-3, JSC-1, and VG-1 cells were infected with lentivirus expressing either a scrambled control shRNA or shRNA targeting TLK2. Cell lysates were collected 96 hours post-infection and Western blots were performed for vIL-6, TLK2, and tubulin. C) Chromatin immunoprecipitation assay. KSHV-293 cells were reverse transfected with 50nM of either a non-targeting control siRNA or a siRNA against TLK2 prior to plating. A ChIP assay was performed 96 hours post-transfection as described in the Experimental Procedures using either an antibody against phosphorylated histone H3 (Ser10) or mouse IgG. The level of phospho-histone H3 bound to the ORF50 promoter was determined by qPCR using promoter-specific primers. Values are given as a relative enrichment compared to the IgG control. Error bars represent standard deviation from the mean. D) Visualization of the qPCR products. PCR products from 5C were run on a 2% agarose gel. Non-specific bands are denoted by an asterisk “*”. See also Supp. Fig. S4.
TLK2 Knockdown Leads to Decreased Phosphorylated Histone H3 Bound to the ORF50 Promoter
The expression and repression of genes, both cellular and viral, is tightly controlled by chromatin structure and modifications. It has been shown that TLKs can phosphorylate histone H3 at the serine 10 position (Li Y, 2001). This histone modification has been shown to modulate transcription (Burkhart et al., 2007, Goto et al., 1999, Van Hooser et al., 1998, Lefebvre, 2002, Mahadevan et al., 1991). In latently infected cells, the KSHV ORF50 promoter is associated with repressive histone, which is released upon reactivation (Gunther and Grundhoff, 2010, Toth et al., 2010). One mechanism by which TLK2 depletion could lead to viral reactivation in KSHV-293 cells is that the histone H3 associated with the ORF50 promoter is dephosphorylated leading to activation and expression of ORF50/RTA.
To determine if depletion of TLK2 led to less phosphorylated histone H3 associated with the ORF50 promoter, we performed a chromatin immunoprecipitation (ChIP) assay. KSHV-293 cells were transfected with either a non-targeting control siRNA or the TLK2 siRNA and incubated for 96 hours. ChIP analysis was performed using an anti-phospho histone H3 (Ser10) or control IgG antibody and the amount of phosphorylated histone H3 bound to the ORF50 promoter was determined by qPCR. Following TLK2 depletion there was an approximate 5.5-fold reduction in the amount of serine 10-phosphorylated histone H3 associated with the ORF50 promoter compared to the control siRNA (Fig. 5C). The qPCR reactions were also run on an agarose gel to visualize the PCR products (Fig. 5D). Since it is possible that the decreased association of phospho-histone H3 with the ORF50 promoter is due to a global decrease in histone H3 phosphorylation, we performed an immunofluorescence assay (IFA) to examine the status of total phosphorylated histone H3 (Ser10). KSHV-293 cells were depleted of TLK2 by siRNA or transfected with a control siRNA and 48 hours later, the cells were stained for pHistone H3 (Ser10). The overall level of phosphorylated histone H3 (Ser10) was unchanged upon TLK2 knockdown (Supp. Fig. 4A). This was confirmed by Western blot as neither total levels of histone H3 nor phospho-histone H3 (Ser10) were altered upon TLK2 knockdown (Supp. Fig. 4B). These data show that the viral reactivation phenotype seen following depletion of TLK2 is associated with less phosphorylated histone H3 bound to the ORF50 promoter and a subsequent increase in expression of ORF50/RTA.
