ABSTRACT
Salmonella and Escherichia coli mannose-binding type 1 fimbriae exhibit highly similar receptor specificities, morphologies, and mechanisms of assembly but are nonorthologous in nature, i.e., not closely related evolutionarily. Their operons differ in chromosomal location, gene arrangement, and regulatory components. In the current study, we performed a comparative genetic and structural analysis of the major structural subunit, FimA, from Salmonella and E. coli and found that FimA pilins undergo diverse evolutionary adaptation in the different species. Whereas the E. coli fimA locus is characterized by high allelic diversity, frequent intragenic recombination, and horizontal movement, Salmonella fimA shows structural diversity that is more than 5-fold lower without strong evidence of gene shuffling or homologous recombination. In contrast to Salmonella FimA, the amino acid substitutions in the E. coli pilin heavily target the protein regions that are predicted to be exposed on the external surface of fimbriae. Altogether, our results suggest that E. coli, but not Salmonella, type 1 fimbriae display a high level of structural diversity consistent with a strong selection for antigenic variation under immune pressure. Thus, type 1 fimbriae in these closely related bacterial species appear to function in distinctly different physiological environments.
IMPORTANCE
E. coli and Salmonella are enteric bacteria that are closely related from an evolutionary perspective. They are both notorious human pathogens, though with somewhat distinct ecologies and virulence mechanisms. Type 1 fimbriae are rod-shaped surface appendages found in most E. coli and Salmonella isolates. In both species, they mediate bacterial adhesion to mannose receptors on host cells and share essentially the same morphology and assembly mechanisms. Here we show that despite the strong resemblances in function and structure, they are exposed to very different natural selection environments. Sequence analysis indicates that E. coli, but not Salmonella, fimbriae are subjected to strong immune pressure, resulting in a high level of major fimbrial protein gene shuffling and interbacterial transfer. Thus, evolutionary analysis tools can provide evidence of divergent physiological roles of functionally similar traits in different bacterial species.
Introduction
Type 1 fimbriae are fibrillar surface appendages that mediate mannose-sensitive bacterial interactions with host cells. They are expressed by many members of the Enterobacteriaceae, including Salmonella enterica and Escherichia coli (1, 2). These adhesive structures are encoded by fim gene clusters and assembled on the bacterial surface via the chaperone/usher pathway (3–5). The shaft of type 1 fimbriae is formed by helically arranged major structural protein FimA subunits (up to 3,000 copies) and distally located minor structural subunits, including a single copy of the tip-associated adhesin FimH. The FimH adhesin is responsible for binding to target receptors, exhibiting specificity for oligosaccharides containing terminal mannose residues (6, 7).
Despite similarities in function, morphology, and biogenesis, Salmonella and E. coli type 1 fimbriae have been found to be nonorthologous and independently acquired by these bacteria (8, 9). Their fim operons are distinctly located on chromosomes and differ in gene organization and composition. The differences apply especially to loci encoding regulatory proteins that, consequently, determine diverse mechanisms controlling type 1 fimbrial phase-variable expression (10–13). It is still unclear why, in these two closely related bacteria, the same adhesive properties are performed by independently acquired fimbrial operons. This is especially puzzling because an ortholog of the Salmonella fim gene cluster designated sfm (Salmonella-like fimbriae) is present at the corresponding location in the chromosome of many E. coli strains. However, sfm fimbriae in E. coli were shown to display no affinity for α-d-mannosides when expressed in a recombinant system (14) and are either nonfunctional or have acquired a different adhesive specificity.
Type 1 fimbriae of both Salmonella and E. coli were demonstrated to contribute to pathogenesis by mediating adhesion to a variety of host cells, including epithelial, endothelial, and lymphoidal cells, and subsequent internalization in these cells (15–20). Recently, interactions of type 1 fimbriae with specific host cell receptors were also shown to play a critical role in initiation and modulation of innate and adaptive immune responses (21–23). Type 1 fimbriae, and particularly FimA, as an abundant surface protein, were shown to be potent targets for host immunity (24–28). Although type 1 fimbriae from both species were shown to elicit a strong immune response, the use of E. coli type 1 fimbriae as vaccine antigens in most cases failed to confer efficient protection against infections (28–30). This was suggested to be due to high antigenic heterogeneity of E. coli FimA. In contrast, considerable antigenic conservation was observed for type 1 fimbriae of many different Salmonella serovars (2, 31, 32). These observations together may indicate that the major structural components of these fimbriae evolve under diverse (host) environmental conditions in different species. However, little direct evidence exists to support this hypothesis.
In the present study, we performed a comparative phylogenetic and structural analysis of the fimA genes from S. enterica and E. coli and investigated possible mechanisms of adaptive evolution in these two major structural subunits.
RESULTS
Nonorthologous nature of fim genes in S. enterica and E. coli.
The fim operons in genome sequences of model strains of Salmonella enterica subsp. I strain LT2 (serovar Typhimurium) and E. coli strain MG1655 (K-12 derivative) were analyzed (Fig. 1). Besides different regulatory gene compositions in fim operons of Salmonella (fimZ-fimW) and E. coli (fimBE), sequence identity between the genes encoding corresponding structural proteins was lower for Salmonella fim and E. coli fim than for Salmonella fim and another set of fimbrial genes in E. coli called the salmonella-like fimbrial (sfm) operon. The protein sequence identity between Salmonella FimA and E. coli FimA for the major pilin subunits was 53%, while it was 65% between Salmonella FimA and SfmA. More importantly, fim operons in Salmonella and E. coli were positioned in different chromosomal locations (see Fig. S1 in the supplemental material) and were flanked by nonhomologous genes (Fig. 1). In contrast, Salmonella fim and E. coli sfm were in essentially the same chromosomal location and flanked by highly homologous genes (with the exception of prophage-related insertions immediately downstream of tRNA-Arg at the 3′-flanking region).
FIG 1 .
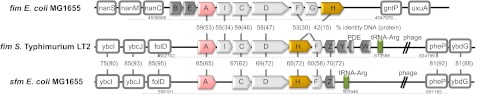
Genetic organization of fim operons in S. enterica serovar Typhimurium and E. coli K-12 and of the sfm operon of E. coli K-12. The fimA (sfmA) and fimH (sfmH) genes are shown in pink and yellow, respectively. The remaining structural fimbrial genes are shown in light gray. The genes involved in regulation are shown in dark gray and tRNA-Arg genes in green. The percent identity for corresponding DNA and protein sequences was determined by pairwise alignment (BioEdit). Chromosomal locations of the operons are designated by numbers below the designations of the neighboring genes of the operons (boxed). “PDE” indicates gene STM0551 encoding phosphodiesterase. The phage-related genes are marked by double slashes (//).
No genes homologous to the E. coli MG1655 fim operon or genes with relative high homology to E. coli fimA could be found in the Salmonella LT2 genome.
Thus, fim operons and their pilin subunits were of a nonorthologous nature in E. coli MG1655 and Salmonella LT2, with the E. coli MG1655 sfm genes being orthologous to the latter, indicating that the fim genes have independent evolutionary histories in these two species.
Sequence diversity of fimA in S. enterica and E. coli strains.
