Abstract
Type 1 diabetes, in human patients and NOD mice, results from immune attack on insulin-producing beta-cells of the pancreas by autoreactive T lymphocytes. In NOD mice, genetically-controlled perturbations in the signaling pathways downstream of the antigen-specific T cell receptor (TCR) may be instrumental in the altered responses of T cells, manifest as inefficient induction of apoptosis after recognition of self-antigens in the thymus, or as perturbed reactivity of mature T cells in peripheral organs. To map this signaling difference(s), we have used mass spectrometry-based quantitative phosphoproteomics to compare the activation of primary CD4+ T cells of diabetes-prone NOD and -resistant B6.H2g7 mice. Immunoprecipitation and IMAC purification of tyrosine-phosphorylated peptides, combined with a stable-isotope iTRAQ labeling, enabled us to identify and quantify over 77 phosphorylation events in 54 different proteins downstream of TCR stimulation of primary CD4+ T cells. This analysis showed a generally higher level of phosphotyrosine in activated NOD cells, as well as several phosphorylation sites that appeared to be differentially regulated in these two strains (involving TXK, CD5, PAG1, and ZAP-70). These data highlight the differences in signaling between CD4+ T cell compartments of NOD and B6g7 mice, and may underlie the dysregulation of T cells in NOD mice.
Keywords: Phosphotyrosine, mass spectrometry, type 1 diabetes, NOD
Introduction
Type-1 diabetes (T1D) is an organ-specific autoimmune disease in which inflammatory cell invasion of the pancreatic islets promotes destruction of the insulin-producing beta cells. The non-obese diabetic (NOD) mouse strain has been used as an important animal model for the study of this disease. In NOD mice, diabetes develops spontaneously, sharing several critical features with the human disorder. As in man, the course of pathology in NOD animals is progressive and diabetes in these animals is primarily T lymphocyte-mediated, although other cell-types such as B cells or macrophages, may also play an important role 1. The genetic determinism of T1D also substantiates the central role of T lymphocytes: the major susceptibility factor maps to the molecules of the Major Histocompatibility Complex (MHC), which control the presentation of antigens and the activation of T lymphocytes via their antigen receptor (TCR); other susceptibility loci influence their phenotypic differentiation and effector functions 2,3.
Although the progression of T1D certainly involves defects in immunoregulatory pathways, such as the control by FoxP3+ Treg cells 4, several lines of evidence have implied that central tolerance pathways that delete maturing T lymphocytes with reactivity to self-antigens may be defective in NOD mice. Abnormal clonal deletion of NOD thymocytes was documented after engagement of TCRs by systemic injection of an anti-CD3 monoclonal antibody 5, a result that was confirmed in a more physiological setting by crossing of a TCR transgenic mouse line with a second line expressing cognate neo-self-Ag in the thymus 6–8. This abnormality proved to be thymocyte-intrinsic, denoting an inappropriate response of the immature thymocytes. Interestingly, it has been proposed that human diabetes patients may have inefficient clonal deletion of thymocytes reactive to insulin, thought to be an important diabetogenic autoantigen 9. In addition, this defective tolerance induction in the thymus of NOD mice is also accompanied by perturbations in alternative pathways of thymic differentiation 10.
The activation of a T cell is initiated by the recognition via the TCR of the complex formed by a peptide bound to a MHC molecule. Thus, a defect in thymic tolerance pathways could be ascribed to perturbations in the signaling pathways downstream of the TCR. Indeed, there is documented evidence that T cells in NOD mice have unusual responses to TCR triggers 11–14. Most recently, our group showed that naïve T cells from NOD mice are over-reactive after activation by antiCD3/28 15. Thus, there is reason to believe that an important element in the pathogenesis of T1D is a genetically determined defect in signal transduction from the TCR, which results in defective induction of self-tolerance as well as lymphocyte over-reactivity.
Signaling via TCR results in the activation of a number of signaling cascades 16. Upon TCR activation, CD4 binds to the MHC molecule, resulting in the proximity of LCK that leads to the phosphorylation of the immunoreceptor tyrosine-based activation motif (ITAM) on the CD3ζ, followed by recruitment and phosphorylation of ZAP-70. This complex phosphorylates tyrosine residues within two key adapter molecules LAT and SLP-76, forming the base of a platform for the recruitment of other signaling molecules which drive the assembly of the calcium initiation complex, cell proliferation, differentiation and immunological response. ZAP-70 can also phosphorylate PLCγ, which activates PKC, leading to NF-κB transactivation, and calcium-dependent pathways. These activities result in transport of NF-AT into the nucleus, where its transcription activation is critical for the development and function of the immune system.
How genetic variation might influence the efficacy of these phosphorylation events during signal transduction in T lymphocytes, directly or indirectly, is thus of great importance to understand the susceptibility to autoimmune deviation. Studying the effect of genetic variation presents several daunting technical challenges, however. First, it is necessary to use primary cells, rather than the more facile cell lines adapted to tissue culture, which are easy to grow in large numbers but present with inevitable phenotypic drift during culture. Second, it is necessary to obtain as broad as possible a perspective on the different signaling cascades. As is often the case, a slight change in a particular phosphorylation site may suffice to bias the outcome of TCR activation, and precise quantification is required.
The challenge of identifying and quantifying protein phosphorylation sites which regulate cellular signal transduction has been recognized for many years. Mass spectrometry allied with stable isotope labeling of peptides has been successfully applied to characterize the dynamic nature of signal transduction. Compared to western blots with phosphorylation-specific antibodies, mass spectrometry offers a much greater dynamic range, more precise quantification, and the ability to quantify many phosphorylation events on multiple proteins simultaneously, in an unbiased manner. To analyze tyrosine phosphorylation mediated signaling networks, enrichment of tyrosine phosphorylated species using anti-phosphotyrosine peptide immunoprecipitation coupled with immobilized metal affinity chromatography (IMAC) and liquid chromatography tandem mass spectrometry (LC-MS/MS) has been demonstrated to give broad coverage of multiple networks in a variety of biological systems 17–19.
To date most phosphoproteomics studies have been dedicated to identification of large numbers of protein phosphorylation sites in cultured cell lines. For instance, analysis of TCR signaling in T lymphoma Jurkat cell lines treated with pervanadate has led to the identification of dozens of tyrosine phosphorylation sites 20–22. More recently, several groups were able to identify and quantify approximately 130 different phosphotyrosine sites in different cell states by stimulating Jurkat cells with anti-CD3 and anti-CD28 or anti-CD3 alone or anti-CD3 and anti-CD4 23–25. T cell phosphoproteomics has also recently been extended to the analysis of primary T cells isolated from peripheral blood. Among the 230 phosphorylation sites identified in this study, only 8 tyrosine phosphorylation sites were identified, with no known function in TCR signaling 26.
In the present work, we have attempted to harness the sensitivity and resolution of MS phosphoproteomics to analyze, in primary T lymphocytes, the impact of genetic variation on tyrosine phosphorylation events downstream of TCR triggering. We compared phosphorylation events in lymphocytes from diabetes-susceptible NOD mice and from diabetes-resistant C57Bl/6.H-2g7 (B6g7) congenic mice. The results provide a useful perspective on signaling pathways in primary T lymphocytes ex vivo, and bring forth several differences between NOD and B6g7 mice in this respect.