TLKs Also Regulate Epstein-Barr Virus Reactivation From Latency
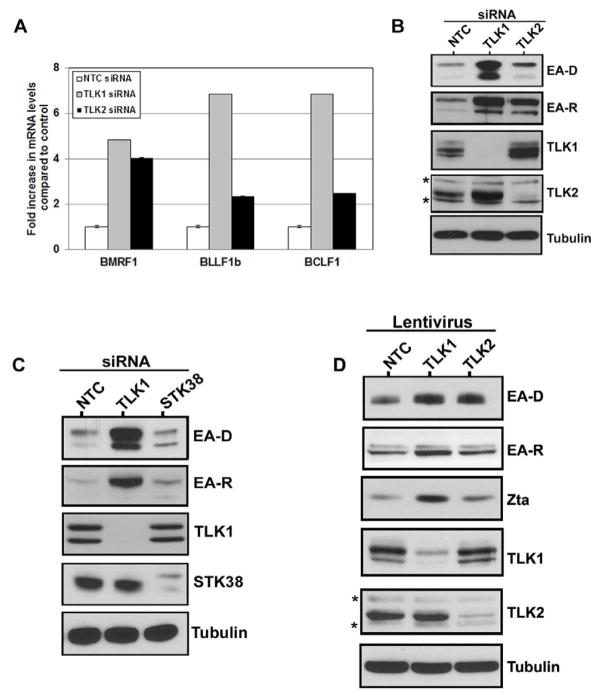
To determine if the actions of the Tousled-like kinases in maintenance of latency are specific to KSHV or if they also modulate reactivation of other related oncogenic gammaherpesviruses, we examined the effects of TLK depletion on the Epstein-Barr virus (EBV) lifecycle. We depleted TLK1 or TLK2 by siRNA in an EBV-infected gastric cancer cell line (AGS-EBV) (Zhou et al., 2005) and measured the expression of signature lytic viral mRNAs and proteins (Figs. 6A and 6B). Interestingly, in the case of EBV, TLK1 appears to have the predominant role in the regulation of EBV reactivation from latency. As can be seen in Fig. 6A, depletion of either TLK1 or TLK2 led to induction of lytic EBV mRNA transcripts, but knockdown of TLK1 resulted in higher levels of all the lytic viral mRNAs tested. A similar effect was seen at the protein level; siRNA-mediated knockdown of either TLK1 or TLK2 led to increased expression of the viral lytic proteins EA-D and EA-R with the highest levels seen following TLK1 depletion (Fig. 6B). To ensure that EBV reactivation in our system is specific to the TLKs, we also knocked down the cellular kinase STK38. Depletion of STK38 in the AGS-EBV cells showed no appreciable reactivation above background (Fig. 6C). We next examined whether the reactivation phenotype observed in the AGS-EBV cells is also seen in the Burkitt’s lymphoma cell line, Akata. Akata-BX1 cells were infected with lentiviruses expressing either a scrambled shRNA, or a shRNA targeting TLK1 or TLK2. Cells were infected for 96 hours at which point cell lysates were harvested and EBV reactivation was measured by expression of the viral lytic proteins EA-D and EA-R, as well as the lytic transactivator protein, Zta. Similar to AGS-EBV cells, knockdown of both TLK1 and TLK2 led to viral reactivation as shown by increased expression of all three lytic proteins (Fig. 6D). Also, cells depleted of TLK1 expressed higher levels of the lytic proteins than those which had TLK2 knocked down, further suggesting that TLK1 is the predominant Tousled-like kinase for maintenance of EBV latency and suppression of reactivation. The levels of viral reactivation in Akata and Akata-BX1 cells following TLK knockdown were also compared to reactivation following human IgG treatment, a known inducing agent of reactivation in Akata cells (Supp. Fig. 5). These data indicate that the Tousled-like kinases not only play a role in KSHV reactivation, but also another gammaherpesvirus, EBV.
Figure 6. Knockdown of TLKs Leads to Reactivation of the Related Gammaherpesvirus, EBV.
A) EBV lytic mRNAs are expressed upon knockdown of the TLKs. AGS-EBV cells were reverse transfected with 50nM of the non-targeting control siRNA or single TLK1 or TLK2 siRNAs. Levels of viral lytic transcripts BMRF1, BLLF1b, and BCLF1 were measured by qPCR at 90 hours post-transfection. Values are normalized to the control siRNA. Error bars represent standard deviation from the mean. B) EBV lytic proteins are induced upon knockdown of the TLKs. AGS-EBV cells were reverse transfected with 50nM of a single siRNA against TLK1, TLK2, or the non-targeting control and cell lysates were harvested 120 hours post-transfection. Western blots were performed for the EBV lytic proteins EA-D and EA-R, as well as TLK1, TLK2, and tubulin. C) Reactivation is specific to the TLKs. AGS-EBV cells were reverse transfected with 50nM of the non-targeting control siRNA or a single siRNA targeting TLK1 or a control kinase, STK38. Cell lysates were harvested 96 hours post-transfection and Western blots were performed for EA-D, EA-R, TLK1, STK38, and tubulin. D) Reactivation in Burkitt’s lymphoma cells. Akata-BX1 cells were infected with lentiviral particles expressing either a non-targeting control (NTC), TLK1, or TLK2 shRNA. 96 hours later cells were harvested and Western blots were performed for the indicated proteins. Asterisks “*” denote non-specific bands. See also Supp. Fig. S5.
