Abstract
Legionella pneumophila proliferates within various protists and metazoan cells, where a cadre of ~300 effectors is injected into the host cell by the defect in organelle trafficking/intracellular multiplication (Dot/Icm) type IVB translocation system. Interkingdom horizontal gene transfer of genes of protists and their subsequent convergent evolution to become translocated effectors has probably enabled L. pneumophila to adapt to the intracellular life within various protists and metazoan cells through exploitation of evolutionarily eukaryotic processes, such as endoplasmic reticulum-to-Golgi vesicle traffic, phosphoinositol metabolism, AMPylation, deAMPylation, prenylation, polyubiquitination, proteasomal degradation and cytosolic amino- and oligo-peptidases. This is highlighted by the ankyrin B (AnkB) F-box effector that exploits multiple conserved eukaryotic machineries to generate high levels of free amino acids as sources of carbon and energy essential for intracellular proliferation in protists and metazoan cells and for manifestation of pulmonary disease in mammals.
Keywords: prenylation, farnesylation, polyubiquitination, phosphatidylinositol, F-box, AnkB, proteasome
Intracellular life cycle of L. pneumophila within mammalian and protozoan cells
L. pneumophila is ubiquitous in aquatic habitats where it survives and replicates within a wide range of protists, such as amoeba and ciliated protozoa [1]. Upon transmission to humans, L. pneumophila replicates in alveolar macrophages leading to Legionnaires’ disease [1,2].
The molecular and cellular aspects of infection by L. pneumophila in both protozoa and mammalian phagocytes are similar [3,4]. When either protozoan hosts or macrophages engulf L. pneumophila, the bacterium evades targeting the Legionella-containing vacuole (LCV) to degradation by the lysosomes [1,5,6]. Instead, mitochondria and endoplasmic reticulum (ER)-derived vesicle are rapidly recruited to the LCV [4–7]. Within a few minutes, the LCV becomes remodeled into an ER-derived compartment [1,6–8]. Within the ER-remodeled LCV, L. pneumophila replicates to high numbers until it disrupts the phagosomal membrane and escapes into the host cell cytosol where the last one to two rounds of replication occurs [9]. This culminates in the expression of various bacterial virulence traits and bacterial escape to the extracellular environment [10,11].
The ability to evade phagocytic killing and to remodel the LCV within both hosts is mediated by the type IVB secretion system known as the Dot/Icm secretion system [6] that injects ~300 effectors into the host cell [12]. This exceptional number of translocated bacterial effectors includes many substrates harboring eukaryotic domains leading to interference or exploitation of various eukaryotic cellular processes and some of the cellular processes are highly conserved through evolution [7,13–16]. This may not be surprising because the molecular and cellular aspects of intracellular proliferation of L. pneumophila within protists and metazoan cells are similar. This review highlights the conserved eukaryotic machineries exploited by L. pneumophila and the translocated bacterial effectors involved.
Exploitation of the eukaryotic prenylation pathway by L. pneumophila
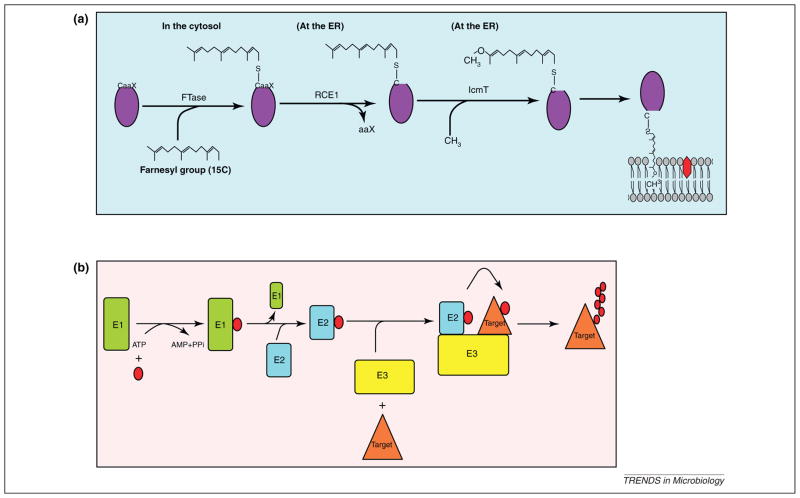
Many of the injected effectors of L. pneumophila are targeted into various host cell membranes and the LCV membrane [13,14,17–20], through exploitation of various cellular processes such as prenylation [21] or lipid phosphatidylinositol (PI) derivatives [22]. Prenylation is an evolutionarily conserved post-translational lipid modification of eukaryotic proteins that increases protein hydrophobicity to enable anchoring a hydrophilic protein to the lipid bilayer of membranes or to associate with other hydrophobic proteins [21]. Prenylation is initiated by covalent linkage of a farnesyl group (15C) or geranyl-geranyl group (20C) to a cysteine residue in a conserved C-terminal CaaX motif of eukaryotic proteins by farnesyl transferase (FTase) or two geranyl-geranyl tranferases (GGTases I and II), respectively [21]. The terminal aaX tripeptide is then cleaved by the endoprotease Ras-converting enzyme-1 (RCE1) followed by carboxyl methylation by the isoprenyl cysteine carboxyl methyl transferase (IcmT) and membrane association of the prenylated proteins (Figure 1a). The enzymatic machinery of prenylation is evolutionarily conserved in eukaryotes but is not present in prokaryotes. Prenylation plays a key role in functional activity of many eukaryotic proteins such as the Ras and Rab proteins, involved in signaling and vesicle fusions, respectively [21].
