Abstract
Therapeutic targeting of histone/protein deacetylase 6 (HDAC6), HDAC9, or the sirtuin-1 (Sirt1) augments the suppressive functions of regulatory T cells (Tregs) that contain the transcription factor Foxp3. However, it is unclear whether distinct mechanisms are involved or whether combined inhibition of these targets would be more beneficial. We compared the suppressive functions of Tregs from wild-type C57BL/6 mice with those from mice with either global (HDAC6−/−, HDAC9−/−, and HDAC6−/−HDAC9−/−), or conditional (fl-Sirt1/CD4-Cre or fl-Sirt1/Foxp3-Cre) HDAC deletion, as well as treatment with isoform-selective HDAC inhibitors. We found that the heat shock response was important for the improvement of Treg suppressive function mediated by HDAC6 inhibition, but not Sirt1 inhibition. Furthermore, although HDAC6, HDAC9, and Sirt1 all deacetylated Foxp3, each protein had diverse effects on transcription factors controlling Foxp3 gene expression. For example, loss of HDAC9 was associated with stabilization of the acetylation of signal transducer and activator of transcription 5 (STAT5) and of its transcriptional activity. Hence, targeting different HDACs increased Treg function by multiple and additive mechanisms, which indicates the therapeutic potential for combinations of HDAC inhibitors in the management of autoimmunity and organ transplantation.
INTRODUCTION
Autoimmune diseases and transplantation require therapeutic suppression of undesired immune responses. Ideally, suppression should be as limited and specific as possible, preserving the host’s ability to fight infections and cancer. Unfortunately, however, most current therapeutic regimens are unable to achieve this, causing an enormous burden of toxicity (1) and impairment of vital host immunity (2). Regulatory T cells (Tregs) constitute a T-cell subset that is important for the attenuation of antigen-specific immune responses (3). Tregs are currently being evaluated for therapeutic applications, either through the direct administration of Tregs that were expanded in numbers ex vivo or through pharmacological measures aimed at selectively strengthening Treg function (4). Tregs are characterized by their expression of Forkhead box P3 (Foxp3), a transcription factor that plays a key role in their development and functions (5, 6). We showed that the suppressive capacity of murine and human Foxp3+ Tregs can be increased by exposure to inhibitors of histone/protein deacetylases (HDACs) (7, 8), as well as by deletion of HDAC9 (8), HDAC6 (9), or the class III HDAC Sirtuin-1 (Sirt1) (10).
Such data suggest the potential for therapeutic targeting of combinations of HDAC enzymes; however, each HDAC deacetylates discrete sets of target proteins, and the mechanisms by which each enzyme regulates Treg function may be quite different (11). For example, a deficiency in HDAC6 markedly augments the heat shock response in Tregs (9), whereas deletion of Sirt1 may dampen the heat shock response in these cells, because Sirt1 is required to stabilize the heat shock factor (HSF)-1 trimer (12). As a result, combined inhibition of Sirt1 and HDAC6 might be counter-productive in efforts to promote the suppressive functions of Tregs. It is also unclear how important the heat shock response is to the increased Treg function observed when HDAC6 (or Sirt1) is targeted, and whether there are heat shock response-independent mechanisms for the enhancement of Treg function by inhibiting HDAC6, such as control of Foxp3 acetylation, as was reported for HDAC9 (8) and Sirt1 (13, 14). Furthermore, deletion of HDAC6, HDAC9, or Sirt1 increases expression of the gene encoding Foxp3 (8–10); however, apart from data showing a role for nuclear factor κB (NF-κB) in the case of Sirt1 deletion in Tregs (10), the transcription factors involved in the enhanced suppressive functions of HDAC6−/− and HDAC9−/− Tregs are unknown. Therefore, we sought to investigate the mechanisms by which isotype-specific inhibition or deletion of HDACs affected Treg function with regards to the heat shock response, Foxp3 acetylation, and transcription factors relevant for Foxp3+ Tregs, as well as to assess whether the combined effects of targeting HDAC6, HDAC9, and Sirt1 on Treg function were additive, in vitro and in vivo. Our findings have implications for the future development of Treg therapies based on HDAC inhibitors for anti-inflammatory and immune-mediated responses.
RESULTS
Activation of the heat shock response is required for efficacy of HDAC6 targeting in Treg cells
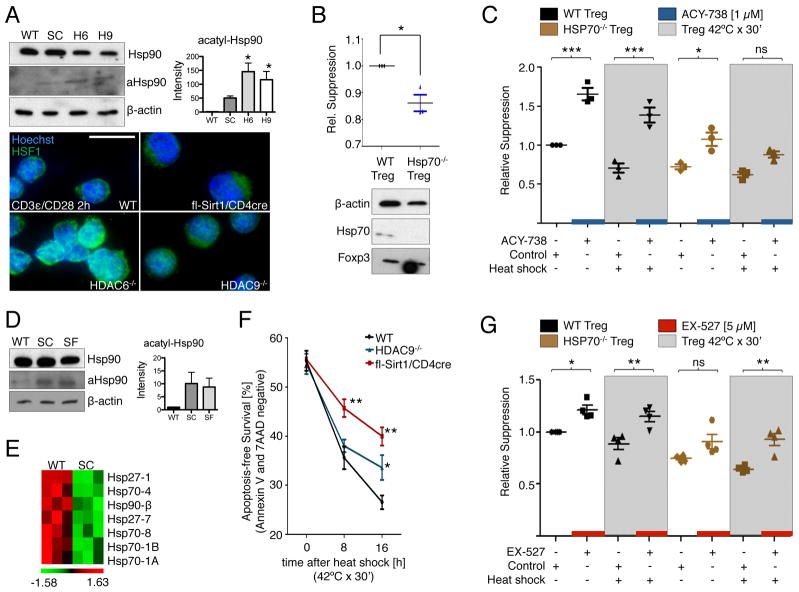
We started our investigation by comparing the involvement and relative importance of the heat shock response in mouse models testing HDAC6 and Sirt1 inhibition to improve Treg function. Gene deletion of HDAC6 or HDAC9 led to hyperacetylation of Hsp90 at Lys294 (K294) in Tregs (Fig. 1A), in association with the nuclear translocation of heat shock factor 1 (HSF-1) (Fig. 1A, lower panel), and the induction of key heat shock response genes, such as those that encode Hsp27 and Hsp70 (9, 15). Although the exact mechanism remains unknown, a functioning heat shock response is considered important for the physiologic suppressive capabilities of Tregs (11). Consistent with this concept, we found that knockout of the gene encoding Hsp70 diminished the ability of wild-type Tregs to constrain T cell proliferation (Fig. 1B).