Discussion
During KSHV infection the virus undergoes two different transcriptional programs, latency and lytic replication. Following primary infection, KSHV can establish latency in most cells, with only a small proportion of infected cells harboring lytically replicating virus (Zhong et al., 1996, Dittmer, 2003). Both the latent and lytic phases of the viral lifecycle have been shown to be important in KSHV-induced malignancies. Essentially all KS, PEL, and MCD lesions harbor KSHV DNA and the vast majority of these are latently infected (Soulier et al., 1995, Dupin et al., 1999, C. Parravinci, 1997, Staskus et al., 1997, Dittmer, 2011). Latent viral products have been shown to transform cells and affect cell survival and proliferation which supports the premise that latency plays a key role in the generation of KSHV-associated diseases (reviewed in (Järviluoma and Ojala, 2006)). Lytic proteins also play a role in tumorigenesis via paracrine methods as they induce the expression of chemokines and cytokines that promote inflammation, angiogenesis, and proliferation of neighboring cells (Wang et al., 2004, Ye et al., 2007a, Cai J, 1994, Chang et al., 2000).
Since both latent and lytic proteins play key roles in gammaherpesvirus-associated diseases, it is important to understand the mechanisms that regulate the two phases of the viral life cycle. Our kinome screen identified the TLKs as important regulators of gammaherpesvirus reactivation. Although we identified other hits in our screen, we have only validated TLK2 and none of the other hits. When the TLKs were depleted in cells harboring latent KSHV or EBV, there was robust reactivation of the virus as illustrated by expression of viral lytic mRNA and protein, and production of infectious progeny virions. In eukaryotic cells, DNA wraps around histones to form nucleosomes. The structure of chromatin is a critical factor in the regulation of gene function. Heterochromatin is tightly packaged chromatin that has low accessibility to the transcriptional and replicative machinery and is regarded as functionally inactive, while euchromatin is more loosely packed which promotes active transcription of DNA. Post-translational modifications of histones, such as acetylation, methylation, and phosphorylation regulate chromatin structure (reviewed in (Bannister and Kouzarides, 2011)). TLK activity is tightly linked to ongoing DNA replication and the TLKs play a role in chromatin assembly through interaction with, and phosphorylation of the human chromatin assembly factors, Asf1a and Asf1b (Silljé and Nigg, 2001, Takahata et al., 2009).
The same rules that apply to human chromosomes hold true for viral episomes. Similar to the host cell DNA, viral DNA is chromatinized and this plays a role in regulating viral gene transcription. Most of the KSHV and EBV latent episomes are associated with nucleosomes (Stedman et al., 2004, Gunther and Grundhoff, 2010, Toth et al., 2010, Chau and Lieberman, 2004, Zhou et al., 2005). Treatment of latently infected cells (either in cell culture or patients) with histone deacetylase inhibitors stimulates lytic gene expression suggesting that heterochromatin promotes maintenance of latency by suppression of viral lytic genes (Zhou et al., 2005, Ye et al., 2007b, Krown et al., 2011). The KSHV ORF50 promoter is partially repressed by the positioning of a nucleosome over a Sp1 binding site and the transcription start site (Lu et al., 2003, Ye et al., 2005). Histone H3 is known to be a phosphorylation target of the TLKs at residue Ser10 (Li Y, 2001), and histone H3 Ser10 phosphorylation has been shown to lead to chromatin condensation (Burkhart et al., 2007, Goto et al., 1999, Van Hooser et al., 1998). Our data shows that knockdown of TLK2 results in decreased association of Ser10-phosphorylated histone H3 with the KSHV ORF50 promoter, which may allow for more “open” and actively transcribed chromatin resulting in the reactivation phenotype observed. It is also possible that knockdown of TLK2 is inducing a more global change in chromatin and this is an area of future study.