Figure 1.
Evolutionarily conserved eukaryotic pathways exploited by Legionella in amoeba and mammals. (a) Farnesylation modification of eukaryotic proteins. Proteins that contain CaaX motif are recognized by the cytosolic farnesyl transferase (FTase), which modifies the protein by the addition of a 15-carbon farnesyl group to the conserved cysteine residue within the C-terminal CaaX motif. The farnesylated proteins are trafficked to the endoplasmic reticulum (ER) where the aaX tripeptide is cleaved by the Ras-converting enzyme-1 (RCE1) protease, followed by methylation of the farnesyl group by the isoprenyl cysteine carboxyl methyl transferase (IcmT) enzyme. The modified protein is subsequently targeted to specific membranes. (b) The eukaryotic ubiquitination pathway. Ubiquitin modification is an ATP-dependent process involving the sequential action of three enzymes (E1–E3). Ubiquitin activating enzyme (E1) activates ubiquitin (red circle) via the generation of a high energy thioester bond between ubiquitin and an E1 cysteine residue. The activated ubiquitin is then transferred to ubiquitin conjugating enzyme (E2). The final step of the ubiquitination is where substrate-specific ligase (E3) binds simultaneously to E2 and the substrate, resulting in transfer of ubiquitin monomer from the E2 enzyme to a target protein. The fate of the modified protein is determined by the lysine linkages utilized in polyubiquitination.
L. pneumophila is the first example of a pathogen that hijacks the eukaryotic prenylation machinery to anchor several bacterial effectors into the pathogen-containing vacuolar membrane and other host membranes [13,14,19–21]. Among these membrane-associated effectors is the well characterized ankyrin B (AnkB), which is found in all sequenced L. pneumophila strains (lpg2144, lpa03071, lpl2072, lpp2082, lpc1593, legAU13) and is present in 211 of 217 tested strains [23–25]. During macrophage and amoeba infection with L. pneumophila, the C-terminal CaaX (CLVC) motif of AnkB is modified by the host farnesylation machinery [13,14,20]. This host-mediated farnesylation is essential for anchoring AnkB to the cytosolic face of the LCV membrane (Figure 2) [13,14,21]. The CaaX (CLVC) motif of AnkB is essential for anchoring ectopically expressed AnkB to the plasma membrane of human cells and amoeba [13,14,21]. A substitution of the conserved cysteine residue in the CaaX motif with alanine (AnkB169C/A) results in redistribution of ectopically expressed AnkB169C/A into the cytosol of human cells and amoeba. Expression of the AnkB169C/A variant by Legionella during infection results in failure to anchor AnkB to the LCV membrane, severe defects in intracellular proliferation in amoeba and macrophages, and attenuation of intrapulmonary proliferation in the mouse model, similar to ablation of ankB [14]. Disruption of the host farnesylation machinery by chemical inhibitors or by silencing FTase by RNAi results in loss of membrane anchoring and biological function of AnkB [14].
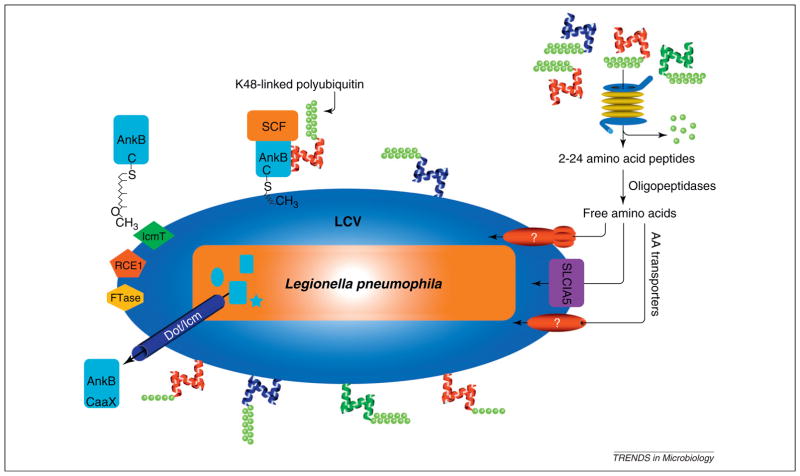
Figure 2.
Ankyrin B (AnkB)-mediated generation of amino acids through promoting proteasomal degradation of amoeba and mammalian hosts. Upon translocation of AnkB by the defect in organelle trafficking/intracellular multiplication (Dot/Icm) type IV secretion system, AnkB is farnesylated by the three host enzymes farnesyl transferase (FTase), Ras-converting enzyme-1 (RCE1) and isoprenyl cysteine carboxyl methyl transferase (IcmT) that are recruited to the Legionella-containing vacuole (LCV) by the Dot/Icm system. Upon farnesylation, AnkB is anchored into the cytosolic face of the LCV membrane, where it interacts with the SCF1 E3 ubiquitin ligase complex and acts as a platform for the docking of K48-linked polyubiquitinated proteins to the LCV. The K48-linked polyubiquitinated proteins are targeted for proteasomal degradation that generates short peptides that are rapidly degraded by cytosolic oligo- and amino-peptidases. This results in generation of high levels of free amino acids that are imported to the LCV by various amino acid (AA) transporters, such as SLC1A5. The whole process is highly conserved and is essential for intracellular proliferation of Legionella in amoeba and human cells and for intrapulmonary proliferation in mice. Adapted from [53].
