ABSTRACT
Environmental pathogens survive and replicate within the outside environment while maintaining the capacity to infect mammalian hosts. For some microorganisms, mammalian infection may be a relatively rare event. Understanding how environmental pathogens retain their ability to cause disease may provide insight into environmental reservoirs of disease and emerging infections. Listeria monocytogenes survives as a saprophyte in soil but is capable of causing serious invasive disease in susceptible individuals. The bacterium secretes virulence factors that promote cell invasion, bacterial replication, and cell-to-cell spread. Recently, an L. monocytogenes chitinase (ChiA) was shown to enhance bacterial infection in mice. Given that mammals do not synthesize chitin, the function of ChiA within infected animals was not clear. Here we have demonstrated that ChiA enhances L. monocytogenes survival in vivo through the suppression of host innate immunity. L. monocytogenes ΔchiA mutants were fully capable of establishing bacterial replication within target organs during the first 48 h of infection. By 72 to 96 h postinfection, however, numbers of ΔchiA bacteria diminished, indicative of an effective immune response to contain infection. The ΔchiA-associated virulence defect could be complemented in trans by wild-type L. monocytogenes, suggesting that secreted ChiA altered a target that resulted in a more permissive host environment for bacterial replication. ChiA secretion resulted in a dramatic decrease in inducible nitric oxide synthase (iNOS) expression, and ΔchiA mutant virulence was restored in NOS2−/− mice lacking iNOS. This work is the first to demonstrate modulation of a specific host innate immune response by a bacterial chitinase.
IMPORTANCE
Bacterial chitinases have traditionally been viewed as enzymes that either hydrolyze chitin as a food source or serve as a defense mechanism against organisms containing structural chitin (such as fungi). Recent evidence indicates that bacterial chitinases and chitin-binding proteins contribute to pathogenesis, primarily via bacterial adherence to chitin-like molecules present on the surface of mammalian cells. In contrast, mammalian chitinases have been linked to immunity via inflammatory immune responses that occur outside the context of infection, and since mammals do not produce chitin, the targets of these mammalian chitinases have remained elusive. This work demonstrates that a Listeria monocytogenes-secreted chitinase has distinct functional roles that include chitin hydrolysis and suppression of host innate immunity. The established link between chitinase and the inhibition of host inducible nitric oxide synthase (iNOS) expression may help clarify the thus far elusive relationship observed between mammalian chitinase enzymes and host inflammatory responses occurring in the absence of infection.
Introduction
Environmental pathogens are organisms that are capable of survival in the outside environment while maintaining the capacity to cause human disease. These organisms represent reservoirs of infection that are widespread and difficult to eradicate. Defining how environmental pathogens develop and maintain the capacity to infect humans is important for understanding emerging infections and for reducing disease transmission under conditions of environmental change. Selective fitness advantages conferred by gene products expressed during host infection could potentially contribute little to survival in environments outside the host, and thus these coding regions might be anticipated to accrue loss-of-function mutations during bacterial replication in the outside environment. Alternatively, it is possible that gene products that contribute to bacterial virulence are functionally diverse, having been adapted for distinct roles in disparate environments.
Listeria monocytogenes is a Gram-positive facultative intracellular bacterium that survives as a saprophyte in soil but upon ingestion can cause serious disease in susceptible individuals (1, 2). Most outbreaks have been associated with the consumption of L. monocytogenes-contaminated food products, and the bacterium is a frequent cause of large and expensive food recalls (3–5). Following ingestion, L. monocytogenes crosses the intestinal barrier and replicates within the liver and spleen (6). The bacterium expresses a number of gene products that enable it to invade host cells, escape from cell phagosomes, replicate within the cytosol, and spread to adjacent cells (7). L. monocytogenes thus expresses a number of gene products that target distinct aspects of eukaryotic cell physiology.
Recent attention has focused on bacterial chitinases and chitin binding proteins in recognition of their contributions to bacterial virulence within infected mammals (8–13). These proteins have historically been thought to contribute to microbial life outside mammalian hosts based on the absence of chitin in mammals and the abundance of the polymer in fungal cell walls, as well as the exoskeletons of mollusks, arthropods, and crustaceans (14, 15). Chitin is a linear polysaccharide consisting of N-acetylglucosamine residues linked by β1,4-glycosidic linkages (16). Hydrolysis of chitin provides an abundant source of carbon and nitrogen for organisms that express chitinases; however, chitinases and chitin binding proteins have now been linked to pathogenesis in mammals for at least four bacterial pathogens, Vibrio cholerae, Serratia marcescens, Legionella pneumophila, and L. monocytogenes (8–13). For V. cholerae, the GbpA chitin binding protein has been demonstrated to contribute to bacterial colonization of the mouse small intestine via binding to mucin (8, 11, 13). The chitin binding protein CBP21 of S. marcescens contributes to bacterial adherence to colonic epithelial cells through its interactions with mammalian chitinase 3-like 1 (CHI3L1) (12). The ChiA chitinase of L. pneumophila contributes to bacterial colonization of the lung through an as yet unidentified mechanism (10).
L. monocytogenes has two chitinase-encoding genes, chiA and chiB, and one gene that encodes a chitin binding protein (lmo2467) (17, 18). The gene products of all three genes have been shown to contribute to bacterial virulence in vivo (9). The most significant defect in virulence was associated with the loss of chiA, which encodes a member of the glycosyl hydrolase family 18 of chitinases (17). ChiA has activity toward chitin as well as the chitin pseudosubstrates p-(GlcNAc)2 and p-(GlcNAc)3 but not toward a pseudocellulose substrate or toward peptidoglycan (17). The expression of chiA is increased in L. monocytogenes within macrophages (19) and is regulated by PrfA (20), a transcriptional activator required for the expression of gene products associated with L. monocytogenes virulence (21). The role of chitinase in L. monocytogenes pathogenesis is not known, nor has it been demonstrated that the chitinase activity of ChiA is required for virulence.
Given that chitin is not produced by mammals, it is intriguing that multiple bacterial chitinases are associated with virulence. Although mammals do not synthesize chitin, the presence of mammalian chitinases has been associated with host inflammatory responses in the apparent absence of microbial infection in diseases such as asthma (22–24). Mammalian chitinases, such as the acidic mammalian chitinase (AMCase) and macrophage differentiation marker YKL-40, both glycosyl hydrolase 18 family members similar to ChiA, may have chitin-like targets that modulate host immunity (25). The direct targets of these chitinases have not yet been identified, but many factors involved in mammalian immune responses are glycosylated with N-acetylglucosamine residues linked by β1,4-glycosidic bonds (14) and thus could potentially serve as substrates for an enzyme with chitinase activity. Endogenous carbohydrates, such as heparan sulfate and hyaluronic acid, that share structural similarities with chitin have been proposed as chitinase substrates; however, proof of this interaction has not been demonstrated (25).