Chromatin structure and histone modifications are significant factors in whether gammaherpesviruses remain in their latent phase, or reactivate and undergo lytic replication. Our discovery that the TLKs play a key role in gammaherpesviral reactivation may potentially be used in a therapeutic strategy to treat malignancies associated with these viruses. The idea of “lytic therapy” involves inducing viral reactivation from latency in combination with antiviral drugs. This was shown for EBV by inducing lytic replication with the chemotherapeutic drugs, doxorubicin and gemcitabine, followed by treatment with anti-herpesviral drugs, such as gancyclovir (Feng et al., 2004), and with bortezomib and radiotherapy for both EBV and KSHV (Fu et al., 2008). Reactivating gammaherpesviruses by depletion of the TLKs in combination with drugs such as gancyclovir could prove to be an effective treatment modality for gammaherpesvirus-associated malignancies.
Experimental Procedures
Cell Culture and Generation of KSHV-293 Cells
HEK-293 and Vero cells were maintained in DMEM media (Cellgro) containing 10% FBS and 1% penicillin-streptomycin (PS). PEL cells were grown in RPMI 1640 media (Cellgro) containing 10% FBS, 1% PS, 0.05mM beta-mercaptoethanol, and 0.007% sodium bicarbonate. AGS-EBV cells were grown in F-12 media (Gibco) containing 10% FBS, 1% PS, and 500 μg/mL G418 as described previously (Marquitz et al., 2012). Akata cells were maintained in RPMI 1640 media (Gibco) containing 10% FBS and 1% PS. Akata-BX1 cells were maintained in RPMI 1640 media containing 1% PS and 0.5 mg/mL G418 as previously described (Molesworth et al., 2000). Induction of EBV reactivation was induced by treatment with 10 μg/mL of anti-human IgG (cat# I5260; Sigma) for 24 hours. KSHV-293 cells were generated by infecting HEK-293 cells with the rKSHV.219 dual reporter virus (a kind gift from J. Vieira)(Vieira and O’Hearn, 2004). KSHV-293 cells were maintained in DMEM media containing 10% FBS, 1% PS, and puromycin (1 μg/mL).
siRNAs and Transfections
All siRNA duplexes were purchased from Dharmacon except the TLK2 siRNA used in Supplemental Figure 2A and 2B which was purchased from Sigma (cat# SIHK2298). The control siRNAs include GAPDH (cat# D-001140-01), non-targeting siRNA #1 (cat# D-001810-01), and UBB (cat# LU-013382). ON-TARGETplus Set of 4 Upgrade pool of siRNAs were used for TLK1 (cat# LU-004174), TLK2 (cat# LU-005389), and STK38 (cat# LU-004674). The target sequences of the siRNAs are listed in Supplemental Table 1. Where indicated, single siRNAs from the set of pooled siRNAs against TLK2 were used. Cells were transfected with siRNAs using either Dharmacon DharmaFECT1 (HEK293, KSHV-293, and AGS-EBV cells) or Invitrogen Lipofectamine RNAiMAX (BCBL-1 cells). Where indicated, fluorescent microscopy images were acquired using a Nikon Eclipse Ti inverted microscope.
shRNA and Lentivirus
The scrambled control and TLK1 (TRCN0000007057) shRNAs are in the pLKO.1-puro background and were purchased from Sigma Aldrich. The TLK2 shRNA was generated by insertion of the hairpin target sequence into the pLKO.1-puro vector by site-directed mutagenesis using the Quikchange II XL kit (Agilent Technologies). The shRNA sequences are listed in Supplemental Table 1. Lentivirus for each of the shRNAs was produced by using the ViraPower Lentiviral Expression System (Invitrogen) per manufacturer’s instructions. Lentiviral infections were performed by addition of lentivirus and polybrene (4 μg/mL) to cells in 15 mL conical tubes and spinoculation at 2500 rpm for 2 hours at 30°C as described previously (West and Damania, 2008).