Covalent linkage of the 15C farnesyl moiety to cysteine within the CaaX motif occurs in the cytoplasm where FTase is localized. Then cleavage of the aaX terminal tripeptide and methylation of the farnesyl group takes place at the cytosolic face of the ER where RCE1 and IcmT are located (Figure 1a) [21]. However, host-mediated farnesylation of AnkB probably occurs locally at the ER-derived LCVs in both hosts, where the three host farnesylation enzymes FTase, RCE1 and IcmT are recruited to the LCV in a Dot/Icm-dependent manner. It is thought that there are specific Dot/Icm effectors involved in recruiting FTase, whereas IcmT and RCE1 are part of the ER [13,14].
Besides AnkB, the LCV is highly enriched with other prenylated proteins [13,14,20]. In silico analyses of four sequenced L. pneumophila genomes revealed the presence of at least 11 genes that encode CaaX motif-containing proteins and those proteins have been designated as prenylated effectors of Legionella (Pel)/CaaX motif protein (CMP) [19,20]. Most of the PelA-K effectors are translocated substrates of the Dot/Icm type IV secretion system, but their functions are unknown. During ectopic expression, Pels/CMPs are modified by the mammalian prenylation machinery and exhibit distinct membrane localization in HEK293 and COS-1 cells [19,20]. The Pels/CMPs exhibit a cytosolic shift in their cellular localization upon chemical inhibition of either FTase or GGTases [19,20]. Although none of the Pels/CMPs have been tested in amoeba, we speculate that some if not all of the Pels/CMPs are modified by the amoeba prenylation machinery because the LCV harboring the ankB null mutant is decorated with farnesylated proteins, which is mediated by the Dot/Icm secretion system [13,14]. The strategy of exploitation of the evolutionarily conserved host prenylation machinery is a remarkable example of pathogenic evolution of Legionella and its host adaptation to infect phylogenetically diverse hosts.
Numerous other bacterial pathogens have the potential to exploit the prenylation machinery to anchor their injected effectors into the pathogen-containing vacuolar membrane or other host membranes [20,21]. The Salmonella effector SifA was the first reported prenylation-dependent membrane-anchored effector that is also modified by host S-palmitoylation [21]. It is possible that prenylation inhibitors might be used as a target for therapeutic agents against Legionella and other pathogens that require the host prenylation machinery to cause disease.
Exploitation of host phosphoinositides to anchor Legionella effectors to the LCV
Many pathogens including Listeria, Shigella, Salmonella, Brucella and Mycobacterium spp. exploit host cell lipid PI derivatives to anchor their effector proteins to various host membranes to promote host cell invasion or establish a suitable replicative niche [26–28]. PI can be phosphorylated/dephosphorylated at the 3, 4 and/or 5 positions on the inositol ring by PI kinases or phosphatases, respectively [22]. The PI derivatives are highly conserved through evolution and play roles in various eukaryotic cellular processes, including cell signaling, actin remodeling and membrane trafficking in amoebae and mammalian cells [29].
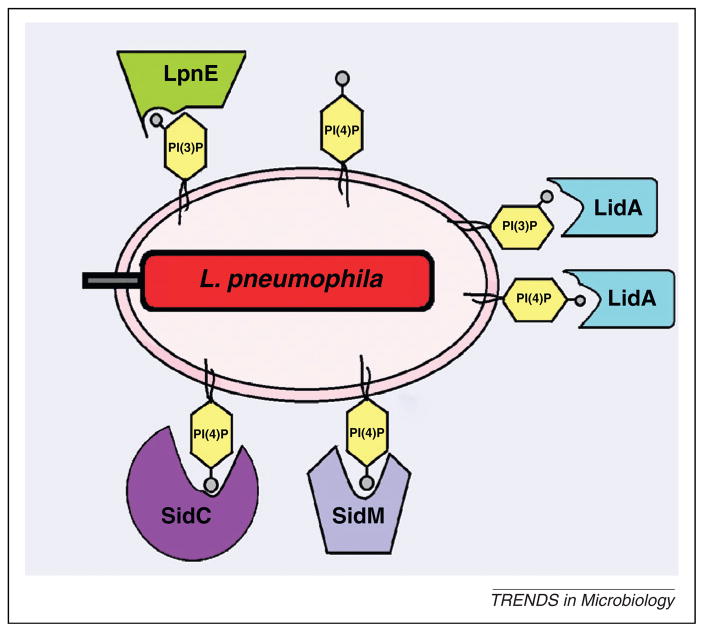
The presence of PI derivatives on the cytosolic side of the LCV membrane allows L. pneumophila to employ them for anchoring some of the translocated effectors on the LCV [22]. The main PI derivative present on the LCVs is phosphatidylinositol (4) phosphate [PI(4)P], which is a Golgi marker that acts as a second messenger to mediate export of early secretory vesicles from ER exit sites [22]. A major PI(4)P-binding effector on the LCVs is SidC and its paralog SdcA [30]. Anchoring those effectors to the LCV membrane through PI(4)P promotes recruitment of ER vesicles to the LCV within amoeba and macrophages [31–33]. The SidM/DrrA effector is a competitor of SidC for free PI(4)P-binding sites on the LCV (Figure 3). Anchoring SidM/DrrA to the LCV via PI(4)P recruits Rab1 to the LCVs. The SidM/DrrA effector modulates the activity of Rab1 through its guanine nucleotide exchange factor (GEF) activity [18], its GDI displacement factor (GDF) activity [34], and its AMPylation and deAMPylation of Rab1 [46–39] to promote recruitment of early secretory vesicles to LCV [5,7]. The GEF activity of SidM is promoted by the LidA effector, which is localized to the LCV. Anchoring LidA to the LCV membrane is achieved by interacting with PI(4)P and with lower affinity to PI(3)P (Figure 3) [38,39]. Interestingly, the Rab1-modulating SidM/DrrA is sufficient to stimulate non-canonical membrane vesicle fusion [31].