Fig. 1.
The heat shock response is required to improve Treg function in response to inhibition of HDAC6, but not Sirt1. (A) Increased Hsp90 K294 acetylation (aHsp90) in HDAC6−/− Tregs (H6) and HDAC9−/− Tregs (H9), and to a lesser degree in fl-Sirt1/CD4cre Tregs (SC). Densitometry data pooled from two experiments. Immunofluorescence of stimulated Tregs showed nuclear translocation of HSF1, mostly in HDAC6−/− cells, whereas fl-Sirt1/CD4cre Tregs exhibited the least amount of intranuclear HSF1. (B) Hsp70−/− Tregs had diminished suppressive function. Data are from three experiments. (C) Hsp70−/− Tregs were less responsive to ACY-738 (HDAC6i) than WT Tregs, especially under conditions of heat shock. Data are from three experiments. (D) Sirt1-deficient Tregs exhibited a trend to increased acetylation of Hsp90 compared to wild-type (WT) Tregs. Densitometry data pooled from two experiments. SF, fl-Sirt1/Foxp3cre. (E) Transcriptional repression of genes involved in the heat shock response in fl-Sirt1/CD4cre Tregs as analyzed by microarrays, with data shown after z-score transformation. (F) Apoptosis-free survival of Tregs after heat shock. (G) The suppressive function of Tregs was improved by inhibition of Sirt1 (with EX-527) in the absence of Hsp70 and under conditions of heat shock. Data are from four experiments. Effector T cells (Teff) and APCs used in the suppression assays were from (B) WT or (C and G) Hsp70−/− mice. WT refers to the appropriate control mouse strain, that is C57BL6 mice for (A), (D), (E), and (F) or B6/129 mice for (B), (C), and (G). *p<0.05, **p<0.01, and ***p<0.001. Scale bar = 10 μm. Additional statistics for (C) and (G) are available in tables S1 and S2.
We next analyzed whether the effects of HDAC6 inhibition on Treg function were also dependent upon the presence of Hsp70 in Tregs, using standard or heat shock conditions. Wild-type and Hsp70−/− Tregs were subjected to a 30-min heat shock (at 42°C), exposed to a previously uncharacterized HDAC6 selective inhibitor (HDAC6i), ACY-738 (fig. S1), and tested for their ability to suppress the proliferation of Hsp70−/− effector T cells that were costimulated with Hsp70−/− antigen-presenting cells (APCs) and monoclonal antibody (mAb) against CD3ε. We found that the HDAC6i enhanced the suppressive function of Tregs under standard conditions and after exposure to heat shock conditions (Fig. 1C). However, loss of Hsp70 substantially decreased the effects of HDAC6 inhibition on Tregs under standard conditions, and Hsp70−/− Tregs subjected to heat shock lost most of their responsiveness to HDAC6 inhibition (Fig. 1C and table S1). We conclude that activation of the heat shock response is required for the effect of HDAC6 inhibition in increasing the suppressive function of Tregs, but it is not the sole mechanism responsible.
Improving Treg function through Sirt1 inhibition does not require activation of the heat shock response
Conditional CD4cre and Foxp3cre mediated deletion of Sirt1 led to a modest increase in K294 acetylation in Hsp90 in Tregs (Fig. 1, A and D). However, in contrast to HDAC6−/− Treg, Sirt1 deletion did not result in nuclear translocation of HSF-1 (Fig. 1A). Consistent with these data, we found the reduced expression of numerous heat shock response genes in Sirt1−/− Tregs compared to those in wild-type Tregs (Fig. 1E). These observations led us to consider whether diminished transcription of genes involved in the heat shock response might decrease the resistance of Sirt1−/− Tregs to stress or affect their suppressive functions. We therefore exposed freshly isolated CD4+CD25+ Tregs from fl-Sirt1/CD4cre mice and wild-type mice to a heat shock similar to the conditions described earlier (Fig. 1C), and tested their ability to survive without initiating apoptosis or cell death, as determined by flow cytometric analysis of the amount of signal from annexin V and 7AAD, respectively. Compared to Tregs from wild-type mice, Tregs from fl-Sirt1/CD4cre mice showed improved survival, despite their lack of activation of genes involved in the heat shock response (Fig. 1F). Given that deletion of Sirt1 seemingly preserved the ability of Tregs to withstand cell stress despite the lack of expression of their heat shock response genes, we hypothesized that improved Treg function as a result of Sirt1 inhibition might be independent of the expression of genes involved in the heat shock response. Indeed, Hsp70−/− Tregs challenged by heat shock still exhibited a marked improvement in their suppressive capability in response to the Sirt1-specific inhibitor, EX-527 (Fig. 1G and table S2). We conclude that activation of the heat shock response is not required to mediate the effect of Sirt1 inhibition on the suppressive functions of Tregs.
