Abstract
Reprogramming to induced pluripotent stem cells (iPSCs) proceeds in a step-wise manner with reprogramming factor binding, transcription, and chromatin states changing during transitions. Evidence is emerging that epigenetic priming events early in the process may be critical for pluripotency induction later. Chromatin and its regulators are important controllers of reprogramming, and reprogramming factor levels, stoichiometry, and extracellular conditions influence the outcome. The rapid progress in characterizing reprogramming is benefiting applications of iPSCs and already enabling the rational design of novel reprogramming factor-cocktails. However, recent studies have uncovered an epigenetic instability of the X-chromosome in human iPSCs that warrants careful consideration.
Decades of research were dedicated to studies of cell fate changes during development and led to the view that, in vivo, differentiated cells are irreversibly committed to their fate. However, reprogramming of somatic cells by transfer into enucleated oocytes pioneered by John Gurdon and colleagues in the 50ies (Gurdon et al., 1958), fusion with other cell partners (Blau et al., 1983), or ectopic transcription factor expression (Davis et al., 1987; Takahashi and Yamanaka, 2006), revealed a remarkable plasticity of the differentiated state. Particularly the exposure to ectopic transcription factors offers a powerful and unexpectedly flexible technique to shift a somatic cell towards alternative somatic identities or pluripotency. The reprogramming field exploded after Takahashi and Yamanaka established a major landmark with the generation of induced pluripotent stem cells (iPSCs) from fibroblasts by simple ectopic expression of Oct4 (O), Sox2 (S), cMyc (M), and Klf4 (K) (Takahashi and Yamanaka, 2006). Aptly, the Nobel Prize, awarded to John Gurdon and Shinya Yamanaka in 2012, symbolizes the extraordinary contribution reprogramming experiments made (and will make) to our understanding of cellular identity and the apparently unlimited practical applications of iPSCs and other reprogrammed cells.
The beauty of transcription factor-induced reprogramming to iPSCs lies in its simplicity and robustness since many different cell types from a wide range of species can be reprogrammed to pluripotency by ectopic expression of OSKM (for a recent summary see (Stadtfeld and Hochedlinger, 2010)). A fundamental feature of the resulting iPSCs is that they are, in their ideal state, functionally indistinguishable from embryonic stem cells (ESCs), which are pluripotent cells derived from pre-implantation embryos, and capable of differentiation into cells of all three germ layers (Bock et al., 2011; Carey et al., 2011) Consequently, reprogramming changes the transcriptome and chromatin state of the somatic cell to that of a pluripotent cell (Chin et al., 2009; Hawkins et al., 2010; Lister et al., 2011; Maherali et al., 2007; Mikkelsen et al., 2008; Okita et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007). Therefore, iPSCs offer an invaluable source of patient-specific pluripotent stem cells for disease modeling, drug screening, toxicology tests, and regenerative medicine (recently reviewed in (Onder and Daley, 2012; Trounson et al., 2012)), and already have been employed to unmask novel insights into human diseases (Koch et al., 2011) .
Despite the extraordinary fidelity of the iPSC technology, the induction of pluripotency upon OSKM expression typically requires an extended latency period of around 1-2 weeks and occurs in less than 1% of the starting cells, even when they are genetically identical and the expression levels of the four transcription factors are similar across all cells in the culture dish (for a review see (Stadtfeld and Hochedlinger, 2010)). Albeit heterogeneity of the starting cell population and differentiation state may affect reprogramming efficiency to a certain degree (Stadtfeld and Hochedlinger, 2010), a key question has therefore been why only a few of a pool of seemingly equivalent OSKM-expressing cells induce pluripotency. Genomic approaches, RNAi screens and simpler genetic methods, as well as emerging single cell analyses, are beginning to provide answers by defining critical reprogramming events as well as regulators and epigenetic properties that promote or hinder specific reprogramming transitions, which we will focus on in the first part of this Review.
Particularly the activation of pluripotency genes appears to present a formidable task for the reprogramming factors. Generally, transcriptional activation begins with the binding of transcription factors to distal enhancer and promoter elements, which initiates the recruitment of co-activators and facilitates the binding of the general transcription machinery and the assembly of the RNA Polymerase II-containing pre-initiation complex (PIC) at the core promoter (Green, 2005). Transcription factors can also promote steps in the transcription process subsequent to PIC assembly (which is of interest for the reprogramming factor cMyc) (Green, 2005). Importantly, the packaging of DNA into nucleosomes affects all aspects of transcription, from transcription factor binding to PIC formation and transcriptional elongation (Beato and Eisfeld, 1997; Li et al., 2007). The ability of transcription factors to bind their recognition elements is further modulated by changes in chromatin structure including DNA methylation, histone modifications, histone variants, or ATP-dependent chromatin remodeling. Chromatin therefore plays a critical role in the establishment of cell type-specific expression patterns and is responsible for the extreme stability of a given cellular identity under physiological conditions, ensuring the stable silencing of lineage-inappropriate genes and restricting transcription factor action to only a subset of their target motifs in the genome (Filion et al., 2010; Gaetz et al., 2012). In differentiated cells, pluripotency loci therefore appear to be in an unfavorable chromatin landscape for binding by most transcription factors. However, we will discuss in this review that the reprogramming factors have a remarkable capability to engage closed chromatin and induce extensive chromatin changes early in reprogramming before any major transcriptional changes take place, unmasking interesting parallels between reprogramming and developmental processes, and highlighting the power of the OSKM reprogramming cocktail. Together, these recent findings have transformed the iPSC system into a powerful model for the dissection of mechanisms underlying cell fate transitions.
The reprogramming process is most scrutinized in the mouse system, but studies of the induced pluripotent state have been extensively performed for both mouse and human iPSCs. Most likely due to the fact that conventional mouse and human iPSCs represent different states of pluripotency, these cells differ epigenetically as highlighted by their X chromosome inactivation state. In the second part of this Review, we will discuss a selection of recent studies that revealed an epigenetic instability of the inactive X chromosome in female human iPSCs, reminiscent of processes in human ESCs, and focus on the implications of these findings for the utility of iPSCs.
The path to pluripotency
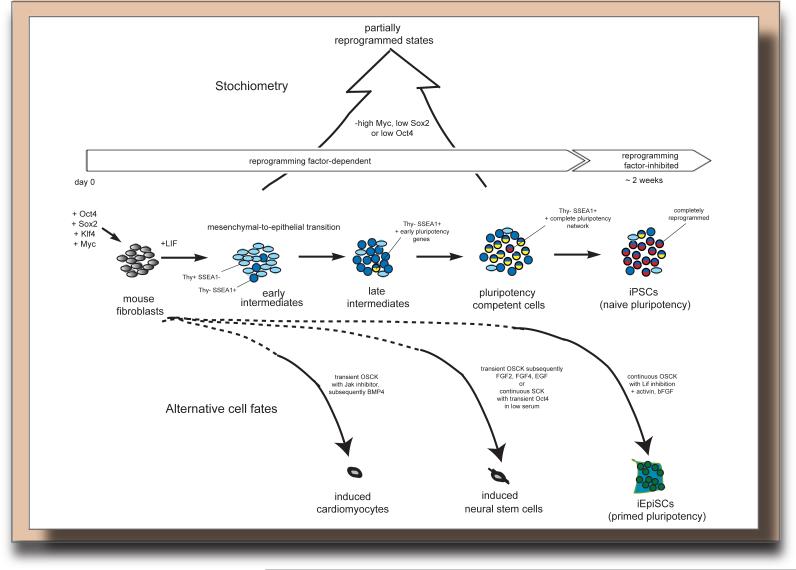
The development of improved reprogramming techniques that include homogeneous and inducible reprogramming factor expression systems (summarized in (Stadtfeld and Hochedlinger, 2010)) has enabled a more detailed view of the mechanism underlying reprogramming despite the fact that only few starting cells become iPSCs. Mouse embryonic fibroblasts are most commonly used as starting cell type for the dissection of the reprogramming process due to the ease of culture and the possibility of derivation from different genetic backgrounds and mouse models. Current evidence argues that reprogramming of these cells to iPSCs requires cell division (Hanna et al., 2009) and is a multi-step process in which the successful induction of the pluripotent state entails the transition through sequential gene expression states (or intermediates) (Figure 1). Failure to transition through any of these steps would lead to a block in reprogramming and account for the low overall reprogramming efficiency. Consistent with this model, it was shown early on by the Jaenisch and Hochedlinger groups that reprogramming cultures represent heterogeneous cell populations that can be resolved based on the expression of cell surface markers (Brambrink et al., 2008; Stadtfeld et al., 2008). Utilizing specific surface marker combinations, cells poised to become iPSCs can be enriched at different times of reprogramming. This knowledge allowed the inference of a reprogramming path, where successfully reprogramming cells first downregulate the fibroblast-associated marker Thy1, then transition to a state positive for the embryonic marker SSEA1, and finally induce the full pluripotency network (Brambrink et al., 2008; Polo et al., 2012; Stadtfeld et al., 2008) (Figure 1). The downregulation of Thy1 occurs in a large fraction of starting cells, the subsequent gain of SSEA1 only in a subset of Thy1-negative cells, and the induction of the pluripotency network in a small subset of SSEA1-positive cells, indicating that transitions between each of these steps occur with low probability (Figure 1). Cells unable to silence Thy1 relatively quickly upon OSKM expression become refractory to the action of the reprogramming factors and can yield iPSCs but with dramatic delay and at much lower efficiency (Polo et al., 2012). Accordingly, a single cell cloning experiment demonstrated that virtually all starting cells have the potential to induce pluripotency in a small subset of their daughter cells when reprogramming is followed over a six months period (Hanna et al., 2009). The intermediate states defined by cell sorting experiments likely represent the most favored possibilities on the path of reprogramming. Further purification of reprogramming intermediates should be feasible and provide insight into whether all reprogramming cells have to pass through the same stages to induce pluripotency. Of interest, SSEA1-positive intermediate cells are still plastic early in reprogramming in that some of these cells can regress to the Thy1-positive (i.e. an earlier) reprogramming state in the presence of reprogramming factor expression. By contrast, later in reprogramming, these cells appear to have matured and become much more committed to progressing to the pluripotent state (Polo et al., 2012), indicating that cellular identity is only stabilized and locked in at the end of the reprogramming process.
Figure 1. The generation of iPSCs is a multistep process that can be modulated by extracellular cues and reprogramming factor levels.