Figure 3.
Exploitation of host cell lipid phosphatidylinositol (PI) derivatives to anchor Legionella pneumophila effectors. Many defects in organelle trafficking/intracellular multiplication (Dot/Icm)-translocated effectors such as SidC, SidM, LidA and LpnE are anchored to the Legionella-containing vacuole (LCV) membrane through PI(4)P or PI(3)P to interfere with signal transduction and promote host vesicle trafficking.
Another effector of L. pneumophila that interacts with PI(3)P is LpnE. The LpnE effector is a Sel1 repeat-containing protein, which is exported independent of both type II and the Dot/Icm type IV secretion systems, and is involved in the entry of L. pneumophila into macrophages and epithelial cells and for virulence in mice [40]. This effector binds exclusively to PI(3)P (Figure 3) [28], and to PI-metabolizing enzymes; the mammalian oculocerebrorenal syndrome of Lowe 1 (OCRL1) and its Dictyostelium homolog Dictyostelium discoideum 5-phosphatase 4 (Dd5P4), which hydrolyze PI(4,5)P2 to yield PI(4)P [22].
Although SidC and its paralog SdcA are the only effectors known to bind PI(4)P within amoeba and macrophages [32,33], it is likely that other L. pneumophila effectors exploit host PI derivatives in a similar strategy within both hosts because the LCVs in both hosts is decorated with PI(4)P that interferes with signal transduction and promotes host vesicle trafficking [22,33]. Therefore, L. pneumophila exploits at least two evolutionary conserved eukaryotic processes, PI metabolism and prenylation, to anchor translocated effectors to the LCV membrane within phylogenetically diverse eukaryotic hosts.
Exploitation of eukaryotic ubiquitination by F-box effectors of Legionella
Ubiquitination is another evolutionarily conserved post-translational modification process that is essential for various eukaryotic processes but it is absent in prokaryotes [41]. Polyubiquitination involves covalent polymerization of the 76-amino acid ubiquitin monomers through one of the seven lysine residues in ubiquitin (K6, K11, K27, K29, K33, K48 or K63) [42]. Polyubiquitination through K48 linkages is a hallmark for degradation by the 26S proteasomes. However, K63 linkages in polyubiquitin modulate the biological activity or location of the polyubiquitinated proteins.
Ubiquitination is mediated by sequential action of three classes of enzymes (E1–E3) [42]. The ubiquitin activating enzyme (E1) transfers ubiquitin to a conjugating enzyme (E2), followed by a substrate-specific E3 ubiquitin ligase that links ubiquitin to the target protein (Figure 1b) [42]. Several classes of E3 ubiquitin ligases have been described based on the presence of U-box or F-box domains [43]. One well-characterized E3 ubiquitin ligase family is the multi-protein SCF1 complex that is composed of SKP1, Cul1 and RBX1 [44]. It is not surprising that many pathogens exploit host ubiquitination [45,46], which is achieved via pathogen-injected effectors that mimic E3 ubiquitin ligases or by bona fide U/F-box effectors [16,45–48].
In silico genomic analyses of Legionella reveals the presence of F- or U-box containing effector encoding genes. The AnkB F-box effector is one of very few Dot/Icm-translocated effectors that plays a major role in intracellular replication within both macrophages and protozoa, and is the only effector known to be required for intrapulmonary proliferation in mice [11,16,23,24,48]. The AnkB effector harbors an F-box domain, two ankyrin (ANK) protein–protein interaction domains in addition to the c-terminal farnesylation CaaX (CLVC) motif [11,16,23,24,49]. In both macrophages and protozoa, AnkB functions as a bona fide F-box protein where it interacts with the host SCF1 complex and functions as a platform for the docking of polyubiquitinated proteins to the LCV membrane in macrophages and amoeba (Figure 2) [13,16,48,49].
In addition to AnkB, the genome sequence of L. pneumophila reveals the presence of at least another four F-box-containing proteins [47]. These include Lpg2224 (PpgA), Lpg2525 and LicA [50]. LegU1 is another F-box effector that interacts with the SCF1 complex, and targets a host protein for ubiquitination. However, in the Philadelphia strain, the mutant that harbors in-frame deletion of the F-box domain of legU1 does not display any defects in either the accumulation of polyubiquitinated proteins on the LCV or intracellular proliferation as compared to the wild type strain in either primary bone marrow-derived mouse macrophages or amoeba [50]. Thus, multiple L. pneumophila F-box proteins hijack the highly conserved SCF1 ubiquitin ligase of amoeba and mammalian cells.