HDAC6 has intranuclear functions and deacetylates Foxp3
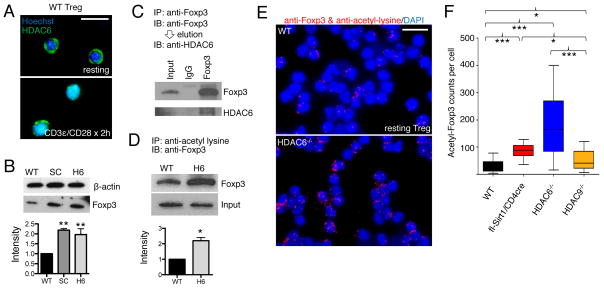
Given our finding that pharmacologic targeting of Sirt1 improved Treg function independently of the heat shock response, we hypothesized that HDAC6 might also have roles in Treg biology that were independent of the heat shock response. Whereas HDAC6 is regarded primarily as a cytosolic protein that regulates the acetylation of cytoskeletal proteins such as α-tubulin, we found that HDAC6 translocated to the nucleus of Tregs activated through CD3ε and the coreceptor CD28 (Fig. 2A). This observation suggested that HDAC6 might have nuclear functions, such that targeting HDAC6 might affect the extent of acetylation of nuclear proteins relevant to Treg function. Indeed, we found that the Foxp3 protein was more abundant in HDAC6−/− Tregs than in wild-type Tregs (Fig. 2B), that it coimmunoprecipitated with HDAC6 (Fig. 2C), and that there was more acetylated Foxp3 in the absence of HDAC6 than in its presence (Fig. 2, D to F). These data suggest that HDAC6 deacetylates Foxp3, and that loss of HDAC6 promotes Foxp3 acetylation, with a consequent increase in the resistance of Foxp3 to proteasomal degradation (14). HDAC6 may also contribute to the regulation of additional transcription factors required for Tregs (fig. S2, and fig. S3), including promoting the nuclear translocation of cAMP response element–binding protein (CREB) phosphorylated at Ser133. We conclude that HDAC6 has important heat shock response-independent functions in Tregs, including regulating the acetylation of Foxp3.
Fig. 2.
Functions of HDAC6 in Foxp3+ Tregs that are independent of the heat shock response. (A) Immunofluorescence showing the translocation of HDAC6 into the nuclei of WT Tregs after stimulation with antibodies against CD3ε and CD28. (B) Western blotting analysis of Treg lysates indicated that Foxp3 is more abundant in HDAC6−/− cells than in WT cells. Densitometry data pooled from of three experiments. (C) Co-immunoprecipitation of HDAC6 and Foxp3 in WT Treg lysates. (D) Immunoprecipitation studies of Treg lysates indicated that Foxp3 is more acetylated in HDAC6−/− Tregs than in WT Tregs. Densitometry data pooled from two experiments. (E) Proximity ligation assays showed more acetylated Foxp3 in HDAC6−/− Tregs than in WT Tregs. (F) Quantification of proximity ligation assays with BlobFinder and statistical analysis with the Kruskal-Wallis test. *P<0.05, ***P<0.001. Data shown as boxplot with the median, 25th, and 75th quartiles; whiskers indicate the 10th and 90th percentiles. Scalebar: 10 μm.
Loss of HDAC9 promotes STAT5 acetylation and Treg phenotype
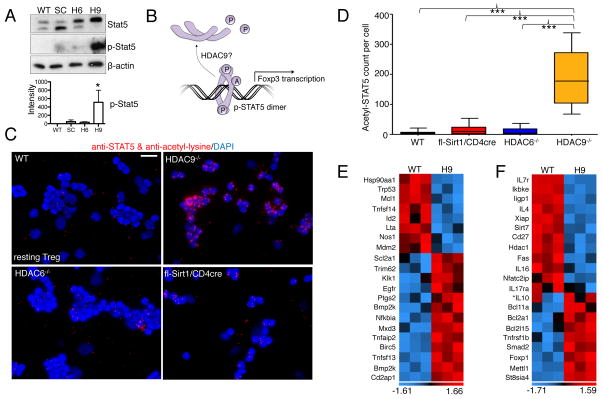
The transcription factor signal transducer and activator of transcription 5 (STAT5), which transduces signals resulting from the binding of interleukin-2 (IL-2) to the IL-2 receptor (IL-2R), is important for the functions of Foxp3+ Tregs (16). We showed that HDAC9−/− CD4+CD25− effector T cells more readily convert into induced Tregs (iTregs) than do the corresponding wild-type effector T cells when stimulated with transforming growth factor–β (TGF-β) (15). Such a predisposition to iTreg conversion does not occur for effector T cells from fl-Sirt1/CD4cre mice (fig. S4), supporting the hypothesis that each HDAC might affect different aspects of Treg biology. Because IL-2 signaling is important in stabilizing iTregs (17), we hypothesized that acetylation of the downstream IL-2 target, STAT5, might determine the differential abilities of HDAC9−/− and Sirt1−/− T-cells to undergo conversion to iTregs (Fig 3B). Indeed, we found increased phosphorylation of STAT5 (Y694) (Fig. 3A) and increased acetylation of STAT5 (Fig. 3C, D) in HDAC9−/− Tregs compared to wild-type Tregs, as was the case to a lesser degree in a comparison of fl-Sirt1/CD4cre Tregs and wild-type Tregs.
Fig. 3.
Deletion of HDAC9 stabilizes the acetylation, phosphorylation, and transcriptional activity of STAT5. (A) Analysis of Treg lysates indicated that STAT5 was relatively more phosphorylated in HDAC9−/− cells than in WT cells. Densitometry data pooled from three experiments. (B) Conceptual model of the acetylation of STAT5, which protects it from losing its transcriptionally active, phosphorylated dimeric form. (C) Proximity ligation assay showed prominent acetylation of STAT5 in HDAC9−/− Tregs compared to that in WT Tregs. (D) Quantification of the data in (C) was performed as described for Fig. 2F. (E) Microarray analysis showed STAT5-dependent signaling in HDAC9−/− Tregs compared to WT Treg (n=3/group) based on previously reported STAT5 targets (19). (F) Further STAT5 signaling as well as other transcriptional alterations promoted a suppressive phenotype in HDAC9−/− Tregs. Data are shown after z-score transformation. *IL-10 not significant. Scale bar: 10 μm. Trp, transformation-related protein; Mcl, myeloid leukemia cell differentiating protein; Tnfsf, tumor necrosis factor superfamily; Id, inhibitor of DNA; Lta, lymphotoxin-α; Nos, NO synthase; Mdm2, an E3 ubiquitin ligase; Scl2a1, GLUT1; Trim, tripartite motif; Klk, Kallikrein; ptgs, post-transcriptional gene silencing; bmp2k, bone morphogenetic protein inducible kinase-2; Cd2ap, CD2-associated protein; Ikbke, Inhibitor of nuclear factor κB kinase ε; Iigp, IFN-inducible GTPase; Xiap, X-linked inhibitor of apoptosis; Fas, CD95; Bcl, B-cell lymphoma; mettl, methyltransferase-like; St8sia, ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase-4.
