Abstract
Glucose-6-phosphate (G-6-P) formation in Staphylococcus aureus is catalysed by glucokinase (glkA) gene under high glucose concentration leading to upregulation of various pathogenic factors; therefore the present study is aimed in the cloning and characterization of glk A gene from S. aureus ATCC12600. The glk A gene was cloned in the Sma I site of pQE 30, sequenced (Accession number: JN645812) and expressed in E. coli DH5α. The recombinant glk A expressed from the resultant glk A 1 clone was purified using nickel metal chelate chromatography, the pure enzyme gave single band in SDS-PAGE with molecular weight of 33kDa. The rglk A showed very high affinity to glucose Km 5.1±0.06mM with Hill coefficient of 1.66±0.032mM. Analysis of glucokinase sequence of S. aureus showed presence of typical ATP binding site and ROK motif CNCGRSGCIE. Sequentially and phylogenetically S. aureus glk A exhibited low identity with other bacterial glk A and 21% homology with human glucokinase (GCK). Functionally, S. aureus glk A showed higher rate of G-6-P formation compared to human GCK which may have profound role in the pathogenesis.
Keywords: glk A, pQE 30, ROK, Km
Background
Staphylococcus aureus is an important nosocomial and community-acquired pathogen and its infections are known to be a leading cause of bacteremia and are associated with severe morbidity and mortality in hospitalized infections [1]. In S. aureus 85% of Glucose is catabolised through EMP pathway and the very first step of the formation of G-6-P is carried out in two pathways one through phosphotransferase (PTS) which usually functions under low glucose concentration and the second PTS independent system catalysed by glucose permease (glk U) and glucokinase (glk A) which functions mostly under high glucose concentration [2]. This G-6-P plays critical role in the pathogen for energy generation in catabolic reactions for the synthesis of all the intermediates for its very survival [3].
Recent findings of cytoplasmic glucokinase is detected in both Gram positive and Gram negative bacteria has 315-321 residues and a monomeric mass of 33-35 kDa, Km values of glucokinase varied from 0.3- 0.8mM for glucose and 0.4-4mM for ATP substrates in both Gram positive and Gram negative bacteria [4]. This enzyme showed greater affinity for glucose than with either mannose or galactose and showed detectable activity with fructose in Gram positive bacteria, while this enzyme showed no affinity for fructose in Gram negative bacteria [5]. Glk A in Gram positive bacteria is composed of higher percentage of alpha-helix compared to β-strand or β-turns and the enzyme exhibited both structural and functional differences with human glucokinase. Glucokinases of bacteria are divided in to 2 groups (1) Glucokinases that belongs to repressor/open reading frames of unknown function/sugar kinases (ROK) family which is characterized by the presence of CXCGX (2)GCXE motifs, and (2) Glucokinases without ROK motifs. However, in Archaea two types of glucokinases are found one ATP-dependent glucokinase and the other ATP-dependent glucokinase having ROK motifs [6]. In multidrug resistant and vancomycin resistant strains of S. aureus increased cell wall biosynthesis and high reductive conditions are the characteristic features where G-6-P formation in such strains may be playing a critical role in drug resistance and pathogenesis [7]. Therefore glk A may be playing a pivotal role, hence the present study is focused on the cloning, expression and characterization of glucokinase from Staphylococcus aureus ATCC12600.
Methodology
Bacterial strains and culture conditions:
Staphylococcus aureus ATCC12600 and Escherichia coli DH5α were obtained from Bangalore Genei Pvt Ltd. S. aureus was grown on modified Baird Parkar media at 37°C. After overnight incubation, a single black shiny colony with distinct zone was picked, inoculated in Brain heart infusion (BHI) broth and incubated at 37°C for overnight. Thus, grown S. aureus ATCC12600 culture was used to characterize Glucokinase and for the isolation of chromosomal DNA for glk A gene amplification. E. coli DH5α was used in the expression of S. aureus glucokinase cloned in pQE 30 vector [8, 9].