siRNA Screen
For the primary screen KSHV-293 cells were reverse transfected using the Dharmacon SMARTpool protein kinase siRNA library (cat #G-003505-10) in a 384-well format purchased by the RNAi Screening Core within the UNC Lineberger Comprehensive Cancer Center. Each well contained a pool of 4 different siRNAs targeting a single gene. The siRNA library consists of 3 “Mother plates” that were each dispensed into triplicate “Daughter plates” using a Biomek FXp Laboratory Automation Workstation (Beckman Coulter). The control siRNAs GAPDH (cat# D-001140-01) and UBB (cat# J-013382-05) were pipetted by hand into the plates. The reverse transfection was performed in the following manner. DMEM media with no phenol red (GIBCO cat# 31053) was added to the wells of the daughter plates using a MicroFlo Select dispenser (BioTek) followed by the addition of the siRNAs so that the final concentration was 25nM/well. DharmaFECT1/DMEM mixture was made and incubated at room temperature for 5 minutes and then added to the wells. The plate was manually mixed by gentle agitation by hand and then spun down and incubated at room temperature for 25 minutes. While the plate was incubating, KSHV-293 cells were trypsinized, counted, and resuspended to reach a concentration of 2500 cells/well. Cells were seeded on top of the DharmaFECT/siRNA mixture using the MicroFlo Select, briefly centrifuged, and incubated at 37°C for 70 hours. At 70 hours post-transfection plates were imaged using a Cellomics ArrayScan VTI HCS Reader (Thermo Scientific) to measure GFP and RFP intensity. For each well, five different fields were imaged and analyzed.
Western Blots
At the indicated times post-transfection, cells were washed with cold PBS and lysed in RIPA buffer (150mM NaCl, 1% NP-40, 50mM Tris pH8, 0.5% sodium deoxycholate, 0.1% SDS) with protease inhibitor cocktail (Roche) for 30 minutes at 4°C. Samples were clarified by centrifugation for 10 minutes at 8.2 × g. Protein amounts were determined by a Bradford assay (BioRad) and equal amounts of protein were resolved on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked for 30 minutes at room temperature in either 5% nonfat dry milk (NFDM) or 5% BSA in a 1X TBS/0.1% Tween (TBST) solution. Membranes were then incubated with primary and secondary antibodies. For antibodies used, see the Supplemental Experimental Procedures section.
Viral Load Assay
For determining intracellular viral load, cells were harvested at the indicated timepoints and DNA was extracted using the Qiagen DNeasy Blood & Tissue Kit according to the manufacturer’s instructions. The ORF57 primer was used for determining viral genomes and genome copies were quantitated by comparison to an ORF57 plasmid-derived standard curve. For viral lytic mRNA transcription, cells were harvested and RNA was isolated using the Qiagen RNeasy Plus Mini Kit followed by treatment with 1 unit of RQ1 DNase (Promega) per μg of RNA for 30 minutes at 37°C. DNase was heat-inactivated at 65°C for 10 minutes. cDNA was reverse transcribed from 100ng of RNA using MMLV reverse transcriptase (Invitrogen) and random hexamer primers (Invitrogen) by incubation at 37°C for 60 minutes followed by heat-inactivation at 95°C for 5 minutes. Gene expression levels were determined by quantitative Real-Time PCR (qPCR) using Power SYBR Green PCR mastermix (Applied Biosystems). The qPCR reactions were run on an ABI 7300 machine. Relative fold calculations were determined by the ΔΔCT method (Dittmer, 2003).
Reverse Transcriptase PCR
Total RNA was extracted and converted into cDNA as described above for the viral load assay. The cDNA was then used as the template in reverse transcriptase PCR (RT-PCR) reactions using Taq polymerase (Qiagen). PCR products were then run out on an agarose gel.
KSHV Infection Assay
KSHV-293 cells were transfected with the indicated siRNAs for 96 hours. Supernatants were collected and spun down at 5,000 rpm for 5 minutes to remove cellular debris. Polybrene (4 μg/mL) was added to the clarified supernatant and incubated for 10 minutes at room temperature. The supernatants were then added to naïve Vero cells (80,000 cells/well) in a 12-well plate and spinoculated at 2,500 rpm for 1.5 hours at 30°C as previously described (West and Damania, 2008). Ninety-six hours later, cells were examined by fluorescent microscopy using the Nikon Eclipse Ti inverted microscope for GFP expression and intracellular KSHV viral loads were determined by qPCR as described above.
