Abstract
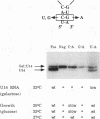
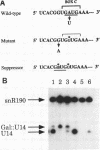
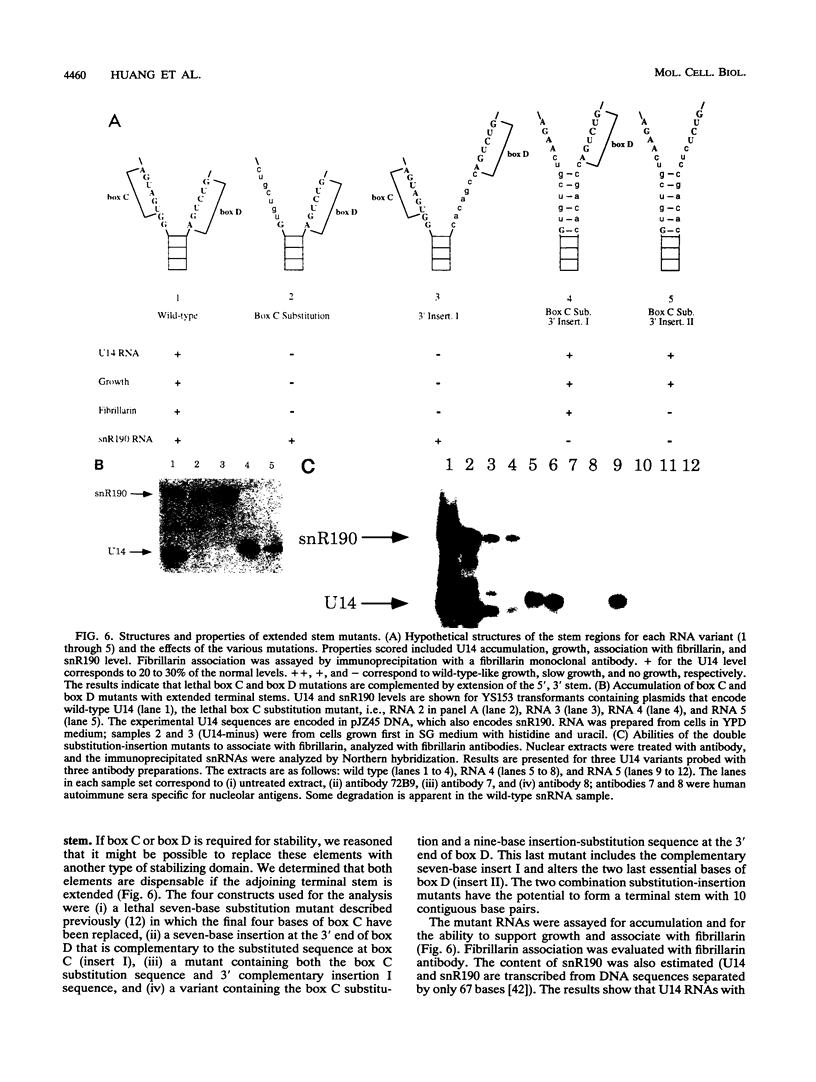
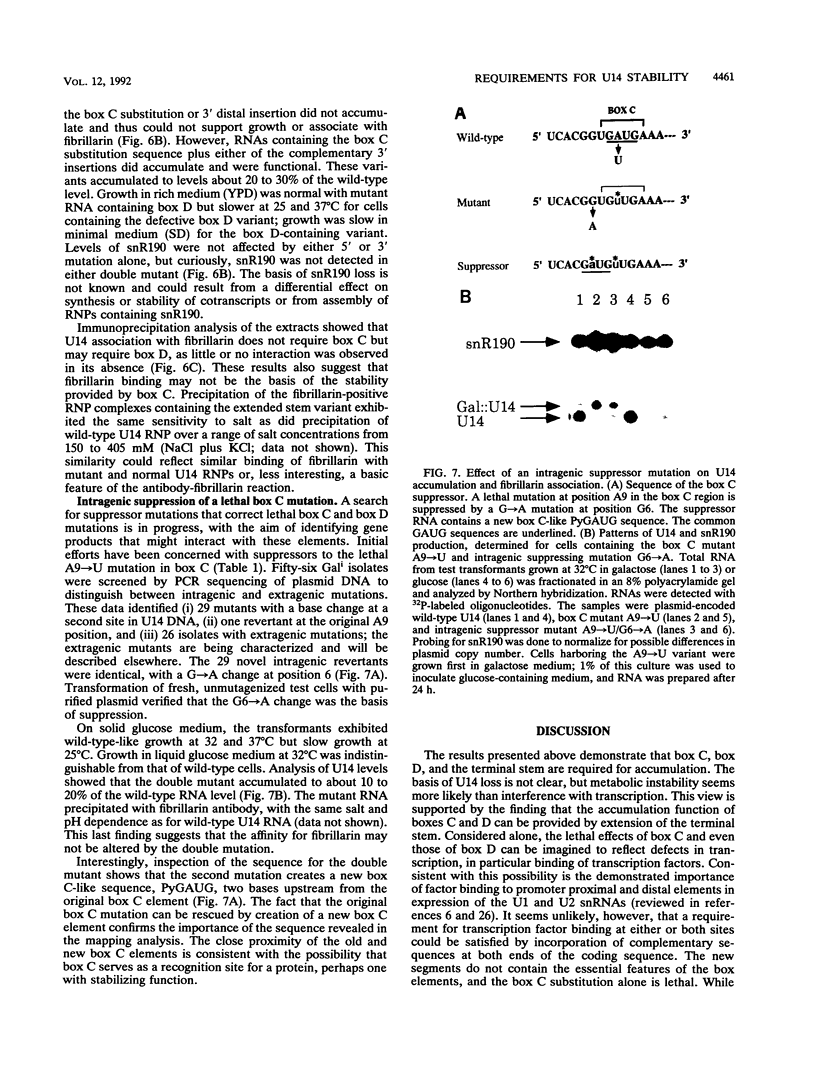
U14 is one of several nucleolar small nuclear RNAs required for normal processing of rRNA. Functional mapping of U14 from Saccharomyces cerevisiae has yielded a number of mutants defective in U14 accumulation or function. In this study, we have further defined three structural elements required for U14 accumulation. The essential elements include the U14-conserved box C and box D sequences and a 5', 3' terminal stem. The box elements are coconserved among several nucleolar small nuclear RNAs and have been implicated in binding of the protein fibrillarin. New mutational results show that the first GA bases of the box C sequence UGAUGA are essential, and two vital bases in box D have also been identified. An intragenic suppressor of a lethal box C mutant has been isolated and shown to contain a new box C-like PyGAUG sequence two bases upstream of normal box C. The importance of the terminal stem was confirmed from new compensatory base changes and the finding that accumulation defects in the box elements can be complemented by extending the terminal stem. The results suggest that the observed defects in accumulation reflect U14 instability and that protein binding to one or more of these elements is required for metabolic stability.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. S., Herrera R. J., Luhrmann R., Lizardi P. M. Isolation and partial characterization of U1-U6 small RNAs from Bombyx mori. Biochemistry. 1985 Jan 1;24(1):117–125. doi: 10.1021/bi00322a017. [DOI] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988 Jul;107(1):17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S. J., Yang X. D., Steitz J. A. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991 Sep;10(9):2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss F. T., Ingrahm J. L. Mutation in Saccharomyces cerevisiae conferring streptomycin and cold sensitivity by affecting ribosome formation and function. J Bacteriol. 1974 May;118(2):319–328. doi: 10.1128/jb.118.2.319-328.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. J., Cannon M. Maturation of ribosomal precursor RNA in Saccharomyces cerevisiae. A mutant with a defect in both the transport and terminal processing of the 20 S species. J Mol Biol. 1980 Oct 25;143(2):179–199. doi: 10.1016/0022-2836(80)90197-7. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis F. L., Parry C. B. Management of causalgia after peripheral nerve injury. Int Disabil Stud. 1989 Jan-Mar;11(1):15–20. doi: 10.3109/02599148909166371. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Oliphant A. R., Struhl K. Mutagenesis with degenerate oligonucleotides: an efficient method for saturating a defined DNA region with base pair substitutions. Methods Enzymol. 1987;155:558–568. doi: 10.1016/0076-6879(87)55036-4. [DOI] [PubMed] [Google Scholar]