Experimental Procedures
Mice, Cell preparation and activation
NOD/LtJ.DOI (NOD) and B6.H2g7 mice were bred under specific pathogen free (SPF) conditions at the Joslin Diabetes Center (IACUC protocol 99-20). CD4+ T cells were isolated from cell suspensions from subcutaneous lymph nodes (cervical, inguinal, axillary, brachial, lumbar, and sciatic) of NOD and B6g7 females (5 weeks of age) by negative magnetic selection (using MACS beads coated with anti-CD8a, anti-B220, anti-CD49b, anti-CD11b and anti-Ter-119; CD4+ T cell isolation kit, Miltenyi), in addition to depletion of pre-activated cells (anti-CD25 and anti-CD69). Purity was confirmed by flow cytometry and CD4+ T cells used in these experiments were regularly >97%. Purified cells were aliquoted at 2.5×106 and pre-incubated with anti-CD3-biotin (3 µg) for 20 min on ice in 30 µL RPMI/ 3% FCS. Cells were further incubated 1 min at 37°C and cross-linked with 1.5ng streptavidin (Jackson Immunochemicals) for 5 minutes at 37°C, after which they were immediately lysed in 500 µL of 8 M urea supplemented with phosphatase inhibitors (1mM Na3VO4, 1mM NaF, 1mM beta-glycerolphosphate). Lysates were pooled to 20×106 cells/condition for mass spectrometry analysis. Control cells were subjected to the same process as activated cells, but without antibody. All three biological replicates were prepared similarly, but on different days.
Sample preparation and peptide immunoprecipitation
Cell lysates were proteolyzed and iTRAQ stable-isotope labeled as described 17. Briefly, samples were reduced with 10 mM DTT for 1 h at 56 °C and alkylated with 55 mM iodoacetamide for 1h at room temperature. Samples were digested with 40 µg of trypsin (Promega) overnight at room temperature and desalted with C18 Sep-Pak Plus Cartridges (Waters). iTRAQ (Applied Biosystems) labeling was performed according to the manufacturer’s directions. Samples labeled with four different isotopic iTRAQ reagents were combined, concentrated to 10 µL and then dissolved in 200 µL of IP buffer (100 mM Tris, 100 mM NaCl, and 1% Nonidet P-40, pH 7.4) and 200 µL of water to a final volume of 400 µL. The mixed sample was incubated with 4 µg of immobilized anti-phosphotyrosine pY100 antibody (Cell Signaling Technologies) for 8h at 4 °C. Protein G agarose beads slurry (20 µL, IP04, Calbiochem) were added to the mixture and incubated overnight at 4°C. Beads were washed three times with rinse buffer (100 mM Tris, 100 mM NaCl, pH 7.4) and retained peptides were eluted from antibody with 70 µL of elution buffer (100 mM glycine pH 2.5) for 1 h at room temperature.
IMAC phosphopeptide purification and LC-MS/MS analysis
Immobilized metal affinity chromatography (IMAC) was performed to enrich for phosphorylated peptides and remove nonspecifically retained nonphosphorylated peptides. IMAC capillary columns were constructed as described 27. Briefly, 100 µm I.D. fused-silica column was packed with POROS 20 MC and activated with 100 mM of FeCl3. Sample was loaded at a flow rate of 1 µL/min, washed with 250 µL (20 min, 12.5 µL/min) of a solution containing 100 mM NaCl (Sigma) in acetonitrile (Mallinckrodt), deionized water, and glacial acetic acid (25:74:1, v/v/v). Organic buffer was removed by rinsing the column with 250 µL of 0.1% acetic acid. Peptides retained on the IMAC column were eluted with 50 µL of 250 mM sodium phosphate (pH 8.0) to a reverse-phase (C18) pre-column (100 µm I.D., packed with 10 cm of 10 µm C18 ODS-A beads (Kanematsu)). Following elution, the pre-column was attached to the HPLC (Agilent) and rinsed with solvent A (H2O/HOAc, 99:1 (v/v)) for 10 minutes at 10 µL/min to remove phosphate salt. After rinsing, the pre-column was attached to an analytical column 28 (50 µm I.D. fused silica capillary packed with 10 cm of 5 µm C18 beads (Millipore)) with an integrated electrospray tip with ~1 µm I.D. orifice. Peptides were eluted using a 100-min gradient with solvent A (H2O/HOAc, 99:1 (v/v)) and B (H2O/MeCN/HOAc, 29:70:1 (v/v/v)): 10 min from 0% to 15% B, 75 min from 15% to 40% B, and 15 min from 40% to 70% B. Eluted peptides were directly electrosprayed into a QqTOF mass spectrometer (QSTAR XL Pro; Applied Biosystems) operated in information-dependent acquisition mode: MS/MS spectra of the five most intense peaks with two to five charges in the MS scan were automatically acquired with previously selected peaks excluded for 40 s.
MS/MS spectra were searched against a rodent (mouse and rat) protein database (NCBI) by using ProQuant (v1.1 Applied Biosystems) and MASCOT (v2.2 Matrix Science) with trypsin as enzyme and allowing up to 3 missed cleavages. Oxidation of M and phosphorylation of S, T, Y were included as variable modifications, while carbamidomethylation of cysteine and iTRAQ modification of –NH2 lysine side chain and the N-terminus were included as fixed modifications. Mass tolerance was set to 2.2 atomic mass units for precursor ions and 0.15 atomic mass units for fragment ions. Peptide sequence validation was further confirmed manually for each of the identified peptides (e.g. those with MASCOT and ProQuant scores greater than 20) by checking the raw MS/MS data for possible mixed spectra, non-assigned abundant peaks, and phosphorylation position. Phosphopeptide quantification was determined via ProQuant by calculating peak area for iTRAQ marker ions (m/z: 114.1, 115.1, 116.1, and 117.1). Quantification results were manually validated and corrected for potential contamination due to peaks at m/z 114.06, 115.06, 116.06 and 117.06. To account for protein loading differences in the four samples, a small fraction (~0.1%) of the supernatant from the tyrosine phosphopeptide immunoprecipitation was analyzed by LC-MS/MS, thereby providing quantification for total non-phosphorylated peptide in each sample. Protein loading quantification was then used to normalize the iTRAQ marker ion data for tyrosine phosphorylated peptides.
Western Blotting, Positive and Negative control of TXK
2.5×106 CD4+ T cells were lysed in 250 µL RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS) with protease inhibitor cocktail tablet (Complete, Roche) and phosphatase inhibitors (0.5 mM Na3VO4, 0.5 mM NaF, 10 mM beta-glycerolphosphate). 60 µg total protein (~105 cells), as determined by bicinchroninic acid protein assay (Thermo Pierce), was applied in each lane of a 10% Tris-HCl acrylamide gel. Following 1-dimensional separation and transfer to PVDF membrane, the membrane was incubated overnight at 4°C with 1:1000 rabbit anti-TXK (Santa Cruz Biotechnology). Immunoreactive bands were visualized by enhanced chemiluminescence, pico-ECL (Thermo Pierce) and the blots were exposed to x-ray XAR film (Kodak). For positive and negative controls of TXK immunoprecipitation, 293FT cells transfected with TXK-IRES-GFP MIG vector or empty vector were used.
Results
Identification and quantification of tyrosine phosphorylation in CD4+ T cells from NOD and B6g7 mice
To determine the influence of genetic differences between the diabetes-susceptible NOD mice and diabetes-resistant C57Bl/6.H-2g7 (B6g7) congenic mice on the TCR signaling network, tyrosine phosphorylation sites were quantified in primary CD4+ T cells from both NOD and B6g7 mice, in resting state and after TCR triggering. We chose activation by crosslinking of bound anti-CD3 antibody, which allows a virtually synchronous activation of all cells in the sample, and a short readout time (5 minutes), a time at which early steps of the signaling cascades are optimally activated. This experimental strategy resulted in four samples (baseline and activated for NOD and B6g7 cells). Control samples were treated in the same manner, but without antiCD3 antibody. Stable isotope (iTRAQ) labeling of cell lysates was used to compare these four samples in the same LC/MS/MS run analysis, schematically represented in Fig. 1. One major challenge in this analysis was the limited sample size (~2×107 cells, corresponding to ~400 µg total protein) for each condition. Given the goal of quantifying tyrosine phosphorylation in primary cells from this limited amount of sample, we applied nano-flow (5–10 nL/min) ESI-LC-MS/MS using a ~1 µm I.D. tip diameter to enhance sensitivity for these analyses.