Known events occurring in early, middle and late phases during the OSKM-mediated reprogramming of mouse embryonic fibroblasts to iPSCs are depicted. During the final emergence of fully reprogrammed iPSCs, the so-called “reprogramming-competent cells” are inhibited by the continued expression of the factors. The reprogramming process can be preferentially trapped in partially reprogrammed states when certain reprogramming factor levels and/or stoichiometry's are employed (top), or can be redirected to a different cell fate, without going through the pluripotent state, by changing culture/growth factor conditions and OSKM expression timing (bottom).
Genome-wide transcriptional profiling was used to further delineate the sequence of events that drive reprogramming. Initially, cells appear to respond relatively homogeneously to the expression of the reprogramming factors (Polo et al., 2012) and robustly silence typical mesenchymal genes expressed in fibroblasts (such as Snai1, Snai2, Zeb1, and Zeb2) (Li et al., 2010; Mikkelsen et al., 2008; Polo et al., 2012; Samavarchi-Tehrani et al., 2010). These events lead to the activation of epithelial markers (such as Cdh1, Epcam, Ocln) in a process called mesenchymal-to-epithelial transition (MET), which seems critical for the early reprogramming phase and is accompanied by morphological changes, increased proliferation, and the formation of cell clusters (Li et al., 2010; Mikkelsen et al., 2008; Samavarchi-Tehrani et al., 2010; Smith et al., 2010). Notably, the aforementioned transition to the SSEA1-positive state appears to correlate with the occurrence of MET (Polo et al., 2012; Samavarchi-Tehrani et al., 2010) (Figure 1). The key characteristic of subsequent reprogramming phase is the gradual activation of pluripotency-associated genes (Brambrink et al., 2008; Buganim et al., 2012; Golipour et al., 2012; Mikkelsen et al., 2008; Polo et al., 2012; Samavarchi-Tehrani et al., 2010; Sridharan et al., 2009; Stadtfeld et al., 2008). For example, the pluripotency loci Nanog and Sall4 are transcriptionally upregulated at a late intermediate stage, while others, such as Utf1 or endogenous Sox2, are induced even later, closely mirroring the acquisition of the full pluripotency expression programming (Figure 1). Albeit detailed time course studies describing these transitions in reprogramming cells still need to be performed at the single cell level, a recent single cell expression study that compared the expression of candidate genes at various reprogramming stages strongly supports a serious of consecutive pluripotency gene activation steps late in the reprogramming process (Buganim et al., 2012). Together, these events culminate in the establishment of the pluripotent state that can be sustained independently of ectopic reprogramming factor expression (Brambrink et al., 2008; Maherali et al., 2007; Okita et al., 2007; Stadtfeld et al., 2008; Wernig et al., 2007).
Fine-tuning reprogramming
Early studies employing inducible reprogramming factor expression systems indicated that reprogramming intermediates are dependent on continued OSKM expression to complete the reprogramming process (Brambrink et al., 2008; Stadtfeld et al., 2008). Evidence is growing that the efficiency of reprogramming is strongly influenced by the levels of the reprogramming factors. For example, fibroblasts engineered to express a higher dose of OSKM in all cells have a dramatically enhanced ability to induce pluripotency (Polo et al., 2012). A peculiar observation is that cells that become refractory to reprogramming early on (and stay Thy1-positive) have dramatically reduced protein levels of the four reprogramming factors compared to cells able to progress towards pluripotency (Polo et al., 2012). Since the RNA levels of the reprogramming factors are similar between these two cell populations, these transcription factors may be prone to increased ubiquitination and degradation specifically in refractory cells (Buckley et al., 2012; Polo et al., 2012). The inability to sustain high reprogramming factor expression contributes strongly to the reprogramming block in refractory cells as a further increase in OSKM expression specifically in these cells induces them to convert to the next reprogramming stage and subsequently to iPSCs more efficiently (Polo et al., 2012). Although continuity of reprogramming factor expression is essential for driving somatic cells towards pluripotency, a recent study pointed out that high levels of ectopic OSKM during the final reprogramming steps may be inhibitory to the efficient induction of the full pluripotency network (Golipour et al., 2012) (Figure 1). This finding is consistent with the observations that retrovirally expressed reprogramming factors are efficiently turned off in faithfully reprogrammed cells (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007), and that the activation of endogenous pluripotency regulators during reprogramming coincides with transgene-independence (Stadtfeld et al., 2008). The reduction of ectopic reprogramming factors at the end of reprogramming may be necessary because even a modest increase in Oct4 levels in ESCs is detrimental to the pluripotent state (Niwa et al., 2000).
Not just overall levels and timing, but also the specific balance of the reprogramming factors relative to each other is critical for the outcome of reprogramming (Figure 1). For example many studies agree that high Oct4 levels and low levels of Sox2 increase the efficiency of reprogramming (Nagamatsu et al., 2012; Tiemann et al., 2011; Yamaguchi et al., 2011). High Sox2 levels have been associated with the stronger induction of developmental markers during reprogramming, which may guide cells away from the path to pluripotency (Yamaguchi et al., 2011). Moreover, even though ectopic expression of cMyc enhances reprogramming, it also leads to emergence of a large fraction of partially reprogrammed ESC-like colonies trapped before the upregulation of the pluripotency program (Nakagawa et al., 2008; Wernig et al., 2008). Remarkably, differences in reprogramming factor stoichiometry appear to have consequences for the epigenetic state and developmental potential of the resulting iPSCs (Carey et al., 2011). This is an interesting result in light of the ongoing debate on epigenetic differences between iPSCs and ESCs (for a recent discussion see (Lowry, 2012)) and suggests that at least some of the observed variations between iPSCs and ESCs are not inherent to the reprogramming process, but due to experimental variables that often are not easy to control, highlighting how a better understanding of the mechanisms underlying reprogramming will benefit the production of safer iPSC lines.
The efficiency of iPSC formation can also be improved by altering media composition and growth factor conditions (Chen et al., 2011; Esteban et al., 2010; Ichida et al., 2009; Li et al., 2010; Samavarchi-Tehrani et al., 2010). While it is likely that downstream effectors of signaling pathways directly alter the transcriptional output of their target genes, specific culture conditions can also modulate the activity and levels of chromatin regulators thereby indirectly affecting OSKM functionality (Chen et al., 2012; Marks et al., 2012; Wang et al., 2011a; Zhu et al., 2013). To mention just one example, vitamin C (ascorbic acid) addition to the media increases reprogramming efficiency and potentially the quality of resulting iPSCs at least in part by influencing the functionality of histone demethylases that depend on iron (Esteban et al., 2010; Stadtfeld et al., 2012; Wang et al., 2011a).
Notably, by supplementing OSKM reprogramming cultures with a growth factor cocktail normally required for the establishment and maintenance of epiblast stem cells (EpiSCs), mouse fibroblasts can be reprogrammed to an EpiSC-like state instead of the ESC-like iPSC state (Han et al., 2011) (Figure 1). Mouse EpiSCs and ESCs capture two different states of pluripotency, which will be discussed in more detail in the second part of this Review. During the last couple years it has also become clear that OSKM (or a subset of these factors) can even prompt the establishment of various somatic cell fates, including cardiomyocytes, blood progenitors, and neural stem cells, when overexpressed temporally and guided by appropriate extracellular cues, without the transition through the pluripotent state (Figure 1) (reviewed in (Sancho-Martinez et al., 2012)). The induction of various developmental regulators at intermediate stages of reprogramming to pluripotency may explain why OSKM can efficiently redirect the reprogramming path to other cell identities upon exposure to suitable signaling cues, and likely reflects a function of Sox2 and Klf4 as critical regulators of various differentiation paths during development (Mikkelsen et al., 2008; Polo et al., 2012; Sridharan et al., 2009). Alternatively, and not mutually exclusive, reprogramming intermediates arising due to OSKM expression may represent normally occurring developmental progenitor states. Though the picture is emerging that signaling cues affect the cell fate choices made during reprogramming and/or lead to the stabilization of particular cell identities that arise during the process, still relatively little is known about the exact role of signaling pathways and their downstream regulators in reprogramming and the intersection with the reprogramming factors. Comparing the molecular dynamics of OSKM-dependent induction of pluripotency and alternative cell fates should demonstrate how cell fate decision processes can be efficiently modulated and facilitate the development of patient-specific somatic cell populations for clinical applications.
Defining the target repertoire of the reprogramming factors
One approach towards a better understanding of the cascade of molecular events underlying the establishment of pluripotency is the definition of reprogramming factor targets at different stages of the reprogramming process. It is generally believed that three of the four reprogramming factors, Oct4, Sox2, and Klf4, are necessary for the induction of pluripotency because they are critical components of an intrinsic, highly stable pluripotency network (Boyer et al., 2005; Chen et al., 2008; Jiang et al., 2008; Kim et al., 2008; Loh et al., 2006; Sridharan et al., 2009). Specifically, Oct4, Sox2, and Klf4 tend to co-localize at many cell type-specific enhancers in ESCs, often together with additional pluripotency transcription factors like Nanog, Esrrb, Klf2, Sall4, Zfp42, and signaling pathway regulators such as Smad1 and Stat3 (Chen et al., 2008; Kim et al., 2008), reinforcing the importance of OSK for the pluripotent state and the view that enhancers are sentinels of cell type-specific gene expression patterns (Visel et al., 2009). The integration of numerous pluripotency transcription factors and signaling cues at these enhancers ensures the expression of many genes with known roles in pluripotency and provides stability to the ESC gene expression program. Another important aspect of the pluripotency network is that many pluripotency transcription factors constitute a transcriptional circuit wired in a feed-forward type of regulation as they induce their own expression and positively regulate each other (Boyer et al., 2005; Chen et al., 2008; Jiang et al., 2008; Kim et al., 2008) (Figure 2A).
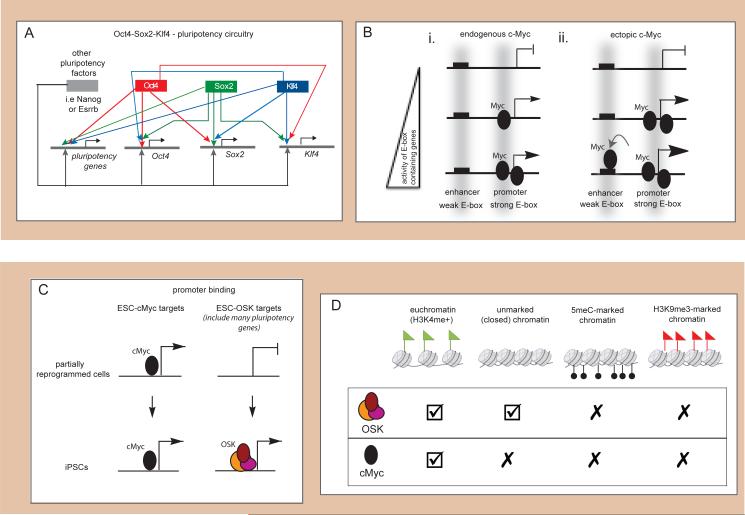
Figure 2. Features of OSKM in ESCs and during reprogramming.