Exploitation of eukaryotic ubiquitination by U-box effectors of Legionella
The Legionella U-box protein (LubX) effector mimics the host E3 ubiquitin ligase activity during mammalian infection [51]. A U-box is a class of eukaryotic E3 ubiquitin ligases that serves as a docking site for E2 ubiquitin conjugating enzymes [52]. LubX harbors two U-box domains where both U-box domains are essential for the LubX biological activity [51]. LubX acts as a negative regulator of the SidH effector by targeting it for proteasomal degradation at later stages of the infection [51]. Interestingly, although LubX triggers ubiquitination of mammalian Cdc2-like kinase 1 (Clk1), its role in the protozoan host has not been reported. It is likely that LubX mimics the host E3 ubiquitin ligase activity in similar fashion during the infection within both evolutionarily distant hosts. There is no doubt that identifying the host targets of U-box and F-box effectors will provide an insight into how Legionella exploits evolutionarily conserved E3 ubiquitin ligases of phylogenetically diverse hosts.
AnkB promotes proteasome-mediated generation of free amino acids essential as energy and carbon sources
Recent studies have shown that the polyubiquitinated proteins assembled by AnkB on the LCV are preferentially enriched for Lys48-linked polyubiquitinated proteins, which is a hallmark for proteasomal degradation, that generate 2–24 amino acid peptides [53]. Substitution of Lys48 to Arg abolishes the decoration of the LCV with polyubiquitinated proteins and blocks intracellular proliferation [53]. This indicates that turnover of Lys48-linked polyubiquitinated proteins is needed for continuous and dynamic remodeling of the LCV or that proteasomal degradation is needed to generate a surplus of amino acids for bacterial proliferation of L. pneumophila. Inhibition of the proteasome, or the host amino- and oligo-peptidases that degrade the short peptides generated by proteasomal degradation [54,55], blocks intracellular proliferation. However, both inhibitions are bypassed by excess amino acid supplementation [53]. Interestingly, failure of wild type L. pneumophila to proliferate in human cells overexpressing ubiquitin variants with Lys48-Arg substitution is accompanied with a strong bacterial starvation response for amino acids, and this response is completely suppressed upon supplementation of the infected cells with excess amino acids, which also rescue intracellular bacterial proliferation [53]. Cells infected by the wild type strain have high levels of free amino acids compared to the ankB mutant-infected cells, which are similar to uninfected cells [53]. Importantly, supplementation of ankB mutant-infected amoeba or macrophages with excess amino acids rescue the mutant for intracellular proliferation and suppress its amino acids starvation response, similar to genetic complementation by the native ankB allele [53]. Screening the 20 amino acids individually for their ability to rescue the ankB mutant for intracellular proliferation in macrophages and amoeba has shown that cysteine or serine supplementation rescue the ankB mutant as efficiently as the mixture of amino acids [53].
Cysteine and serine are converted by Legionella into pyruvate that feeds the tricarboxylic acid (TCA) cycle, and L. pneumophila relies on amino acids as the major sources of carbon and energy production through the TCA cycle. Interestingly, supplementation of amoeba or macrophages with pyruvate or citrate rescues the ankB mutant for intracellular proliferation within amoeba and macrophages and for its starvation response, similar to the supplementation by the mixture of amino acids. Remarkably, injection of mice with the mixture of amino acids or with cysteine specifically rescues the ankB mutant for intrapulmonary proliferation, similar to genetic complementation [53]. These findings indicate that the ultimate goal of exploitation of multiple evolutionarily conserved eukaryotic processes by AnkB is to generate amino acids by promoting host proteasomal degradation of the K48-linked polyubiquitinated proteins. The generated amino acids are required as sources of carbon and energy for intracellular growth of Legionella (Figure 2). Because either cysteine or serine are sufficient to rescue the ankB mutant, it is most likely that L. pneumophila has sufficient levels of cellular amino acids with the exception of high levels of a major source of carbon and energy to feed the TCA cycle. Interestingly, cysteine and serine are non-essential amino acids for Acanthamoeba or human cells. In addition, Legionella is auxotrophic for Arg, Cys, Ile, Leu, Met, Val and Thr, which include all the amino acids that Acanthamoeba is auxotrophic for (Arg, Ile, Leu, Met and Val) [56]. With the exception of Cys, human cells are auxotrophic for all the amino acids that Legionella is auxotrophic for, but Cys is relatively scarce in human cells. This indicates a tremendous patho-adaptation of Legionella to the intracellular life within Acanthamoeba and human cells and amoeba. The mechanism of generation of a surplus of amino acids through promoting host proteasomal degradation by an F-box effector Legionella is the first example of a microbial strategy dedicated to generate sources of carbon and energy needed for microbial proliferation in vivo. Remarkably, the amino acids essential for Legionella, due to deficiency of de novo synthesis, are also essential for the two evolutionarily distant hosts, with the exception of Cys, which is scarce in human cells. Whether other organisms exploit similar, or idiosyncratic, strategies to access host sources of carbon and energy in vivo remains to be explored [57].
Evolution of Legionella translocated effectors
The high number of L. pneumophila eukaryotic-like proteins that exhibit structural and functional mimicry of eukaryotic proteins may reflect the diversity of eukaryotic pathways that are exploited by L. pneumophila during infection of phylogenetically diverse eukaryotic hosts [13,16,23,58]. Even though ~10% of the Legionella genome is dedicated to injecting effectors into host cells, most of these effectors do not play critical roles in intracellular proliferation in macrophages, suggesting potential host tropism for redundant effectors that are tailored to exploit a certain host in the environment. In silico analysis of the L. pneumophila genome identified several genomic islands that were indispensable for bacterial growth within protists but not in macrophages or nutrient-rich media [59]. Interestingly, the necessity of individual genomic islands for intracellular proliferation varies among the amoeba species. This indicates that Legionella is a generalist pathogen, which distinguishes this bacterial species from most other specialist pathogens [59].