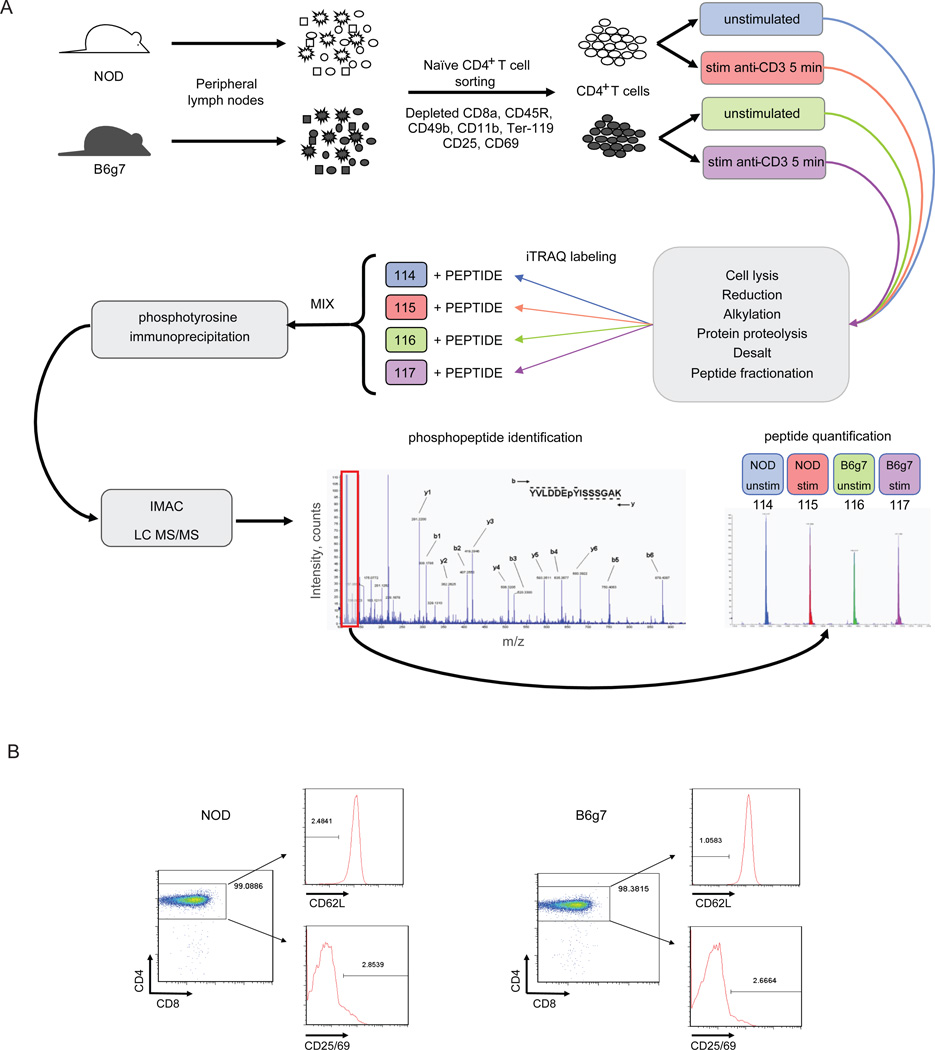
Figure 1.
A. Experimental scheme: subcutaneous lymph nodes were isolated from NOD and B6g7 mice and CD4+ T cells were magnetically separated by negative selection, thereby depleting non-CD4+ T cells using anti-CD8a, anti-CD45R, anti-CD49b, anti-CD11b and anti-Ter-119 to deplete CD8+ T cells, B cells, NK cells, macrophages, and erythroid cells, respectively. In addition preactivated or regulatory T cells were also depleted using anti-CD25 and anti-CD69 antibody. Half of the cells were left unstimulated and the other half was stimulated with anti-CD3 antibody for 5 min. Lysed cells were enzymatically digested, labeled separately with iTRAQ and combined. Upon phosphotyrosine enrichment by phosphotyrosine immunoprecipitation and IMAC, sample was subjected to LC-MS/MS analysis. Identification of each peptide was manually confirmed and quantification was performed by the analysis of the area from iTRAQ marker ions present in the MS/MS analysis. B. Assessment of CD4+ T cell separation analysis from both NOD and B6g7 subcutaneous lymph nodes by flow cytometry. CD4+ T cells separation were essentially >97%.
Naïve CD4+ T cells were isolated from peripheral lymph nodes by negative magnetic selection (i.e. using antibodies that bound to cells other than the desired CD4+ cells) to avoid possible interfering signals from antibodies bound to the CD4 co-receptor. The depleting cocktail included antibodies to CD69 and CD25 to deplete preactivated or regulatory cells. Purity of the resulting CD4+ T cell preparations were assessed by flow cytometry (routinely >97% purity), and essentially all were CD62Lhi, indicating naïve status (Fig. 1b). Isolated CD4+ T cells were stimulated with anti-CD3 for 5 min, and immediately lysed. Cell lysates were digested with trypsin, and the resulting peptides labeled with one of the four isoforms of the iTRAQ reagent (one for each of the four different conditions). The labeled extracts were combined, and phosphotyrosine peptides were enriched sequentially by immunoprecipitation with anti-phosphotyrosine antibody and by immobilized metal affinity chromatography (IMAC) before LC-MS/MS analysis. The identification of the peptide sequence from each of the MS/MS spectra were performed using ProQuant and MASCOT against a rodent protein database (NCBI). Identified phosphorylated peptides were manually validated by checking the MS/MS spectra for unassigned ions with greater than 10% abundance (relative to base peak in the MS/MS spectrum), contamination within the isolation window, and for the position of the phosphorylation site 29 (Supplemental Figure 1). Phosphopeptides were quantified across the four different conditions by comparing the peak area for each of the four iTRAQ labels (m/z: 114, 115, 116, and 117). To correct for possible variations in the starting amounts of sample for each condition, peak areas were normalized for each condition relative to the total value of nonphosphorylated peptides in the supernatant of the immunoprecipitation step.
Three independent experiments were performed in this manner, which from approximately 2000 spectra altogether identified 77 tyrosine-phosphorylated peptides in 54 different proteins, providing a broad perspective of signaling pathways downstream of T cell receptor stimulation (the complete data are presented in Supplementary Table 1). These included 10 of the 12 known immunoreceptor tyrosine-based activation motif (ITAM) sites on the CD3 subunits of the TCR complex. We also detected proteins known to be directly connected to the response to TCR stimulation, ZAP-70 (pY290 and pY491) and LCK (pY192, pY505 and likely pY394 (the tryptic peptide containing pY394 (LIEDNEpYTAR) could not be unambiguously assigned to LCK, SRC, FYN or YES because its sequence is identical among these Src kinase family members)). Downstream of ZAP-70 and LCK, several other protein kinases were identified, including TXK (pY91, pY148, and pY420), PI3K (pY199), as well as several phosphatases: PTPN6 (a.k.a. SHP1) (pY536), PTPN11 (a.k.a. SHP2) (pY62), and PTPRA (pY825). In addition, phosphopeptides were identified in downstream signaling proteins such as PLCγ (pY771), WASP (pY293) and SLAP-130 (pY559) as well as nuclear proteins like Histone-4 (H4 pY170) and GSK3 (pY279). In addition, of the 77 phosphopeptides observed, only 3 phosphopeptides were observed that were not found in current phosphopeptide databases (KIAA1462 pY320, LIME1 pY207, and TXK pY214), suggesting that, at least at this level of sensitivity, we may be approaching the full spectrum of phosphotyrosine peptides linked to TCR signaling in naïve T lymphocytes.