(A) In ESCs, Oct4, Sox2, and Klf4 bind their own and each others promoters and enhancers, as well as those of many additional ESC-specific (pluripotency) genes. Further contributing to the pluripotency circuitry, many of these ESC-specific genes are also bound by various additional pluripotency regulators including Nanog and Esrrb, such that ESC-specific enhancers represent hotspots of pluripotency transcription factor binding. (B) (i) In ESCs (and many other cell types), cMyc targets most actively transcribed genes at the core promoter by binding high-affinity E-box sequences and functions by enhancing transcriptional elongation. Expression levels correlate with cMyc occupancy. (ii) Upon overexpression, cMyc does not appear to regulate new target genes, but amplifies the existing gene expression pattern by binding the same genes but now at elevated levels and occupying additional, low affinity E-box-like sequences in both the core promoter and enhancer regions of these genes. (C) Scheme illustrating different contributions of the reprogramming factors to the late phase of reprogramming, highlighting separable engagement of OSK and cMyc during reprogramming. Many genes occupied by cMyc in ESCs/iPSCs are already bound by this transcription factor and expressed in partially reprogrammed cells, which represent a late reprogramming intermediate. By contrast, OSK bind the promoter regions of many of their ESC-specific target genes only late in reprogramming, accompanying their transcriptional upregulation. This is particularly obvious for those genes that are co-bound by OSK in their promoter region in ESCs. (D) Chromatin can affect the ability of transcription factors to bind to their DNA motifs, which is thought to explain why most transcription factors bind to only a small subset of their recognition motifs in the genome. Here, we summarize the chromatin preferences of the four reprogramming factors early in reprogramming.
By contrast, cMyc is unique among the reprogramming factors, as it is neither a component of the core pluripotency network (Chen et al., 2008; Kim et al., 2010) nor absolutely necessary for reprogramming to iPSCs (Nakagawa et al., 2008; Wernig et al., 2008). Indeed, cMyc is a central player in many diverse biological processes, including cell growth and differentiation. Two recent reports (Lin et al., 2012; Nie et al., 2012) strongly support a model in which cMyc is not a transcription factor that is responsible for OFF/ON switches of its target genes such as OSK. Instead, cMyc is a non-linear amplifier of transcriptional outputs that acts universally on active genes containing the E-box DNA motif. Mechanistically, cMyc promotes transcription by regulating RNA polymerase II pause-release and increasing the rate of transcriptional elongation (Rahl et al., 2010). Therefore, cMyc occupies the core promoter regions of many active genes in ESCs/iPSCs and is typically not present at enhancers (Chen et al., 2008; Kim et al., 2010; Nie et al., 2012; Soufi et al., 2012; Sridharan et al., 2009) (Figure 2Bi). Analysis of cMyc binding across different inducible expression levels in tumor cells demonstrated that cMyc predominantly binds high-affinity E-box sites at core promoters of almost all active genes when expressed at low levels, but spills over to weaker E-box sites within enhancers of the same active genes upon higher expression, likely because promoter sites become saturated (Figure 2Bii) (Lin et al., 2012; Nie et al., 2012). Thus, the target repertoire of cMyc does not appear to change when cMyc is strongly expressed, but transcriptional output is increased. The significant differences between OSK and cMyc have important implications for the reprogramming process: Oct4, Sox2, and Klf4 are probably crucial for specifying cell fate change in reprogramming, while cMyc may simply act by amplifying arising expression changes due to OSK action at genes that contain E-boxes, potentially helping to trap genes in the ON state.
The low efficiency of reprogramming makes the application of genome-wide analysis techniques of reprogramming factor binding, such as chromatin immunoprecipitation combined with massive parallel sequencing (ChIP-Seq), challenging for cells at intermediate stages of the reprogramming process. To circumvent this problem, our lab initially mapped reprogramming factor binding within promoter regions in iPSCs and in partially reprogrammed cells, which represent a clonal, trapped late reprogramming intermediate expanded from ESC-like colonies that arise in reprogramming cultures and fail to express pluripotency regulators, and compared occupancy data with gene expression patterns (Sridharan et al., 2009). In both cell types, genes co-occupied by the reprogramming factors are highly expressed, indicating that an intrinsic property of reprogramming factor co-binding is to activate genes. Interestingly, genes more highly expressed in partially reprogrammed cells than in ESCs are often more efficiently targeted by the OSKM factors in the intermediate state than in ESCs, while genes more highly expressed in ESCs are generally less bound in partially reprogrammed cells than in ESCs. Thus, many genes are more strongly expressed in partially reprogrammed cells compared to ESCs due to targeting of the four factors to promoter regions that they do not normally bind in ESCs, and, conversely, the failure to activate ESC-specific genes appears to result from the inability of the factors to bind these genes in the intermediate state. These findings are consistent with the reprogramming factors being directly responsible for the “ectopic” expression of developmental genes in reprogramming intermediates, which is known to hinder reprogramming (Mikkelsen et al., 2008). Notably, the widespread lack of ESC-specific promoter binding in partially reprogrammed cells impinges more dramatically on Oct4, Sox2, and Klf4 than on cMyc and particularly affects many pluripotency-related genes that are co-occupied by combinations of Oct4, Sox2, and Klf4 in ESCs (Figure 2C). In the case of these genes, it appears that the OSK promoter engagement occurs only towards the very end of the reprogramming process and is likely required for their transcriptional activation (Figure 2C). These findings not only demonstrate a separable contribution of cMyc and OSK to the activation of various pluripotency loci and a change in the reprogramming factor target repertoire during the reprogramming process, but also indicate that the promoter engagement of key pluripotency genes is a critical task for reprogramming.
Recently, Zaret and colleagues obtained a picture of the initial chromatin engagement of the reprogramming factors by performing ChIP-seq 48 hours after the induction of reprogramming factor expression in human fibroblasts (Soufi et al., 2012), when most cells still undergo very similar expression changes (see above) (Polo et al., 2012). Comparing OSKM binding patterns between the early reprogramming stage and the pluripotent state, Zaret and colleagues made two interesting observations (Soufi et al., 2012). First, many more genes are bound by all four factors early in reprogramming than in the pluripotent state, which could be due to the high expression levels of the induced factors. However, OSKM binding of apoptosis-regulating genes early in the process suggests that the extensive cell death apparent in reprogramming cultures (reviewed in (Plath and Lowry, 2011)) is a direct consequence of reprogramming factor binding, potentially representing a general cellular defense mechanism to ectopic transcription factor expression (Soufi et al., 2012). Furthermore, initial target genes of the reprogramming factors are significantly enriched for regulators of the mesenchymal-to-epithelial transition, the critical early reprogramming event discussed above, while pluripotency loci such as NANOG and DPPA4 are not yet bound, corroborating that a redistribution of OSKM binding occurs as cells move along the reprogramming path and suggesting that the reprogramming factors directly target at least some of the genes that transcriptionally change early in the process. The second, more surprising finding is that the reprogramming factors interact extensively with distal genomic sites including some known enhancers. Indeed, 85% of all initial binding events occur distal to promoter regions (Soufi et al., 2012). Since it appears that in the pluripotent state the transcription factors have shifted to a binding pattern that includes promoter regions much more strongly, Zaret and colleagues proposed that the binding of the reprogramming factors to distal elements is an early step in reprogramming that precedes promoter binding and transcriptional activation of many target genes (Soufi et al., 2012).
Reprogramming factors as pioneers
The next question then is which features anticipate the recruitment of ectopically expressed OSKM? The DNA motifs of the four factors are enriched at their respective binding sites indicating that the factors are recruited directly through their sequence motifs rather than randomly targeting or scanning the genome (Soufi et al., 2012; Sridharan et al., 2009). However, transcription factors work in a concentration-dependent manner and will, at higher concentration, also occupy DNA sites of lower affinity, which may be important for reprogramming, where very high levels of ectopic OSKM are expressed (Lin et al., 2012; Nie et al., 2012; Soufi et al., 2012) (Figure 2Bii). Lineage-specification factors present in the starting cell type may contribute to the targeting of the reprogramming factors to a subset of their DNA motifs. For example, during lineage development, Sox transcription factors often occupy sites pre-marked by other Sox proteins, which were expressed in the previous developmental stage (Bergsland et al., 2011). If such lineage-specific factors are involved in the initial targeting of the reprogramming factors, one might predict that reprogramming factors will target different genomic locations in different starting cell types.
Importantly, chromatin is thought to strongly affect the ability of transcription factors to bind their cognate DNA motifs, and certain chromatin states, characterized for example by the presence of specific combinations of histone modifications, may be especially conducive to DNA binding by specific transcription factors (Filion et al., 2010). As expected, binding of the reprogramming factors does occur in open and accessible chromatin, marked by active histone modifications such as H3K4 methylation (Soufi et al., 2012; Sridharan et al., 2009) (Figure 2D). Among the reprogramming factors, cMYC binding is much more strictly associated with a pre-existing active chromatin state than that of OSK (Soufi et al., 2012; Sridharan et al., 2009), consistent with active chromatin being a pre-requisite for the binding of cMyc (Guccione et al., 2006) (Figure 2D). An astonishing observation by Zaret and colleagues is that the vast majority (around 70%) of reprogramming factor binding events early in human fibroblast reprogramming occurs within genomic regions that display a closed chromatin state in the starting fibroblasts characterized by the absence of DNAse hypersensitivity and, surprisingly, any histone modifications (Soufi et al., 2012). Thus, the reprogramming factors can efficiently access their target sequences within genomic regions that are packed with nucleosomes and probably even further condensed into higher-order structures. This is particularly true for OSK and to a much lesser extent for cMYC (Soufi et al., 2012) (Figure 2D). Indeed, the ability of cMYC to access target sites in closed chromatin is dependent on OSK occupancy (Soufi et al., 2012). OSK can occupy these sites in the absence of ectopic cMYC, but cMYC cannot bind when overexpressed in the absence of ectopic OSK. In turn, ectopic cMYC enhances the initial binding of OSK to these sites. These data are in agreement with cMyc potentiating the action of the other three reprogramming factors rather than initiating these events.