Luciferase Assay
To assay ORF50 activity, 120,000 HEK-293 cells/well of a 12-well plate were reverse transfected with siRNAs against GAPDH, TLK1, or TLK2 at a final concentration of 50nM and incubated at 37°C for 24 hours. Cells were then tr ansfected with 2 μg of a KSHV ORF50 promoter luciferase reporter plasmid and incubated for 48 hours at 37°C. Cells were harvested and luciferase activity was measured according to manufacturer’s instructions (Promega).
KSHV Viral Array
KSHV-293 cells were either transfected with the indicated siRNAs or treated with 0.1mM sodium butyrate and incubated for 96 hours at 37°C. Total RNA was extracted and treated with DNase as described above for the viral load assay. cDNA was reverse transcribed from 10 μg of RNA using the Superscript II system (Invitrogen) and random hexamer primers (Invitrogen). The reaction mixture was sequentially incubated at 30°C for 15 minutes, 42°C for 15 minutes, 52°C for 15 minutes, and 70°C for 10 minutes. Following reverse transcription, the remaining RNA was digested by treatment with 1 U of RNaseH for 30 minutes at 37°C and the reaction was quenched by heating to 95°C for 5 minutes. The dup licate samples were subjected to a KSHV real-time PCR viral array as described previously (Dittmer, 2003) with the addition of redundant primers, such that more than one primer pair was used to measure each known viral transcript.
Chromatin Immunoprecipitation (ChIP)
KSHV-293 cells were plated at 2 × 106 cells per 10-cm dish and transfected with 50nM of either a non-targeting control siRNA or a siRNA targeting TLK2 and cells were incubated for 96 hours post-transfection. The ChIP assay was performed using the EZ-ChIP kit (Millipore cat#17-371) per manufacturer’s instructions. Immunoprecipitations were performed by adding either 2 μg of normal mouse IgG or anti-phospho histone H3 (Ser10) (Millipore cat#CS200553) and incubating overnight at 4°C. The amount of serine 10 phosphor ylated histone H3 bound to the ORF50 promoter was determined by qPCR using ORF50 promoter-specific primers (Fwd: GGTACCGAATGCCACAATCTGTGCCCT; Rev: TTGTGGCTGCCTGGACAGTATTCTCAC).
For all the above assays, additional details and primer sequences can be found in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
A kinome screen identified Tousled-like kinases (TLKs) as regulators of KSHV reactivation
TLK depletion activated lytic gene expression, including the viral lytic switch protein
TLK knockdown in latently-infected cells led to complete KSHV reactivation
Knockdown of TLKs also reactivated the related gammaherpesvirus, EBV, from latency
Acknowledgments
We thank Brian Golitz and Noah Sciaky (University of North Carolina, Chapel Hill, NC) for technical and analytical assistance and the members of the Damania and Dittmer labs for helpful discussions. We thank Lindsey Hutt-Fletcher for the gift of the AGS-EBV and Akata-BX1 cell lines. PJD was supported by the LCCC training grant, T32CA009156. BD is supported by grants CA096500 and CA163217, and BD, DD and NRT are supported by grant CA019014. BD is a Leukemia & Lymphoma Society Scholar and a Burroughs Wellcome Fund Investigator in Infectious Disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart BA, et al. Osmotic Stress-dependent Repression Is Mediated by Histone H3 Phosphorylation and Chromatin Structure. Journal of Biological Chemistry. 2007;282:4400–4407. doi: 10.1074/jbc.M609041200. [DOI] [PubMed] [Google Scholar]
- Parravinci C, MC, Paulli M, Magrini U, Lazzarino M, Moore PS, Chang Y. Expression of a virus-derived cytokine, KSHV vIL-6, in HIV-seronegative Castleman’s disease. The American Journal of Pathology. 1997;151:1517–1522. [PMC free article] [PubMed] [Google Scholar]
- Cai JGP, Masood R, Chandrasoma P, Jung B, Law Re, Radka Sf. Oncostatin-M is an autocrine growth factor in Kaposi’s sarcoma. The American Journal of Pathology. 1994;145:74–79. [PMC free article] [PubMed] [Google Scholar]
- Canfield CRJ, De Benedetti A. TLK1B promotes repair of DSBs via its interaction with Rad9 and Asf1. BMC Molecular Biology. 2009:10. doi: 10.1186/1471-2199-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, et al. Kaposi’s Sarcoma–Associated Herpesvirus-Like DNA Sequences in AIDS-Related Body-Cavity–Based Lymphomas. New England Journal of Medicine. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chang J, et al. Inflammatory Cytokines and the Reactivation of Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. Virology. 2000;266:17–25. doi: 10.1006/viro.1999.0077. [DOI] [PubMed] [Google Scholar]
- Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chau CM, Lieberman PM. Dynamic chromatin boundaries delineate a latency control region of Epstein-Barr virus. J Virol. 2004;78:12308–19. doi: 10.1128/JVI.78.22.12308-12319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, et al. KSHV Reactivation from Latency Requires Pim-1 and Pim-3 Kinases to Inactivate the Latency-Associated Nuclear Antigen LANA. PLoS Pathog. 2009;5:e1000324. doi: 10.1371/journal.ppat.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B, et al. Comparison of the Rta/Orf50 Transactivator Proteins of Gamma-2-Herpesviruses. Journal of Virology. 2004;78:5491–5499. doi: 10.1128/JVI.78.10.5491-5499.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer DP. Transcription Profile of Kaposi’s Sarcoma-associated Herpesvirus in Primary Kaposi’s Sarcoma Lesions as Determined by Real-Time PCR Arrays. Cancer Research. 2003;63:2010–2015. [PubMed] [Google Scholar]
- Dittmer DP. Restricted Kaposi’s sarcoma (KS) herpesvirus transcription in KS lesions from patients on successful antiretroviral therapy. MBio. 2011;2:e00138–11. doi: 10.1128/mBio.00138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin N, et al. Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease, and primary effusion lymphoma. Proceedings of the National Academy of Sciences. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng WH, et al. Lytic Induction Therapy for Epstein-Barr Virus-Positive B-Cell Lymphomas. Journal of Virology. 2004;78:1893–1902. doi: 10.1128/JVI.78.4.1893-1902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DX, et al. Bortezomib-induced enzyme-targeted radiation therapy in herpesvirus-associated tumors. Nat Med. 2008;14:1118–22. doi: 10.1038/nm.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, et al. Identification of a Novel Phosphorylation Site on Histone H3 Coupled with Mitotic Chromosome Condensation. Journal of Biological Chemistry. 1999;274:25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- Gregory SM, et al. Toll-like receptor signaling controls reactivation of KSHV from latency. Proceedings of the National Academy of Sciences. 2009;106:11725–11730. doi: 10.1073/pnas.0905316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Ganem D. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. The Journal of Clinical Investigation. 2004;113:124–136. doi: 10.1172/JCI200417803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, et al. PKU-β/TLK1 regulates myosin II activities, and is required for accurate equaled chromosome segregation. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2008;657:63–67. doi: 10.1016/j.mrgentox.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Järviluoma A, Ojala PM. Cell signaling pathways engaged by KSHV. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2006;1766:140–158. doi: 10.1016/j.bbcan.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Krown SE, et al. Pilot study of oral valganciclovir therapy in patients with classic Kaposi sarcoma. J Infect Dis. 2011;203:1082–6. doi: 10.1093/infdis/jiq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Ozato K, Lefebvre P. Phosphorylation of histone H3 is functionally linked to retinoic acid receptor beta promoter activation. EMBO Rep. 2002;3:335–340. doi: 10.1093/embo-reports/kvf066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YDR, Anthony C, Sunavala G, De Benedetti A. A translationally regulated Tousled kinase phosphorylates histone H3 and confers radioresistance when overexpressed. Oncogene. 2001;20:726–738. doi: 10.1038/sj.onc.1204147. [DOI] [PubMed] [Google Scholar]
- Lu F, et al. Chromatin Remodeling of the Kaposi’s Sarcoma-Associated Herpesvirus ORF50 Promoter Correlates with Reactivation from Latency. Journal of Virology. 2003;77:11425–11435. doi: 10.1128/JVI.77.21.11425-11435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan LC, et al. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- Marquitz AR, et al. Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc Natl Acad Sci U S A. 2012;109:9593–8. doi: 10.1073/pnas.1202910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molesworth SJ, et al. Epstein-Barr Virus gH Is Essential for Penetration of B Cells but Also Plays a Role in Attachment of Virus to Epithelial Cells. Journal of Virology. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet JL, et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–13. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- Roe JL, et al. TOUSLED Participates in Apical Tissue Formation during Gynoecium Development in Arabidopsis. The Plant Cell Online. 1997;9:335–353. doi: 10.1105/tpc.9.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JL, et al. The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell. 1993;75:939–950. doi: 10.1016/0092-8674(93)90537-z. [DOI] [PubMed] [Google Scholar]