The variability of complete sequences of the fimA locus from 55 strains of S. enterica subsp. I was analyzed and compared to that of internal regions of their housekeeping genes (aroC, thrA, and hisD) that are commonly used as part of multilocus sequence typing (MLST) schemes. In Salmonella subsp. I, sequence analysis revealed 24 unique alleles of fimA with average pairwise diversity π = 1.4%, which was comparable to the nucleotide diversity of three concatenated Salmonella housekeeping genes (Table 1). When fimA and housekeeping loci from 11 strains of other five subspecies (II, IIIa, IIIb, IV, and VI) were included in the analysis, the diversity of fimA increased almost 3-fold (to π = 3.9 ± 0.12), but it correlated with an increase of the diversity in the housekeeping genes (to π = 3.2 ± 0.08) (Table 1).
TABLE 1 .
Nucleotide diversity of fimA and 3-locus MLST in S. enterica and E. coli
| Gene category |
S. entericaa |
E. colia |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subsp. I (55) |
Subsp. I–VI (66) |
Entire species (53) |
B2 group (38) |
|||||||||
| π (%) | dS | dN | π (%) | dS | dN | π (%) | dS | dN | π (%) | dS | dN | |
| 3-locus housekeeping genesb | 1.4 ± 0.02 | 0.055 | 0.001 | 3.2 ± 0.08 | 0.132 | 0.004 | 1.7 ± 0.05 | 0.075 | 0.001 | 0.5 ± 0.02 | 0.019 | 0.001 |
| fimA | 1.4 ± 0.04 | 0.035 | 0.007 | 3.9 ± 0.12 | 0.125 | 0.015 | 7.9 ± 0.14 | 0.222 | 0.041 | 7.8 ± 0.14 | 0.215 | 0.041 |
Numbers in parentheses represent numbers of analyzed sequences.
Data represent the results determined for concatenated sequences of internal (450- to 500-bp) fragments of the housekeeping genes from S. enterica (thrA, aroC, and hisD) and E. coli (adk, fumC, and gyrB) MLST schemes.
The nucleotide diversity of fimA in 53 strains of E. coli (comprising representative strains of the entire species) was on average four times higher (π = 7.9%) than the diversity of housekeeping genes from the E. coli MLST scheme (adk, gyrB, and fumC). While the housekeeping gene diversity of E. coli (1.7%) was only slightly higher than that of S. enterica subsp. I (1.4%), the E. coli fimA diversity was as much as 5-fold higher (Table 1). Importantly, while the housekeeping gene diversity of all S. enterica subspecies was twice as high as in E. coli, the fimA diversity of the former was half that of the latter. In addition, E. coli fimA was characterized by the highest rates of synonymous (dS) and nonsynonymous (dN) values compared to E. coli and Salmonella genes (Table 1). Interestingly, when a subgroup of E. coli strains belonging to phylogenetic B2 group was analyzed, the average nucleotide diversity of their housekeeping genes was much lower than that seen even in S. enterica subsp. I strains (π = 0.5%) but fimA diversity remained as high as in the entire E. coli species (7.8%).
Thus, while fimA diversity in Salmonella is on par with the diversity of housekeeping genes, E. coli fimA is significantly more diverse and its diversity appears to be independent of the level of housekeeping gene diversity within individual phylogenetic groups of E. coli.
Interstrain movement of fimA in both species.
The fact that the fimA diversity of phylogenetic group B2 strains of E. coli was the same as in the entire species prompted us to assess the horizontal movement (homologous exchange) of fimA within both E. coli and S. enterica. Maximum-likelihood (ML) trees were constructed based on aligned fimA and concatenated 3-locus MLST sequences of Salmonella and E. coli followed by the analysis of their congruence. Based on the three housekeeping loci used, a total of 40 different MLST sequence types (STs) were identified among Salmonella study strains, and at least 12 of the STs were represented by at least two strains (Fig. 2A). In general, Salmonella strains with the same ST represented the same serovar. Comparison of the STs and the fimA trees in S. enterica (Fig. 2B) showed that though the topologies of these trees are not identical, strains of the same ST had the same fimA allele. This was observed for all 12 ST/fimA clades, indicating limited movement of S. enterica subsp. I fimA between different strains. Also, no signature of the fimA horizontal movement between strains of different S. enterica subspecies (I to VI) was observed, as strains of the same subspecies clustered on both MLST tree and the corresponding fimA tree.
FIG 2 .
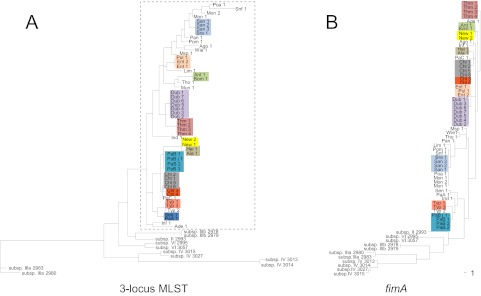
Maximum-likelihood DNA phylograms of concatenated 3-locus MLST and fimA sequences of S. enterica. The MLST (A) and fimA (B) trees were constructed based on an alignment of 66 sequences obtained for S. enterica subsp. I to VI. The colored boxes mark strain clades with identical sequence types (STs). The S. enterica subsp. I clade is boxed (gray dashed line). The scale at the bottom of the phylograms indicates phylogenetic distance and corresponds to a 1-nucleotide difference.
In E. coli, in contrast, where 25 STs were found, with 9 represented by two or more strains, (Fig. 3A), fimA alleles from strains of the same ST were distributed in different clades on the corresponding fimA tree (Fig. 3B). This was observed for 4 of 9 STs (P = 0.02), indicating that, unlike Salmonella, the E. coli fimA locus frequently moves horizontally between phylogenetically distinct isolates. Importantly, horizontal movement of highly diverse fimA alleles was specifically notable in phylogenetic group B2 strains clustered into a single clade on the E. coli MLST tree (boxed in Fig. 3A).
FIG 3 .
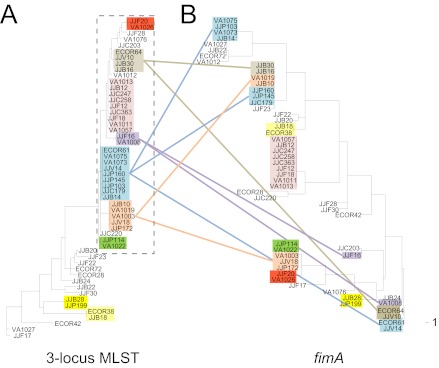
Maximum-likelihood DNA phylograms of concatenated 3-locus MLST and fimA sequences of E. coli. The MLST (A) and fimA (B) trees were constructed based on an alignment of 53 sequences obtained for E. coli. The colored boxes mark strain clades with identical sequence types (STs). Cross-connecting lines indicate corresponding MLST strain clades that carry different fimA haplotypes. The E. coli clade of strains with very low MLST sequence diversity (the major clade in the MLST tree) is boxed (gray dashed line). The scale at the bottom of the phylograms indicates phylogenetic distance and corresponds to a 1-nucleotide difference.
Thus, E. coli fimA moves frequently within clonally related strains, while Salmonella fimA tends to stay within the clonally related strain groups, without evidence of horizontal movement.
Whole-genome sequence analysis of fimA diversity in S. enterica and E. coli.