We then assessed whether increases in STAT5 phosphorylation and acetylation were associated with decreased methylation of the Treg-specific demethylated region (TSDR) in effector T cells, which has been proposed as a methylation sensitive region controlling Foxp3 expression (18). However, pyrosequencing of six key CpG islands within the TSDR did not show any difference in methylation between wild-type Tregs and Tregs deficient in HDAC6, HDAC9, or Sirt1 (fig. S5 and fig. S6). Upon checking the expression of known STAT5-dependent transcripts by microarray analysis, we noted that HDAC9−/− Tregs had a pattern of differential gene expression that was opposite to that of a STAT5 knockdown model reported by Liao et al. (19), supporting our concept of the increased transcriptional activity of STAT5 in HDAC9−/− Treg cells (Fig. 3E). Additional transcripts were altered in HDAC9−/− Tregs showing increased expression of several transcripts associated with the Treg phenotype (Fig. 3F). We conclude that loss of HDAC9 promotes STAT5 signaling in Tregs. Together, our data suggest that HDAC6, HDAC9, and Sirt1 have shared and individual mechanisms of action in increasing the suppressive activities of Tregs. We continued our investigation by combining the targeting of these HDACs with the goal to optimize the suppressive capacity of Tregs.
Combined loss of HDAC6, HDAC9, and Sirt1 augments Treg function
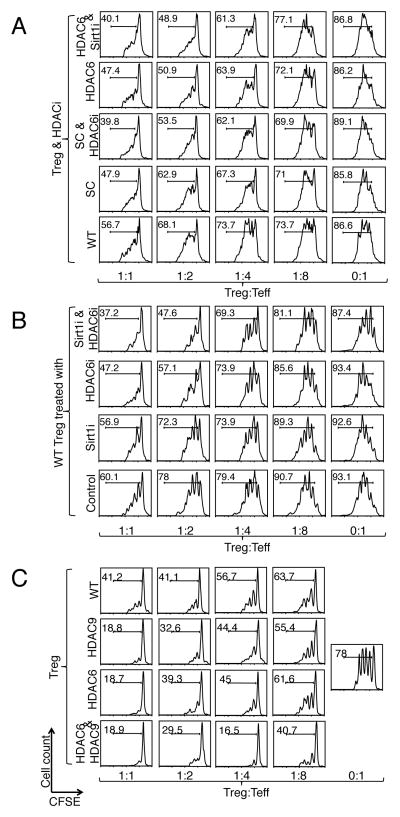
The clinical use of pharmacologic inhibitors of HDACs has primarily been limited to oncologic applications because of the toxicities and side-effects of existing pan-HDAC inhibitors (20). Since we wished to investigate the usefulness of simultaneously targeting specific HDAC proteins in Tregs, and there are, as yet, no specific HDAC9 inhibitors, we combined genetic and pharmacologic approaches with HDAC6−/− and Sirt1−/− mice and the isotype-specific HDAC inhibitors ACY-738 (HDAC6) and EX-527 (Sirt1). We found that the suppressive functions of Tregs from HDAC6−/− or fl-Sirt1/CD4cre mice were augmented by the addition of the complementary HDAC inhibitor (Fig. 4A). Furthermore, this additive benefit was also observed in wild-type Tregs treated with combined HDAC6 and Sirt1 inhibition simultaneously, which achieved better suppression than did treatment with either HDAC inhibitor alone (Fig. 4B). Similarly, we developed HDAC6 and HDAC9 double-knockout mice, and found that Treg function was further strengthened compared to those of Tregs from either single knockout or wild-type mice (Fig. 4C). Combining the targeting of HDAC9 and Sirt1 led to only a marginal improvement in the suppressive function of Tregs (fig. S7). Furthermore, loss of three HDACs (which was achieved by treating HDAC9−/− mice with both HDAC6 and Sirt1 inhibition) did not further augment Treg function compared to dual HDAC6 and Sirt1 inhibition alone (fig. S8). Therefore, we focused our present investigation on the effects of targeting both HDAC6 and HDAC9 or both Sirt1 and HDAC6.
Fig. 4.
Combined targeting of HDACs can further augment the suppressive function of Tregs. (A) Treg suppression assays with WT effector T cells and APCs, as well as with WT or HDAC-deficient Tregs with or without one or two specific HDAC inhibitors. We used EX-527 (5 μM) and ACY-738 (1 μM) to inhibit Sirt1 and HDAC6, respectively. (B) WT Tregs treated with Sirt1i, HDCA6i, or both showed increased suppressive function compared to Tregs treated with single HDAC6i or Sirt1i, or vehicle control treatment. (C) HDAC6−/− HDAC9−/− double knockout Tregs showed stronger suppressive function compared with WT and single knockout Tregs. Data are representative of three independent experiments.
Combined inhibition of Sirt1 and HDAC6 has an additive effect in improving Treg function in vivo
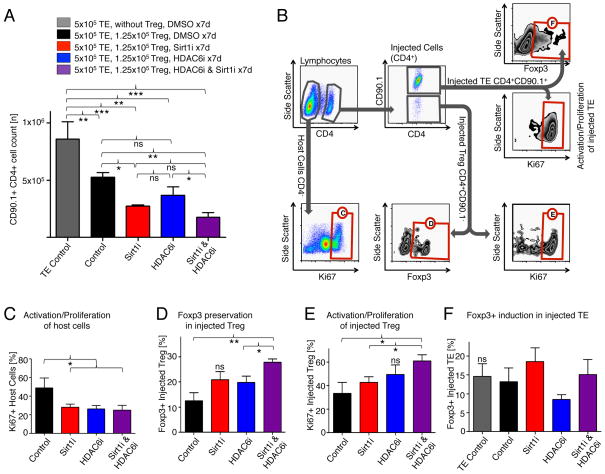
To extend our findings of combined pharmacologic inhibition of HDAC6 and Sirt1 inhibition in vivo, we adoptively transferred Thy1.1 (CD90.1+) CD4+CD25− effector T cells into B6/Rag1−/− mice and administered wild-type CD4+CD25+ Tregs at a 1:4 ratio. In this model, Thy1.1 effector T cells are injected to immunocompromised mice lacking host T and B cells. The injected effector T cells proliferate rapidly, but are limited in their expansion by co-administered Tregs. The mice were then treated with DMSO as a vehicle control or with Sirt1 and HDAC6, inhibitors, both alone or in combination. Co-treatment with Sirt1 and HDAC6 inhibitors was most effective at suppressing the proliferation of the adoptively transferred effector T cells, and was stronger than was the inhibition of either Sirt1 or HDAC6 alone (Fig. 5A). This was associated with less proliferation of host non-CD4 cells (Fig. 5B, C), improved preservation of Foxp3+ Treg, and the increased proliferation of the injected Treg population (Fig. 5D, E). Treatment with Sirt1i, HDAC6i, or both did not affect the induction of Foxp3+ Tregs from the injected Thy1.1 effector T cells (Fig. 5F), and did not the proliferation of effector T cells (fig. S9). We conclude that combining the targeting of Sirt1 and HDAC6 produces an additive increase in Treg suppressive function in vivo.