In this study, we have further investigated the role of ChiA in L. monocytogenes pathogenesis. ChiA chitinase activity was found to be important for the suppression of host innate immune responses that serve to limit bacterial replication within the livers and spleens of infected mice. Mice infected with L. monocytogenes strains lacking chiA exhibited increased levels of NOS2 expression, and bacteria were rapidly cleared from target organs. The virulence of ΔchiA strains was restored in mice lacking iNOS. These data thus establish the first functional link between a bacterial chitinase and direct modulation of host innate immunity.
RESULTS
Loss of ChiA enhances immune clearance of L. monocytogenes from target organs.
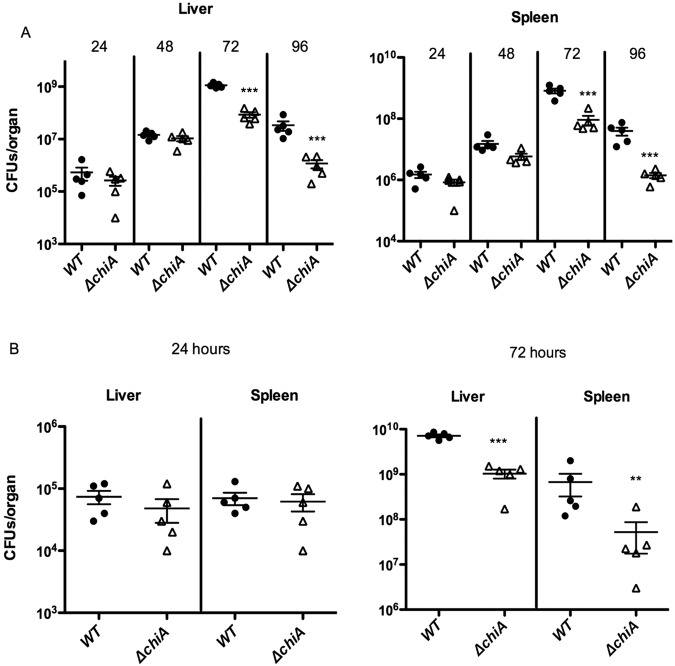
Our laboratory has previously shown that mice intravenously infected with L. monocytogenes mutants lacking chiA have an approximately 20-fold reduction in bacterial burdens present in the liver and spleen at 72 h postinfection (9). It was not determined, however, whether ChiA was required at the initiation of infection or whether the protein was required subsequent to L. monocytogenes colonization of the liver and spleen. To determine the point during infection at which the requirement for ChiA becomes evident, 7- to 8-week-old Swiss Webster mice were infected with a sublethal dose (2 × 104 CFU) of either wild-type L. monocytogenes 10403S or the ΔchiA mutant. At 24, 48, 72, and 96 h postinfection, the livers and spleens of infected mice were harvested to enumerate bacterial burdens. The number of L. monocytogenes CFU recovered from the livers and spleens of animals infected with either the wild type or the ΔchiA mutant strain appeared to be very similar at 24 and 48 h postinfection, indicating that the ΔchiA mutant was fully capable of colonizing and replicating within these target organs. However, by 72 h postinfection, the numbers of bacteria recovered from the livers and spleens of animals infected with the ΔchiA mutant were 13-fold and 9-fold lower, respectively, than in the organs of mice infected with wild-type L. monocytogenes (Fig. 1A). By 96 h postinfection, bacterial burdens in the livers and spleens of animals infected with either bacterium began to drop, reflecting immune clearance of infecting bacteria (Fig. 1A). Mice infected with the ΔchiA mutant exhibited significantly lower (>30-fold) numbers of bacteria in target organs, indicating that the mutant strain was more susceptible to immune clearance. ChiA thus does not contribute to the initial establishment of L. monocytogenes infection in mice, but the protein is required for enhanced bacterial survival in the face of an effective host innate immune response.
FIG 1 .
L. monocytogenes mutants lacking chiA exhibit a virulence defect late in infection. (A) Intravenous infection of mice with the ΔchiA strain. Swiss Webster mice were intravenously infected with 2 × 104 CFU L. monocytogenes wild type or ΔchiA mutant bacteria. The scatter plot shows CFU obtained from the livers and spleens of five individual mice at 24, 38, 72, or 96 h postinfection; the data are representative of two independent experiments. Solid lines and brackets represent the means and standard errors of the means, respectively, for the data points in each group. (B) Intragastric [i.g.] infection of mice with ΔchiA bacteria. C57BL6 mice were intragastrically infected with 1 × 108 CFU L. monocytogenes wild type or ΔchiA bacteria. The scatter plot shows CFU obtained from the livers and spleens of five individual mice at 24 and 72 h postinfection; the data are representative of two independent experiments. Solid lines and brackets represent the means and standard errors of the mean, respectively, for the data points in each group. Statistically significant P values (***, P < 0.0001; **, P < 0.001) for the ΔchiA strain in both the liver and the spleen after 72 and 96 h of infection (intravenous [i.v.] infection) or after 72 h (i.g. infection) compared to results for the wild type were determined using a one-way analysis of variance with Dunnett’s posttest (GraphPad V.5.0A software program).
ChiA is not required for bacterial translocation across the host intestinal barrier.
Thus far, the most definitive roles reported for the contributions of bacterial chitinases or chitin binding proteins to infection have been the enhancement of bacterial adherence to intestinal or colonic epithelial cells (8, 11–13). Given that the natural route of L. monocytogenes infection occurs via bacterial translocation across the intestinal epithelial barrier (26), the potential contributions of ChiA to intragastric infection of mice were examined. Although mice are generally less susceptible to L. monocytogenes oral infection due to amino acid variations present in their E-cadherin, the intestinal epithelial cell receptor recognized by the L. monocytogenes surface protein InlA (27, 28), Wollert et al. (29) have recently described mutations within inlA that increase InlA binding affinity for E-cadherin and facilitate murine oral infection models. The S192N and Y376S mutations encoded by inlA (inlA S192N and Y376S) were thus introduced via homologous recombination into both the wild-type and ΔchiA strains, followed by intragastric inoculation of the strains into mice. At 24 h postinfection, there was no significant difference in the numbers of bacteria recovered from the livers or spleens of mice infected with either the wild type or the ΔchiA strain. By 72 h postinfection, mice infected with the ΔchiA strain exhibited approximately 10-fold-lower numbers of bacteria in both liver and spleen, (Fig. 1B), a difference similar to the fold reduction in bacterial CFUs observed following intravenous infection (Fig. 1A). ChiA therefore does not appear to significantly influence the ability of L. monocytogenes to cross the intestinal epithelium.