The observations made by comparing fimA and three housekeeping loci were validated by a comparative analysis of a broader set of genes using publicly available fully assembled genome sequences of 44 E. coli and 24 S. enterica subsp. I strains (see Fig. S2A and B in the supplemental material). We compared diversities of each species by using a combination of 6 housekeeping loci. We found that the 6-locus-based phylogenies generally corresponded to the 3-locus-based phylogenies (see Fig. S2A and B in the supplemental material), with the E. coli B2 group strains remaining clustered together (see Fig. S2A in the supplemental material). Also, according to the 6-locus phylogeny, fimA alleles move between clonally distinct strains in E. coli more frequently than in Salmonella (see Fig. S2A and B in the supplemental material). Interestingly, the 6-locus-based nucleotide diversity of S. enterica subsp. I strains did not change relative to the 3-locus diversity, while the diversity of E. coli in general and, specifically, of the B2 strains increased (Table 2). Still, the E. coli fimA diversity remained significantly higher than that of the housekeeping genes, both overall and in the B2 strains. Furthermore, we analyzed the diversity of another gene in the corresponding fim operons—fimC, coding for the molecular chaperone of FimA and other structural subunits—and found it to be at the same level as that of the housekeeping genes in both species (Table 2).
TABLE 2 .
Nucleotide diversity of fim and sfm in publicly available fully assembled genome sequences of S. enterica and E. coli
| Gene category |
S. enterica subsp. I (24)a |
E. colia |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Entire species (44) |
B2 group (15) |
||||||||
| π (%) | dS | dN | π (%) | dS | dN | π (%) | dS | dN | |
| 3-locus housekeeping genesb |
1.3 ± 0.05 | 0.049 | 0.001 | 1.8 ± 0.05 | 0.077 | 0.001 | 0.5 ± 0.04 | 0.02 | 0.001 |
| 6-locus housekeeping genesc |
1.2 ± 0.04 | 0.047 | 0.001 | 2.7 ± 0.05 | 0.117 | 0.002 | 1.3 ± 0.07 | 0.053 | 0.002 |
| fimA | 1.2 ± 0.07 | 0.031 | 0.006 | 7.8 ± 0.21 | 0.217 | 0.041 | 8.6 ± 0.62 | 0.246 | 0.045 |
| fimC | 1.2 ± 0.05 | 0.032 | 0.006 | 1.3 ± 0.03 | 0.042 | 0.004 | 1.1 ± 0.01 | 0.034 | 0.004 |
| sfmAd | Not applicable | 0.7 ± 0.06 | 0.025 | 0.002 | Not available | ||||
| sfmCe | Not applicable | 1.5 ± 0.06 | 0.047 | 0.005 | Not available | ||||
Numbers in parentheses represent numbers of analyzed sequences.
Data represent the results determined for concatenated sequences of internal (450- to 500-bp) fragments of the housekeeping genes (thrA, aroC, and hisD for S. enterica and adk, fumC, and gyrB for E. coli).
Data represent the results determined for concatenated sequences of internal (450- to 500-bp) fragments of the housekeeping genes (thrA, aroC, hisD, adk, fumC, and gyrB for both S. enterica and E. coli).
Data represent the results for 29 sfmA sequences determined in 44 E. coli strains.
Data represent the results for 28 sfmC sequences determined in 44 E. coli strains.
Also, based on the genome sequences of E. coli strains, we analyzed the diversity of E. coli sfmA, which is evolutionarily more closely related to Salmonella fimA than to E. coli fimA. sfmA was present in the genomes of 29 (65%) E. coli strains, but, interestingly, none of them belonged to the B2 group (see Fig. S2A in the supplemental material). Also, all Shigella strains carried sfmA but were missing sfmC or/and sfmH, homologous to Salmonella fimC and fimH (coding for the molecular chaperone and adhesive subunit, respectively), suggesting that the sfm operon in these strains is not complete (see Fig. S2A in the supplemental material). The nucleotide diversity of sfmA was even lower than that of fimA in S. enterica subsp. I (π = 0.72% ± 0.025), while the nucleotide diversity of sfmC (where available) was comparable to that of fimC in both species (Table 2). Also, the phylogenetic analysis of sfmA showed limited horizontal movement of sfmA between different E. coli strains (data not shown).
Considering the patchy distribution of sfm operon in E. coli, we also looked throughout the publicly available S. enterica genomes for possible homologues of the E. coli-like fim operon. Based on sequence identity, operon structure, and chromosomal position, no complete or partial presence of an E. coli-like fim operon was detected in any of the 24 S. enterica subsp. I genomes or in the only available genome for other subspecies—that of S. enterica subsp. IIIa strain RKS 2980 (not shown).
Thus, based on the expanded set of genes from strains with whole-genome sequence available, we confirmed the much higher diversity and more frequent horizontal movement of E. coli fimA relative to Salmonella fimA and E. coli sfmA.
Nonsynonymous variability and recombinational shuffling of fimA.
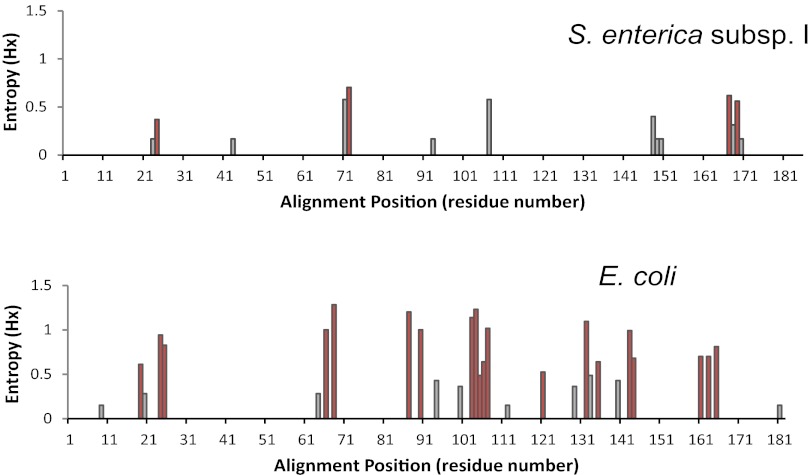
We next compared the levels of nonsynonymous variability of fimA in both species in detail by analyzing the distribution of variable amino acid positions across the protein sequences of S. enterica subsp. I and E. coli FimA (Fig. 4). There was a significant difference between Salmonella and E. coli in the numbers of variable positions in FimA (14 versus 29) and in the level of amino acid variability in these positions. A large majority of polymorphic sites in E. coli and only a minority in Salmonella FimA represented hot spots, indicating multiple independently acquired amino acid substitutions at these positions (Fig. 4, red bars). Also, though the polymorphic sites were in general equally distributed along entire length of FimA from each species, some local clustering of polymorphic sites was observed. Of note, single-amino-acid deletions (in positions 168 and 169) in Salmonella and two-amino-acid insertions (between residues 26 and 27) in E. coli were also present.
FIG 4 .
Distribution of amino acid variability along FimA sequence alignments of S. enterica subsp. I and E. coli. The amino acid variability across the FimA sequence is represented by the Shannon entropy plot as determined by BioEdit software. The entropy values refer to the complexity in amino acid composition at the position in the sequence alignment, taking into account the numbers and frequencies of different amino acids observed for each position. Red bars indicate sites of hot spot mutations. For Salmonella and E. coli, 24 and 28 FimA structural variants were analyzed, respectively.