In comparison to naked DNA, nucleosomal DNA is less accessible for DNA binding factors (Beato and Eisfeld, 1997), and the majority of transcription factors cannot bind their cognate sites when sequestered within a nucleosome and need a structural change in the associated nucleosome or a nucleosome-free-region for binding (Wallrath et al., 1994), highlighting an important functionality of OSK. Cooperative binding or simultaneous engagement of neighboring binding sites could explain the ability of OSK to interact with nucleosomal binding sites (Adams and Workman, 1995). For instance, binding of one factor might partially destabilize a nucleosome, allowing the other transcription factor(s) to access sites that were previously buried. However, each of the OSK reprogramming factors alone can also target sites in closed chromatin, i.e. without the other two factors being detected at those sites (Soufi et al., 2012). Therefore, Zaret and colleagues proposed that Oct4, Sox2, and Klf4 each can act as pioneer factors able to access closed chromatin on their own without the help of additional transcription factors (Soufi et al., 2012). There is additional evidence in support for this idea: First, based on 3D structures, Oct4, Sox2, and Klf4, but not cMyc, interact with one side of the DNA helix when bound to DNA, potentially allowing them to bind DNA in the context of the nucleosome (Beato and Eisfeld, 1997; Soufi et al., 2012). Second, a comparison of nucleosome occupancy with binding of Oct4 and Sox2 in ESCs genome-wide suggests that Oct4 and Sox2 can, at least in part, interact with nucleosomal DNA (Teif et al., 2012). Third, Sp1, a transcription factor belonging to the same family of highly related transcription factors as Klf4, can bind nucleosomal DNA in vitro, making it reasonable to anticipate that Klf4 will share SP1's capacity (Li et al., 1994). Fourth, it was found that pre-existing nucleosomes at the enhancer and promoter regions of the OCT4 and NANOG gene loci are displaced when OCT4 is ectopically expressed in differentiated cells (ie. in the absence of any other reprogramming factors) (You et al., 2011). This chromatin re-organization coincided with Oct4 binding, suggesting that Oct4 is able to directly access DNA sites that are internal to a nucleosome and establish a nucleosome-depleted region (You et al., 2011).
The idea of OSK acting as pioneer factors in reprogramming is exciting because it is reminiscent of early developmental progenitor cells that are marked by pioneer factor binding at enhancers (Gualdi et al., 1996). The efficient activation of lineage-specific genes during development often requires a cascade of DNA-transcription factor interactions and chromatin changes at their enhancer and promoter regions that begins long before these genes are transcribed (Zaret and Carroll, 2011). Pioneer transcription factors initiate this series of events by accessing tissue-specific enhancers already at a very early developmental stage and inducing chromatin decondensation, remodeling and/or a change in local chromatin modifications, thereby priming enhancer and promoter regions for binding by additional transcription factors and transcriptional activation at a later stage of development. Thus, pioneer factors are initiator factors that make regulatory regions competent for activation in response to the right stimulus.
In the context of reprogramming, the binding of OSK to closed chromatin early in reprogramming could therefore be a crucial step for events that happen later in the process, particularly considering that some of these distal binding events overlap with known enhancers. One may speculate that Oct4, Sox2, and Klf4 can engage at least some ESC-specific enhancers early in reprogramming even though they are locked up in closed chromatin in the starting fibroblasts, poising them for promoter binding and transcriptional activation later in the process. In the next section, we will provide additional evidence in support of such epigenetic priming by focusing on chromatin changes that occur early in the reprogramming process.
Promoter and enhancer priming early in reprogramming
An analysis of the initial transcriptional and chromatin changes early in mouse cell reprogramming (i.e. 24-72 hours after induction of the reprogramming factors) revealed striking parallels to the initial reprogramming factor binding pattern (Koche et al., 2011). First, gene expression changes, both up and down, are largely confined to genes with promoter regions carrying active chromatin marks in the starting fibroblasts (i.e. in regions marked by enrichment of H3K4me3, a modification associated with the transcriptional start sites of active and poised genes) (Koche et al., 2011). The restriction of expression changes to genes that are already in an open and accessible chromatin configuration is consistent with the fact that the perturbation of the somatic gene expression program is the major response early in the reprogramming process (Koche et al., 2011; Mikkelsen et al., 2008; Polo et al., 2012; Samavarchi-Tehrani et al., 2010; Sridharan et al., 2009).
Unexpectedly, changes in histone modifications are much more widespread than initial changes in gene expression indicating that an extensive genome-wide chromatin remodeling takes place as immediate response to reprogramming factor expression (Koche et al., 2011). In addition to chromatin changes associated with gene expression switches, H3K4me2 (a histone mark associated with active or poised promoters and enhancers) rapidly emerges de novo in many promoter regions in the absence of transcriptional changes and even before any cell division has taken place (Figure 3). Many of these promoters belong to genes that are transcriptionally activated later in reprogramming, including various pluripotency regulators like Sall4, Pecam1, FoxD3, and Lin28. The gain of H3K4me2 is not accompanied by simultaneous accumulation of the H3K4me3 mark and often occurs on a nucleosome that covers the transcriptional start site. Since nucleosomes at transcriptional start sites are incompatible with the assembly of the basic transcriptional machinery (Lorch et al., 1987), nucleosome depletion must be one of the subsequent steps that allows transactivation of these genes later in reprogramming. Interestingly, promoters with H3K4me2 gain early in reprogramming often display a high CpG density and are enriched for CpG islands (Koche et al., 2011) (Figure 3), which may obviate the need for extensive chromatin remodeling and facilitate quick changes in chromatin structure due to lower nucleosome occupancy (Ramirez-Carrozzi et al., 2009) .
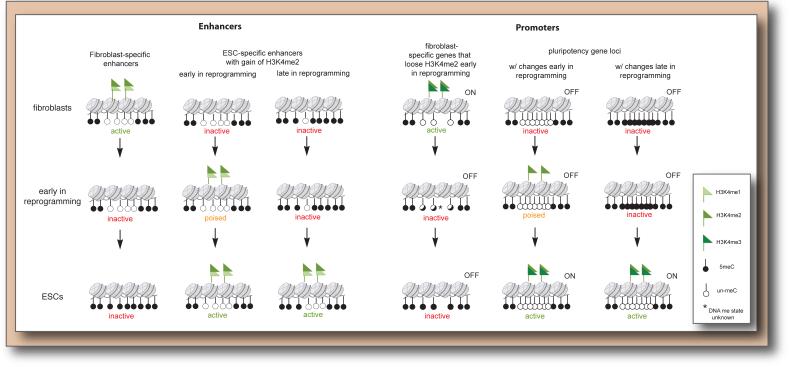
Figure 3. Chromatin dynamics during reprogramming.
Many fibroblast-specific promoters and enhancers are decommissioned early in reprogramming (after 24-48 hours of reprogramming factor expression) by loss of active H3K4 methylation marks but appear to gain DNA methylation only late in reprogramming. ESC-specific enhancers and promoters can be divided into at least two groups – those with dramatic changes in histone modifications already early in reprogramming, long before their transcriptional activation, and those that undergo histone modification changes only much later in the process. One key difference between these groups appears to be the DNA methylation state. For example, the first group includes many pluripotency genes with CpG-dense promoter elements (indicated by higher density of circles) that are hypomethylated in fibroblasts.
Compared to promoters, chromatin changes at enhancers are even more prominent early in the reprogramming (Koche et al., 2011), which is consistent with the observations that many enhancers are active in only a single cell type, and that the chromatin state of enhancers is more variable across cells types than that of promoters (Heintzman et al., 2009). The systematic mapping of enhancers is now possible genome-wide because specific enhancer-associated chromatin signatures have been identified that even reveal the activity of the enhancer (Creyghton et al., 2010; Heintzman et al., 2009; Koche et al., 2011; Rada-Iglesias et al., 2011). In the active state (ie when associated with an actively transcribed gene), enhancer elements are demarcated by domains of H3K27Ac and H3K4me1/me2, but not H3K4me3. In association with inactive genes, enhancers can be in one of two states: unmarked (i.e completely inactive), lacking all of the features that are associated with the active enhancer state; or poised, carrying H3K4me1/me2 in the absence of H3K27ac. It is thought that poised enhancers are important for the plasticity of developmental decisions as a subset can acquire the signature of active enhancers upon change in external stimuli. The specific enhancer state therefore appears to strongly influence the ability of the cell to respond to environmental or developmental stimuli. For example, immediate transcriptional changes to a new signaling cue are often restricted to genes with active and/or poised enhancers, while inactive genes with unmarked (inactive) enhancers remain refractory (Ghisletti et al., 2010; Heintzman et al., 2009).
Importantly, switches in enhancer states occur very rapidly and extensively, even before the first cell division of reprogramming, highlighting an extremely quick departure from the somatic cell identity (Koche et al., 2011). These changes go in both directions: more than 60% of fibroblast-specific enhancers are decommissioned and at least one thousand ESC-specific enhancers are established de novo within the first 24 hours of reprogramming factor expression, based on loss or gain of H3K4me1/2, respectively (Figure 3). Although H3K4me1/2 on its own does not allow one to distinguish between active and poised enhancer states, it is likely that many of the newly marked ESC-specific enhancers are in a poised state that will be activated at later stages of reprogramming. Thus, extensive chromatin remodeling at ESC-specific promoters and enhancers precedes the transcriptional activation of many pluripotency genes.
Together, these chromatin dynamics are likely crucial for the shutdown of the somatic expression program and the transition towards pluripotency. During differentiation, pluripotency genes acquire a silent state that is associated with a repressive chromatin environment that can include DNA methylation, histone variants, covalent histone modifications, chromatin regulatory proteins, and occupancy of regulatory regions by nucleosomes (Feldman et al., 2006; Mikkelsen et al., 2008; You et al., 2011). To activate pluripotency genes it seems that the reprogramming factors must surmount at least two separable obstacles: the binding block at upstream regulatory regions (i.e distal enhancer and promoter elements), and a block in the transactivation of the core promoter, which prevents the assembly and activation of the RNA-polymerase II-containing basal transcription machinery. Therefore, it may not be too surprising that the activation of pluripotency genes in reprogramming is relatively slow and potentially requires a cascade of events. The findings described above suggest that the formation of poised ESC-specific enhancers early in reprogramming may be a critical first step to orchestrate the productive engagement of the core promoter and transcriptional activation of ESC-specific genes later in the process when proper signals are available (Taberlay et al., 2011). This likely requires further chromatin remodeling and/or additional transcriptional and signaling regulators that are unavailable early in reprogramming (for more discussion see the transition section below). Importantly, this epigenetic priming does not affect all pluripotency genes early on as many only gain an active/poised chromatin signature at their enhancer and promoter regions late in the process (Polo et al., 2012; Sridharan et al., 2009) (Figure 3) (see below). Understanding the regulation of enhancer/promoter pairs of pluripotency genes during reprogramming will be an important task for the future that will increase our general knowledge about the dynamics of promoter and enhancer interactions (Taberlay et al., 2011).