Fig. 5.
Combined pharmacologic targeting of HDAC6 and Sirt1 is additive in improving the suppressive function of Tregs and preserving Foxp3 in vivo. (A) Homeostatic proliferation assays showed increased suppressive function of Tregs in mice after individual treatment with either Sirt1i (EX-527, 1 mg/kg/d i.p.) or HDAC6i (Tubastatin, 40 mg/kg/d i.p.) as well as further improvement with combined treatment. (B) Gating strategy to identify cell populations of interest in spleens harvested from B6/Rag1−/− mice; combined treatment sample shown. Red labeled boxes identify populations of cells subsequently analyzed in the indicated panels. (C) Ki67 is reduced in host non-CD4+ cells treated with Sirt1i or HDAC6i. (D) Foxp3+ was better preserved in Tregs from mice treated with Sirt1i and HDAC6i, especially in combination, than in untreated mice. (E) Tregs from mice treated with Sirt1i, HDAC6i, and combined treatment groups had a more activated phenotype (Ki67) than did untreated cells. (F) Neither Sirt1i nor HDAC6i improves the induction of Tregs from effector T cells. Data are from three mice per group (N=15). *P < 0.05, **P < 0.01, and ***P < 0.001.
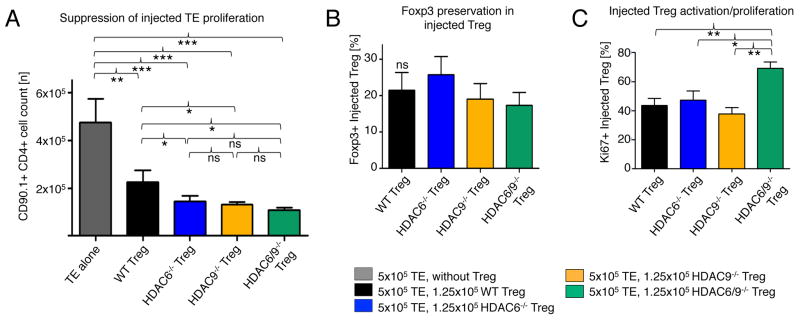
Next, we conducted a similar homeostatic proliferation experiment to compare HDAC6−/−, HDAC9−/−, HDAC6−/− HDAC9−/− double-knockout Tregs and wild-type controls. Unlike the case with pharmacologic targeting of Sirt1 and HDAC6, the double-knockout HDAC6−/− HDAC9−/− Tregs did not further suppress effector cell proliferation anymore than did either single knockout in vivo (Fig. 6A). In contrast to the combined inhibition of Sirt1 and HDAC6, Foxp3 preservation within the injected Tregs was not altered (Fig. 6B), although we observed a trend with HDAC6−/− cells that was consistent with a similar change in cells treated with HDAC6i (Fig. 5D). Lastly, we noted that the abundance of the proliferation marker Ki67 was higher on HDAC6−/− HDAC9−/− double-knockout Tregs than on single HDAC6 or HDAC9 knockout or wild type Treg. (Fig. 6C). Thus, we conclude that the combined targeting of HDAC6 and Sirt1 is so far the most promising lead to achieve additive improvement in Treg function.
Fig. 6.
Combined genetic targeting of HDAC6 and HDAC9 does not substantially improve the suppressive function of Tregs in vivo. (A) Homeostatic proliferation assays showed improved suppression of effector T cells by Tregs with either individual deficiency in either HDAC6 or HDAC9, but no added benefit from combined deletion. (B) Preservation of Foxp3 was not changed by combined knockout of HDAC6 and HDAC9. (C) Ki67 abundance was increased in HDAC6−/− HDAC9−/− double knockout Treg compared to that in single knockout and WT cells. Data are from three to four mice per group (N=18). The gating strategy used was similar to that in Fig. 5B. *P < 0.05, **P < 0.01, and ***P <0.001.
DISCUSSION
To move beyond the limitations of contemporary immunotherapies, the induction and maintenance of tolerance will need to become central components of future immunosuppressive regimens. To that end, direct administration of Tregs and the use of pharmacological agents that support Treg function hold substantial promise (20). However, current regimens for increasing the numbers of Tregs ex vivo face a number of challenges, among them the relatively short-lived persistence of the Treg phenotype in vivo (21). Augmenting the number or efficiency of endogenous Tregs through the use of HDAC inhibitors is an attractive alternative to administration of ex vivo expanded or induced Treg, and may also be relevant to stabilizing their phenotype after administration to the patient (22). Here, we identified multiple HDACs that are suitable targets for inhibition, and have tested several HDAC inhibitors, both class- and isotype-specific with in vitro and in vivo models. However, because the network of HDAC interactions and their roles in Treg biology remain ill-defined, and because suppression of certain HDAC isoforms may even worsen Treg function, a strategy that focuses on the inhibition of Sirt1, HDAC6, and HDAC9 is both logical and feasible (11). Our findings support the concept that these three HDACs share common mechanisms (for example, the deacetylation of Foxp3), but they also have distinctly different effects on Treg biology (for example, transcription factors controlling Foxp3 expression, heat shock response). Our studies showed that HDAC6 can regulate Foxp3 acetylation, which is a signature mechanism of action of several HDACs in Tregs (8, 10, 13, 23, 24). Acetylation of Foxp3 is an important posttranslational mechanism that affects Foxp3 abundance, because it protects Foxp3 from K48-mediated polyubiquitination and proteasomal degradation (13). In addition to extending the half-life of the Foxp3 protein, acetylation increases its DNA-binding capability, which leads to enhanced suppressive function of Tregs (25).