Tyrosine phosphorylation differentially regulated in CD4+ T cells from NOD or B6g7
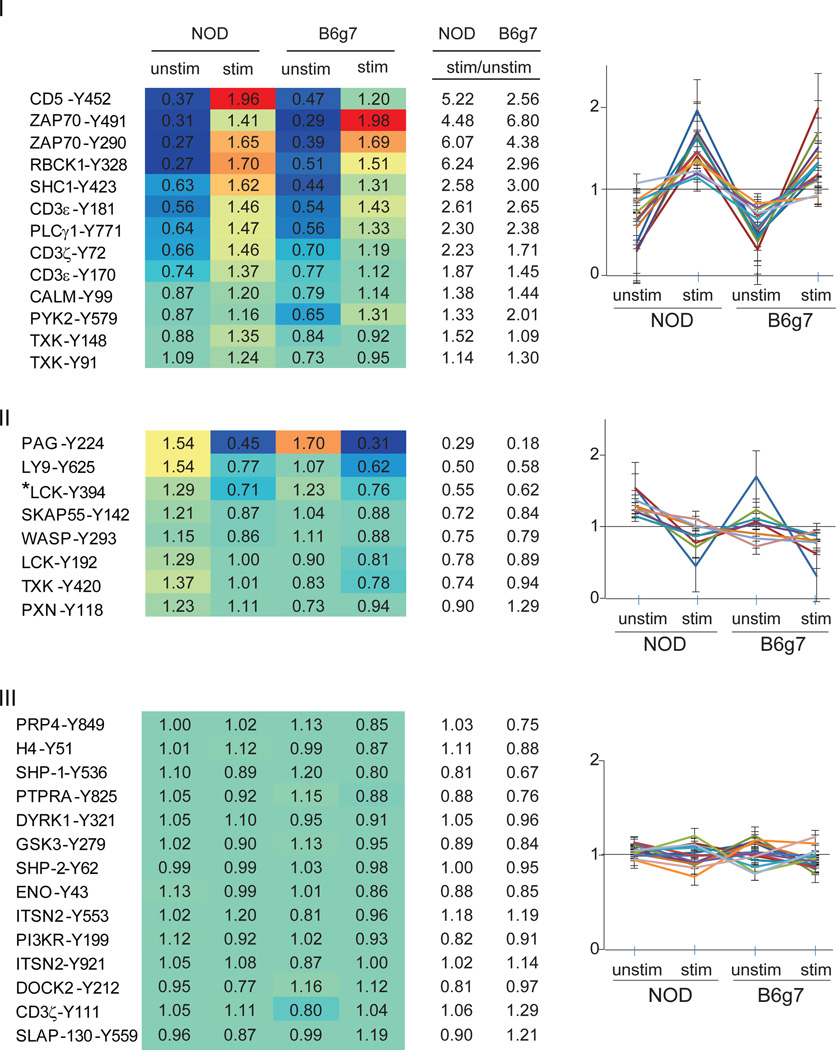
Among the 77 tyrosine phosphorylation sites identified, 35 peptides were detected in at least two separate experiments, and we focused on these phosphopeptides for which quantification was deemed more reliable. Clustering tools showed that they could be grouped into three categories (Fig. 2; values were normalized, for each phosphopeptide, to the mean value of the 4 conditions). The first category includes peptides whose phosphorylation increased upon activation. Many of these proteins and phosphorylation sites are well-characterized members of the canonical TCR signaling network, including several of the CD3 and ZAP-70 phosphopeptides. This increase in phosphorylation was generally present in CD4+ T cells from both NOD and B6g7, indicating that activation signals proximal to the receptor are generally shared, as one would expect. This response to activation is also visible in the plot of Fig. 3, which compares the responses to activation by showing the resting/stimulated cells in NOD and B6g7. Conversely, the second group of phosphopeptides had decreased phosphorylation in activated cells relative to non-stimulated cells, and this was also generally true in cells from both mouse strains. This group included peptides from PAG1 (a.k.a. CBP) pY224, and LY9 (a.k.a. SLAMf3) pY625. In T cells, PAG1 is involved in the negative regulation of T-cell activation by inactivation of Src-family kinases via its association with CSK, and the inhibitory effect of PAG1 on T-cell activation is dependent on its tyrosine phosphorylation 30–32. PAG1-Y224 is dephosphorylated upon stimulation, presumably relieving this inhibition. Finally, the third group of peptides showed little or no change during activation, either because unrelated to TCR signaling, or because affected only later in the process, and not yet activated at the five-minute time point.
Figure 2.
Heatmap and graphical clustering analysis of phosphotyrosine sites in unstimulated and stimulated CD4+ T cells from NOD and B6g7 mice. Clusters were classified into three major different categories: I. increased phosphorylation, II. decreased phosphorylation in both NOD and B6g7 upon treatment with anti-CD3 antibody, and III. peptides with minimal change during activation. Values in the table are normalized row-wise by the mean of all four conditions (unstimulated, stimulated; NOD and B6g7 mice) for each of the peptide. Heatmap color graduation varies from low (blue) to high (red) tyrosine phosphorylation. * The tryptic peptide containing pY394 could not be unambiguously assigned to LCK, SRC, FYN or YES because its sequence is identical among these Src kinase family members.
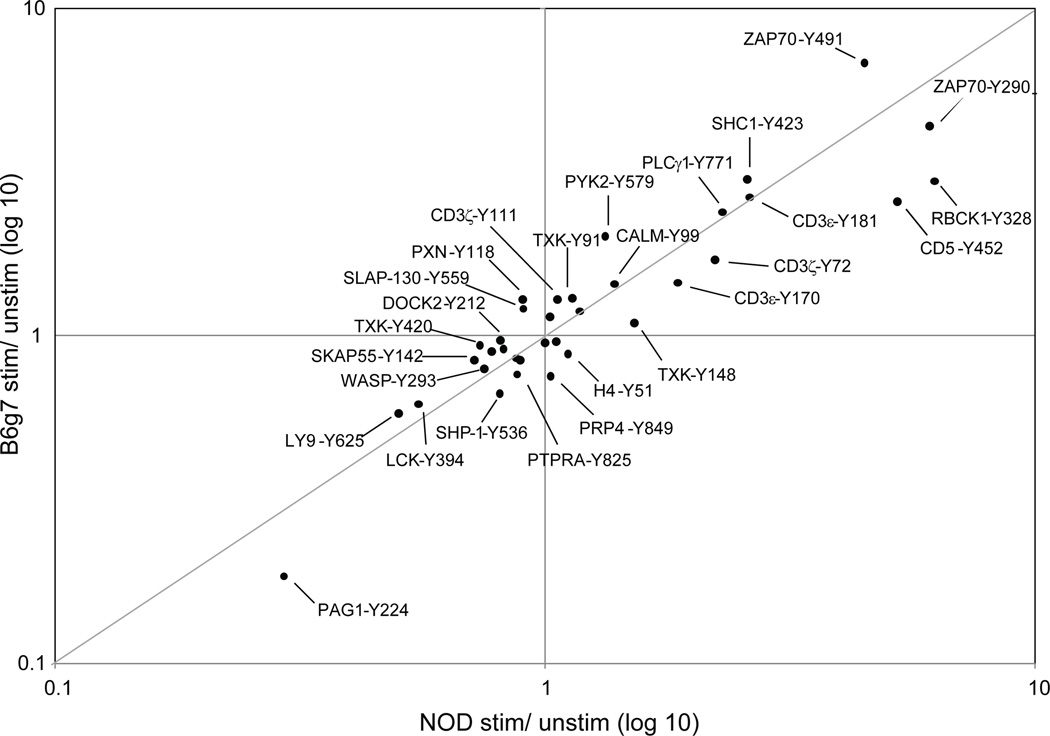
Figure 3.
Quantification of tyrosine phosphorylation in B6g7 and NOD CD4+ T cells stimulated with anti-CD3 for 5 minutes. For each phosphorylated peptide, fold-change phosphorylation level was calculated for stimulated to unstimulated CD4+ T cells. Fold-change for B6g7 cells was then compared to fold-change for NOD cells to obtain the plot.