The virulence defect of the ΔChiA mutant can be rescued in trans during coinfection with wild-type L. monocytogenes.
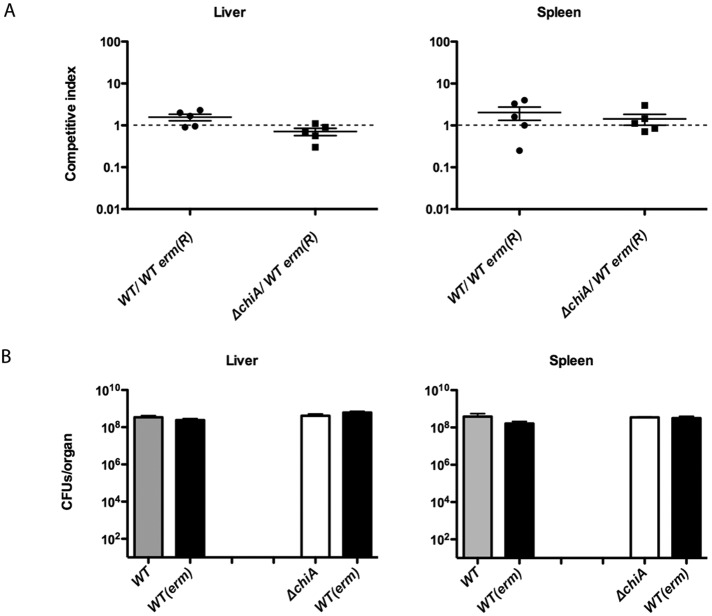
ChiA is a secreted protein, and thus we determined if the defect conferred by the absence of ChiA during infection could be rescued by secreted ChiA provided in trans by wild-type L. monocytogenes. Mice were intravenously injected with a 1:1 ratio of the ΔchiA strain and a wild-type derivative of L. monocytogenes that contains a chromosomal erythromycin resistance gene cassette that has been shown not to impact bacterial virulence (30). After 72 h of infection, the livers and spleens were harvested and assessed for bacterial burdens on solid medium with and without erythromycin. If the presence of wild-type L. monocytogenes was capable of complementing the virulence defect of the ΔchiA strain the 1:1 ratio of the wild-type strain to the ΔchiA strain should not change significantly during the course of infection. However, if the presence of wild-type L. monocytogenes did not restore ΔchiA strain virulence, then the wild-type strain should demonstrate a competitive advantage over the ΔchiA strain, thereby altering the ratio of the wild type to the ΔchiA strain to a value greater than 1. Despite the original 10- to 20-fold decrease in recovery of the ΔchiA mutant from livers and spleens of monoculture-infected animals in comparison to results for the wild type at 72 h postinfection (Fig. 1), mice infected with mixtures of the wild type and the ΔchiA strain maintained approximately equivalent ratios of the two strains after 72 h (Fig. 2A). The competitive index (CI) value reflecting the numbers of ΔchiA bacteria recovered in comparison to the wild type was not significantly different from 1 (P > 0.1), indicative of equivalent bacterial fitness levels (Fig. 2A). In addition, the absolute numbers of wild-type bacteria present in the liver and spleen were similar in mixed infections with either the ΔchiA strain or the wild-type erythromycin-resistant strain, indicating that the ΔchiA mutant did not negatively impact wild-type growth (Fig. 2B). These data indicate that the virulence defect of the ΔchiA mutant can be rescued in trans by the presence of wild-type L. monocytogenes, suggesting that secreted ChiA broadly influences some aspect of host physiology to promote ΔchiA replication during the course of infection.
FIG 2 .
Virulence defects associated with ΔchiA are complemented in trans by the presence of wild-type L. monocytogenes. (A) Wild-type and ΔchiA strain mixed infections in mice. Swiss Webster mice were intravenously infected with 2 × 104 total CFUs of a 1:1 mixture of the L. monocytogenes DP-L3903 (10403S with a silent Tn917-lac insertion conferring erythromycin resistance) and ΔchiA strains. After 72 h of infection, livers and spleens of infected mice were harvested and bacteria were enumerated. As a control, groups of mice were infected with a 1:1 mixture of wild-type L. monocytogenes strains with the DP-L3903 erythromycin-resistant strain. Each data point represents the competitive index or the ratio of ΔchiA CFU recovered to that of wild-type L. monocytogenes. Data shown are representative of at least 2 independent experiments. (B) Total CFU of wild-type and ΔchiA mutant strains recovered from each organ. Data shown are representative of at least 2 independent experiments.
Chitinase activity appears to be important for ChiA-dependent enhancement of L. monocytogenes virulence.
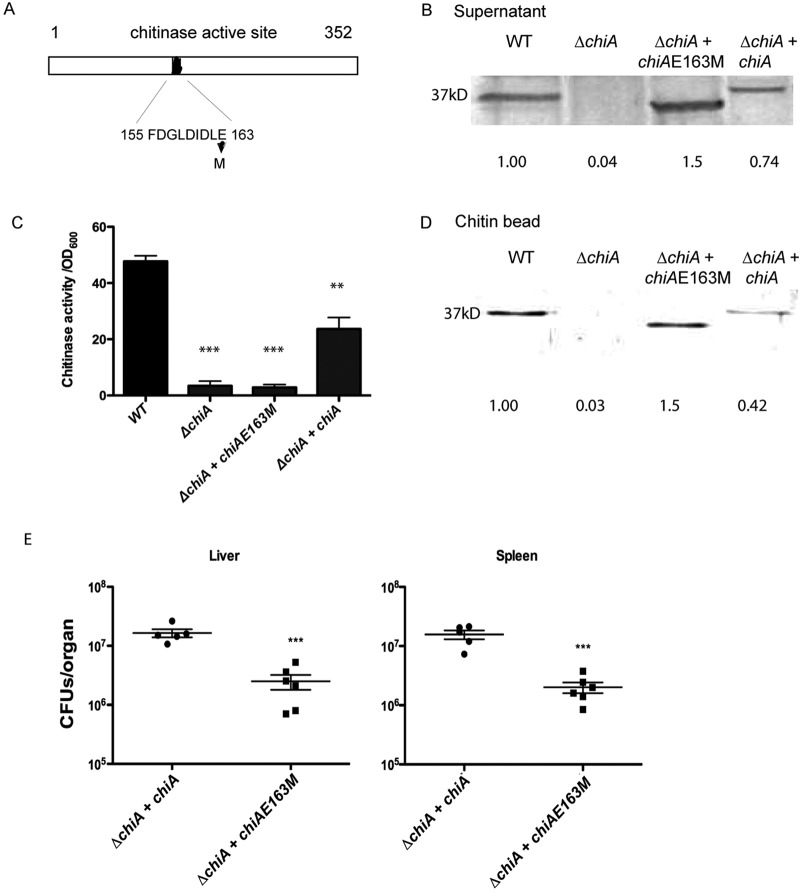
ChiA contains a highly conserved chitinase catalytic region consisting of 10 amino acids (FDGLDIDDLE); the glutamic acid residue at position 163 within this sequence is required for chitin hydrolysis (31, 32) (Fig. 3A). The substitution of methionine for the catalytic glutamate residue in the related ChiA chitinase of Vibrio harveyi eliminates chitinase activity without affecting chitin binding (31). To determine if the chitinase and/or the chitin binding activity of L. monocytogenes ChiA was important for bacterial virulence, a similar chiA E163M (E163M amino acid change encoded by chiA) mutation was introduced into the chiA gene carried on the integrative plasmid vector pPL2 (33) (Fig. 3A). The pPL2-chiA and pPL2-chiA E163M plasmids were stably introduced in single copy into the chromosome of L. monocytogenes ΔchiA, and the resulting strains (the ΔchiA + pPL2-chiA and ΔchiA + pPL2-chiA E163M mutants, respectively) were assessed for secreted ChiA, chitin binding, and chitinase activity.