Important differences in the actions of the individual HDACs in Tregs are also apparent from our studies. The most important mechanism for the enhancement of Treg function as a result of HDAC6 inhibition appears to be the induction of the heat shock response. Activation of the heat shock response enables the production of chaperone proteins, such as Hsp27 and Hsp70, which protect the cell from stresses such as hypoxia or hyperthermia, and could give Tregs with an improved heat shock response a “survival advantage” under inflammatory conditions. Indeed, we found that Hsp70-containing Tregs had decreased suppressive function compared to that of wild type control Tregs. Although DNA binding by the HSF1 trimer is stabilized by Sirt1 (12), and deletion of Sirt1 led to a diminished heat shock response, this effect did not impair the survival of Sirt1−/− Tregs under heat shock. We hypothesized that Sirt1 might deacetylate Hsp70 and thereby influence the cellular heat shock response at a posttranslational level, because acetylation of Hsp70 increases its chaperone activity (26); however, we could not detect increased Hsp70 acetylation in cells lacking Sirt1. Even if Hsp70 were hyperacetylated in the absence of Sirt1, the importance of Hsp70 for the functional improvement of Tregs in which Sirt1 is inhibited appeared quite minimal. This is a central point, because it provides a rationale for combining inhibition of HDAC6 with that of Sirt1, despite their apparently contrary effects on the heat shock response, to achieve optimized inhibition of Tregs.
Diversity in the effects of HDACs was also apparent at the level of Foxp3 transcription factors. Although the net result of deleting HDAC6, HDAC9, or Sirt1 is a similar increase in the expression of the gene encoding Foxp3 (8–10), the transcription factors involved are different. Loss of Sirt1 stabilizes the acetylation of K310 of p65 (10), which increases its transcriptional activation (27), and is consistent with the nuclear translocation of p65 in fl-Sirt1/CD4cre Tregs that we observed (fig. S2). On the other hand, loss of HDAC9 favored the acetylation of STAT5. STAT5 has three reported acetylation sites (K694, K701, and K359), which, when acetylated, stabilize the transcriptionally active STAT5 dimer (28). Thus, the abundance of STAT5 Y694 phosphorylation, which is responsible for STAT5 nuclear translocation (29), may be secondary because of the stabilized STAT5 dimer being protected from dephosphorylation. Lastly, our study demonstrated the usefulness of combined targeting of HDACs by genetic deletion, the use of two isotype-specific HDAC inhibitors, or both approaches. Hence, targeting selected HDACs in combination is a promising strategy to augment Treg function, and with potentially less toxicity than occurs through the use of one or more pan-HDAC inhibitors. Because the combined pharmacologic inhibition of HDAC6 and Sirt1 was most effective in our in vivo studies, and because isotype-selective inhibitors are commercially available, this combination is, for the time being, the best lead to achieve this goal. Our study provides a rationale for further testing of combinations of isotype-specific HDAC inhibitors with relevant in vivo models, as well as in vitro studies with human cells.
MATERIAL AND METHODS
Mice
C57BL/6 and B6/Rag1−/− (Jackson Laboratory), B6/129 (Taconic), HDAC6−/− (30), HDAC9−/− (31), fl-Sirt1/CD4cre (10), fl-Sirt1/Foxp3cre (10), and Hsp70−/− (32) mice, all on an H-2b background, were housed under specific-pathogen-free conditions and were studied under a protocol approved by the Institutional Animal Care and Use Committee of the Children’s Hospital of Philadelphia.
Antibodies and small molecules
For flow cytometric analysis, we purchased mAbs against murine CD4 (conjugated to APC-eFluor 780, PE), CD8 (Pe-Cy7), CD25 (PE-Cy5), and Foxp3 (APC, PE) from eBioscience and against CD4 (Pacific Blue) and CD90.1 (PE Cy7) from BD Pharmingen. For Western blotting and fluorescence studies, we purchased antibodies against Sirt1, Hsp90, acetyl-lysine, STAT5 (recognizing both α and β forms), phosphorylated STAT5 (pSTAT5, Y694), Smad3, pSmad3 (S423/425), pCREB (S133), and CREB from Cell Signaling. We also purchased antibodies against acetyl-Hsp90 (K294, Rockland), Foxp3 (eBioscience), and β-actin from Santa Cruz. EX-527 was purchased from Tocris Bioscience, and the HDAC6 inhibitor compounds, Tubastatin and ACY-738, were provided by Dr. Alan Kozikowski (University of Illinois at Chicago) and Acetylon Pharmaceuticals, respectively. Small molecules were dissolved in DMSO, and DMSO alone was used as a control.
Cell isolation, flow cytometry, and pyrosequencing
Spleens and somatic lymph nodes were harvested from mice and processed to a single-cell suspension of lymphocytes. We used magnetic beads (Miltenyi Biotec) for the isolation of CD4+CD25− effector T cells, CD4+CD25+ Tregs, CD8+ T cells, and Thy1.2- APCs. Cells of interest were analyzed for their surface markers. To detect Foxp3, cells that had been stained for surface markers were fixed, permeabilized, and labeled with Foxp3-specific mAb (33). Data were analyzed with Flowjo 6.4.7 software (Treestar). Pyrosequencing of TSDR methylation in isolated effector T cells was performed by EpigenDx (assay-ID: ADS443FS2).
Analysis of T cell function
For Treg suppression assays, purified effector T cells were labeled with carboxyfluorescein succinimidyl ester (CFSE, Molecular Probes) and stimulated with irradiated APCs and antibody against CD3ε (1 μg/ml, BD Pharmingen). After 72 hours, cells were labeled with fluorescent mAb against CD4 and analyzed by flow cytometry. For quantification, we normalized relative suppression and calculated areas under the curve as previously reported (7). To measure the extent of conversion of effector T cells into Foxp3+ Tregs, we incubated effector T cells with TGF-β (3 ng/ml) and IL-2 (25 U/ml) for 4 days, and then assessed the cells by flow cytometry for the presence of Foxp3+ (15). For in vivo Treg suppression assays, we adoptively transferred Thy1.1+ CD4+ effector T cells in the presence or absence of Tregs into B6/RAG1−/− mice at a 4:1 ratio (8). Lymphocytes were isolated after one week, and the increase in numbers of cells identified by the markers Thy1.1, CD4, Foxp3, and Ki67 was assessed by flow cytometry.