Kinetic study of Glucokinase:
The enzyme assay was performed on a Cyber lab spectrophotometer (USA). Glucokinase is exclusively present in cytoplasm of bacteria therefore cytoplasmic fraction was isolated from the bacteria [10]. 2ml of reaction mixture contains the following components 60Mm Tris-HCl buffer pH-7.5, 0.5 mM MgCl2, 0.2M ATP, 0.9mM NAD, 1unit of Glucose-6- phosphate dehydrogenase, 12mM Glucose (substrate) and 10µl of crude (isolated from S. aureus ATCC 12600) or pure enzyme and incubated for 30 minutes at 37°C. The absorbance was measured at 340 nm against blank. Enzyme activity was expressed as the micro moles of NADH formed per ml. per minute which is equivalent to the amount of G-6-P formed [11].
Calculation of Hills coefficient:
The Hills coefficient was calculated by plotting the graph with log [Vi / Vmax – Vi] on Y-Axis and log [S] on X- axis where Vi is the velocity at different substrate concentrations, Vmax is the maximum velocity of the enzyme at which the enzyme is fully saturated with the substrate concentration [11].
glk A gene Amplification and sequencing from Staphylococcus aureus ATCC12600:
glk A gene was amplified from S. aureus chromosomal DNA using the primers glk A -F: 51-CGCT GAT GTA GGC GGG ACG 31 and glk A -R: 51-GGT CTT GAT TAA TCC 31 which were designed from the Glucokinase gene sequence of S. aureus Mu 50 strain [12]. The reaction mixture contained in a final volume of 50µl which consisted of 100ρmoles of each primers, 100µmol of dNTPS mix, 10mM Tris- HCl (pH 8.8), 1.5mM MgCl2, 1U of hot start Taq DNA Polymerase (Bangalore Genei pvt ltd) and 0.25µg of chromosomal DNA. Amplification parameters included an initial denaturation step for 10 min at 94°C; 35 cycles of 94°C for 60 seconds of denaturation, 60 seconds of annealing at 460C and 100 seconds of amplification at 72°C which was followed by a final extension step at 72°C for 5 min in a Mastercycler gradient Thermocycler (Eppendorf). Amplified products were purified with NP-PCR Purification kit, Taurus Scientific, USA and were sequenced by dye terminating method at MWG Biotech India Ltd. Thus, obtained glk A gene sequence was deposited at Gen Bank (http://www.ncbi.nlm.nih.gov/nuccore/JN645812).
Cloning, expression and purification of Glucokinase:
glk A gene was cloned in the Sma I site of pQE 30 and transformed into E. coli DH5α, the resultant clone was named as glk A1. The insert in the clone was sequenced and after ascertaining the sequence of glk A was over expressed with 1mM IPTG. The rglk A was purified from the cytosolic fraction of glk A1 clone by passing through nickel metal chelate agarose column (by following QIA express expression system protocol) and protein was eluted using 300mM immidazole hydrochloride. The product was analysed on 10% SDS-PAGE [8]. The enzyme kinetics of purified rglk A was performed as described in earlier section.
Sequence Analysis of glk A:
The glk A sequences were retrieved for Staphylococcus aureus (AEY84930.1), Bacillus subtilis (YP_004877992.1), Bacillus cereus (AAU16254.1), Bacillus megaterium (ADF41316.1), Corynebacterium glutamicum (AAF80161.1), Renibacterium salmoninarum (YP_001625658.1) from Genbank. All these structures were scanned against PROSITE data base to predict the structures of ROK motif. The predicted ROK motifs of all these glk A sequences were aligned using clustalX tool [13] to find out the ROK signature and dissimilarities among them.