- Hughes J. M., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991 Dec;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. M., Konings D. A., Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987 Jul;6(7):2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Zagorski J., Li H. V., Fournier M. J. Identification of essential elements in U14 RNA of Saccharomyces cerevisiae. EMBO J. 1990 Dec;9(13):4503–4509. doi: 10.1002/j.1460-2075.1990.tb07901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Stebbins-Boaz B., Gerbi S. A. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988 Mar 25;16(5):2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Guthrie C. Unexpected flexibility in an evolutionarily conserved protein-RNA interaction: genetic analysis of the Sm binding site. EMBO J. 1990 Aug;9(8):2555–2561. doi: 10.1002/j.1460-2075.1990.tb07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Solymosy F. Molecular analysis of a U3 RNA gene locus in tomato: transcription signals, the coding region, expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res. 1990 Apr 25;18(8):1941–1949. doi: 10.1093/nar/18.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Tóth M., Solymosy F. Plant small nuclear RNAs. Nucleolar U3 snRNA is present in plants: partial characterization. Eur J Biochem. 1985 Oct 15;152(2):259–266. doi: 10.1111/j.1432-1033.1985.tb09192.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Fournier M. J. U14 function in Saccharomyces cerevisiae can be provided by large deletion variants of yeast U14 and hybrid mouse-yeast U14 RNAs. EMBO J. 1992 Feb;11(2):683–689. doi: 10.1002/j.1460-2075.1992.tb05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischwe M. A., Ochs R. L., Reddy R., Cook R. G., Yeoman L. C., Tan E. M., Reichlin M., Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J Biol Chem. 1985 Nov 15;260(26):14304–14310. [PubMed] [Google Scholar]
- Liu J., Maxwell E. S. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res. 1990 Nov 25;18(22):6565–6571. doi: 10.1093/nar/18.22.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima A., Nakayama N., Miyajima I., Arai N., Okayama H., Arai K. Analysis of full-length cDNA clones carrying GAL1 of Saccharomyces cerevisiae: a model system for cDNA expression. Nucleic Acids Res. 1984 Aug 24;12(16):6397–6414. doi: 10.1093/nar/12.16.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K. A., Steitz J. A. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987 Aug;7(8):2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Simmons T., Shuster E. O., Siliciano P. G., Guthrie C. Genetic analysis of small nuclear RNAs in Saccharomyces cerevisiae: viable sextuple mutant. Mol Cell Biol. 1988 Aug;8(8):3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter G. L., Brennwald P. J., Holm K. A., Wise J. A. The sequence of U3 from Schizosaccharomyces pombe suggests structural divergence of this snRNA between metazoans and unicellular eukaryotes. Nucleic Acids Res. 1988 Nov 11;16(21):10131–10152. doi: 10.1093/nar/16.21.10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Henning D., Busch H. Nucleotide sequence of nucleolar U3B RNA. J Biol Chem. 1979 Nov 10;254(21):11097–11105. [PubMed] [Google Scholar]
- Reddy R., Henning D., Busch H. Primary and secondary structure of U8 small nuclear RNA. J Biol Chem. 1985 Sep 15;260(20):10930–10935. [PubMed] [Google Scholar]
- Reimer G., Pollard K. M., Penning C. A., Ochs R. L., Lischwe M. A., Busch H., Tan E. M. Monoclonal autoantibody from a (New Zealand black x New Zealand white)F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987 Jul;30(7):793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Schimmang T., Tollervey D., Kern H., Frank R., Hurt E. C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989 Dec 20;8(13):4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanab G. M., Maxwell E. S. Proposed secondary structure of eukaryotic U14 snRNA. Nucleic Acids Res. 1991 Sep 25;19(18):4891–4894. doi: 10.1093/nar/19.18.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A. Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Hurt E. C. The role of small nucleolar ribonucleoproteins in ribosome synthesis. Mol Biol Rep. 1990;14(2-3):103–106. doi: 10.1007/BF00360433. [DOI] [PubMed] [Google Scholar]
- Trinh-Rohlik Q., Maxwell E. S. Homologous genes for mouse 4.5S hybRNA are found in all eukaryotes and their low molecular weight RNA transcripts intermolecularly hybridize with eukaryotic 18S ribosomal RNAs. Nucleic Acids Res. 1988 Jul 11;16(13):6041–6056. doi: 10.1093/nar/16.13.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K., Steitz J. A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989 Oct;8(10):3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990 Jun;2(3):521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Wise J. A., Weiner A. M. Dictyostelium small nuclear RNA D2 is homologous to rat nucleolar RNA U3 and is encoded by a dispersed multigene family. Cell. 1980 Nov;22(1 Pt 1):109–118. doi: 10.1016/0092-8674(80)90159-2. [DOI] [PubMed] [Google Scholar]
- Zagorski J., Tollervey D., Fournier M. J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible and essential small nuclear RNAs. Mol Cell Biol. 1988 Aug;8(8):3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko G. S., Mitchell A. P., Jones E. W. Mapping of the proteinase b structural gene PRB1, in Saccharomyces cerevisiae and identification of nonsense alleles within the locus. Genetics. 1980 Sep;96(1):137–146. doi: 10.1093/genetics/96.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]