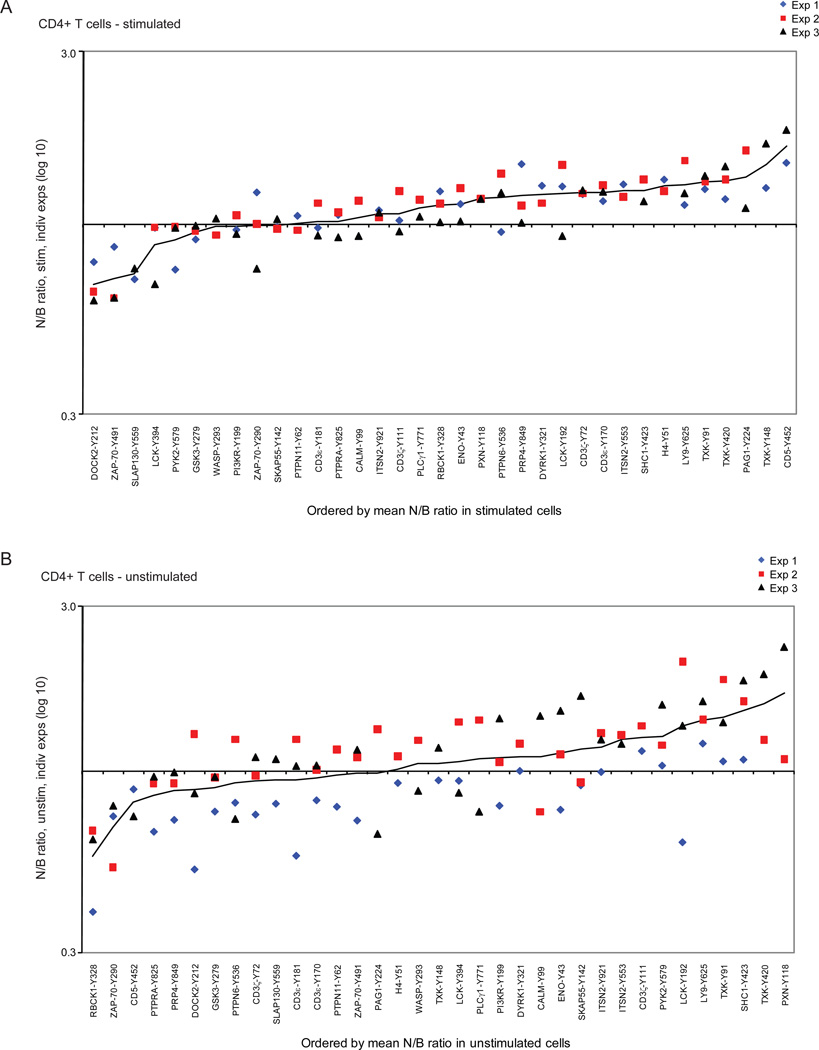
Although the profiles were generally similar in CD4+ T cells from NOD or B6g7 mice, some quantitative differences were present, reflected in the color scheme of Fig. 2 or by off-diagonal points in Fig. 3. For instance, the patterns were quantitatively different for activation phosphopeptides CD5-Y452 or ZAP-70-Y491, or for the downregulated phosphorylation of LY9-Y625. To better analyze these differences, the NOD/B6g7 ratio for each tyrosine phosphopeptide was calculated for stimulated and unstimulated cells, and plotted according to ranking of the mean NOD/B6g7 ratio (Fig. 4). This analysis showed several tyrosine phosphorylation sites from several proteins to be differentially regulated in these two strains, and this was generally consistent in biological replicates, particularly for the more extreme deviations. For stimulated cells (Fig. 4a), tyrosine phosphorylation in NOD CD4+ T cells was overall higher than in B6g7 CD4+ T cells, with 26 of the 36 phosphopeptides featuring a mean NOD/B6g7 ratio above 1. Several peptides also stood out at the extreme of the distribution: in particular, tyrosine phosphorylation sites at CD5-pY452, PAG1-pY224 and three different phosphotyrosines in the kinase TXK (pY91, pY148 and pY420) had the highest phosphorylation levels in NOD relative to B6g7 T cells. At the other extreme, ZAP-70-pY491, DOCK2-pY212 and SLAP-130-pY559 presented higher phosphorylation levels in T cells from B6g7 mice compared to NOD. The higher levels of TXK or CD5-Y452 phosphopeptides in stimulated NOD T cells might result from a higher responsiveness, or from higher baseline levels. Examination of the profile in unstimulated cells (Fig. 4b) showed that TXK phosphopeptides were already over-represented prior to TCR engagement, thus reflecting a difference in baseline levels rather than inducibility (which is modest for these phosphorylation sites in any case – Fig. 3). In contrast, the CD5-Y452 phosphopeptide was actually under-represented in resting NOD cells, and thus it is a stronger response to TCR engagement that leads to greater phosphorylation in stimulated NOD T cells relative to stimulated B6g7 T cells.
Figure 4.
Comparison of tyrosine-phosphorylation in peripheral CD4+ T cells from diabetes-susceptible NOD vs. –resistant B6g7 mice (a) stimulated or (b) unstimulated with anti-CD3 antibody. Proteins plotted by order of the mean of NOD (N) over B6g7 (B) ratio. Different colors represent different experiments. Protein tyrosine-phosphorylation sites are represented by Y-position of phosphorylation.
Although these trends appeared generally reproducible, particularly for the more extreme deviations, it was important to ascertain the statistical significance of the conclusions. These experiments with primary cells exhibited significantly more variation in phosphopeptide composition than experiments run in the same time period in our laboratory with tissue culture cells (data not shown). As would be expected from the distribution of the data points in Fig. 4A, none of the differences between NOD and B6g7 for individual phosphopeptides were statistically significant when their relative abundance was compared individually by a Student’s t.test, particularly when an appropriate Bonferroni correction for multiple sampling was applied. Rather than estimating the significance of individual points, however, it was important to estimate the significance of the global deviations we observe, which was performed with a data permutation strategy. Simulated datasets were generated under the null hypothesis that all differences observed between NOD and B6g7 cells were merely random fluctuations around a common mean; this mean and its variance were calculated for each phosphopeptide, and 1,000 iterations of the same experimental design (3 replicates, same distribution of missing values) were simulated computationally by random number generation following a normal distribution with these means and variances. The NOD vs B6g7 differences were tabulated in these simulated datasets. As expected, some differences were found for individual phosphopeptides in the simulated datasets, but in only 7 of 1000 permutations did we observe as many concordant deviations (>1.4-fold in either direction), with an average of 1.48 deviations per simulated dataset, vs 5 in the real dataset. This analysis thus confirms that the differential profiles we observed denote statistically significant differences in responses between CD4+ T cells from NOD and B6g7 mice.
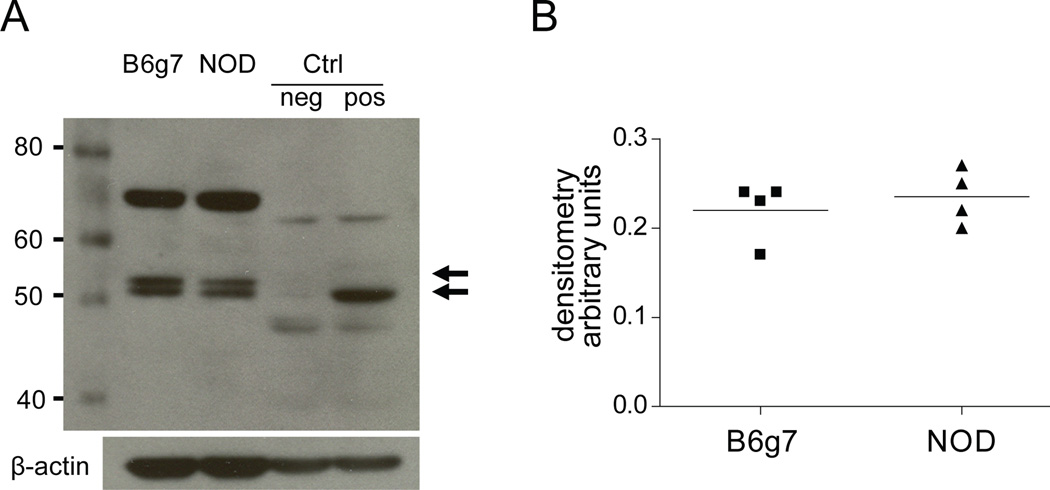
Given the presence of several overly abundant TXK phosphopeptides in NOD CD4+ T cells, it seemed plausible that the difference might reflect a different level of TXK protein. To verify this point, total TXK protein was quantitated by immunoblotting in extracts of B6g7 and NOD CD4+ T cells. This analysis showed no significant difference in TXK protein expression between the two strains (Fig. 5). Similarly, microarray analysis of splenic CD4+ T cells from B6g7 and NOD also showed no difference in the corresponding mRNA (H. Suwanai, personal communication). This result indicated that phosphorylation differences observed in NOD vs B6g7 were not due to overall quantity of the protein, but instead are indicative of altered phosphorylation stoichiometry between the strains.