FIG 3 .
ChiA chitinase activity appears to contribute to L. monocytogenes virulence. (A) Schematic diagram of ChiA showing the conserved region of 10 amino acids within the active site of the chitinase. The substitution of methionine (M) 163 for glutamate (E) has been demonstrated to eliminate chitinase activity without affecting chitin binding in other closely related ChiA family members (31). (B) Detection of secreted ChiA by Western blot analysis. Bacterial supernatant proteins from overnight cultures of wild-type, ΔchiA, ΔchiA + pPL2-chiA, and ΔchiA + pPL2-chiA E163M bacteria were isolated using trichloroacetic acid (TCA) precipitation. Sample volumes were adjusted to reflect equivalent bacterial cell densities. Secreted ChiA was detected using rabbit polyclonal antibody directed against ChiA. The amount of protein detected for each sample in comparison to that in the wild-type lane (set at 1.0) as determined by densitometry is indicated at the bottom of each panel. Data shown are representative of at least 3 independent experiments. (C) Assessment of chitinase activity in bacterial supernatants. Supernatants derived from overnight cultures of wild-type, ΔchiA, ΔchiA + pPL2-chiA, and ΔchiA + pPL2-chiA E163M were assessed for chitinase activity using the colorimetric substrate nitrophenyl N,N′--diacetyl-β-d-chitobioside. Chitinase activity was reflected by substrate hydrolysis as measured by absorbance at an optical density at 405 nm (OD405) as a function of bacterial cell density. Data shown represent the means ± standard errors for three independent experiments. Statistically significant differences as determined by one-way analysis of variance with Tukey’s multiple-comparison test are indicated (*, P < 0.01; **, P < 0.001; ***, P < 0.0001) (GraphPad V.5.0). (D) Detection of chitin binding by the ChiA wild-type and mutant proteins. Bacterial supernatants derived from overnight cultures of wild-type, ΔchiA, ΔchiA + pPL2-chiA, and ΔchiA + pPL2-chiA E163M bacteria were adjusted to control for equivalent cell densities, concentrated 50-fold, and incubated with chitin beads for 1 h at 4°C. Beads were washed with PBS, and bound protein was recovered following boiling of the beads in SDS-PAGE sample buffer. ChiA bound to chitin was detected by Western blot analysis using rabbit polyclonal antibody directed against ChiA. The amount of protein detected for each sample in comparison to that in the wild type lane (set at 1.0) as determined by densitometry is indicated at the bottom of each panel. Data shown are representative of at least 3 independent experiments. (E) Assessment of bacterial virulence in mice. Swiss Webster mice were intravenously infected with 2 × 104 CFU of either ΔchiA + pPL2-chiA or ΔchiA + pPL2-chiA E163M bacteria. The scatter plot shows CFU obtained from the livers and spleens of five individual mice at 72 h postinfection; the data are representative of two independent experiments. Solid lines and brackets represent the means and standard errors of the means, respectively, for the data points in each group. ***, statistically significant value (P < 0.0001) for ΔchiA + pPL2-chiA E163M results in both the liver and the spleen after 72 h of infection compared to ΔchiA + pPL2-chiA results using a one-way analysis of variance with Dunnett’s posttest (GraphPad V.5.0A).
ChiA and ChiA E163M were detected as secreted products in the supernatants of both pPL2-chiA- and pPL2-chiA E163M-complemented ΔchiA strains as determined by Western blot analysis of bacterial supernatants (Fig. 3B). The ChiA E163M protein was observed to migrate as a smaller-molecular-mass species (approximately 1 kDa smaller based on molecular size standards); this appeared to reflect a proteolytic cleavage event, since a similarly sized lower-molecular-mass band could also occasionally be detected migrating below wild-type ChiA (see a faint band detectable in Fig. 3B). The ΔchiA + pPL2-chiA E163M mutant was deficient in secreted chitinase activity as detected by an in vitro chitinase assay of bacterial supernatants (Fig. 3C). However, both wild-type ChiA and ChiA E163M were able to bind chitin, as demonstrated by the binding and retention of the proteins on chitin beads (Fig. 3D). The chiA E163M mutation therefore results in the elimination of detectable chitinase activity while not affecting chitin binding.
When the plasmid-complemented ΔchiA strains were tested for bacterial virulence in mouse intravenous infection models, the ΔchiA + pPL2-chiA E163M mutant remained nearly as attenuated as strains lacking chiA entirely (Fig. 3E). Mice infected with ΔchiA + pPL2-chiA E163M strains had a greater than 6-fold reduction in bacterial CFUs recovered from the liver and spleen in comparison to results for animals infected with ΔchiA + pPL2-chiA strains at 72 h postinfection (Fig. 3E). Although the ChiA E163M protein appears more susceptible to proteolytic cleavage, the undiminished chitin binding activity of the mutant suggests that the chitin binding activity of ChiA alone is not sufficient to enhance L. monocytogenes virulence.