Immunofluorescence staining
We centrifuged freshly isolated CD4+CD25+ Tregs or CD4+CD25+ Tregs that had been treated for 2 hours with antibodies against CD3ε and CD28 onto cytospin slides (Fisherfinest; Thermo Fisher Scientific). We permeabilized the cells with 10% normal goat serum and 1% Triton X. Double-stranded DNA was visualized with Hoechst 33342 (Invitrogen). Images were obtained with an Olympus BX51 Bright-field microscope (Olympus) at 100x magnifications. All raw images were universally processed with Adobe Photoshop 7.0 with the auto-contrast feature and were merged with Image-J 1.44 (National Institutes of Health, http://rsbweb.nih.gov/ij/) as shown in fig. S10A.
Western blotting analysis
After cell lysis in Radio-Immunoprecipitation Assay buffer with Halt™ protease inhibitor (Thermo Fisher Scientific) and the determination of protein concentration (DU640, Beckman-Coulter), samples were mixed with Laemmli sample buffer containing 2-mercaptoethanol (BioRad) and boiled for 5 min. Samples were loaded onto Mini-PROTEAN TGXT 4–15% gradient gels (BioRad), and after separation (at 200 V for 30 min), proteins were transferred to PolyScree PVDF Hybridization Transfer membranes (Perkin Elmer). Membranes were cut according to the molecular weights of the proteins of interest (as indicated with Precision Plus Protein Dual Color, BioRad), and incubated with primary and horseradish peroxidase (HRP)-conjugated secondary antibodies. We used SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific) and X-OMAT Blue Film XB (Kodak) for image development. Depending on the background, films were scanned or photographed and then processed with Adobe Photoshop 7.0 (grayscale conversion, and auto-contrast function, see fig. S10B). Densitometry was analyzed using ImageJ 1.44.
Proximity ligation assays
We prepared cytospin slides of CD4+CD25+ Tregs and used primary antibodies as described earlier. Antibodies against acetyl-lysine and either STAT5 or Foxp3 were chosen to be matching mouse- and rabbit-based antibodies (that is, mouse Foxp3 mAb was combined with a rabbit acetyl-lysine antibody, and the rabbit STAT5 mAb was matched with a mouse acetyl-lysine antibody). Subsequently, we used a Duolink in situ proximity ligation assay kit “Orange” (Olink Bioscience), with corresponding secondary antibodies (PLUS-rabbit, MINUS-mouse), according to the manufacturer’s instructions. We quantified the data with BlobFinder software by Allalou and Wahlby (Centre for Image Analysis, Uppsala, Sweden, http://www.cb.uu.se/~amin/BlobFinder/) (34).
Microarrays
RNA was extracted with RNeasy kits (Qiagen), and RNA integrity and quantity were analyzed by photometry (DU640; Beckman-Coulter). Microarray experiments were performed with whole-mouse-genome oligoarrays (Mouse430a; Affymetrix), and array data were analyzed with MAYDAY 2.12 software (35). Array data were subjected to robust multiarray average normalization. Normalized data were used for calculating fold changes of genes that were increased or decreased in expression with the Student’s t test, and data with >2-fold differential expression (p<0.05 with Storey’s FDR<0.1) were included in the analysis. Data underwent z-score transformation for display.
Statistical analysis
Data were analyzed with GraphPad Prism 5.0d software. All normally distributed data were displayed as means ± standard deviation (SD). Measurements between two groups were performed with a Student’s t test or Mann-Whitney U test. Groups of three or more were analyzed by one-way analysis of variance (ANOVA) or the Kruskal-Wallis test.
Supplementary Material
Fig. S1. ACY-738 and Tubastatin are HDAC6 inhibitors.
Fig. S2. Immunofluorescence analysis of Tregs to assess the cytosolic and nuclear localization of Foxp3 related transcription factors.
Fig. S3. Western blotting analysis of Treg lysates to compare Foxp3 transcription factors.
Fig. S4. Loss of Sirt1 does not affect conversion to iTregs.
Fig. S5. Loss of HDAC9 does not alter TSDR methylation.
Fig. S6. Purity control of the effector T cells used for the pyrosequencing methylation assay (fig. S5).
Fig. S7. Combined deletion of Sirt1 and HDAC9 produces minor improvements in Treg function.
Fig. S8. Targeting of three HDACs does not improve Treg function any more than does use of dual HDACi.
Fig. S9. Proliferation of effector T cells is not affected by HDAC inhibitors.
Fig. S10. Examples of image processing.
Table S1. Statistical analysis for Fig. 1C.
Table S2. Statistical analysis for Fig. 1G.
Acknowledgments
Funding: The project described was supported by Award Number K08AI095353 (to U.H.B.), AI073489, and AI095276 (to W.W.H.) from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Author contributions: U.H.B. and W.W.H. conceived the study and designed the experiments. U.H.B. and L.W. did most of the experiments. R.H. prepared immunofluorescence staining. T.A. and Y.L. assisted with homeostatic proliferation assays and advised with flow cytometry and protein biochemistry assays. U.H.B., L.W., T.A., Y.L., and W.W.H. analyzed data. U.H.B. wrote the manuscript and W.W.H. edited the manuscript.
Data accessibility: We deposited our data in the NCBI Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under accession numbers GSE26425 and GSE36095.
Competing interests: None.