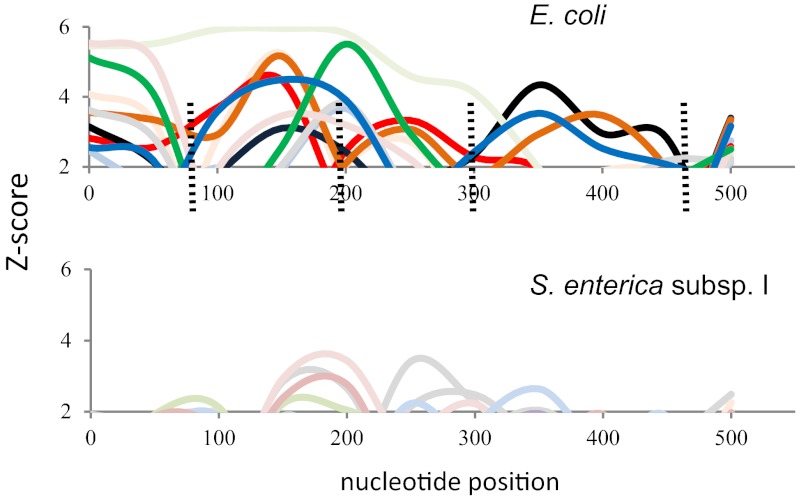
To analyze to what extent the structural polymorphism in pilin sequences results from mutation versus recombination (intragenic shuffling) events, we tested the presence of intragenic recombination regions in fimA using the MaxChi statistic (33). The analysis showed strong signals of recombination in E. coli fimA (MaxChi P = 0.001) but failed to indicate any evidence of recombination in S. enterica subsp. I fimA (MaxChi P = 0.95). In addition, we randomly selected 10 sets of triplet sequences (see Materials and Methods for details) for both E. coli and Salmonella fimA. On each triplet set, we applied MaxChi and SiScan (34) for detection of recombination. There was a clear (P < 0.05) signal of recombination in 5 of the 10 randomly chosen triplet sets from E. coli (Fig. 5; see also Fig. S3A in the supplemental material). In contrast, none of the Salmonella triplet sets (Fig. 5; see also Fig. S3B in the supplemental material) provided significant signals of recombination (P < 0.01).
FIG 5 .
Intragenic recombination in E. coli and S. enterica subsp. I fimA as determined by SiScan and MaxChi tests. Combined Z score plots (showing values higher than the threshold score of 1.96, i.e., P < 0.05) obtained for 10 sets of randomly chosen triplet sequences of E. coli and Salmonella are shown. The sequence triplets with recombination signal are shown in bold colors (red, orange, green, blue, and black). The sequence triplets without recombination signal are shown in light colors. Dashed lines indicate frequent recombination breakpoints. Graphs presenting Z score plots for each sequence triplet separately are shown in Fig. S3A and B in the supplemental material. The colors used here correspond to those used in the supplemental figures for these triplets.
Thus, the variability of the Salmonella FimA primary structure appears to result primarily from acquired mutations, whereas in E. coli FimA polymorphism is due to both mutations and intragenic shuffling.
Distribution of amino acid changes in the tertiary structure of fimA.
While the E. coli FimA structure has been resolved (Protein Data Bank [PDB] 2JTY) (35), the structure of Salmonella FimA has not been reported. A prediction of the Salmonella FimA three-dimensional (3D) structure was obtained for the S. Typhimurium LT2 sequence (including residues 23 to 185) by homology modeling using the protein fold recognition server Phyre2 (36). The Phyre2 results indicated that the E. coli FimA structure (PDB 2JTY) is the template ranked best, with 52% sequence identity and 98% sequence coverage. The confidence level for the matching was 100%. The solved E. coli FimA structure represents a self-complemented FimA protein variant (scFimA) in which the C terminus is fused with an additional donor strand. As scFimA mimics the state of wild-type FimA in the context of the quaternary structure of the fimbrial rod, Salmonella FimA was modeled accordingly (see Materials and Methods). The resulting query-template primary sequence alignment with the secondary-structure motifs and predicted Salmonella FimA structural model is presented in Fig. 6. As shown in the alignment, the predicted secondary folds for Salmonella are remarkably conserved and fit closely overall to the solved secondary structure of E. coli.
FIG 6 .
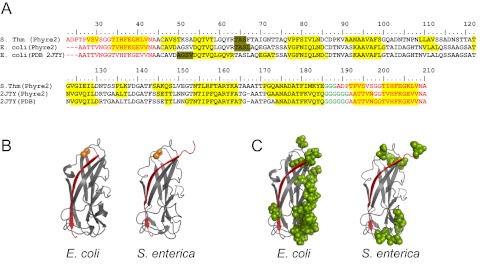
Primary, secondary, and tertiary structures of Salmonella and E. coli FimA. (A) Primary sequence alignment with predicted (Phyre2) and solved (PDB 2JTY) secondary motifs of S. Typhimurium (S. Thm) and E. coli FimA. Beta strands are indicated as yellow highlights; α helices are marked in light brown. Cysteine residues are shown in blue. The sequences of N-terminal extensions and added donor strands are in red, and sequences of glycine linkers used to construct the self-complemented subunits are in green. (B) Ribbon representation of the 3D structure of E. coli FimA (PDB 2JTY) and the predicted Salmonella FimA model. The orange spheres represent C-α atoms of cysteine residues. (C) Distribution of structural mutations in tertiary structures of E. coli and Salmonella FimA. The amino acids at variable sites are shown as green spheres.
As viewed by PyMol (Fig. 6B), the predicted Salmonella FimA model retains a complete Ig fold, where the added donor strand (red) occupies the hydrophobic groove as observed in E. coli FimA 2JTY. Moreover, similar to those in E. coli, cysteine residues in the Salmonella model are juxtaposed in a position suitable for the formation of a stabilizing disulfide bond (Fig. 6B, orange spheres).
Using the E. coli FimA structure and Salmonella FimA model, we next compared spatial distributions of variable positions in the major subunits from these two species (Fig. 6C). The analysis revealed that, in E. coli, the amino acid substitutions predominantly target one side of FimA molecule opposite the position of the complementing donor strand (red). According to the previously reported fimbrial rod model, the donor strand faces the inside core of the fimbrial rod (37). Thus, the highly polymorphic loops are predicted to be exposed on the outer surface of the fimbrial rod (Fig. 6C). In contrast, in Salmonella, the variable positions tend to cluster on the top (residues 23, 24, 43, 93, 148, 149, and 150) and the bottom (residues 167, 168, 169, and 170) of the immunoglobulin-like fold, with only sporadic occurrence (residues 71, 72, and 107) in external loops.
The helical structure of the type 1 fimbriae with its extensive subunit-subunit interactions raises the possibility that mutations in FimA residues might occur in pairs and affect residues that interact with one another in the intersubunit interface, representing compensatory intragenic suppression. We determined the number of events where mutations were acquired in pairs by E. coli and Salmonella fimA per total number of mutation events using Zonal phylogeny (ZP) software (38) that provides branch-specific information on nonsynonymous substitutions in the phylogenetic tree. For Salmonella fimA we detected that 2 of 34 mutations and, similarly, for E. coli we detected that 2 of 44 mutations represented events where mutations were likely acquired in pairs, indicating that intergenic suppression does not play a significant role in the structural diversification of FimA in either species.