Relating the extensive binding of OSK to distal sites in unmarked, closed chromatin early in human cell reprogramming (Soufi et al., 2012) to the epigenetic priming of many ESC-specific enhancers early in mouse reprogramming (Koche et al., 2011) implies that the reprogramming factors may cause at least some of these initial epigenetic priming events directly. To test this hypothesis, simultaneous analysis of transcription factor binding, chromatin and transcription states is required, and detailed studies, both in vitro and in vivo, need to address if Oct4, Sox2, and Klf4 can indeed bind regulatory DNA sites packaged in nucleosomes and change chromatin structure. The ability of the reprogramming factors to engage regulatory genomic elements in closed (silent) chromatin may be a critical feature and explain why OSK are such potent inducers of pluripotency, and that in many different somatic cell types.
DNA methylation and H3K9 methylation influence reprogramming factor binding
Given that OSK appear to be able to efficiently engage closed chromatin regions already early in reprogramming, it may be surprising that many regulatory regions bound by OSKM in the pluripotent state are not occupied early in the process (Soufi et al., 2012; Sridharan et al., 2009). What then are the impediments to reprogramming factor binding and action? DNA methylation has arisen as an important factor in restricting early reprogramming events. ESC-specific promoters and enhancers that gain active chromatin modifications only late in reprogramming tend to be hypermethylated in the starting fibroblasts and become demethylated only late in reprogramming (Koche et al., 2011) (Figure 3). For example, hypermethylation of key pluripotency gene promoters, including those of Nanog and Oct4, is observed until late in reprogramming (Mikkelsen et al., 2008; Polo et al., 2012), suggesting that demethylation of these promoters is a rate-limiting step. By contrast, promoters and enhancers that already gain active chromatin marks (H3K4me2) early in reprogramming exhibit hypomethylation throughout the entire reprogramming process (Koche et al., 2011) (Figure 3). Thus, DNA methylation appears to limit where initial histone modification changes can occur. Furthermore, Oct4 expression can establish a nucleosome-depleted region at the distal enhancers of OCT4 and the proximal promoter of NANOG in somatic cells, but only if these regions are unmethylated (You et al., 2011), indicating that DNA methylation can prevent the recruitment of the reprogramming factors (Figure 3D). In case of Oct4, DNA methylation must affect binding indirectly as its DNA motif does not contain a CpG. Jones and colleagues proposed that DNA methylation in flanking sequences may stabilize the nucleosome and prevent binding (You et al., 2011). Similarly, binding of cMyc is inhibited by CpG methylation within its CACGTG target site (Prendergast and Ziff, 1991). However, the binding of other transcription factors, such as the Klf4 related transcription factor SP1 is not affected by DNA methylation (Harrington et al., 1988), suggesting that the reprogramming factors may be differentially affected by DNA methylation. Importantly, DNA methylation is recognized as a feature that limits reprogramming to pluripotency because interference with Dnmt1, the enzyme responsible for the maintenance of DNA methylation (Mikkelsen et al., 2008) promotes iPSC formation (Table 1).
Interestingly, somatic enhancers that are inactivated quickly upon reprogramming factor expression and are typically methylated in the pluripotent state, only gain hypermethylation later in the reprogramming process (Koche et al., 2011) (Figure 3). Thus, both the methylation of somatic genes and the demethylation of some critical pluripotency genes appear to occur only late in reprogramming, establishing the DNA methylation pattern characteristic of the pluripotent state, which is in contrast to the more gradual changes in histone modifications and transcriptional states throughout reprogramming (Koche et al., 2011; Polo et al., 2012). This may explain at least in part why reprogramming intermediates are instable when the reprogramming factors are withdrawn as DNA methylation may be required to permanently lock in a gene expression pattern and cell identity (Koche et al., 2011). However, it needs to be noted that reprogramming occurs normally even upon the genetic ablation of the de novo DNA methyltransferases Dnmt3a and Dnmt3b, indicating that the gain of DNA methylation in somatic promoters and enhancers may not be essential (Pawlak and Jaenisch, 2011) (Table 1). In any case, it will be interesting to elucidate the mechanisms underlying these bidirectional changes of DNA methylation late in the reprogramming process.
In addition to DNA methylation, other repressive chromatin marks affect the ability of the reprogramming factors to engage their target sites. Indeed, Zaret and colleagues uncovered hundreds of large regions of megabase scale that exclude reprogramming factor binding early in human cell reprogramming even though the same regions are bound extensively by the factors in ESCs (Soufi et al., 2012). Albeit gene-poor, these regions contain various well-known pluripotency genes such as NANOG, SOX2, and PRDM14, and almost perfectly overlap with regions of extended H3K9me3 in the starting fibroblasts that are in close contact with the nuclear lamina (Soufi et al., 2012). Importantly, during reprogramming these broad H3K9me3 domains are erased, consistent with their absence in human ESCs (Hawkins et al., 2010; Soufi et al., 2012; Zhu et al., 2013), raising the possibility that the lack of OSKM binding in these large contiguous genomic regions early in reprogramming could be caused by the presence of H3K9me3.
There is currently some debate as to whether the H3K9me3 domains arise during lineage specification or are triggered in differentiated cells in response to specific culture conditions in vitro (Hawkins et al., 2010; Zhu et al., 2013). Regardless, the H3K9 methyltransferase SUV39H1 is required for the maintenance of these H3K9me3 domains and inhibition of TGFβ-signaling lowers the H3K9me3 domain signal (Soufi et al., 2012; Zhu et al., 2013). Notably, both the suppression of SUV39H1 and the inhibition of TGFβ-signaling enhance reprogramming to pluripotency (Ichida et al., 2009; Onder et al., 2012; Soufi et al., 2012) (Table 1), and inhibition of SUV39H1/2 early in human cell reprogramming increases the access of OSKM to sites within H3K9me3 domains (Soufi et al., 2012). Thus, H3K9 methylation represents a barrier to the induction of pluripotency, at least in part by blocking reprogramming factor access (Figure 2D). This conclusion is supported further by the finding that various other H3K9 methyltransferases and H3K9 demethylases control reprogramming efficiency (Chen et al., 2012; Onder et al., 2012; Soufi et al., 2012) (Table 1). In a fascinating twist, the same regions that display a shift from a broad H3K9me3 pattern to OSKM binding during reprogramming encompass nearly all of the 20 hot spots of aberrant epigenetic reprogramming, which exhibit aberrant DNA methylation patterns in human iPSCs compared to ESCs (Lister et al., 2011; Soufi et al., 2012). Thus, the loss of H3K9me3 from these regions may be a very inefficient process that could additionally be influenced by the exact culture conditions used for reprogramming (Zhu et al., 2013).
Transitioning between reprogramming steps
An important question is what exactly the rate-limiting transition steps at various reprogramming stages are. How do reprogramming cells transition from one step to the next? While the field is defining molecules that positively and negatively influence the reprogramming process, this question is still very difficult to address due to the inefficiency of the process. Rate-limiting transitions are likely linked to fluctuations or inherent noise of gene expression, chromatin state, and transcription factor binding, and further influenced by cell-cell contacts or extrinsic signals. Single cell gene expression studies have shown that early reprogramming cultures and intermediate reprogramming populations both display heterogeneity with considerable variation in gene expression between cells (Buganim et al., 2012; Polo et al., 2012), suggesting that stochastic gene activation events could be an important contributor to reprogramming transitions. Some of these expression differences are likely essential for progression towards pluripotency, while others may not have any impact on the reprogramming process or even be inhibitory (Buganim et al., 2012; Polo et al., 2012).
Oct4 physically interacts with various active and repressive chromatin complexes (Pardo et al., 2010; van den Berg et al., 2010), raising the question of whether the activator or repressor function of Oct4 and the other reprogramming factors is more important for reprogramming. Recent reports in which reprogramming factors were fused to strong transcriptional activation domains (TADs) or repressor proteins indicate that activator but not repressor fusions promote reprogramming (Hammachi et al., 2012; Hirai et al., 2011; Wang et al., 2011c), suggesting that transcriptional activation is the main action of the reprogramming factors in reprogramming and may be rate-limiting. However, not all TADs can enhance the induction of pluripotency: TADs of MyoD and VP16 but not those of Mef2C and Gata4 increase iPSC formation when fused to Oct4 (Hammachi et al., 2012; Hirai et al., 2011; Hirai et al., 2012; Wang et al., 2011c). Since TADs serve as scaffold to recruit other transcription factors, co-activators, and specific chromatin modifiers required for transcriptional activation, these findings suggest the need for specific co-regulatory proteins in pluripotency induction. A strong transcriptional activator may bypass the requirement for extensive chromatin remodeling at the promoter for recruitment of the basic transcriptional machinery and pre-initiation complex assembly (Koutroubas et al., 2008). Notably, the ectopic tethering of a strong transcriptional activator (the VP16 TAD) to the silent Oct4 gene in somatic cells is capable of activating this allele within 48 hours. However, this activation only happens in a small number of cells, highlighting the need for additional regulatory events (Hathaway et al., 2012).
Given that the reprogramming factors may act predominantly as transcriptional activators, it may be surprising that the initial transcriptional response includes the silencing of the somatic expression program. However, transcriptional activators could amplify or induce the expression of other transcriptional activators as well as repressors, which in turn could secondarily affect gene expression patterns via emergent feed-forward and feed-back circuitries and thereby contribute to the cell fate change of reprogramming. High levels of strong transcription factors may also contribute indirectly to the repression of other genes by competing for binding at common sites on the basic transcriptional machinery in a process referred to as squelching (Gill and Ptashne, 1988). Additionally, not only coding genes but also miRNAs are dynamically regulated during reprogramming and have been implicated in the control of the reprogramming process, even allowing for the induction of pluripotency without the ectopic expression of any transcription factor (Anokye-Danso et al., 2011; Judson et al., 2009). miRNA expression inversely correlates with target gene expression during reprogramming (Polo et al., 2012), suggesting that miRNAs may be critically contributing to the silencing of the somatic gene expression program and subsequent reprogramming steps. For example, an increase of miR-130 and miR-301 early in reprogramming enhances reprogramming by repressing the developmental regulator Meox2 (Pfaff et al., 2011), and miRNAs of the miR-200 family are induced early and contribute to the repression of the fibroblast regulators Zeb1 and Zeb2 (Samavarchi-Tehrani et al., 2010). The experimental depletion of pre-existing lineage factors also promotes reprogramming (Hanna et al., 2008) likely by facilitating the decommissioning of somatic enhancers, thereby enabling the transition to the next reprogramming stage.