ChiA reduces the expression of iNOS, and ΔchiA strains are fully virulent in NOS2−/− mice.
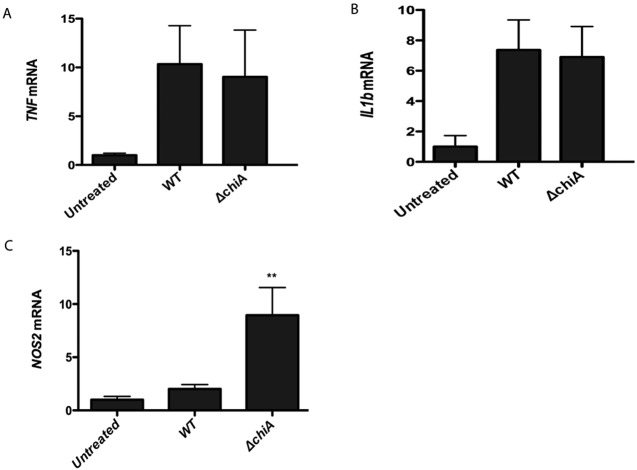
Based on the ability of the ΔchiA mutants to be complemented for virulence in trans by the presence of wild-type L. monocytogenes secreting ChiA, we investigated whether the secretion of ChiA enhanced L. monocytogenes replication within host tissues by modulating host immune responses. The expression of the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin 1β (IL-1β) and of the inducible nitric oxide synthase (iNOS) was assessed in the livers of mice infected with either wild-type or ΔchiA strains at 48 h postinfection, prior to the 72-h time point for which ΔchiA strain numbers can be observed to decrease in comparison to those for the wild type (Fig. 1). At 48 h postinfection, no significance difference was observed in the levels of either TNF-α or IL-1β expression for mice infected with either the wild type or the ΔchiA mutant (Fig. 4A and B). In contrast, expression of iNOS was approximately 4-fold higher in animals infected with the ΔchiA strain than in those infected with the wild-type strain (Fig. 4C).
FIG 4 .
Mice infected with L. monocytogenes ΔchiA strains exhibit increased levels of iNOS expression. Seven- to eight-week-old Swiss Webster mice were infected via the tail vein with 2 × 104 CFU of wild-type or ΔchiA L. monocytogenes for 48 h, followed by analysis of mRNA expression in liver. Data are normalized to gapdh expression levels, with the average for untreated samples set at 1. Data shown are from at least two independent experiments. Statistically significant differences as determined by one-way analysis of variance with Tukey’s multiple-comparison test are indicated (**, P < 0.001) (GraphPad V.5.0).
The significant increase observed in the induction of iNOS in mice infected with the ΔchiA mutant strains suggests that a potential in vivo function of L. monocytogenes ChiA is the suppression of iNOS expression and activity. iNOS activity is an important facet of the host innate immune response that limits L. monocytogenes survival and proliferation (34–36). NOS2−/− mice lacking iNOS have been shown to be approximately 100-fold more susceptible to L. monocytogenes infection than wild-type mice (36). If ChiA is important for reducing iNOS activity, then the enhanced virulence observed for the wild-type strains versus ΔchiA mutants should not be apparent in mice lacking iNOS. C57BL/6 wild-type and NOS2−/− mice were infected with wild-type or ΔchiA L. monocytogenes, and bacterial burdens were monitored in the livers and spleens at 72 h postinfection (Fig. 5). Given the 100-fold increase in susceptibility of NOS2−/− mice to L. monocytogenes infection (36), these animals were inoculated with 100-fold fewer bacteria (2 × 102 CFU) than wild-type C57BL/6 mice. As shown in Fig. 5A, the virulence defect observed for ΔchiA strains in wild-type C57BL/6 mice was similar to that observed for the mutant following the infection of Swiss Webster mice, with the mutant strain exhibiting approximately 10-fold and 7-fold decreases in bacterial CFUs recovered from the liver and spleen, respectively, at 72 h postinfection. Strikingly, this ΔchiA-associated virulence defect was not detectable in NOS2−/− animals (Fig. 5B). These results, when combined with the observed increase in iNOS expression in mice infected with ΔchiA strains, represent the first example of a pathogen-encoded chitinase functioning to enhance virulence through suppression of iNOS expression, a critical host defense mechanism.
FIG 5 .
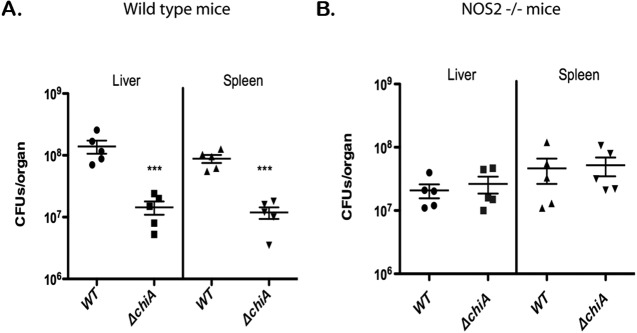
The virulence of L. monocytogenes ΔchiA mutants is restored in mice lacking iNOS. (A) L. monocytogenes intravenous infection. C57BL/6 mice were intravenously infected with 2 × 104 CFU L. monocytogenes wild-type or ΔchiA mutant bacteria. The scatter plot shows CFU obtained from the livers and spleens of five individual mice at 72 h postinfection; the data are representative of two independent experiments. Solid lines and brackets represent the means and standard errors of the means, respectively, for the data points in each group. ***, statistically significant value (P < 0.0001) for ΔchiA bacteria in both the liver and the spleen after 72 of infection compared to results for the wild type using a one-way analysis of variance with Dunnett’s posttest (GraphPad V.5.0A). (B) Intravenous infection of C57BL/6 NOS2−/− mice lacking iNOS. C57BL/6 NOS2−/− mice were intravenously infected with 2 × 102 CFU L. monocytogenes wild-type or ΔchiA mutant bacteria [the infectious dose was adjusted to reflect the ~100-fold increase in susceptibility to L. monocytogenes infection for mice lacking iNOS (36)]. The scatter plot shows CFU obtained from the livers and spleens of five individual mice at 72 h postinfection; the data are representative of two independent experiments. Solid lines and brackets represent the means and standard errors of the means, respectively, for the data points in each group. No significant statistical difference was detected between the bacterial burdens of mice infected with ΔchiA bacteria for either organ (P > 0.05) compared to findings for the wild type using a one-way analysis of variance with Dunnett’s posttest (GraphPad V.5.0A).