Thus, there is a distinct structural pattern of the amino acid changes in the E. coli FimA and the Salmonella FimA, with the variable residues in E. coli FimA concentrated in the outer surface-exposed epitopes of the subunit, i.e., putative antigenic epitopes.
DISCUSSION
FimA, as the major structural component of type 1 fimbriae, is abundantly expressed on the bacterial surface. This protein is thus expected to be a major antigen and evolve under strong selective pressure from the host immune system. In E. coli FimA, the footprint of such selection can be seen in the great allelic variation of the fimA locus evolving under strong diversification selection (8, 39). In this report, however, we show that fimA from a closely related pathogen, S. enterica, evolves under a different adaptive selection pressure with much lower diversity, indicating that fimbriated Salmonella are not subjected to strong immune pressure to vary the fimbrial structure.
In both species, type 1 fimbriae were shown to bind to mannosylated receptors on target cells, facilitating bacterial adhesion and invasion, and triggering and modulating the host immune response (22–24, 40). The mannose-specific interactions involve very similar molecular mechanisms. Despite very low (15%) sequence identity, the fimbrial tip adhesive protein of E. coli and Salmonella, FimH, has highly homologous, two-domain tertiary structures and mediates shear-dependent binding to mannose via an allosteric catch-bond mechanism (41, 42). Both adhesins evolve under positive selection for accumulation of structural mutations that greatly affect their adhesive properties, with markedly similar distribution patterns of functional mutations in the corresponding tertiary structures (43–46). In particular, mutations that enhance mannose binding under static conditions (and are common among uropathogenic E. coli and systemically invasive Salmonella serovars) are primarily localized to the interdomain interface of FimH, exerting an allosteric effect on binding pocket affinity (46–49).
In contrast to the high structural and functional similarities of the adhesive subunits, we found that FimA, the major structural subunit from Salmonella and E. coli, displays distinct adaptive patterns. The E. coli fimA locus is characterized by a high level of allelic variation, with strong signals for intragenic recombination and frequent horizontal movement. In contrast, the Salmonella fimA locus exhibits relatively low diversity (on par with that of neutrally evolving housekeeping loci) without any strong evidence of intragenic shuffling or interstrain movement. The difference is especially obvious in comparisons of the S. enterica subsp. I and E. coli group B2 strains that were overrepresented in our analysis because of their medical significance. Both represent phylogenetic clades within the corresponding species, combining relatively closely related strains with distinct virulence characteristics. S. enterica subsp. I is comprised of serovars that cause gastroenteritis and/or systemic infections in humans, while E. coli group B2 strains are notorious for their ability to cause urinary tract infections, sepsis, meningitis, and other extraintestinal infections. From the perspective of general genomic characteristics, there is a similarity between these subspecies groups, as shown in our previous genome-wide comparative study of Salmonella and E. coli (38). It was shown that S. enterica subsp. I and the E. coli B2 group strains are very similar in regard to the level of nucleotide diversity of core genes (1% in both species) and the prevalence of mosaic versus core genes (33% versus 63% in S. enterica subsp. I and 25% versus 64% of the total genes in E. coli group B2 strains).
In the data set analyzed here, based on MLST housekeeping gene analyses, B2 E. coli strains had levels of genetic diversity either lower than or similar to those of S. enterica subsp. I. However, the average diversity of fimA alleles in E. coli was 5- to 6-fold higher than that of fimA in the Salmonella isolates, with a distinctively higher rate of horizontal allelic exchange in the former. This indicates that the lower diversity of S. enterica subsp. I fimA does not simply result from the overall lower genetic diversity of S. enterica subsp. I strains.
In E. coli, the functional relevance of the diversifying selection in fimA locus was verified by structural studies. Using a predicted model of type 1 fimbriae, it has been demonstrated that the variable amino acid residues of FimA are predominantly located on the external surface of the fimbrial rod (37). In our study, to compare the spatial distributions of polymorphic sites of Salmonella FimA and E. coli FimA, we used 3D structures of the pilin monomers that included the recently solved structure of E. coli FimA (PDB 2JTY) (35) and we obtained here by homology modeling a putative structure of Salmonella FimA. This analysis revealed that the majority of the amino acid substitutions in E. coli targeted loops on one side of FimA molecule which, according to the previously reported model (37), are exposed on the external surface of the fimbrial rod, thereby strongly supporting the view of antigenic variation in E. coli FimA. In Salmonella, in contrast, the variable positions were detected on the opposite poles of the immunoglobulin-like fold, i.e., on the top and the bottom of FimA, which are more likely to be positioned in the intersubunit interface than surface exposed. In two proposed models of the type 1 fimbrial structure (37, 50), the fimbrial shaft is formed by a helically coiled string of FimA subunits that are connected to each other “head to tail.” Although at this point we still cannot exclude the possibility of involvement of these mutations in epitope diversification, it is clear that there is a significant difference in the structural variabilities of corresponding FimA segments, suggesting that Salmonella FimA evolves under weaker selective pressure for antigenic diversification than E. coli FimA.
Salmonella and E. coli are known to inhabit many diverse niches (hosts, tissues, cellular environments). It is thus possible that immune pressure could act differently on type 1 fimbriae in different niches. However, it is equally likely that the differences in the antigenic diversifications of these two pilins may result from distinct mechanisms of regulation of type 1 fimbrial expression in these bacteria. Both Salmonella and E. coli have been shown to switch between sparsely and highly fimbriated states in response to various environmental signals, in part to escape host immunity. In E. coli, on/off switching of fimbrial expression is determined by the orientation of the promoter-containing DNA region (fimS) (10, 12), where the inversion of the fimS DNA segment is catalyzed by two site-specific recombinases (encoded by fimB and fimE) (51). In Salmonella, in contrast, the promoter is not invertible, but its transcriptional activity is controlled by regulatory proteins encoded by fimZ, fimY, and fimW located downstream of the operon (13, 52–54). It could be speculated that the differences in the regulatory mechanisms of type 1 fimbrial expression in these two bacteria may determine the different efficiencies of phase switching and thus provide different levels of protection against host immune defenses. In this context, the distribution pattern of structural mutations observed for Salmonella FimA may suggest that mutations found at the top and the bottom of the pilin subunit (and thus in the subunit contact interface) could affect FimA-FimA interactions during fimbrial polymer formation and consequently be functionally adaptive for efficient “phase” switching.
On the other hand, it has been demonstrated that in E. coli, the FimA polymer has the ability to coil and uncoil under the influence of mechanical forces (55). Since bacterial adhesion usually occurs in the presence of flowing bodily fluids that create drag forces on bacteria and their adhesins, the mechanical properties of fimbrial shaft exert a significant effect on bacterial adhesion. In this aspect, the mutations located in the intersubunit interface of Salmonella FimA (as well as in that of E. coli FimA) could modulate mechanical properties of the fimbrial shaft under the influence of shear forces and consequently affect mannose-specific adhesion of the bacteria under flow conditions. Although this hypothesis requires experimental verification, a similar observation for enterotoxigenic E. coli (ETEC) class 5 fimbria-mediated adhesion was previously reported. It was shown that point mutations that are localized to the intersubunit interface of the major (nonadhesive) subunit of those fimbriae significantly reduced adhesion under flow conditions (56).