It is likely that horizontal gene transfer (HGT) and convergent evolution are two key mechanisms for the evolution of eukaryotic-like effectors during long term co-evolution of L. pneumophila with diverse primitive eukaryotic hosts (Box 1). The HGT theory is supported by several lines of evidences. First, L. pneumophila is naturally competent for DNA uptake and the Dot/Icm translocation system can exchange DNA between bacteria through conjugation [60]. Second, the difference of G + C content of the eukaryotic-domain encoding genes compared to the rest of the genome suggests more recent acquisition of those genes [61]. Subsequent multiple convergent evolutions of the host gene acquired through HGT is essential for the encoded protein to become a translocated effector functional in the host (Box 1).
Box 1. Obstacles for a eukaryotic gene to encode a translocated functional bacterial effector.
The long close association between Legionella and protozoa in the environment provides the opportunity for HGT to occur. The acquired gene must undergo a number of changes, probably through convergent evolution, to become a translocated functional effector. This involves multiple processes such as removal of the introns, acquisition of a prokaryotic promoter along with regulatory elements and a ribosomal binding site. To become an effector recognized and translocated by the Icm/Dot secretion system, a translocation signal must be acquired along with domains that interact with chaperones needed for effector stability and recognition by the translocation machinery [71]. To become biologically functional, some effectors must also acquire eukaryotic post-translational modification motifs such as prenylation or theS-palmitoylation motif. Convergent evolution of the genes acquired through interkingdom HGT is probably the main mechanism of long term evolution to result in translocated effectors that exhibit biological function in the host cytosol.
It seems more likely that AnkB has been acquired by L. pneumophila through interkingdom HGT as a whole eukaryotic gene or even as fragments during the long intimate association with primitive unicellular eukaryotes such as amoeba or even indirectly from another intracellular bacterium or endosymbiont of unicellular eukaryotes [47,72]. The domain architecture of AnkB of L. pneumophila more closely resembles the F-box proteins of unicellular eukaryotes, where the protein–protein interaction domains of F-box proteins are ankyrin domains (ANK) [47]. By contrast, ANK domains are absent from F-box proteins of metazoans such as mammals. Instead, mammalian F-box proteins have WD repeats or leucine-rich repeats (LRR) as protein–protein interaction domains [73]. Interestingly, all eukaryotic F-box proteins are cytosolic, whereas AnkB is a membrane-anchored protein. The CaaX motif of AnkB could have gradually evolved through convergent evolution to be anchored to the LCV membrane. Nevertheless, acquisition of the CaaX motif through horizontal gene transfer cannot be excluded [13,14]. There is a growing list of L. pneumophila effectors that have probably been acquired by HGT from protozoan host, such as the LegK1 and LegK3, which exhibit similarity to amoeba protein kinases [72]. Moreover, the L. pneumophila LegS2 effector is highly homologous to the protozoan sphingosine-1-phosphate lyase (SPL) [71]. Evidence shows that many genes of Legionella have been acquired from other organisms that reside within amoeba such as Lpg2416, which is an ankyrin-containing protein that has homologs only in Acanthamoeba polyphaga mimivirus [74].
Patho-adaptation of L. pneumophila to metazoan hosts
The accumulation of acquired genetic materials through HGT helps L. pneumophila to increase its fitness and adaption to diverse hosts, which provides selective pressure for maintaining the newly acquired genes and expanding the host range [59]. With the presence of high numbers of translocated bacterial effectors, functional redundancy is observed, where mutation in a gene does not have any phenotype in either macrophages or protozoa. It is likely that the eukaryotic-like proteins that exploit conserved eukaryotic processes initially evolved to manipulate diverse protozoan hosts. However, the presence of conserved targets in mammalian cells enables L. pneumophila to exploit conserved pathways in mammalian cells via the same effector such as AnkB [13,14,16,48]. However, Legionella modulates mammalian-specific processes that are absent from unicellular eukaryotes [62]. One example is modulation of programmed cell death by L. pneumophila, where it is thought that L. pneumophila generates a delicate balance between triggering both pro-apoptotic and anti-apoptotic pathways to render mammalian cells permissive for intracellular proliferation [12,62]. Even though caspase-3 is activated in L. pneumophila-infected human macrophages, L. pneumophila inhibits apoptosis by triggering anti-apoptotic pathways via SdhA and SidF effectors [12] until later stages of infection, giving enough time for L. pneumophila to replicate [4,63–65]. However, L. pneumophila is incapable of inducing apoptosis in amoebae [66] because amoeba has a primitive program of cell death that lacks apoptosis regulatory proteins such as the Bcl2 and caspase families of proteins [67]. In addition, modulation of activation of the inflammasomes and nuclear factor-kappaB (NF-κB) by Legionella is also specific to mammals and is absent from unicellular eukaryotes [62,68]. The Dot/Icm-mediated translocation of at least four kinases (LegK1–LegK4) to modulate host phosphorylation activities by L. pneumophila is a clear indication of interference with host signaling by L. pneumophila. Some of this interference with host signaling may be limited to the mammalian host, such as phosphorylation and activation of NF-κB by LegK1 [10,68–70]. It may also be limited to the protozoan host, such as the kinase activity of LegK2 is essential for remodeling the LCV by the ER, and for intracellular proliferation in Acanthamoeba [68]. Whether LegK2 is functionally active and is required for intracellular proliferation in mammalian cells is still to be determined. Therefore, it is most likely that Legionella required a lengthy period of interaction with metazoan hosts, such as mammals, before pathogenesis of disease manifestation was exhibited.