REFERENCES AND NOTES
- 1.Naesens M, Kuypers DR, Sarwal M. Calcineurin inhibitor nephrotoxicity. Clin J Am Soc Nephrol. 2009;4:481–508. doi: 10.2215/CJN.04800908. [DOI] [PubMed] [Google Scholar]
- 2.Fishman JA. Infection in solid-organ transplant recipients. N Engl J Med. 2007;357:2601–2614. doi: 10.1056/NEJMra064928. [DOI] [PubMed] [Google Scholar]
- 3.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009;10:689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 4.McMurchy AN, Bushell A, Levings MK, Wood KJ. Moving to tolerance: Clinical application of T regulatory cells. Semin Immunol. 2011;23:304–313. doi: 10.1016/j.smim.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 6.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 7.Akimova T, Ge G, Golovina T, Mikheeva T, Wang L, Riley JL, Hancock WW. Histone/protein deacetylase inhibitors increase suppressive functions of human FOXP3+ Tregs. Clin Immunol. 2010;136:348–363. doi: 10.1016/j.clim.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 9.de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, Bradner JE, Mazitschek R, Kozikowski AP, Matthias P, Hancock WW. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol. 2011;31:2066–2078. doi: 10.1128/MCB.05155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beier UH, Wang L, Bhatti TR, Liu Y, Han R, Ge G, Hancock WW. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol Cell Biol. 2011;31:1022–1029. doi: 10.1128/MCB.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beier UH, Akimova T, Liu Y, Wang L, Hancock WW. Histone/protein deacetylases control Foxp3 expression and the heat shock response of T-regulatory cells. Curr Opin Immunol. 2011;23:670–678. doi: 10.1016/j.coi.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Loosdregt J, Brunen D, Fleskens V, Pals CE, Lam EW, Coffer PJ. Rapid temporal control of Foxp3 protein degradation by sirtuin-1. PLoS ONE. 2011;6:e19047. doi: 10.1371/journal.pone.0019047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, Brenkman AB, Hijnen DJ, Mutis T, Kalkhoven E, Prakken BJ, Coffer PJ. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 15.de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010;138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, Bellucci R, Raderschall E, Canning C, Soiffer RJ, Frank DA, Ritz J. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q, Kim YC, Laurence A, Punkosdy GA, Shevach EM. IL-2 controls the stability of Foxp3 expression in TGF-beta-induced Foxp3+ T cells in vivo. J Immunol. 2011;186:6329–6337. doi: 10.4049/jimmunol.1100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 19.Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunstein CG, Miller JS, Cao Q, McKenna DH, Hippen KL, Curtsinger J, Defor T, Levine BL, June CH, Rubinstein P, McGlave PB, Blazar BR, Wagner JE. Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood: safety profile and detection kinetics. Blood. 2011;117:1061–1070. doi: 10.1182/blood-2010-07-293795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye J, Su X, Hsueh EC, Zhang Y, Koenig JM, Hoft DF, Peng G. Human tumor-infiltrating Th17 cells have the capacity to differentiate into IFN-gamma+ and FOXP3+ T cells with potent suppressive function. Eur J Immunol. 2011;41:936–951. doi: 10.1002/eji.201040682. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, Basu S, Riley JL, Hancock WW, Shen Y, Saouaf SJ, Greene MI. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao R, de Zoeten EF, Ozkaynak E, Wang L, Li B, Greene MI, Wells AD, Hancock WW. Histone deacetylase inhibitors and transplantation. Curr Opin Immunol. 2007;19:589–595. doi: 10.1016/j.coi.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y, Li B, Zhou Z, Hancock WW, Zhang H, Greene MI. Histone acetyltransferase mediated regulation of FOXP3 acetylation and Treg function. Curr Opin Immunol. 2010;22:583–591. doi: 10.1016/j.coi.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Park JH, Jung Y, Kim JH, Jong HS, Kim TY, Bang YJ. Histone deacetylase inhibitor enhances 5-fluorouracil cytotoxicity by down-regulating thymidylate synthase in human cancer cells. Mol Cancer Ther. 2006;5:3085–3095. doi: 10.1158/1535-7163.MCT-06-0419. [DOI] [PubMed] [Google Scholar]
- 27.Ghizzoni M, Haisma HJ, Maarsingh H, Dekker FJ. Histone acetyltransferases are crucial regulators in NF-kappaB mediated inflammation. Drug Discov Today. 2011;16:504–511. doi: 10.1016/j.drudis.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L, Gao JS, Guan Y, Shi X, Zhang H, Ayrapetov MK, Zhang Z, Xu L, Hyun YM, Kim M, Zhuang S, Chin YE. Acetylation modulates prolactin receptor dimerization. Proc Natl Acad Sci U S A. 2010;107:19314–19319. doi: 10.1073/pnas.1010253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng R, Aoki Y, Yoshida M, Arai K, Watanabe S. Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J Immunol. 2002;168:4567–4575. doi: 10.4049/jimmunol.168.9.4567. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, Lombard D, Mizeracki A, Matthias G, Alt FW, Khochbin S, Matthias P. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao R, Wang L, Han R, Wang T, Ye Q, Honjo T, Murphy TL, Murphy KM, Hancock WW. Differential effects of B and T lymphocyte attenuator and programmed death-1 on acceptance of partially versus fully MHC-mismatched cardiac allografts. J Immunol. 2005;175:5774–5782. doi: 10.4049/jimmunol.175.9.5774. [DOI] [PubMed] [Google Scholar]
- 34.Allalou A, Wahlby C. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput Methods Programs Biomed. 2009;94:58–65. doi: 10.1016/j.cmpb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Battke F, Symons S, Nieselt K. Mayday--integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. ACY-738 and Tubastatin are HDAC6 inhibitors.
Fig. S2. Immunofluorescence analysis of Tregs to assess the cytosolic and nuclear localization of Foxp3 related transcription factors.
Fig. S3. Western blotting analysis of Treg lysates to compare Foxp3 transcription factors.
Fig. S4. Loss of Sirt1 does not affect conversion to iTregs.
Fig. S5. Loss of HDAC9 does not alter TSDR methylation.
Fig. S6. Purity control of the effector T cells used for the pyrosequencing methylation assay (fig. S5).
Fig. S7. Combined deletion of Sirt1 and HDAC9 produces minor improvements in Treg function.
Fig. S8. Targeting of three HDACs does not improve Treg function any more than does use of dual HDACi.
Fig. S9. Proliferation of effector T cells is not affected by HDAC inhibitors.
Fig. S10. Examples of image processing.
Table S1. Statistical analysis for Fig. 1C.
Table S2. Statistical analysis for Fig. 1G.