- Silljé HHW, Nigg EA. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Current Biology. 2001;11:1068–1073. doi: 10.1016/s0960-9822(01)00298-6. [DOI] [PubMed] [Google Scholar]
- Sodhi A, et al. Does dysregulated expression of a deregulated viral GPCR trigger Kaposi’s sarcomagenesis? Faseb J. 2004;18:422–7. doi: 10.1096/fj.03-1035hyp. [DOI] [PubMed] [Google Scholar]
- Songyang Z, et al. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–85. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease [see comments] Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- Staskus KA, et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. Journal of Virology. 1997;71:715–9. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, et al. ORC, MCM, and Histone Hyperacetylation at the Kaposi’s Sarcoma-Associated Herpesvirus Latent Replication Origin. Journal of Virology. 2004;78:12566–12575. doi: 10.1128/JVI.78.22.12566-12575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stürzl M, et al. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi’s sarcoma. International Journal of Cancer. 1997;72:68–71. doi: 10.1002/(sici)1097-0215(19970703)72:1<68::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, et al. The radioresistance kinase TLK1B protects the cells by promoting repair of double strand breaks. BMC Molecular Biology. 2005;6:19. doi: 10.1186/1471-2199-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, et al. A dominant negative mutant of TLK1 causes chromosome missegregation and aneuploidy in normal breast epithelial cells. BMC Cell Biology. 2003;4:16. doi: 10.1186/1471-2121-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, et al. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 2009;28:3378–89. doi: 10.1038/emboj.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team., R. D. C. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. URL http://www.R-project.org. [Google Scholar]
- Tiedemann RE, et al. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 2010;115:1594–1604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, et al. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 2010;6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser A, et al. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. Journal of Cell Science. 1998;111:3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- Vieira J, O’hearn PM. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325:225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- Wang HW, et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–693. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. The protein kinase TOUSLED is required for maintenance of transcriptional gene silencing in Arabidopsis. EMBO Rep. 2007;8:77–83. doi: 10.1038/sj.embor.7400852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J, Damania B. Upregulation of the TLR3 Pathway by Kaposi’s Sarcoma-Associated Herpesvirus during Primary Infection. Journal of Virology. 2008;82:5440–5449. doi: 10.1128/JVI.02590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, et al. Kaposi’s Sarcoma-Associated Herpesvirus Promotes Angiogenesis by Inducing Angiopoietin-2 Expression via AP-1 and Ets1. Journal of Virology. 2007a;81:3980–3991. doi: 10.1128/JVI.02089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, et al. De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi’s sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J Virol. 2007b;81:9279–91. doi: 10.1128/JVI.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, et al. An Sp1 Response Element in the Kaposi’s Sarcoma-Associated Herpesvirus Open Reading Frame 50 Promoter Mediates Lytic Cycle Induction by Butyrate. Journal of Virology. 2005;79:1397–1408. doi: 10.1128/JVI.79.3.1397-1408.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, et al. Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus. PLoS Pathog. 2007;3:e44. doi: 10.1371/journal.ppat.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, et al. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proceedings of the National Academy of Sciences. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, et al. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 2005;24:1406–17. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.