Concluding remarks
Approximately 10% of the coding capacity of the L. pneumophila genome is dedicated to translocation of ~300 effectors, which is the largest cadre of effectors relative to any other known pathogen. This exceptional number of effectors is likely to have equipped L. pneumophila with the capacity to exploit multiple protist hosts with effectors tailored to certain hosts. Many host pathways are exploited, and the few discovered so far are undoubtedly the tip of the iceberg of this bacteria–host interaction. Much of the host exploitation is achieved by eukaryotic-like effectors that mimic functions of eukaryotic proteins. Genes encoding effectors have probably been acquired by HGT through long adaptation and co-evolution with unicellular eukaryotes in the aquatic environment and by subsequent convergent evolution such as the AnkB, LegK3 and LegS2 effectors that exhibit high similarity to amoeba proteins. There is no doubt that protozoa have been a productive ‘training’ ground for intracellular proliferation of L. pneumophila, which has a remarkable capacity to exploit the structural and molecular conservation of cellular processes in unicellular eukaryotes to emerge and infect more evolved mammalian cells. The AnkB effector is a fascinating example of exploitation of multiple evolutionarily conserved processes, and its biological function is ultimately dedicated to generate a surplus of host amino acids as sources of carbon and energy needed for intracellular proliferation within protists and metazoan hosts. Many new exciting studies on the role of SidM/DrrA in modulating Rab1 activity in vesicle traffic in metazoan cells have not examined the role in protists [5,7,18,34–39]. However, considering the high conservation of vesicle traffic through evolution, we speculate that the role of SidM/DrrA in modulating Rab1 activity to intercept vesicle traffic from the ER [5,7] is also exhibited in amoeba [18,34–39].
However, the ability of Legionella to exploit many highly evolved eukaryotic processes, such as the inflammasomes, pro- and anti-apoptotic pathways that are absent in protozoa suggests long term co-evolution of Legionella with more evolved eukaryotic hosts. Regardless of the origin of the acquired gene by HGT, the gene must undergo long term evolution before the encoded proteins becomes an injected effector that modulates cellular processes. It is possible that many Legionella eukaryotic-like proteins that are not translocated are still undergoing evolution of acquisition of various motifs needed for translocation. Further understanding of the molecular aspects of Legionella–protist interaction will continue to fascinate us and educate us about a remarkable evolution of a bacterial pathogen from interacting with protists to its patho-adaptation to metazoans.
Acknowledgments
Y.A.K. is supported by Public Health Service Awards R01AI43965 and R01AI069321 from NIAID and by the commonwealth of Kentucky Research Challenge Trust Fund.
Footnotes
The authors declare no competing financial interests.
References
- 1.Molmeret M, et al. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harb OS, et al. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ Microbiol. 2000;2:251–265. doi: 10.1046/j.1462-2920.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- 3.Gao LY, et al. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, et al. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo ZQ. Legionella secreted effectors and innate immune responses. Cell Microbiol. 2012;14:19–27. doi: 10.1111/j.1462-5822.2011.01713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isberg RR, et al. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge J, Shao F. Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cell Microbiol. 2011;13:1870–1880. doi: 10.1111/j.1462-5822.2011.01710.x. [DOI] [PubMed] [Google Scholar]
- 8.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 9.Molmeret M, et al. Disruption of the phagosomal membrane and egress of Legionella pneumophila into the cytoplasm during the last stages of intracellular infection of macrophages and Acanthamoeba polyphaga. Infect Immun. 2004;72:4040–4051. doi: 10.1128/IAI.72.7.4040-4051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molmeret M, et al. Temporal and spatial trigger of post-exponential virulence-associated regulatory cascades by Legionella pneumophila after bacterial escape into the host cell cytosol. Environ Microbiol. 2010;12:704–715. doi: 10.1111/j.1462-2920.2009.02114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Khodor S, et al. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 2010;18:132–139. doi: 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Z-Q. Striking a balance: modulation of host cell death pathways by Legionella pneumophila. Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Quadan T, Abu Kwaik Y. Molecular characterization of exploitation of the polyubiquitination and farnesylation machineries of Dictyostelium discoideum by the AnkB F-box effector of Legionella pnemophila. Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price CT, et al. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med. 2010;207:1713–1726. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolery AR, et al. AMPylation: Something old is new again. Front Microbiol. 2010;1 doi: 10.3389/fmicb.2010.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price CT, et al. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conover GM, et al. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 18.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov SS, et al. Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J Biol Chem. 2010;285:34686–34698. doi: 10.1074/jbc.M110.170746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price C, et al. Host-mediated post-translational prenylation of novel Dot/Icm-translocated effectors of Legionella pneumophila. Front Microbiol. 2010;1 doi: 10.3389/fmicb.2010.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Quadan T, et al. Anchoring bacterial effectors to host membranes through host-mediated lipidation by prenylation: A common paradigm. Trends Microbiol. 2011;19:573–579. doi: 10.1016/j.tim.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Hilbi H, et al. Anchors for effectors: subversion of phosphoinositide lipids by Legionella. Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Khodor S, et al. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol Microbiol. 2008;70:908–923. doi: 10.1111/j.1365-2958.2008.06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habyarimana F, et al. Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol. 2008;10:1460–1474. doi: 10.1111/j.1462-2920.2007.01560.x. [DOI] [PubMed] [Google Scholar]