DISCUSSION
Environmental pathogens survive and replicate in disparate habitats that include the outside environment as well as inside human cells. For many of these organisms, humans may be infrequently encountered hosts, and thus to remain pathogens, these organisms must be capable of maintaining a repertoire of virulence factors that allow host colonization. Gene products that function in multiple contexts would provide a fitness advantage to pathogens in diverse habitats while reducing the genetic repertoire that must be maintained. Bacterial chitinases appear to represent such a class of multifunctional virulence factors. These enzymes promote bacterial survival in the environment by facilitating the use of chitin as an energy source and/or by providing a competitive advantage over organisms, such as fungi, that contain chitin as a structural component (37–39). It is now apparent that for some pathogens, chitinases function to enhance bacterial fitness within mammalian hosts either by promoting colonization (8, 10–13) or, as shown in this study, by modulating host innate immune responses. Here we have provided evidence that the secreted ChiA chitinase of L. monocytogenes enhances bacterial survival within host tissues through the downregulation of iNOS activity. To our knowledge, this is the first demonstration of a bacterial chitinase influencing a mammalian host innate immune response.
Given that humans and other mammals do not produce chitin, the target of the L. monocytogenes ChiA enzyme remains unclear. While it is possible that chitin or organisms producing chitin may be found within the human gastrointestinal tract or within oral and nasal passages, L. monocytogenes ChiA must play a role in mammalian pathogenesis that is distinct from the hydrolysis of microbe- or arthropod-synthesized chitin. The virulence defect of the L. monocytogenes ΔchiA mutant was evident following intravenous inoculation based on bacterial replication within the liver and spleen, organs not anticipated to harbor exogenous sources of chitin. Molecules similar to chitin can be found associated with glycoproteins and glycolipids present on the cell surfaces of a variety of mammalian cell types (40, 41). Studies have also identified a Xenopus chitin oligosaccharide synthase that is conserved in mice and that, when expressed from a plasmid in mouse 3T3 cells, stimulated hyaluronic acid synthesis (42). A number of key molecules involved in mammalian immune responses and signal transduction cascades are glycoproteins containing (β1,4) N-acetyl glucosamine residues (43–45), and it is possible that such a target resides within a NOS2 induction pathway. Elucidation of the specific mammalian target for ChiA is a very challenging task, perhaps best illustrated by the longtime recognition of the association of mammalian chitinases with sterile inflammation and the continuing failure to identify a specific mammalian target for these enzymes. It will thus be interesting to ascertain if similar associations between chitinase activity and reductions in iNOS expression can be established for other bacterial and mammalian chitinase enzymes.
The ability of secreted ChiA to rescue the defect of L. monocytogenes ΔchiA mutant strains during mixed infections is consistent with ChiA modification of a target that influences host immunity. ΔchiA mutants have no detectable defects in host cell adherence or invasion, vacuole escape, intracellular replication, or cell-to-cell spread (9). ΔchiA mutants also exhibited robust bacterial replication in NOS2−/− mice (Fig. 5B), indicating that the mutants were not metabolically limited as the result of the loss of chitinase-dependent hydrolysis of a substrate needed for growth. It is anticipated that ChiA-dependent suppression of iNOS activity would impact multiple facets of host immunity. iNOS contributes to host resistance to L. monocytogenes (34–36, 46, 47), although it has recently been implicated in increasing the susceptibility of activated macrophages to L. monocytogenes spread from adjacent cells (48). iNOS catalyzes the synthesis of nitric oxide (NO) from arginine (49), and induced NO plays several important roles in mediating clearance of bacterial infection (34–36, 46, 47). NO is capable of causing significant membrane and protein damage, and it also functions as a mammalian signaling molecule (50–52). Prolific secretion of NO by TNF- and iNOS-producing dendritic cells (TipDCs) at sites of infection has been shown to enhance macrophage activation and bacterial clearance (53, 54). In general, the ability of L. monocytogenes ChiA to downregulate NOS2 expression would appear consistent with enhanced bacterial survival within host tissues.
Mammalian chitinases and chitin binding proteins are expressed in macrophages and in epithelial cells of the lung and digestive tract, where they provide a first line of defense against exogenous agents, including chitin-containing pathogens (55). As mentioned previously, the expression of mammalian chitinases has been associated with a number of chronic inflammatory and tissue-remodeling disorders in the absence of infection, including asthma, chronic obstructive pulmonary disease (COPD), Sjögren’s syndrome, and Alzheimer’s disease (24, 25, 55–57); however, the specific targets of mammalian chitinases in the context of these diseases have not yet been determined. Mammalian chitinases have been reported to augment Th2 inflammatory responses (25), and thus it is tempting to speculate that one function of L. monocytogenes ChiA may be to skew the host immune response from a Th1 to a Th2 inflammatory pathway, leading to the downregulation of iNOS and enhancing L. monocytogenes survival within host tissues. It will be interesting to determine if the chitinase enzymes produced by other intracellular bacterial pathogens contribute similar roles in the modulation of host immunity so as to promote bacterial infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains are listed in Table 1. The ΔchiA and inlA S192N Y369S strains were constructed using allelic exchange as previously described (9). Bacterial cultures were grown in bovine heart infusion (BHI) medium prior to in vivo experiments or in Luria broth (LB) supplemented with 0.1% N-acetylglucosamine (20).
TABLE 1 .
Bacterial strains used in this study
| Strain | Descriptiona | Genotype(s)b | Reference |
|---|---|---|---|
| NF-L100 | Wild type 10403S parent strain | 59 | |
| NF-L1593 | In-frame deletion of chiA in NF-L100 | ΔchiA | 9 |
| NF-L1772 | 10403S with inlA S192N Y376S | inlA S192N Y376S | This work |
| NF-L1980 | NF-L1593 with inlA S192N Y376S | ΔchiA inlA S192N Y376S | This work |
| NF-L1801 | NF-L1593 with pPL2-chiA | ΔchiA + pPL2-chiA | 9 |
| NFL-1941 | NF-L1593 with pPL2-chiA E163M | ΔchiA + pPL2-chiA E163M | This work |
| DP-L3903 | 10403S with silent Tn917 insertion | WT Ermrc | 30 |
All strains are derived from L. monocytogenes 10403S.
Description of strains as provided within the text.
Erythromycin-resistant strain.
Construction of pPL2-chiA E163M plasmid.
Site-directed mutagenesis was used to introduce a methionine residue in place of glutamate at position 163 of ChiA using plasmid pPL2-chiA (9) and the Change-IT multiple-mutation site-directed mutagenesis kit (USB) with primer E163M (5′ GGATTAGACATCGACTTAATGCAAAGTGCGATTACCGCGGGA 3′).