Distinct adaptive patterns of fimA in Salmonella and E. coli raise questions about evolutionary trajectories of these distinct types of type 1 fimbriae. The lack of E. coli-like fim genes in S. enterica indicates that this operon was either lost from or not acquired by the latter. At this point, based on the patchy distribution of either Salmonella-like or E. coli-like fim genes in other enterobacterial species (see Table S1 in the supplemental material), it is difficult to be definitive about that and further analysis is required to address the interesting issue about the evolutionary interplay of the different types of mannose-specific fimbriae in members of the family Enterobacteriaceae. Though E. coli has both fimbrial types, it appears that the Salmonella-like fimbrial operon (sfm) in E. coli either is nonfunctional or has acquired functions distinct from those of E. coli and Salmonella fim operons. Our analysis of sfm genes indicates that they are present in 65% of the E. coli genomes, with none found in strains representing the B2 group and partial inactivation in some of the other E. coli groups. Even when most of the operon is present (as in E. coli strain MG1655), the sfm genes appear to diverge more significantly from the corresponding Salmonella fim genes than the orthologous genes on the flanks. Also, we found that all sfmA genes in E. coli carry a 5′ deletion relative to the Salmonella fimA corresponding to a deletion of 5 amino acid residues at position 2 to position 6 of the amino acid sequence that is part of the donor strand crucial for FimA-FimA polymerization by beta-strand complementation. Such a structural defect could significantly affect fimbrial assembly or even abrogate it. No data with regard to natural expression of the Sfm fimbriae have been reported to date. Though it was possible to express the sfm fimbriae of E. coli K-12 from an artificial promoter, the only small afimbrial structures that were observed were structures whose functionality was not established (14). Thus, taken together, these data indicate that fim-encoded type 1 fimbriae are the only mannose-specific organelles in the E. coli species. It remains to be determined, however, whether the fim operon was acquired later than sfm and functionally replaced Salmonella-like fimbrial genes or, alternatively, whether the two traits shared a long evolutionary history in E. coli and sfm genes functionally diverged over time.
In summary, by using microevolutionary analytical tools, we demonstrate here that surface organelles that presumably perform essentially identical adhesive functions in closely related bacterial pathogens can be under highly dissimilar types of selective pressure. While the exact basis of this difference remains to be elucidated, this indicates possibly distinct ecological and/or pathogenic environments in which these organelles are functioning. Despite the genes appearing to be under relatively weak immune pressure to diversify, our results also suggest the possibility that type 1 fimbria-based vaccines may be more successful for treatment of infections by Salmonella, considering the structurally conserved nature of the major subunit. Finally and foremost, this report shows the potential power of comparative population genetic analysis in determining adaptive and, thus, functional peculiarities of specific bacterial traits in different species. This should assist us in unraveling the physiological significance and possibly pathogenic roles of the traits, especially in the age of continuous accumulation of genomic data of a large number of individual strains from the same species.
MATERIALS AND METHODS
Bacterial strains.
S. enterica and E. coli strains used in this study are listed in Tables S2 and S3 in the supplemental material, respectively. The S. enterica collection included 53 isolates of subspecies I representing 30 different serovars, including 23 strains of systemic and 30 strains of different intestinal serovars, and 11 strains of other subspecies (subspecies II to VI). The E. coli collection consisted of 53 isolates representing convenient samples of diverse pathotypes and nonpathogenic isolates. Bacteria were routinely grown overnight in LB medium at 37°C without shaking.
Gene amplification and sequencing.
Genomic DNA was isolated from S. enterica and E. coli strains using a DNeasy Blood & Tissue kit (Qiagen). The fimA was PCR amplified from the genomic DNA using the following pairs of primers: primer pair fimASe F (5′GGATGCCGAAACCGGGTG3′) and fimASe R (5′CTGTGGCGACAGCGCAGCC3′) (for S. enterica fimA) and primer pair fimAEc F (5′ACGTTTCTGTGGCTCGACGCATCT3′) and fimAEc R (5′ACGTCCCTGAACCTGGGTAGGTTA3′) (for E. coli fimA). S. enterica and E. coli housekeeping locus fragments (from aroC, hisD, and thrA and from fumC, adk, and gyrB, respectively) were amplified in accordance with the protocols available at the MLST database (http://mlst.ucc.ie/mlst/dbs/Senterica/documents/primersEnterica_html and http://mlst.ucc.ie/mlst/dbs/Ecoli/documents/primersColi_html). The PCR products were purified using ExoSAP-IT reagent (Affymetrix) in accordance with the manufacturer’s instructions and subjected to sequencing by the GENEWIZ sequencing service (Genewiz Seattle, Seattle, WA).
Phylogenetic analysis.
The nucleotide sequences were aligned using ClustalW with default settings (57). Zonal phylogeny (ZP) analysis and associated statistics were performed using Zonal Phylogeny Software (ZPS) (38). The maximum-likelihood (ML) phylograms as implemented in ZPS were generated by PAUP* 4.0 b using the general time-reversible (GTP) substitution model with codon-position-specific estimated base frequencies (58). Sequence diversity was measured as the average pairwise diversity index (π) and the rates of nonsynonymous (dN) and synonymous (dS) mutations (59) using MEGA version 4 (60). Analysis of statistical significance was performed using the z test for π and dN/dS values (61). The presence of structural hot spot mutations was determined using ZPS.
Intragenic recombination detection.
MaxChi (33) was used to provide a summary statistic for the detection of recombination, where a gene data set showing MaxChi statistic P < 0.05 was considered to have recombinant sequence(s). For three-sequence-based (triplet) analysis of recombination, we applied MaxChi along with SiScan (34), which depicts probable recombinant and parent sequences. In SiScan, Z scores were plotted based on identities of all three sequence pairs of a triplet set across the gene length using a sliding window of 100 nucleotides, a step size of 50 nucleotides, and 100 randomizations for Monte Carlo sampling. The data set that showed identity of two of the three sequence pairs with Z scores > 1.96 (i.e., significance at P < 0.05) at considerably nonoverlapping stretches across the gene length indicated putative intragenic recombination with the presence of both parents in the data set.
An in-house perl script was used to develop a random sequence number generator. In the aligned fimA allelic variant sets of E. coli and Salmonella, we assigned numbers to each gene sequence. Since Salmonella had lower number of unique sequences (i.e., 24 alleles), the program generated sets of 3 numbers (i.e., triplets) ranging from 1 to 24 sets. We incorporated the constraint that the numbers within any triplet set would differ from one another by a value of at least 5 in order to avoid considering sequences in a triplet that were phylogenetically too close. We generated 10 random triplet sets of numbers and used identical sets for the two species to choose strain sequences corresponding to each number in the aligned datasets.
FimA structural analysis and modeling.
The distribution of amino acid variability in the sequence alignment was computed by the use of BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and is represented by Shannon entropy plots (62), where the entropy data refer to the complexity in the amino acid composition at each position (taking into account the numbers and frequencies of different amino acids at the position) in the sequence alignment.