- 25.Cazalet C, et al. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res. 2008;18:431–441. doi: 10.1101/gr.7229808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizarro-Cerda J, Cossart P. Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat Cell Biol. 2004;6:1026–1033. doi: 10.1038/ncb1104-1026. [DOI] [PubMed] [Google Scholar]
- 27.Hilbi H. Modulation of phosphoinositide metabolism by pathogenic bacteria. Cell Microbiol. 2006;8:1697–1706. doi: 10.1111/j.1462-5822.2006.00793.x. [DOI] [PubMed] [Google Scholar]
- 28.Weber SS, et al. Pathogen trafficking pathways and host phosphoinositide metabolism. Mol Microbiol. 2009;71:1341–1352. doi: 10.1111/j.1365-2958.2009.06608.x. [DOI] [PubMed] [Google Scholar]
- 29.Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 30.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arasaki K, et al. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe. 2012;11:46–57. doi: 10.1016/j.chom.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ragaz C, et al. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 2008;10:2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 33.Weber SS, et al. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2006;2:e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh HY, et al. Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. EMBO J. 2010;29:496–504. doi: 10.1038/emboj.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan Y, et al. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci USA. 2011;108:21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan Y, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 38.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Brombacher E, et al. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton HJ, et al. Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect Immun. 2007;75:5575–5585. doi: 10.1128/IAI.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grillari J, et al. Post-translational modification of cellular proteins by ubiquitin and ubiquitin-like molecules: role in cellular senescence and aging. Adv Exp Med Biol. 2010;694:172–196. doi: 10.1007/978-1-4419-7002-2_13. [DOI] [PubMed] [Google Scholar]
- 42.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 43.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 45.Perrett CA, et al. Interactions of bacterial proteins with host eukaryotic ubiquitin pathways. Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Angot A, et al. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price C, Abu Kwaik Y. Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front Microbiol. 2010;1 doi: 10.3389/fmicb.2010.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lomma M, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12:1271–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 49.Cazalet C, et al. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 2010;6:e1000851. doi: 10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ensminger AW, Isberg RR. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun. 2010;78:3905–3919. doi: 10.1128/IAI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubori T, et al. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 52.Hatakeyama S, Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302:635–645. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 53.Price C, et al. Bacterial F-box effector exploits eukaryotic K48-linked polyubiquitination and proteasome machineries to generate amino acids essential for intracellular growth in diverse hosts. Science. 2011;334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 54.Saric T, et al. Pathway for degradation of peptides generated by proteasomes: a key role for thimet oligopeptidase and other metallopeptidases. J Biol Chem. 2004;279:46723–46732. doi: 10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- 55.Reits E, et al. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 56.Ingalls CS, Brent MM. Defined minimal growth medium for Acanthamoeba polyphaga. J Protozool. 1983;30:606–608. doi: 10.1111/j.1550-7408.1983.tb01430.x. [DOI] [PubMed] [Google Scholar]
- 57.Rohmer L, et al. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011;19:341–348. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price CT, et al. Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect Immun. 2010;78:2079–2088. doi: 10.1128/IAI.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Connor TJ, et al. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci USA. 2011;108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stone BJ, Kwaik YA. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franco IS, et al. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11:1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 62.Amer AO. Modulation of caspases and their non-apoptotic functions by Legionella pneumophila. Cell Microbiol. 2010;12:140–147. doi: 10.1111/j.1462-5822.2009.01401.x. [DOI] [PubMed] [Google Scholar]
- 63.Molmeret M, et al. Activation of caspase-3 by the Dot/Icm virulence system is essential for arrested biogenesis of the Legionella-containing phagosome. Cell Microbiol. 2004;6:33–48. doi: 10.1046/j.1462-5822.2003.00335.x. [DOI] [PubMed] [Google Scholar]
- 64.Abu-Zant A, et al. Incomplete activation of macrophage apoptosis during intracellular replication of Legionella pneumophila. Infect Immun. 2005;73:5339–5349. doi: 10.1128/IAI.73.9.5339-5349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Losick VP, Isberg RR. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med. 2006;203:2177–2189. doi: 10.1084/jem.20060766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao LY, Abu Kwaik Y. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect Immun. 1999;67:862–870. doi: 10.1128/iai.67.2.862-870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haenssler E, Isberg RR. Control of host cell phosphorylation by Legionella pneumophila. Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ge J, et al. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci USA. 2009;106:13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Losick VP, et al. LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol. 2010;12:1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Degtyar E, et al. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol. 2009;11:1219–1235. doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 72.Hervet E, et al. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun. 2011;79:1936–1950. doi: 10.1128/IAI.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith TF, et al. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 74.Lurie-Weinberger MN, et al. The origins of eukaryotic-like proteins in Legionella pneumophila. Int J Med Microbiol. 2010;300:470–481. doi: 10.1016/j.ijmm.2010.04.016. [DOI] [PubMed] [Google Scholar]