Figure 5.
a. One dimension western blotting analysis with anti-TXK antibody (1:1000) of B6g7 and NOD CD4+ T cell lysates and positive (+) and negative (−) controls (lysates of 293FT cell lines expressing or not TXK protein, respectively). Arrows show the two isoforms present in the mouse cell lysate. b. Densitometry quantification of the bands in B6g7 and NOD. Both 52 kDa and 57 kDa were merged.
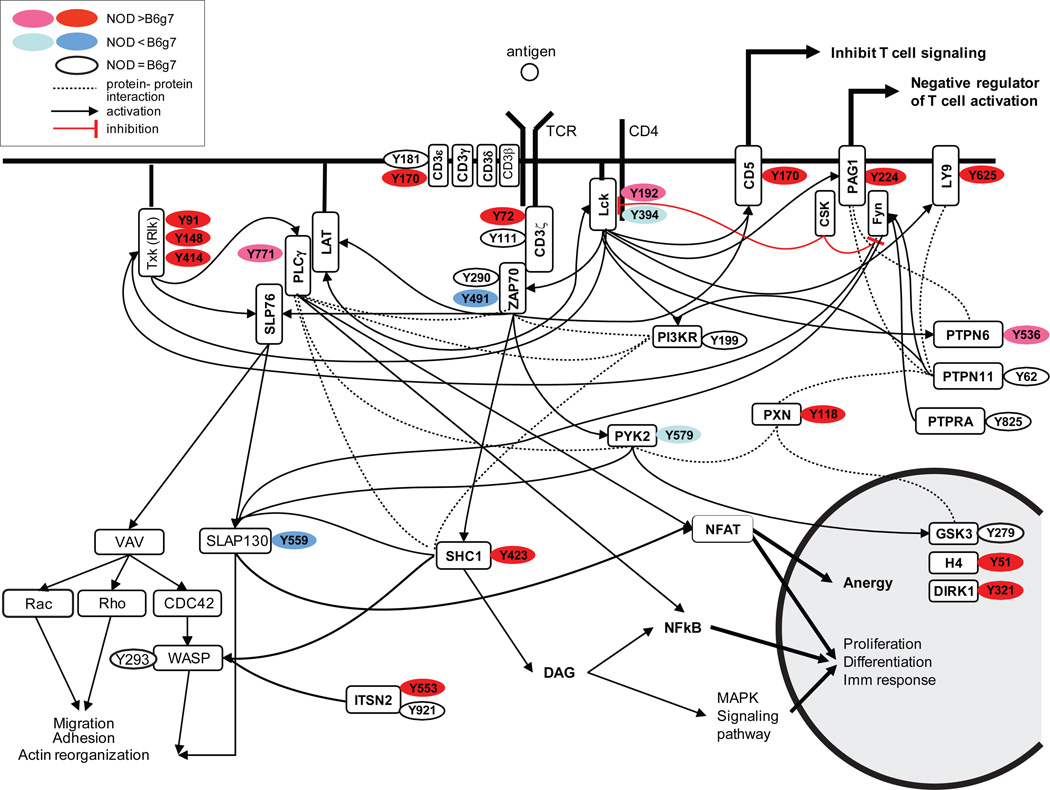
To help in the identification of TCR signaling pathways that might be differentially activated in NOD and B6g7 CD4+ T cells, the comparative abundance in stimulated cells is represented graphically in Fig. 6, overlayed on the connectivity map of molecules downstream from the TCR (recompiled data from KEGG Pathway database and Refs 33, and 34). The differential phosphorylation observed in activated NOD CD4+ T cells did not bring forth any immediately obvious pathway, as the corresponding proteins mapped to different areas of the schema.
Figure 6.
T cell signaling pathways and protein phosphorylation sites identified to be differential between NOD and B6g7 mice in stimulated cells. In red: high tyrosine-phosphorylation upregulated in NOD; pink: low tyrosine-phosphorylation in NOD; blue: high tyrosine phosphorylation upregulated in B6g7; light blue: low tyrosine phosphorylation upregulated in B6g7; in white: tyrosine-phosphorylation equal in NOD and B6g7 (recompiled from KEGG Pathway database, Refs 33 and 34).
Discussion
The susceptibility of NOD mice to autoimmune diabetes originates from several mechanistic levels (defective T cell tolerance induction in the thymus, over-reactive T cells in the periphery, deficient control of the autoreactive effector T cells by Treg cells) all of which could potentially be ascribed to biased signaling downstream from the TCR. The work presented here quantified the tyrosine phosphoproteome of T cell signaling pathways in NOD relative to B6g7 mice, with the goal of defining the key differences in signaling between these two strains. We used a mass spectrometry strategy using iTRAQ labeling of phosphopeptides, initially developed by our laboratory to elucidate tyrosine phosphorylation signaling networks in human HMEC and Jurkat cell lines, to quantitatively measure protein tyrosine phosphorylation in primary CD4+ T cells from NOD and B6g7 mice. To our knowledge, this is the first application of tyrosine phosphoproteome screening to primary T cells, and to the comparative analysis of genetic variants.
Overall, 77 different tyrosine phosphorylation sites in 54 different proteins were identified in CD4+ T cells, these sites were quantified in both the unstimulated cells and in those cells stimulated with anti-CD3 antibody. Several of the phosphorylation sites are homologous to previously reported phosphopeptides in the human Jurkat cell line 21,23,25(Supplemental Table 1), reinforcing the confidence that we are indeed observing significant peptides. Indeed, the response pattern of several phosphorylation sites in the Jurkat cell line, including those on CD3 family members and ZAP-70, were similar compared to our observations in primary CD4+ T cells. In addition, the phosphoproteins identified in the current analysis include all the “usual suspects” of TCR-initiated signaling pathways 35.
The phosphopeptides presenting the highest induction upon stimulation in control B6g7 cells belong to ZAP-70, which is perhaps not surprising since this molecule is at the very apex of the TCR signaling cascade. Y290 (corresponding to position Y292 in human ZAP-70) is located at the interdomain B and was reported as tied to negative feedback, possibly by promoting ligand-induced TCR internalization 36. Y491 is located in the putative regulatory loop of ZAP-70 kinase domain and a Y>F mutation in the Jurkat cell line (corresponding to position pY492 in human) inhibited TCR-induced activation of NFAT by interfering with both intracellular calcium increase and Ras-regulated activation of Erk pathway kinases 37. Interestingly, ZAP-70-pY491 is one of the exceptions whose phosphorylation decreases in NOD T cells relative to B6g7 (Fig. 2), unlike pY290, which responds equivalently in both strains. One might speculate that pY491 reflects biased signaling in NOD cells, although the origin and specific effect of this decreased phosphorylation remains to be elucidated.