Modeling of the S. enterica FimA 3D structure was performed using Phyre2 fold recognition and template modeling software (36). Briefly, the amino acid sequence of S. Typhimurium SL1344 FimA (including residues 23 to 185) was submitted to the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The structure of E. coli FimA (PDB code 2JTY) (35) was selected as the best-ranked template for modeling of S. Typhimurium FimA, with 52% sequence identity, 100% confidence, and 98% sequence coverage. As E. coli FimA (template) represents a self-complemented variant of the protein, the C terminus of the S. Typhimurium SL1344 FimA sequence was completed with the corresponding self-donor strand sequence (ADPTPVSVSGGTIHFEGKLVNA) via the use of the (Gly)3 linker and resubmitted to the Phyre2 server. The resulting structures of the S. enterica FimA model and E. coli FimA (PDB code 2JTY) were viewed and analyzed using the molecular visualization system PyMOL.
Nucleotide sequence accession numbers.
The DNA sequences of the new S. enterica and E. coli fimA alleles have been deposited in GenBank under accession numbers KC405503 through KC405538. Accession numbers of all S. enterica and E. coli fimA alleles of the study are presented in Tables S2 and S3 in the supplemental material.
SUPPLEMENTAL MATERIAL
Location of fim and sfm operons on S. enterica serovar Typhimurium LT2 and E. coli K-12 chromosomes. The positions of fim and sfm operons (red) are shown relative to the positions of the origins of replication (ORI) and housekeeping genes (adk, hisD. thrA, fumC, gyrB, and aroC) in the genome sequences of S. Typhimurium strain LT2 (GenBank accession no. NC_003197) and E. coli K-12 strain MG1655 (GenBank accession no. U00096). Download
Maximum-likelihood DNA phylograms of concatenated 3- and 6-locus MLST and fimA sequences of E. coli and S. enterica. (A) The MLST and fimA trees were constructed based on sequences from 44 E. coli strains for which fully assembled genomes are publicly available. The colored boxes mark strain clades with identical sequence types. Cross-connecting lines indicate the same fimA allele carried by different MLST type strains (black boxes). The E. coli clade of B2 strains is boxed (gray dashed line). The red asterisk indicates the presence of sfmA, and the blue asterisk indicates that the sfm operon is not complete. The scale at the bottom of the phylograms indicates phylogenetic distance and corresponds to a one-nucleotide difference. (B) Maximum-likelihood DNA phylograms of concatenated 3- and 6-locus MLST and fimA sequences of S. enterica. The MLST and fimA trees were constructed based on sequences of 24 S. enterica subsp. I strains for which fully assembled genomes are publicly available. The colored boxes mark strain clades with identical sequence types. The scale at the bottom of the phylograms indicates phylogenetic distance and corresponds to a one-nucleotide difference. Download
Z score plot by SiScan test of recombination for 10 sets of randomly chosen triplet sequences of E. coli (A) and Salmonella (B). A particular set (1 through 10) for each species includes identical randomly generated sequence numbers (see Materials and Methods for details). In each plot, the black dotted line represents the significant threshold of P = 0.05 (for Z score = 1.96). For each triplet sequences, that MaxChi statistic P value is provided. Sets 1, 2, 5, 7, and 8 of E. coli show significant recombination signals via both MaxChi and SiScan statistics. Download
BLAST analysis of fimA in different enterobacteria.
List of Salmonella enterica strains used in this study.
List of Escherichia coli strains used in this study.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant RO1 GM084318.
We gratefully acknowledge Steve Moseley (University of Washington) for critical reading of the manuscript and helpful advice. We also thank to following individuals for providing Salmonella and E. coli strains: Ferric Fang, Stephen Libby, Mansour Samadpour, Maciej Ugorski, Roderick I. Mackie, Howard Ochman, and James R. Johnson.
Footnotes
Citation Kisiela DI, Chattopadhyay S, Tchesnokova V, Paul S, Weissman SJ, Medenica I, Clegg S, Sokurenko EV. 2013. Evolutionary analysis points to divergent physiological roles of type 1 fimbriae in Salmonella and Escherichia coli. mBio 4(2):e00625-12. doi:10.1128/mBio.00625-12.
REFERENCES
- 1. Duguid JP, Anderson ES, Campbell I. 1966. Fimbriae and adhesive properties in salmonellae. J. Pathol. Bacteriol. 92:107–138 [DOI] [PubMed] [Google Scholar]
- 2. Duguid JP, Campbell I. 1969. Antigens of the type-1 fimbriae of salmonellae and other enterobacteria. J. Med. Microbiol. 2:535–553 [DOI] [PubMed] [Google Scholar]
- 3. Klemm P, Jørgensen BJ, van Die I, de Ree H, Bergmans H. 1985. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol. Gen. Genet. 199:410–414 [DOI] [PubMed] [Google Scholar]
- 4. Gerlach GF, Clegg S, Ness NJ, Swenson DL, Allen BL, Nichols WA. 1989. Expression of type 1 fimbriae and mannose-sensitive hemagglutinin by recombinant plasmids. Infect. Immun. 57:764–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waksman G, Hultgren SJ. 2009. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7:765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thankavel K, Shah AH, Cohen MS, Ikeda T, Lorenz RG, Curtiss R, III, Abraham SN. 1999. Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J. Biol. Chem. 274:5797–5809 [DOI] [PubMed] [Google Scholar]
- 7. Jones CH, Pinkner JS, Roth R, Heuser J, Nicholes AV, Abraham SN, Hultgren SJ. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. U. S. A. 92:2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyd EF, Hartl DL. 1999. Analysis of the type 1 pilin gene cluster fim in salmonella: its distinct evolutionary histories in the 5′ and 3′ regions. J. Bacteriol. 181:1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nuccio SP, Bäumler AJ. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71:551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abraham JM, Freitag CS, Clements JR, Eisenstein BI. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 82:5724–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freitag CS, Abraham JM, Clements JR, Eisenstein BI. 1985. Genetic analysis of the phase variation control of expression of type 1 fimbriae in Escherichia coli. J. Bacteriol. 162:668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eisenstein BI. 1981. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214:337–339 [DOI] [PubMed] [Google Scholar]
- 13. Clegg S, Hancox LS, Yeh KS. 1996. Salmonella typhimurium fimbrial phase variation and FimA expression. J. Bacteriol. 178:542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Korea CG, Badouraly R, Prevost MC, Ghigo JM, Beloin C. 2010. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 12:1957–1977 [DOI] [PubMed] [Google Scholar]
- 15. Guo A, Lasaro MA, Sirard JC, Kraehenbühl JP, Schifferli DM. 2007. Adhesin-dependent binding and uptake of Salmonella enterica serovar typhimurium by dendritic cells. Microbiology 153:1059–1069 [DOI] [PubMed] [Google Scholar]
- 16. Abraham S, Shin J, Malaviya R. 2001. Type 1 fimbriated Escherichia coli-mast cell interactions in cystitis. J. Infect. Dis. 183(Suppl 1):S51–S55 [DOI] [PubMed] [Google Scholar]
- 17. Ponniah S, Abraham SN, Dockter ME, Wall CD, Endres RO. 1989. Mitogenic stimulation of human B lymphocytes by the mannose-specific adhesin on Escherichia coli type 1 fimbriae. J. Immunol. 142:992–998 [PubMed] [Google Scholar]