Results
Cloning, expression and characterization of glucokinase gene:
In the present study glk A gene was cloned, expressed and characterized from S. aureus ATCC 12600. glk A gene was amplified from the chromosomal DNA of S. aureus ATCC 12600 and the PCR product of (0.987kb) (Figure 1A) was cloned in the Sma I site of pQE 30 vector and transformed in E. coli DH5α and the clone was named as glk A1. The insert in the clone was confirmed by PCR using the same primers and the PCR product was sequenced (Accession number: JN645812) and the BLAST results resembled with all the glk A gene sequence reported for other strains of S. aureus. The PCR product obtained was cloned in “-1” frame and successfully induced with 1mM IPTG for glk A expression in clone glk A1. The 10% SDS PAGE results clearly indicated the expression of rglk A with a molecular weight 33 KDa corresponding to the insert cloned (Figure 1B). The enzyme kinetics of pure recombinant glk A exhibited in the cytosol was 0.1053±0.01mM of NADPH/ml/min and Km 5.22± 0.17mM with Hill coefficient of 1.71± 0.025mM which is close to the native glk A of S. aureus ATCC12600 (0.20817±0.04mM of NADPH/ml/min and Km 5.1±0.06mM with Hill coefficient of 1.66±0.032mM). The close Km and Hill coefficient values of native glk A and rglk A corroborated the sequence results indicating presence of only one kind of glk A in the S. aureus Table 1 (see supplementary material). These results also explain that both native glk A and rglk A showed positive cooperativity towards glucose. The above results also reflected on the functional properties of the glk A, with human GCK showing very high Km compared with S. aureus Km suggesting lower affinity of substrate for the enzyme Table 2 (see supplementary material). From the results it can be concluded that S. aureus glk A and human GCK are dissimilar enzymes.
Figure 1.
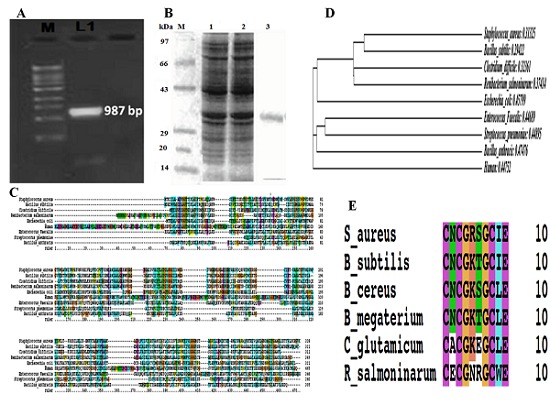
A) PCR amplification of glk A gene (0.987kb) from clone glk A1; B) SDS-PAGE analysis of analysis glk A protein from recombinant clone glk A 1; C) Sequence alignment of bacterial glk A sequences and human glk A sequence; D) Phylogenetic analysis of S. aureus with other Gram positive & Gram negative bacterial glk A; E) Multiple sequence alignment of S. aureus with other Gram positive and Gram negative bacterial glk A which is having ROK CXCGX(2)GCXE motif.
Sequence analysis of S. aureus glk A:
The glk A protein sequence of S. aureus ATCC12600 showed highly conserved regions with both Gram negative (E. coli) and positive (Bacillus subtilis) bacteria, while very low identity (21%) was observed with human GCK gene (Figure 1C). A clear distinction between S. aureus glk A and other glucokinases of both Gram positive and Gram negative pathogens was observed in multiple sequence alignments and phylogenetic analysis (Figure 1D) which showed that phylogenetically not very close relationship occurs among bacteria and humans. Further, the glk A sequence of S. aureus showed typical ATP binding site and ROK motif which is indicated by three conserved C residues in the motif CNCGRSGCIE (Figure 1E) such ATP dependent glk A having ROK motif is the property of glk A of Arachae. Therefore, it was exciting to identify the role of the amino acid sequences that are uniquely conserved in glk A of S. aureus which is having very low homology with other glk A of Gram positive and Gram negative bacteria indicating variable rate of G-6-P formation which also reflects on variability in the pathogenesis.
Discussion
In Staphylococcus aureus the glk A functions optimally at higher glucose concentration which probably leads to increased reductive conditions in the pathogen probably resulting in upregulation of butanediol pathway, cell wall biosynthesis, Polysaccharide Intracellular Adhesion (PIA) molecules, exopolysaccharide synthesis and dTMP levels, these are the characteristic features Small Colony Variants (SCV) which are usually observed in relapsed episodes of S. aureus infections in humans [14, 15].