What leads to the hierarchical pluripotency gene activation late in reprogramming? As discussed before, their efficient transcription requires the combinatorial and synergistic action of multiple activators bound to the enhancer and/or distal promoter. Enhancers can be modular, where each transcription factor contributes to the transcriptional output, or be non-modular, where each transcription factor is essential such that the target gene is only turned on when all transcription factors are present. Particularly considering that many ESC-specific enhancers are bound by a large number of pluripotency transcription factors in ESCs (Figure 2A), the presence of OSKM alone is likely not sufficient for efficient binding and/or transactivation. One of the factors that needs to act alongside OSK appears to be the pluripotency transcription factor Nanog. Nanog co-occupies many pluripotency genes together with OSK in ESCs and targets promoter regions that fail to bind OSK until the end of the reprogramming process (Sridharan et al., 2009) (Figure 2). Intriguingly, Nanog is essential for the establishment of iPSCs (Silva et al., 2009) and becomes expressed before many other pluripotency genes during the reprogramming process (Golipour et al., 2012), suggesting that it could be required for their activation. Overexpression of Esrrb, another pluripotency factor, can rescue OSKM-induced reprogramming in the absence of endogenous Nanog (Festuccia et al., 2012). Fitting in the concept of hierarchical pluripotency activation, Esrrb is a direct target of Nanog in ESCs (Festuccia et al., 2012). Therefore, a critical function of Nanog in reprogramming may be to activate Esrrb, which in turn directly interacts with the general transcriptional machinery and also co-occupies many pluripotency loci with OSK and Nanog (Percharde et al., 2012; van den Berg et al., 2010). Interestingly, a recent RNAi screen identified various chromatin regulators including Morc1 as regulators of the final reprogramming steps, which have not yet directly been implicated in the maintenance of pluripotency (Golipour et al., 2012), indicating that in addition to transcriptional activation an extensive chromatin remodeling may be required at the late reprogramming stage.
Today, we are just beginning to discover how chromatin limits but also guides reprogramming factors and how the factors overcome chromatin barriers. Direct interactions of the reprogramming factors with chromatin regulators may be important. For example, Oct4 can interact with subunits of the BAF chromatin-remodeling complex (Pardo et al., 2010; van den Berg et al., 2010), which enhances reprogramming and could stimulate the binding of transcription factors to nucleosomal sites (Singhal et al., 2010; Utley et al., 1997). Similarly, the activity of the reprogramming factors can be modulated by post-translational modifications such as O-GlcNAc, which in the case of Oct4 is required for activation of target genes in ESCs and for Oct4's full functionality in reprogramming (Jang et al., 2012). Recent studies have identified additional chromatin regulators that are essential for the process (for a summary see Table 1). For example, the H3K27me demethylase Utx also interacts with OSK and is critical for the removal of this repressive H3K37me3 from pluripotency loci (Mansour et al., 2012). Similarly, decreasing the levels of histone marks associated with transcriptional elongation promotes the downregulation of the somatic gene expression program and suppression of senescence regulators (Liang et al., 2012; Onder et al., 2012; Wang et al., 2011a). While it is likely that appropriate regulatory factors need to be co-expressed and function alongside OSKM to gain access to pluripotency genes that are locked into repressive chromatin (Doege et al., 2012), such an opportunity may normally arise during every cell division, immediately following DNA replication before nucleosome assembly (Wolffe, 1991). It remains to be determined whether replication (i.e. cell proliferation) is required for changing gene expression patterns at all stages of the reprogramming process.
X chromosome state in mouse cell differentiation and reprogramming
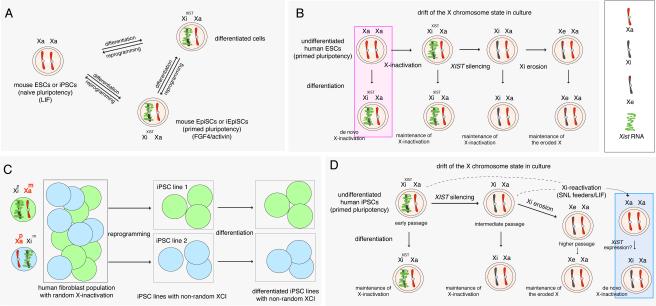
In the remaining sections of this Review, we will focus on the characterization of the induced pluripotent state considering both mouse and human iPSCs, highlighting differences and parallels between these two cell types with an emphasis on the epigenetic state of the X chromosome. In mammals, X-chromosome inactivation (XCI) leads to the transcriptional silencing of one X chromosome in female (XX) cells, equalizing gene dosage to XY males. This process involves several non-coding RNAs and a dramatic reorganization of chromatin with various epigenetic layers of regulation such as DNA methylation, histone modifications, and late replication in S-phase (reviewed in (Wutz, 2011)). In the mouse, X chromosome silencing is established very early in embryonic development in the epiblast cells of the implanting blastocyst, which will give rise to the embryo proper and represent the in vivo counterpart of mouse ESCs. XCI can therefore be recapitulated in vitro in differentiating mouse ESCs. Differentiation induces expression of the non-coding RNA Xist, which then quickly spreads to coat the chromosome in cis, mediating silencing of X-linked genes and inducing a repressive chromatin character along the entire chromosome (Wutz, 2011) (Figure 4A). This process is random such that the paternally and maternally inherited X chromosome (Xp and Xm, respectively) become silenced with equal chance. However, in the mouse system, two states of pluripotency exist in vivo and in vitro. ESCs and the epiblast cells of the pre-implantation blastocyst represent the naïve pluripotent state. By contrast, primed pluripotent cells are isolated from the epithelialized epiblast of the post-implantation embryo as mouse epiblast stem cells (EpiSC) and represent a developmentally advanced pluripotent state (Brons et al., 2007; Tesar et al., 2007). Consequently, EpiSCs are distinct from ESCs in gene expression, growth factor dependence, morphology, and the ability to contribute to blastocyst chimeras, although various core pluripotency regulators are present in both mouse ESCs and EpiSCs, and both cell types are capable of multi-lineage differentiation in vitro (reviewed in (Nichols and Smith, 2009)). Importantly, EpiSCs are post X-inactivation, i.e. are XiXISTXa, mirroring the state of the epithelialized epiblast in vivo (Pasque et al., 2011) (Figure 4A). Therefore, in the mouse system, the XaXa state appears to be a hallmark specifically of naïve pluripotency
Figure 4. X chromosome states in mouse and human pluripotent cells.
(A) X chromosome inactivation and reactivation cycle in the mouse system, highlighting the relationship between naïve and primed pluripotency and the associated X-inactivation states. Xa=active X chromosome, Xi=inactive X chromosome. (B) Hierarchy of X chromosome states in female human ESCs during long-term culture. Xe=eroded Xi. The box marks the only X-chromosome state that allows de-novo X-inactivation upon induction of differentiation. (C) Xi-reactivation does not occur when female human somatic cells are reprogrammed to primed iPSCs. While fibroblasts are mosaic for which X is inactivated (Xp=paternal X; Xm=maternal X), each early passage iPSC line carries the X-inactivation state of the differentiated cell that initiated the reprogramming event. This state is subsequently maintained upon differentiation. (D) As in (B), but for the hierarchy of X chromosome states in female human iPSCs during long-term culture.
Since XCI represents one of the most dramatic events of facultative heterochromatin formation in mammalian development, the question arises of how the somatically silent X chromosome is regulated during reprogramming. In the mouse system, the typical reprogramming experiment establishes naïve pluripotency, i.e. iPSCs that are equivalent to LIF-dependent, naïve ESCs. Our lab demonstrated that female mouse iPSCs, like female mouse ESCs, carry two active X chromosomes (XaXa) indicating that the Xi is reactivated during reprogramming to naïve pluripotency (Maherali et al., 2007) (Figure 4A). The activation of genes on the Xi is accompanied by the loss of all known heterochromatic chromatin marks, the silencing of Xist, enabling random X-inactivation upon induction of differentiation, indicating that there is no epigenetic memory for the prior Xi left behind (Maherali et al., 2007). Xi-reactivation occurs very late in the reprogramming process at around the time of pluripotency gene expression (Stadtfeld et al., 2008). In contrast to iPSCs, induced EpiSCs (iEpiSCs), generated by OSKM expression and culture conditions required for support of the primed pluripotent state (bFGF/activin), are XiXISTXa (Han et al., 2011) (Figure 4A). EpiSCs can be reprogrammed to the ESC-like state with various transcription factors and a switch in culture environment, establishing the XaXa state (Nichols and Smith, 2009) (Figure 4A). Together, these findings establish the X chromosome state as a sensitive indicator of the developmental state in the mouse system, both in differentiation and reprogramming processes, and demonstrate that the XaXa state is indisputably only associated with the naïve state of pluripotency.
X chromosome status in human ESCs and iPSCs
The analysis of human ESCs led to the puzzling observation that various ESC lines differ in their X chromosome status (Hoffman et al., 2005; Shen et al., 2008; Silva et al., 2008) (Figure 4B): (i) They can be XaXa and undergo XCI upon differentiation, comparable to mouse ESCs. (ii) Some human ESC lines have already undergone XCI and display a heterochromatic Xi with XIST RNA coating the undifferentiated state (XiXISTXa). (iii) The majority of human ESCs has a silent Xi that lacks XIST expression (Xiw/oXISTXa). Currently it is thought that newly derived human ESCs start in the XaXa state and subsequently drift towards XCI and later lose of XIST RNA with additional time in culture (Figure 4B). The strongest support for this model comes from the fact that the XaXa state can be stabilized in newly derived ESCs under physiological oxygen conditions, while chronic exposure to atmospheric oxygen concentrations irreversibly induces XCI (Lengner et al., 2010). Regardless of the X chromosome state, human ESCs generally share more features with the primed pluripotent state of the mouse than with mouse ESCs (Nichols and Smith, 2009). Therefore, the XaXa state is not restricted to naïve pluripotency in the human system and can also mark in the primed pluripotent state. To date, the occurrence of the XaXa state and the instability of the X have not been described for mouse EpiSCs and, in fact, for any other cell type.