Overall, however, we observed a higher level of tyrosine phosphorylation for a majority of phosphopeptides in NOD T cells, which is consistent with the over-reactivity of NOD CD4+ T cells observed upon in vitro stimulation 15. Our data also highlight tyrosine phosphorylation at three sites of the TXK kinase. TXK is a member of the Tec kinase family which is comprised of TXK, TEC, and ITK. The exact biological role of the TXK kinase and functional significance of the identified phosphorylation sites are not well known. Current data suggests partial redundancy between Tec family kinases in modulating and amplifying the efficiency of signal transduction downstream from the TCR. Specifically, mutation of TXK does not prevent T cell development nor block TCR signaling but instead reduces TCR signaling in a manner that can alter thymocyte development and mature T cell differentiation and function 34. Kashiwakura and colleagues 38, showed that Y91 of TXK is an autophosphorylation site, important for the promotion of IFN-gamma synthesis, suggesting that phosphorylation of Y91 plays a positive regulatory role in TXK function. Phosphorylation at Y420 was identified as a transphosphorylation substrate for FYN 39, but the other sites have yet to be characterized. The origin and significance of increased phosphorylation of TXK in NOD T cells relative to B6g7 T cells remains to be elucidated, but it is interesting to note that all three sites show the same bias, suggesting that they might be phosphorylated by the same kinase complex.
Several proteins demonstrated decreased phosphorylation in NOD compared to B6g7, including ZAP-70-Y491, DOCK2-Y212 and SLAP-130-Y559. DOCK2 is an adaptor protein, a member of the hematopoietic cell-specific CDM family that is essential for lymphocyte chemotaxis; it plays a role in TCR-induced activation of Rac2 and IL-2 transcription, at least in part through interactions with RAC, CRKL, and CD3-zeta 40. SLAP-130 (a.k.a. FYB) is an adapter protein for FYN and SLP76 in T cells 41,42. TCR ligation leads to slightly increased tyrosine phosphorylation of SLAP-130 at Y559, and this phosphorylation event allows SLAP-130 to dampen SLP76 activity; yet this does not happen in NOD T cells, where SLAP-130 phosphorylation at Y559 is slightly decreased, possibly contributing to over-responsiveness of NOD T cells.
In summary, by using mass spectrometry analysis to precisely quantify the differences in the dynamics of the phosphorylation in the T cell signaling events, we were able to identify several tyrosine phosphorylation events that have increased phosphorylation in stimulated NOD CD4+ T cells compared to B6g7 cells. Differences in the ratio of the characterized phosphorylation sites were not very high, which explains why we were unable to detect remarkable differences in tyrosine phosphorylation in NOD vs B6g7 mice by conventional biochemical analyses. The heightened level of basal activation of the signaling network in NOD T cells might unbalance the response to TCR stimulation, thereby contributing to their over-reactivity. We can hypothesize that slight modifications in several phosphorylation events of the T cell signaling pathway in NOD T cells, at baseline and after stimulation, as well as their lower responsiveness at several inhibitory sites, might contribute to a final over-responsiveness of the Teff cells.
Supplementary Material
Protein list of tyrosine phosphorylation sites identified and quantified derived from unstimulated and stimulated CD4+ T cell lysate from diabetes-prone NOD and -resistant B6g7 mice in three independent experiments. 2×107 CD4+ T cells isolated from subcutaneous lymph nodes for each of the conditions were used.
Values correspond to relative areas in each peptide normalized to the non-phosphorylated peptides. Most values correspond to the area-under-the-curve directly obtained from the ProQuant data analyzer. In a few cases, these ProQuant values were deemed not reliable because of the existence of a pre-peak at 116.07 amu, which ProQuant could not distinguish from the main iTRAQ peak at 116.1 amu; for those cases, the area was measured manually from the spectra, and span a lower scale (shown as bold) . Position of phosho in human indicates the phosphorylation position in the human protein when different from mouse. A, B and C indicates phosphorylation previously described in Jurkat T lymphoma cell line by A. Ref 21 B. Ref 23; and C. Ref 25. * corresponds to novel tyrosine phosphorylation sites not identified in current phosphopeptide databases. *** The tryptic peptide containing pY394 (LIEDNEpYTAR) could not be unambiguously assigned to LCK, SRC, FYN or YES because its sequence is identical among these Src kinase family members.
Manually verified MS/MS spectra of each phosphopeptide identified from the CD4+ T cells reported in this article. Serine, Threonine and Tyrosine phosphorylated amino acids were indicated as B, O and Z or pS, pT or pY, respectively. –NH2 side chain iTRAQ labeled Lysine was indicated as J.
Acknowledgements
We thank K. Hattori for assistance with mice and Prof. Andre Veillette for useful discussion. This work was supported by a grant from the NIH (R33 AI 065354) to DM, CB and FW; LKI was supported by a mentor-based postdoctoral fellowship from the American Diabetes Association.
Abbreviations used in this paper
- NOD
non obese diabetic
- T1D
type 1 diabetes
- TCR
T cell receptor
- Teff
T effector cells
- Treg
T regulatory cells
- Ag
antigen
Footnotes
Supporting Information
Supporting Information Available: This material is available free of charge via the internet at http://pubs.acs.org.
Bibliography
- 1.Kikutani H, Makino S. The murine autoimmune diabetes model: NOD and related strains. Adv. Immunol. 1992;51:285–322. doi: 10.1016/s0065-2776(08)60490-3. [DOI] [PubMed] [Google Scholar]
- 2.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N. Engl. J. Med. 2009;360(16):1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 3.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009 doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluestone JA, Tang Q, Sedwick CE. T Regulatory Cells in Autoimmune Diabetes: Past Challenges, Future Prospects. J Clin Immunol. 2008 doi: 10.1007/s10875-008-9242-z. [DOI] [PubMed] [Google Scholar]
- 5.Kishimoto H, Sprent J. A defect in central tolerance in NOD mice. Nat. Immunol. 2001;2(11):1025–1031. doi: 10.1038/ni726. [DOI] [PubMed] [Google Scholar]
- 6.Lesage S, Hartley SB, Akkaraju S, Wilson J, Townsend M, Goodnow CC. Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J Exp Med. 2002;196(9):1175–1188. doi: 10.1084/jem.20020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liston A, Lesage S, Gray DH, O'Reilly LA, Strasser A, Fahrer AM, Boyd RL, Wilson J, Baxter AG, Gallo EM, Crabtree GR, Peng K, Wilson SR, Goodnow CC. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity. 2004;21(6):817–830. doi: 10.1016/j.immuni.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Zucchelli S, Holler P, Yamagata T, Roy M, Benoist C, Mathis D. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity. 2005;22:385–396. doi: 10.1016/j.immuni.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Pugliese A, Miceli D. The insulin gene in diabetes. Diabetes Metab. Res. Rev. 2002;18:13–25. doi: 10.1002/dmrr.261. [DOI] [PubMed] [Google Scholar]
- 10.Feuerer M, Jiang W, Holler PD, Satpathy A, Campbell C, Bogue M, Mathis D, Benoist C. Enhanced thymic selection of FoxP3+ regulatory T cells in the NOD mouse model of autoimmune diabetes. Proc Natl Acad Sci U S A. 2007;104(46):18181–18186. doi: 10.1073/pnas.0708899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zipris D, Lazarus AH, Crow AR, Hadzija M, Delovitch TL. Defective thymic T cell activation by concanavalin A and anti-CD3 in autoimmune nonobese diabetic mice. Evidence for thymic T cell anergy that correlates with the onset of insulitis. J. Immunol. 1991;146(11):3763–3771. [PubMed] [Google Scholar]