Mouse infection models.
Swiss Webster, C57BL/6, and C57BL/6 NOS2−/− mice were obtained from Harlan Labs (Swiss Webster) or Jackson Laboratory (C57BL/6 strains). Oral and intravenous infections of 7- to 8-week-old mice were carried out as described elsewhere (58). Mixed infections were carried out as described previously (59) with a 1:1 mixture of the wild-type DP-L3903 (30) and ΔchiA strains (total bacterial CFU = 2 × 104) in 200 µl phosphate-buffered saline (PBS) injected via the tail vein.
Chitinase and chitin binding assays.
The chitinase and chitin binding assays were carried out using bacterial culture supernatants derived from overnight cultures as described elsewhere (17).
RNA analysis.
Mice (7- to 8-week-old Swiss Webster) were intravenously infected as described previously (58) for 48 h. Livers were removed and homogenized in the presence of Trizol (Invitrogen) and processed as directed by the manufacturer. cDNA synthesis was performed with Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) and oligo(dT) (Invitrogen) as directed. Quantitative PCR (qPCR) was performed with SYBR Advantage mix (Clontech) using oligonucleotides specific for gapdh, nos2, IL-1β, and tnfa (IDT).
ACKNOWLEDGMENTS
We thank Omar Jawald for help with the construction of the chiA E163M mutation.
This work was supported by Public Health Service grants AI076693 (to N.P.C. and N.E.F.) and AI083241 (to N.E.F.) from NIAID.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the funding sources.
Footnotes
Citation Chaudhuri S, Gantner BN, Ye RD, Cianciotto NP, Freitag NE. 2013. The Listeria monocytogenes ChiA chitinase enhances virulence through suppression of host innate immunity. mBio 4(2):e00617-12. doi:10.1128/mBio.00617-12.
REFERENCES
- 1. Czuprynski CJ. 2005. Listeria monocytogenes: silage, sandwiches and science. Anim. Health Res. Rev. 6:211–217 [DOI] [PubMed] [Google Scholar]
- 2. Xayarath B, Freitag NE. 2012. Optimizing the balance between host and environmental survival skills: lessions learned from Listeria monocytogenes. Future Microbiol. 7:839–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. CDC 2003. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—selected sites, United States. MMWR Morb. Mortal. Wkly. Rep. 53:338–343 [PubMed] [Google Scholar]
- 4. CDC 2009. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states. MMWR Morb. Mortal. Wkly. Rep. 58:333–337 [PubMed] [Google Scholar]
- 5. CDC 2011. Multistate outbreak of listeriosis associated with Jensen farms cantaloupe—United States, August–September 2011. MMWR Morb. Mortal. Wkly. Rep. 60:1357–1358 [PubMed] [Google Scholar]
- 6. Cossart P, Toledo-Arana A. 2008. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10:1041–1050 [DOI] [PubMed] [Google Scholar]
- 7. Cossart P. 2011. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 108:19484–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. 2008. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect. Immun. 76:4968–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaudhuri S, Bruno JC, Alonzo F, III, Xayarath B, Cianciotto NP, Freitag NE. 2010. Contribution of chitinases to Listeria monocytogenes pathogenesis. Appl. Environ. Microbiol. 76:7302–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DebRoy S, Dao J, Söderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U. S. A. 103:19146–19151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jude BA, Martinez RM, Skorupski K, Taylor RK. 2009. Levels of the secreted Vibrio cholerae attachment factor GbpA are modulated by quorum-sensing-induced proteolysis. J. Bacteriol. 191:6911–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawada M, Chen CC, Arihiro A, Nagatani K, Watanabe T, Mizoguchi E. 2008. Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab. Invest. 88:883–895 [DOI] [PubMed] [Google Scholar]
- 13. Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866 [DOI] [PubMed] [Google Scholar]
- 14. Cohen-Kupiec R, Chet I. 1998. The molecular biology of chitin digestion. Curr. Opin. Biotechnol. 9:270–277 [DOI] [PubMed] [Google Scholar]
- 15. Dahiya N, Tewari R, Hoondal GS. 2006. Biotechnological aspects of chitinolytic enzymes: a review. Appl. Microbiol. Biotechnol. 71:773–782 [DOI] [PubMed] [Google Scholar]
- 16. Bhattacharya D, Nagpure A, Gupta RK. 2007. Bacterial chitinases: properties and potential. Crit. Rev. Biotechnol. 27:21–28 [DOI] [PubMed] [Google Scholar]
- 17. Leisner JJ, Larsen MH, Ingmer H, Petersen BO, Duus JO, Palcic MM. 2009. Cloning and comparison of phylogenetically related chitinases from Listeria monocytogenes EGD and Enterococcus faecalis V583. J. Appl. Microbiol. 107:2080–2087 [DOI] [PubMed] [Google Scholar]
- 18. Leisner JJ, Larsen MH, Jorgensen RL, Brondsted L, Thomsen LE, Ingmer H. 2008. Chitin hydrolysis by Listeria spp., including L. monocytogenes. Appl. Environ. Microbiol. 74:3823–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Larsen MH, Leisner JJ, Ingmer H. 2010. The chitinolytic activity of Listeria monocytogenes EGD is regulated by carbohydrates but also by the virulence regulator PrfA. Appl. Environ. Microbiol. 76:6470–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de las Heras A, Cain RJ, Bielecka MK, Vázquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14:118–127 [DOI] [PubMed] [Google Scholar]
- 22. Eurich K, Segawa M, Toei-Shimizu S, Mizoguchi E. 2009. Potential role of chitinase 3-like-1 in inflammation-associated carcinogenic changes of epithelial cells. World J. Gastroenterol. 15:5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawada M, Hachiya Y, Arihiro A, Mizoguchi E. 2007. Role of mammalian chitinases in inflammatory conditions. Keio J. Med. 56:21–27 [DOI] [PubMed] [Google Scholar]
- 24. Ober C, Chupp GL. 2009. The chitinase and chitinase-like proteins: a review of genetic and functional studies in asthma and immune-mediated diseases. Curr. Opin. Allergy Clin. Immunol. 9:401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, Elias JA. 2011. Role of chitin and chitnase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu. Rev. Physiol. 73:479–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lecuit M. 2005. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin. Microbiol. Infect. 11:430–436 [DOI] [PubMed] [Google Scholar]
- 27. Lecuit M. 2007. Human listeriosis and animal models. Microbes Infect. 9:1216–1225 [DOI] [PubMed] [Google Scholar]
- 28. Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722–1725 [DOI] [PubMed] [Google Scholar]