Given the different states of the X in human ESCs an interesting question was whether reprogramming of female human cells to iPSCs, which recapitulate the primed pluripotent state of human ESCs, would result in Xi-reactivation. Originally, our group demonstrated that female human iPSC lines carry an XIST RNA-coated Xi (XiXISTXa) when they are first derived (Tchieu et al., 2010) (Figure 4C). In contrast to somatic cell populations, which are mosaic with respect to which X chromosome is inactivated, iPSC lines display a nonrandom pattern of XCI that is maintained upon induction of differentiation (Tchieu et al., 2010). As a result, two types of iPSC lines can be derived - those expressing only the Xp (XmiXpa) and those expressing only the Xm (XmaXpi) (Tchieu et al., 2010) (Figure 4C). Therefore, reprogramming to human iPSCs does not elicit Xi reactivation, and iPSCs inherit the Xi of the particular somatic cell in the culture dish that underwent a successful reprogramming event (Pomp et al., 2011; Tchieu et al., 2010). Although subsequent reports confirmed this conclusion (Cheung et al., 2011; Pomp et al., 2011), other groups obtained conflicting results and argued that Xi-reactivation is prevalent in iPSCs (Kim et al., 2011; Marchetto et al., 2010).
Recent reports help to reconcile these apparently contradictory conclusions, and confirm that the silent state of the X is faithfully maintained through the reprogramming process but unravels with the time iPSCs spend in culture (Anguera et al., 2012; Mekhoubad et al., 2012; Nazor et al., 2012; Tchieu et al., 2010; Tomoda et al., 2012). Similar to human ESCs, human iPSCs are prone to undergo XIST silencing upon prolonged passaging, yielding Xiw/oXISTXa lines and accordingly losing all XIST RNA-dependent repressive chromatin marks such as H3K27me3 (Pomp et al., 2011; Tchieu et al., 2010) (Figure 4D). Reprogramming experiments with female fibroblasts heterozygous for a mutation of the X-linked gene HPRT combined with an elegant drug selection system that can distinguish between the expression of wildtype or mutant HPRT, revealed that spontaneous loss of XIST RNA coating coincides with re-expression of the HPRT allele from the Xi (Mekhoubad et al., 2012). Thus, XiHPRTwtXaHPRTmut iPSCs express only the mutant HPRT allele at early passage but activate the wildtype HPRT allele upon XIST RNA loss. Importantly, the activation of Xi-linked genes is not limited to this one gene but appears to affect the Xi more broadly as demonstrated by global expression and DNA methylation profiles of female iPSC lines (Anguera et al., 2012; Mekhoubad et al., 2012; Nazor et al., 2012). Specifically, in early passage XiXISTXa iPSCs, X-linked genes are expressed at the level of male (XaY) iPSCs and display DNA methylation in promoters of Xi-linked genes (Mekhoubad et al., 2012; Nazor et al., 2012). By contrast, higher passage female iPSCs with no XIST RNA (Xiw/oXISTXa) are often characterized by higher expression of various X-linked genes and hypomethylation of a subset of Xi-linked promoters, suggesting that the loss of DNA methylation contributes to the activation of Xi-linked genes.
Importantly, the activation of X-linked genes does not appear to affect the entire X chromosome. Eggan and colleagues coined the partial reactivation of the Xi “erosion of dosage compensation” yielding an eroded Xi, the Xe (Mekhoubad et al., 2012) (Figure 4D). Even with long-term culturing, none of the female human iPSC lines reach the low DNA methylation level along the entire X that is typical for male iPSCs (with their single Xa), indicating that even in the worst case the activation of genes on the Xi is limited in range (Nazor et al., 2012). Across many female human iPSC lines the X chromosome is affected to varying degrees, but the loss of DNA methylation appears to target similar large, non-contiguous regions of the X chromosome, indicating that certain parts of the X can effectively maintain proper silencing while others are more prone to reactivation (Nazor et al., 2012). The patchy erasure of DNA methylation along the X, along with loss of gene silencing and XIST RNA coating, cannot be corrected upon differentiation, nor upon a repeated round of reprogramming (Mekhoubad et al., 2012; Nazor et al., 2012). Together, these findings are most consistent with a model in which reprogramming sustains the XiXISTXa state, but continued passaging of iPSCs results in XIST silencing (Xiw/oXISTXa), which then triggers partial reactivation of the Xi (Xew/oXISTXa) (Figure 4D). Notably, one could argue that these X-related events are a consequence of continued reprogramming processes, particularly given that continuous passaging of iPSCs reduces gene expression differences compared to ESCs (Chin et al., 2010; Polo et al., 2010). However, the erosion of the X has also been recently observed in many human ESC lines upon XIST RNA loss, very similar in extent to iPSCs (Nazor et al., 2012) (Figure 4C). Importantly, iPSCs with an eroded Xi still depend on FGF/Activin signaling to maintain pluripotency (Mekhoubad et al., 2012), confirming that the erosion of the X chromosome occurs in the context of primed pluripotency and is likely not associated with a change in cell identity to naïve pluripotency. Thus, for human pluripotent cells, iPSCs and ESCs, dosage compensation erosion appears to be a problem of cell culture, particularly given that it remains a feature of the differentiated progeny, necessitating the development of improved culturing methods (see below).
Why are XIST expression and the silent state of the X unstable upon long-term culturing? A few relevant observations have been made: iPSC lines obtained from the same reprogramming experiment (i.e. the same fibroblast population) typically display widely different X-states at the same passage, with some lines being able to maintain the XiXISTXa state and others being on the path of erosion (Anguera et al., 2012; Kim et al., 2011; Mekhoubad et al., 2012; Nazor et al., 2012; Tchieu et al., 2010). Similarly, any given iPSC and ESC line can be heterogeneous regarding its X chromosome state (Anguera et al., 2012; Mekhoubad et al., 2012; Silva et al., 2008; Tchieu et al., 2010; Tomoda et al., 2012). These findings, combined with the fact that no genomic abnormalities were found in iPSC lines with an eroded Xi, suggest that epigenetic but not genetic changes are responsible for the instability of the X chromosome (Anguera et al., 2012; Mekhoubad et al., 2012). Consistently, complete methylation of the XIST promoter correlates with the loss of the RNA in iPSCs (Tchieu et al., 2010), implying that de novo methylation contributes to its silencing. Interestingly, in mouse fibroblasts, experimentally induced loss of Xist by itself does not induce the reactivation of candidate X-linked genes (Csankovszki et al., 2001). However, when Xist loss is combined with the deletion of Dnmt1 and loss of DNA methylation, a dramatic reactivation of the Xi occurs in mouse somatic cells (Csankovszki et al., 2001). This parallels what happens when the Xi erodes in human iPSCs (XIST and DNA methylation loss), suggesting that deregulation of the DNA methylation machinery may directly contribute to this process.
An interesting observation is that the propagation of XiXISTXa iPSCs in media containing bFGF and IGF2 and on feeder cells expressing leukemia inhibitory factor (LIF) predictably induces XIST RNA loss and activates genes of the Xi after only a few passages. In this case, silencing is re-initiated upon differentiation, suggesting that complete Xi-reactivation occurred, establishing an XaXa state in human iPSCs, rather than an erosion of the X as discussed above (Tomoda et al., 2012) (Figure 4D). Based on cell morphology, it appears that the XaXa cells still maintain the primed pluripotent state under these conditions (Tomoda et al., 2012). A somewhat surprising observation is that XIST RNA was not detected at the endpoint of differentiation (Tomoda et al., 2012). More work will be needed to test whether XIST is upregulated earlier in the differentiation process, as X-inactivation without XIST expression would be a highly unexpected possibility (Figure 4D). In any case, this study re-emphasizes that culture conditions can have a dramatic impact on the epigenetic state of the X in human iPSCs.
A comparison of the X states in female human ESCs and iPSCs highlights two key differences. The XaXa state appears to be the most “immature” state for primed human ESCs (Lengner et al., 2010) (Figure 4B, boxed), but it is a downstream state in the hierarchy of X states in primed, human iPSCs (Tomoda et al., 2012) (Figure 4D, boxed). Hypoxic conditions or the addition of HDAC inhibitors, which appear to promote the generation and maintenance of XaXa hESCs (Lengner et al., 2010; Ware et al., 2009), do not enhance the establishment of XeXa or XaXa iPSCs (Anguera et al., 2012; Kim et al., 2011; Mekhoubad et al., 2012; Pomp et al., 2011; Tchieu et al., 2010). One reason for the difference in X-state hierarchies between human iPSCs and ESCs may be that the cells are of very different origin - iPSCs are derived from somatic XiXa cells and ESCs from XaXa cells of the female human blastocyst (Okamoto et al., 2011). Understanding the behavior of the human X in ESCs and iPSCs will be an important contribution to the ongoing debate about potential transcriptional, epigenetic, and genetic differences between various iPSC and ESC lines and their relevance (Lowry, 2012).
It is important to realize that human pluripotent cells that resemble the naïve, mouse ESC state, can be established in vitro via transcription factor-induced reprogramming methods. For example, the overexpression of OCT4 and KLF4 or KLF4 and KLF2 in primed human ESCs/iPSCs, or OSKM in fibroblasts, combined with the specific culture condition that support the naïve state, allows the establishment of human naive iPSCs (Hanna et al., 2010). However, currently the naïve state is still relatively difficult to establish and maintain (Hanna et al., 2010; Pomp et al., 2011; Wang et al., 2011b). When derived from XiXISTXa iPSCs, naïve human pluripotent cells become XIST-negative but display XIST RNA coating in virtually all cells upon differentiation (Hanna et al., 2010). Despite the fact that the analysis of the X chromosome state in naïve human cells is still in its infancy, these data argue strongly that the mouse ESC-like XaXa state, which allows XIST-dependent induction of X-inactivation during differentiation, can be established in human cells upon reprogramming to the naïve state. Naïve human pluripotent cells may therefore represent an excellent model to study the regulation of human XCI and may get around problems associated with the instability of the X in primed pluripotent cells. However, the existence of human naïve (mouse ESC-like) pluripotent cells in vivo remains unclear and their derivation from pre-implantation embryos has not been accomplished yet (Kuijk et al., 2012; Roode et al., 2012).