The kinetic results of the present study were closed to native glk A and exhibited higher affinity for glucose compared to other pathogenic bacteria (Table 2) these results correlated with the multiple sequence alignment and phylogenetic analysis of glk A (Figure 1D) indicating the rate of G-6-P formation is different in all these organisms. The pure glucokinase was obtained from glk A1 [8, 9] which exhibited molecular weight of 33 kDa similar to native protein [4]. The sequence analysis of S. aureus glk A showed typical ATP binding site and ROK motifs similar to the glk A present in Archaea [6] which is different in both Gram positive and negative pathogenic bacteria indicating the glk A in S. aureus is a regulated enzyme.
The human GCK counter part of S. aureus glk A showed absence of ROK motif and exhibited higher Km (Table 2) these differences between glk A of S. aureus and human GCK probably suggests that in S. aureus glucose is rapidly phosphorylated and used more in the anabolic pathways such as increased nucleotide biosynthesis, cell wall biosynthesis resulting in rapid proliferation and formation of SCV which are one of the key pathogenic factors of this pathogen [16–21].
Supplementary material
Acknowledgments
This paper forms a part of Ph.D thesis work going to be submitted to JNT University, Hyderabad- 500085, A.P, India.
Footnotes
Citation:Lakshmi et al, Bioinformation 9(4): 169-173 (2013)
References
- 1.Frederiksen MS, et al. Pediatr Infect Dis J. 2007;26:398. doi: 10.1097/01.inf.0000261112.53035.4c. [DOI] [PubMed] [Google Scholar]
- 2.Friedrich götz, et al. Prokaryotes. 2006;4:5. [Google Scholar]
- 3.Blumenthal HJ, et al. Ann N Y Acad Sci. 1974;236:105. doi: 10.1111/j.1749-6632.1974.tb41485.x. [DOI] [PubMed] [Google Scholar]
- 4.Strasters KC, et al. J Gen Microbiol. 1963;33:213. doi: 10.1099/00221287-33-2-213. [DOI] [PubMed] [Google Scholar]
- 5.Meyer D, et al. J Bacteriol. 1997;179:1298. doi: 10.1128/jb.179.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kengen SW, et al. J Biol Chem. 1995;270:30453. doi: 10.1074/jbc.270.51.30453. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Y, et al. Infection and Immunity. 2009;77:4256. doi: 10.1128/IAI.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prasad UV, et al. Appl Biochem Biotechnol. 2013;169:862. doi: 10.1007/s12010-012-0027-8. DOI 10.1007/s12010-012-0027-8. [DOI] [PubMed] [Google Scholar]
- 9.Hari Prasad O. Protein J. 2012;31:345. doi: 10.1007/s10930-012-9410-0. [DOI] [PubMed] [Google Scholar]
- 10.Cardenas ML, et al. J Biochem. 1984;145:163. doi: 10.1111/j.1432-1033.1984.tb08536.x. [DOI] [PubMed] [Google Scholar]
- 11.Patni NJ. J Bacteriol. 1971;105:220. doi: 10.1128/jb.105.1.220-225.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohta T. DNA Res. 2004;11:51. doi: 10.1093/dnares/11.1.51. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, et al. Nucleic Acids Res. 1997;25:4876. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauffman MG, et al. Mol Cell Biol. 1991;11:2538. doi: 10.1128/mcb.11.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zander J, et al. Infect Immun. 2008;76:1333. doi: 10.1128/IAI.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer D, et al. J Bacteriol. 1997;179:1298. doi: 10.1128/jb.179.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierre skarlatos, et al. J Bacteriol. 1998;180:3222. doi: 10.1128/jb.180.12.3222-3226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu, et al. J Mol Biol. 1967;24:213. doi: 10.1016/0022-2836(67)90327-0. [DOI] [PubMed] [Google Scholar]
- 19.Concha MI, et al. FEMS Microbiol Lett. 2000;186:97. doi: 10.1111/j.1574-6968.2000.tb09088.x. [DOI] [PubMed] [Google Scholar]
- 20.Han B, et al. Appl Environ Microbiol. 2007;73:3581. doi: 10.1128/AEM.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathnakar Reddi KVN, et al. Journal of pharmacy research. 2013;205:9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