- 29. Wollert T, Pasche B, Rochon M, Deppenmeier S, van den Heuvel J, Gruber AD, Heinz DW, Lengeling A, Schubert WD. 2007. Extending the host range of Listeria monocytogenes by rational protein design. Cell 129:891–902 [DOI] [PubMed] [Google Scholar]
- 30. Auerbuch V, Lenz LL, Portnoy DA. 2001. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect. Immun. 69:5953–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Songsiriritthigul C, Pantoom S, Aguda AH, Robinson RC, Suginta W. 2008. Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism. J. Struct. Biol. 162:491–499 [DOI] [PubMed] [Google Scholar]
- 32. Svitil AL, Kirchman DL. 1998. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for the ecology and evolution of 1,4-beta-glycanases. Microbiology 144:1299–1308 [DOI] [PubMed] [Google Scholar]
- 33. Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Endres R, Luz A, Schulze H, Neubauer H, Fütterer A, Holland SM, Wagner H, Pfeffer K. 1997. Listeriosis in p47(phox−/−) and TRp55−/− mice: protection despite absence of ROI and susceptibility despite presence of RNI. Immunity 7:419–432 [DOI] [PubMed] [Google Scholar]
- 35. Myers JT, Tsang AW, Swanson JA. 2003. Localized reactive oxygen and nitrogen intermediates inhibit escape of Listeria monocytogenes from vacuoles in activated macrophages. J. Immunol. 171:5447–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shiloh MU, MacMicking JD, Nicholson S, Brause JE, Potter S, Marino M, Fang F, Dinauer M, Nathan C. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10:29–38 [DOI] [PubMed] [Google Scholar]
- 37. Bassler BL, Yu C, Lee YC, Roseman S. 1991. Chitin utilization by marine bacteria: degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276–24286 [PubMed] [Google Scholar]
- 38. Inbar J, Chet I. 1991. Evidence that chitinase produced by Aeromonas caviae is involved in biological control of soil borne plant pathogens by this bacterium. Soil Biol. Biochem. 23:973–978 [Google Scholar]
- 39. Wang S, Shih I, Liang T, Wang C. 2002. Purification and characterization of two antifungal chitinases extracellularly produced by Bacillus amyloliquefaciens V656 in a SCSP medium. J. Agric. Food Chem. 50:2241–2248 [DOI] [PubMed] [Google Scholar]
- 40. Björk S, Breimer ME, Hansson GC, Karlsson KA, Leffler H. 1987. Structures of blood group glycosphingolipids of human small intestine. A relation between the expression of fucolipids of epithelial cells and the ABO, Le and Se phenotype of the donor. J. Biol. Chem. 262:6758–6765 [PubMed] [Google Scholar]
- 41. Liu Z, Masuko S, Solakyildirim K, Pu D, Linhardt RJ, Zhang F. 2010. Glycosaminoglycans of the porcine central nervous system. Biochemistry 49:9839–9845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semino CE, Specht CA, Raimondi A, Robbins PW. 1996. Homologs of the Xenopus developmental gene DG42 are present in zebrafish and mouse and are involved in the synthesis of Nod-like chitin oligosaccharides during early embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 93:4548–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mortier A, Van Damme J, Proost P. 2008. Regulation of chemokine activity by posttranslational modification. Pharmacol. Ther. 120:197–217 [DOI] [PubMed] [Google Scholar]
- 44. Recny MA, Luther MA, Knoppers MH, Neidhardt EA, Khandekar SS, Concino MF, Schimke PA, Francis MA, Moebius U, Reinhold BB, Reinhold VN, Reinherz EL. 1992. N-glycosylation is required for human CD2 immunoadhesion functions. J. Biol. Chem. 267:22428–22434 [PubMed] [Google Scholar]
- 45. Rudd PM, Wormald MR, Stanfield RL, Huang M, Mattsson N, Speir JA, DiGennaro JA, Fetrow JS, Dwek RA, Wilson IA. 1999. Roles for glycosylation of cell surface receptors involved in cellular immune recognition. J. Mol. Biol. 293:351–366 [DOI] [PubMed] [Google Scholar]
- 46. Boockvar KS, Granger DL, Poston RM, Maybodi M, Washington MK, Hibbs JB, Kurlander RL. 1994. Nitric oxide produced during murine listeriosis is protective. Infect. Immun. 62:1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nathan CF, Hibbs JB., Jr. 1991. Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr. Opin. Immunol. 3:65–70 [DOI] [PubMed] [Google Scholar]
- 48. Cole C, Thomas S, Filak H, Henson PM, Lenz LL. 2012. Nitric oxide increases susceptibility of Toll-like receptor-activated macrophages to spreading Listeria monocytogenes. Immunity 36:807–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. MacMicking J, Xie QW, Nathan C. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323–350 [DOI] [PubMed] [Google Scholar]
- 50. Fang FC. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 99:2818–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820–832 [DOI] [PubMed] [Google Scholar]
- 52. Fang FC, Vazquez-Torres A. 2002. Nitric oxide production by human macrophages: there’s NO doubt about it. Am. J. Physiol. Lung Cell. Mol. Physiol. 282:L941–L943 [DOI] [PubMed] [Google Scholar]
- 53. Serbina NV, Jia T, Hohl TM, Pamer EG. 2008. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 26:421–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi C, Pamer EG. 2011. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11:762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee CG. 2009. Chitin, chitinases and chitinase-like proteins in allergic inflammation and tissue remodeling. Yonsei Med. J. 50:22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Greenwell-Wild T, Moutsopoulos NM, Gliozzi M, Kapsogeorgou E, Rangel Z, Munson PJ, Moutsopoulos HM, Wahl SM. 2011. Chitinases in the salivary glands and circulation of patients with Sjögren’s syndrome: macrophage harbingers of disease severity. Arthritis Rheum. 63:3103–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Watabe-Rudolph M, Song Z, Lausser L, Schnack C, Begus-Nahrmann Y, Scheithauer MO, Rettinger G, Otto M, Tumani H, Thal DR, Attems J, Jellinger KA, Kestler HA, von Arnim CA, Rudolph KL. 2012. Chitinase enzyme activity in CSF is a powerful biomarker of Alzheimer disease. Neurology 78:569–577 [DOI] [PubMed] [Google Scholar]
- 58. Alonzo F, III, Port GC, Cao M, Freitag NE. 2009. The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect. Immun. 77:2612–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bruno JC, Jr, Freitag NE. 2010. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One 5:e15138 http://dx.doi.org/10.1371/journal.pone.0015138 [DOI] [PMC free article] [PubMed] [Google Scholar]