- 12.Rapoport MJ, Lazarus AH, Jaramillo A, Speck E, Delovitch TL. Thymic T cell anergy in autoimmune nonobese diabetic mice is mediated by deficient T cell receptor regulation of the pathway of p21ras activation. J. Exp. Med. 1993;177(4):1221–1226. doi: 10.1084/jem.177.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salojin K, Zhang J, Cameron M, Gill B, Arreaza G, Ochi A, Delovitch TL. Impaired plasma membrane targeting of Grb2-murine son of sevenless (mSOS) complex and differential activation of the Fyn-T cell receptor (TCR)-zeta-Cbl pathway mediate T cell hyporesponsiveness in autoimmune nonobese diabetic mice. J. Exp. Med. 1997;186(6):887–897. doi: 10.1084/jem.186.6.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Salojin K, Delovitch TL. Sequestration of CD4-associated Lck from the TCR complex may elicit T cell hyporesponsiveness in nonobese diabetic mice. J. Immunol. 1998;160(3):1148–1157. [PubMed] [Google Scholar]
- 15.D'Alise AM, Auyeung V, Feuerer M, Nishio J, Fontenot J, Benoist C, Mathis D. The defect in T-cell regulation in NOD mice is an effect on the T-cell effectors. Proc Natl. Acad Sci U S. A. 2008;105(50):19857–19862. doi: 10.1073/pnas.0810713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu. Rev. Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell Proteomics. 2005;4(9):1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes. 2006;55(8):2171–2179. doi: 10.2337/db06-0148. [DOI] [PubMed] [Google Scholar]
- 19.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. U. S. A. 2007;104(31):12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salomon AR, Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, Sauer K, Brock A, Horn DM, Schultz PG, Peters EC. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2003;100(2):443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal. Chem. 2004;76(10):2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 22.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat. Methods. 2005;2(8):591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 23.Kim JE, White FM. Quantitative analysis of phosphotyrosine signaling networks triggered by CD3 and CD28 costimulation in Jurkat cells. J. Immunol. 2006;176(5):2833–2843. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto M, Oyamada K, Takahashi H, Sato T, Hatakeyama S, Nakayama KI. Large-scale proteomic analysis of tyrosine-phosphorylation induced by T-cell receptor or B-cell receptor activation reveals new signaling pathways. Proteomics. 2009;9(13):3549–3563. doi: 10.1002/pmic.200900011. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen V, Cao L, Lin JT, Hung N, Ritz A, Yu K, Jianu R, Ulin SP, Raphael BJ, Laidlaw DH, Brossay L, Salomon AR. A new approach for quantitative phosphoproteomic dissection of signaling pathways applied to T cell receptor activation. Mol. Cell Proteomics. 2009;8(11):2418–2431. doi: 10.1074/mcp.M800307-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrascal M, Ovelleiro D, Casas V, Gay M, Abian J. Phosphorylation analysis of primary human T lymphocytes using sequential IMAC and titanium oxide enrichment. J. Proteome. Res. 2008;7(12):5167–5176. doi: 10.1021/pr800500r. [DOI] [PubMed] [Google Scholar]
- 27.Zarling AL, Ficarro SB, White FM, Shabanowitz J, Hunt DF, Engelhard VH. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J. Exp. Med. 2000;192(12):1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2000;72(18):4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 29.Nichols AM, White FM. Manual validation of peptide sequence and sites of tyrosine phosphorylation from MS/MS spectra. Methods Mol. Biol. 2009;492:143–160. doi: 10.1007/978-1-59745-493-3_8. [DOI] [PubMed] [Google Scholar]
- 30.Davidson D, Bakinowski M, Thomas ML, Horejsi V, Veillette A. Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol. Cell Biol. 2003;23(6):2017–2028. doi: 10.1128/MCB.23.6.2017-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janssen E, Zhang W. Adaptor proteins in lymphocyte activation. Curr. Opin. Immunol. 2003;15(3):269–276. doi: 10.1016/s0952-7915(03)00044-x. [DOI] [PubMed] [Google Scholar]
- 32.Smida M, Posevitz-Fejfar A, Horejsi V, Schraven B, Lindquist JA. A novel negative regulatory function of the phosphoprotein associated with glycosphingolipid-enriched microdomains: blocking Ras activation. Blood. 2007;110(2):596–615. doi: 10.1182/blood-2006-07-038752. [DOI] [PubMed] [Google Scholar]
- 33.Lin J, Weiss A. T cell receptor signalling. J. Cell Sci. 2001;114(Pt 2):243–244. doi: 10.1242/jcs.114.2.243. [DOI] [PubMed] [Google Scholar]
- 34.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu. Rev. Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 35.Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu. Rev. Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- 36.Magnan A, Di BV, Mura AM, Boyer C, Richelme M, Lin YL, Roure A, Gillet A, Arrieumerlou C, Acuto O, Malissen B, Malissen M. T cell development and T cell responses in mice with mutations affecting tyrosines 292 or 315 of the ZAP-70 protein tyrosine kinase. J. Exp. Med. 2001;194(4):491–505. doi: 10.1084/jem.194.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mege D, Di BV, Germain V, Tuosto L, Michel F, Acuto O. Mutation of tyrosines 492/493 in the kinase domain of ZAP-70 affects multiple T-cell receptor signaling pathways. J. Biol. Chem. 1996;271(51):32644–32652. doi: 10.1074/jbc.271.51.32644. [DOI] [PubMed] [Google Scholar]
- 38.Kashiwakura J, Suzuki N, Takeno M, Itoh S, Oku T, Sakane T, Nakajin S, Toyoshima S. Evidence of autophosphorylation in Txk: Y91 is an autophosphorylation site. Biol. Pharm. Bull. 2002;25(6):718–721. doi: 10.1248/bpb.25.718. [DOI] [PubMed] [Google Scholar]
- 39.Chamorro M, Czar MJ, Debnath J, Cheng G, Lenardo MJ, Varmus HE, Schwartzberg PL. Requirements for activation and RAFT localization of the T-lymphocyte kinase Rlk/Txk. BMC. Immunol. 2001;2:3. doi: 10.1186/1471-2172-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukui Y, Hashimoto O, Sanui T, Oono T, Koga H, Abe M, Inayoshi A, Noda M, Oike M, Shirai T, Sasazuki T. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412(6849):826–831. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- 41.Boerth NJ, Judd BA, Koretzky GA. Functional association between SLAP-130 and SLP-76 in Jurkat T cells. J. Biol. Chem. 2000;275(7):5143–5152. doi: 10.1074/jbc.275.7.5143. [DOI] [PubMed] [Google Scholar]
- 42.Veale M, Raab M, Li Z, da Silva AJ, Kraeft SK, Weremowicz S, Morton CC, Rudd CE. Novel isoform of lymphoid adaptor FYN-T-binding protein (FYB-130) interacts with SLP-76 and up-regulates interleukin 2 production. J. Biol. Chem. 1999;274(40):28427–28435. doi: 10.1074/jbc.274.40.28427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein list of tyrosine phosphorylation sites identified and quantified derived from unstimulated and stimulated CD4+ T cell lysate from diabetes-prone NOD and -resistant B6g7 mice in three independent experiments. 2×107 CD4+ T cells isolated from subcutaneous lymph nodes for each of the conditions were used.
Values correspond to relative areas in each peptide normalized to the non-phosphorylated peptides. Most values correspond to the area-under-the-curve directly obtained from the ProQuant data analyzer. In a few cases, these ProQuant values were deemed not reliable because of the existence of a pre-peak at 116.07 amu, which ProQuant could not distinguish from the main iTRAQ peak at 116.1 amu; for those cases, the area was measured manually from the spectra, and span a lower scale (shown as bold) . Position of phosho in human indicates the phosphorylation position in the human protein when different from mouse. A, B and C indicates phosphorylation previously described in Jurkat T lymphoma cell line by A. Ref 21 B. Ref 23; and C. Ref 25. * corresponds to novel tyrosine phosphorylation sites not identified in current phosphopeptide databases. *** The tryptic peptide containing pY394 (LIEDNEpYTAR) could not be unambiguously assigned to LCK, SRC, FYN or YES because its sequence is identical among these Src kinase family members.
Manually verified MS/MS spectra of each phosphopeptide identified from the CD4+ T cells reported in this article. Serine, Threonine and Tyrosine phosphorylated amino acids were indicated as B, O and Z or pS, pT or pY, respectively. –NH2 side chain iTRAQ labeled Lysine was indicated as J.