Instability of the human X, differentiation, and disease modeling
iPSCs can be derived for specific diseases and can differentiate into any cell type of the human body. Therefore, they offer an unprecedented opportunity to examine disease states and develop novel drugs (Onder and Daley, 2012; Trounson et al., 2012). The nonrandom X-inactivation in early-passage XiXISTXa iPSCs has an interesting consequence for the modeling of X-linked diseases. Considering females heterozygous for a mutation in an X-linked gene, iPSCs can be derived that express either the wildtype or the mutant form of the protein, which represent an ideal experimental system for the investigation of disease phenotypes, as both wildtype and mutant cell lines are on the same genetic background (Tchieu et al., 2010) (Figure 5). To date, X-linked diseases such as Rett Syndrome and Lesch-Nyhan Syndrome (LNS) have been modeled by such matched iPSCs (Cheung et al., 2011; Kim et al., 2011; Mekhoubad et al., 2012). For example, mutations in the X-linked gene HPRT cause LNS, which leads to behavioral and neurological symptoms in males but is typically non-symptomatic in heterozygous females because of random X-inactivation (Figure 5). From these heterozygous females, XiXIST RNA/HPRTwtXaHPRTmut iPSCs can be obtained that, at early passage, exhibit the LNS phenotype upon differentiation into neurons in vitro, while iPSCs with the opposite X-inactivation pattern (XiXIST RNA/HPRTmutXaHPRTwt) behave normally (Mekhoubad et al., 2012). However, at higher passage, erosion of the Xi in XiHPRT wtXaHPRTmut iPSCs leads to the expression of the wildtype HPRT allele and loss of the disease phenotype (Mekhoubad et al., 2012) (Figure 5). The interpretation of X-linked disease studies therefore requires caution and a careful assessment of the X-state.
Figure 5. Effects of X chromosome instability on disease modeling.
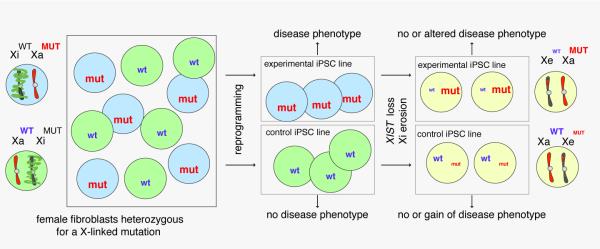
Reprogramming of differentiated cells from females heterozygous for an X-linked mutation results in iPSC lines that either express the mutant or the wildtype allele from the Xa at early passage due to non-random X-inactivation. These cell lines represent pairs of experimental and control cells ideal for modeling X-linked disease on an isogenic background. However, upon XIST loss and Xi erosion, the allele from the Xi can become re-expressed, resulting in the loss or modulation of the disease phenotype.
Problems caused by the erosion of the Xi in human iPSCs and ESCs do not only apply to studies of X-linked diseases, but should also be taken seriously for the modeling of autosomal diseases or, in fact, any differentiation process, as the erosion of the Xi in long-term culture can also alter the expression of some autosomal genes in addition to increasing X-linked gene expression (Anguera et al., 2012). Furthermore, female iPSC lines without XIST expression grow faster in culture, survive better in routine culturing, and appear to form only poorly differentiating teratomas, which may be associated with the upregulation of several X-linked oncogenes (Anguera et al., 2012), indicating that the erosion of the X affects the behavior of female iPSCs and ESCs more broadly. Importantly, all recent studies agree that loss of XIST RNA coating is closely associated with the erosion of the Xi under conventional culture conditions (Anguera et al., 2012; Mekhoubad et al., 2012; Nazor et al., 2012; Tchieu et al., 2010; Tomoda et al., 2012). Thus, currently female human iPSCs with XIST RNA coating should be preferentially used for any downstream application as these cells are in the well-defined XiXa state. Accordingly, XIST RNA coating of the Xi and the accumulation of XIST-dependent chromatin marks such as H3K27me3 can be considered biomarkers as they appear to directly identify the stable XiXa state (Anguera et al., 2012).
Outlook
The improved mechanistic understanding of the path to pluripotency has already enabled the establishment of non-OSK-containing reprogramming cocktails (Buganim et al., 2012; Mansour et al., 2012) and allowed for the replacement of essential endogenous proteins by downstream targets (Festuccia et al., 2012). Currently, we are learning only by analyzing a few snapshots of the reprogramming process. However, more and more snapshots will eventually become a continuous epigenetic movie of cell fate changes, where we can virtually watch how the epigenetic landscape is reset. The 2006 era showcased the potency of diverse transcription factors in converting cell fates. It now seems likely that it may eventually be possible to generate any cell types by forced expression of the appropriate transcription factor(s). Continued dissection of the reprogramming process holds the promise that, at some point in the future, we will be able to predict exactly which transcription factors are most potent as reprogramming factors. Finally, other fields such as tumor biology will benefit from the insight gained through reprogramming studies given for example mutations that avoid senescence have been shown to increase both reprogramming and tumor development.
Acknowledgements
Our work is supported by the NIH (DP2OD001686 and P01 GM099134), CIRM (RN1-00564 and RB3-05080), and by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA. We apologize to all authors whose work could not be cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly Efficient miRNA-Mediated Reprogramming of Mouse and Human Somatic Cells to Pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Wu Y, Guo L, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2012;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu J, Chen Y, Yang J, Liu H, Zhao X, Mo K, Song H, Guo L, Chu S, et al. Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res. 2011;21:884–894. doi: 10.1038/cr.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb Is a Direct Nanog Target Gene that Can Substitute for Nanog Function in Pluripotent Cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J, Clift KL, Fernandes CJ, Mao FF, Lee JH, Zhang L, Baker SW, Looney TJ, Foshay KM, Yu WH, et al. Evidence for a critical role of gene occlusion in cell fate restriction. Cell Res. 2012;22:848–858. doi: 10.1038/cr.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Green MR. Eukaryotic transcription activation: right on target. Mol Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Hammachi F, Morrison GM, Sharov AA, Livigni A, Narayan S, Papapetrou EP, O'Malley J, Kaji K, Ko MS, Ptashne M, et al. Transcriptional activation by Oct4 is sufficient for the maintenance and induction of pluripotency. Cell Rep. 2012;1:99–109. doi: 10.1016/j.celrep.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DW, Greber B, Wu G, Tapia N, Arauzo-Bravo MJ, Ko K, Bernemann C, Stehling M, Scholer HR. Direct reprogramming of fibroblasts into epiblast stem cells. Nat Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington MA, Jones PA, Imagawa M, Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci U S A. 1988;85:2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Katoku-Kikyo N, Karian P, Firpo M, Kikyo N. Efficient iPS cell production with the MyoD transactivation domain in serum-free culture. PLoS One. 2012;7:e34149. doi: 10.1371/journal.pone.0034149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Tani T, Katoku-Kikyo N, Kellner S, Karian P, Firpo M, Kikyo N. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells. 2011;29:1349–1361. doi: 10.1002/stem.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Hall L, Batten JL, Young H, Pardasani D, Baetge EE, Lawrence J, Carpenter MK. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, Kwon YW, Cho EJ, Youn HD. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, Poppe D, Doerr J, Ladewig J, Mertens J, Tuting T, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutroubas G, Merika M, Thanos D. Bypassing the requirements for epigenetic modifications in gene transcription by increasing enhancer strength. Mol Cell Biol. 2008;28:926–938. doi: 10.1128/MCB.01344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijk EW, van Tol LT, Van de Velde H, Wubbolts R, Welling M, Geijsen N, Roelen BA. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. 2012;139:871–882. doi: 10.1242/dev.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Li B, Adams CC, Workman JL. Nucleosome binding by the constitutive transcription factor Sp1. J Biol Chem. 1994;269:7756–7763. [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat Cell Biol. 2012;14:457–466. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285:25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lowry WE. Does transcription factor induced pluripotency accurately mimic embryo derived pluripotency? Curr Opin Genet Dev. 2012;22:429–434. doi: 10.1016/j.gde.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu G, Saito S, Kosaka T, Takubo K, Kinoshita T, Oya M, Horimoto K, Suda T. Optimal ratio of transcription factors for somatic cell reprogramming. J Biol Chem. 2012;287:36273–36282. doi: 10.1074/jbc.M112.380683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Onder TT, Daley GQ. New lessons learned from disease modeling with induced pluripotent stem cells. Curr Opin Genet Dev. 2012;22:500–508. doi: 10.1016/j.gde.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 2011;30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V, Radzisheuskaya A, Gillich A, Halley-Stott RP, Panamarova M, Zernicka-Goetz M, Surani MA, Silva JC. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J Cell Sci. 2012 doi: 10.1242/jcs.113019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak M, Jaenisch R. De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state. Genes Dev. 2011;25:1035–1040. doi: 10.1101/gad.2039011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lavial F, Ng JH, Kumar V, Tomaz RA, Martin N, Yeo JC, Gil J, Prabhakar S, Ng HH, et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012;26:2286–2298. doi: 10.1101/gad.195545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff N, Fiedler J, Holzmann A, Schambach A, Moritz T, Cantz T, Thum T. miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep. 2011;12:1153–1159. doi: 10.1038/embor.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPS Cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomp O, Dreesen O, Leong DF, Meller-Pomp O, Tan TT, Zhou F, Colman A. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roode M, Blair K, Snell P, Elder K, Marchant S, Smith A, Nichols J. Human hypoblast formation is not dependent on FGF signalling. Dev Biol. 2012;361:358–363. doi: 10.1016/j.ydbio.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Sancho-Martinez I, Baek SH, Izpisua Belmonte JC. Lineage conversion methodologies meet the reprogramming toolbox. Nat Cell Biol. 2012;14:892–899. doi: 10.1038/ncb2567. [DOI] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SS, Kim SY, Bestor TH, Shioda T, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405. S391–392. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Hofer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Tiemann U, Sgodda M, Warlich E, Ballmaier M, Scholer HR, Schambach A, Cantz T. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A. 2011;79:426–435. doi: 10.1002/cyto.a.21072. [DOI] [PubMed] [Google Scholar]
- Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, Eilertson KE, Tsang P, Baba S, White MP, et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A, Shepard KA, DeWitt ND. Human disease modeling with induced pluripotent stem cells. Curr Opin Genet Dev. 2012;22:509–516. doi: 10.1016/j.gde.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Utley RT, Cote J, Owen-Hughes T, Workman JL. SWI/SNF stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem. 1997;272:12642–12649. doi: 10.1074/jbc.272.19.12642. [DOI] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Lu Q, Granok H, Elgin SC. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011a;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci U S A. 2011b;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen J, Hu JL, Wei XX, Qin D, Gao J, Zhang L, Jiang J, Li JS, Liu J, et al. Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 2011c;12:373–378. doi: 10.1038/embor.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Implications of DNA replication for eukaryotic gene expression. J Cell Sci. 1991;99(Pt 2):201–206. doi: 10.1242/jcs.99.2.201. [DOI] [PubMed] [Google Scholar]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Hirano K, Nagata S, Tada T. Sox2 expression effects on direct reprogramming efficiency as determined by alternative somatic cell fate. Stem Cell Res. 2011;6:177–186. doi: 10.1016/j.scr.2010.09.004. [DOI] [PubMed] [Google Scholar]
- You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci U S A. 2011;108:14497–14502. doi: 10.1073/pnas.1111309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]