- 18. Althouse C, Patterson S, Fedorka-Cray P, Isaacson RE. 2003. Type 1 fimbriae of Salmonella enterica serovar typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71:6446–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baorto DM, Gao Z, Malaviya R, Dustin ML, van der Merwe A, Lublin DM, Abraham SN. 1997. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature 389:636–639 [DOI] [PubMed] [Google Scholar]
- 20. Khan NA, Kim Y, Shin S, Kim KS. 2007. FimH-mediated Escherichia coli K1 invasion of human brain microvascular endothelial cells. Cell. Microbiol. 9:169–178 [DOI] [PubMed] [Google Scholar]
- 21. Hase K, Kawano K, Nochi T, Pontes GS, Fukuda S, Ebisawa M, Kadokura K, Tobe T, Fujimura Y, Kawano S, Yabashi A, Waguri S, Nakato G, Kimura S, Murakami T, Iimura M, Hamura K, Fukuoka S, Lowe AW, Itoh K, Kiyono H, Ohno H. 2009. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature 462:226–230 [DOI] [PubMed] [Google Scholar]
- 22. Ashkar AA, Mossman KL, Coombes BK, Gyles CL, Mackenzie R. 2008. FimH adhesin of type 1 fimbriae is a potent inducer of innate antimicrobial responses which requires TLR4 and type 1 interferon signalling. PLoS Pathog. 4:e1000233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mossman KL, Mian MF, Lauzon NM, Gyles CL, Lichty B, Mackenzie R, Gill N, Ashkar AA. 2008. Cutting edge: FimH adhesin of type 1 fimbriae is a novel TLR4 ligand. J. Immunol. 181:6702–6706 [DOI] [PubMed] [Google Scholar]
- 24. Ochoa-Repáraz J, Sesma B, Alvarez M, Jesús Renedo M, Irache JM, Gamazo C. 2004. Humoral immune response in hens naturally infected with salmonella enteritidis against outer membrane proteins and other surface structural antigens. Vet. Res. 35:291–298 [DOI] [PubMed] [Google Scholar]
- 25. Kassaify ZG, Banat G, Baydoun E, Barbour EK. 2008. Quantitative assessment of fimbriae-specific serum and egg yolk antibodies induced in chicken layers by a newly developed live salmonella enteritidis vaccine and relationship to infection. Int. J. Appl. Res. Vet. Med. 6:111–120 [Google Scholar]
- 26. Abraham SN, Beachey EH. 1987. Assembly of a chemically synthesized peptide of Escherichia coli type 1 fimbriae into fimbria-like antigenic structures. J. Bacteriol. 169:2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eisenstein BI, Clements JR, Dodd DC. 1983. Isolation and characterization of a monoclonal antibody directed against type 1 fimbriae organelles from Escherichia coli. Infect. Immun. 42:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levine MM, Black RE, Brinton CC, Jr, Clements ML, Fusco P, Hughes TP, O’Donnell S, Robins-Browne R, Wood S, Young CR. 1982. Reactogenicity, immunogenicity and efficacy studies of Escherichia coli type 1 somatic pili parenteral vaccine in man. Scand. J. Infect. Dis. Suppl. 33:83–95 [PubMed] [Google Scholar]
- 29. To SC, Moon HW, Runnels PL. 1984. Type 1 pili (F1) of porcine enterotoxigenic Escherichia coli: vaccine trial and tests for production in the small intestine during disease. Infect. Immun. 43:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guerina NG, Woodson K, Hirshfeld D, Goldmann DA. 1989. Heterologous protection against invasive Escherichia coli K1 disease in newborn rats by maternal immunization with purified mannose-sensitive pili. Infect. Immun. 57:1568–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crichton PB, Yakubu DE, Old DC, Clegg S. 1989. Immunological and genetical relatedness of type-1 and type-2 fimbriae in salmonellas of serotypes gallinarum, pullorum and typhimurium. J. Appl. Bacteriol. 67:283–291 [DOI] [PubMed] [Google Scholar]
- 32. Thorns CJ. 1995. Salmonella fimbriae: novel antigens in the detection and control of salmonella infections. Br. Vet. J. 151:643–658 [DOI] [PubMed] [Google Scholar]
- 33. Smith JM. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129 [DOI] [PubMed] [Google Scholar]
- 34. Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582 [DOI] [PubMed] [Google Scholar]
- 35. Puorger C, Vetsch M, Wider G, Glockshuber R. 2011. Structure, folding and stability of FimA, the main structural subunit of type 1 pili from uropathogenic Escherichia coli strains. J. Mol. Biol. 412:520–535 [DOI] [PubMed] [Google Scholar]
- 36. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 37. Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061–1066 [DOI] [PubMed] [Google Scholar]
- 38. Chattopadhyay S, Dykhuizen DE, Sokurenko EV. 2007. ZPS: visualization of recent adaptive evolution of proteins. BMC Bioinformatics 8:187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peek AS, Souza V, Eguiarte LE, Gaut BS. 2001. The interaction of protein structure, selection, and recombination on the evolution of the type-1 fimbrial major subunit (fimA) from Escherichia coli. J. Mol. Evol. 52:193–204 [DOI] [PubMed] [Google Scholar]
- 40. Ohno H, Hase K. 2010. Glycoprotein 2 (GP2): grabbing the FimH bacteria into M cells for mucosal immunity. Gut Microbes 1:407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas WE. 2008. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J. Biol. Chem. 283:11596–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kisiela DI, Kramer JJ, Tchesnokova V, Aprikian P, Yarov-Yarovoy V, Clegg S, Sokurenko EV. 2011. Allosteric catch bond properties of the FimH adhesin from Salmonella enterica serovar typhimurium. J. Biol. Chem. 286:38136–38147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sokurenko EV, Chesnokova V, Doyle RJ, Hasty DL. 1997. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J. Biol. Chem. 272:17880–17886 [DOI] [PubMed] [Google Scholar]
- 44. Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu XR, Krogfelt KA, Struve C, Schembri MA, Hasty DL. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. U. S. A. 95:8922–8926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, Chattopadhyay S, Johnson JR, Dykhuizen DE. 2004. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol. Biol. Evol. 21:1373–1383 [DOI] [PubMed] [Google Scholar]
- 46. Kisiela DI, Chattopadhyay S, Libby SJ, Karlinsey JE, Fang FC, Tchesnokova V, Kramer JJ, Beskhlebnaya V, Samadpour M, Grzymajlo K, Ugorski M, Lankau EW, Mackie RI, Clegg S, Sokurenko EV. 2012. Evolution of Salmonella enterica virulence via point mutations in the fimbrial adhesin. PLoS Pathog. 8:e1002733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sokurenko EV, Courtney HS, Maslow J, Siitonen A, Hasty DL. 1995. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J. Bacteriol. 177:3680–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109:913–923 [DOI] [PubMed] [Google Scholar]
- 49. Aprikian P, Tchesnokova V, Kidd B, Yakovenko O, Yarov-Yarovoy V, Trinchina E, Vogel V, Thomas W, Sokurenko E. 2007. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J. Biol. Chem. 282:23437–23446 [DOI] [PubMed] [Google Scholar]
- 50. Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Häner M, Taschner N, Burkhard P, Aebi U, Müller SA. 2002. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 323:845–857 [DOI] [PubMed] [Google Scholar]
- 51. Klemm P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5:1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yeh KS, Hancox LS, Clegg S. 1995. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J. Bacteriol. 177:6861–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tinker JK, Hancox LS, Clegg S. 2001. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar typhimurium. J. Bacteriol. 183:435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saini S, Pearl JA, Rao CV. 2009. Role of FimW, FimY, and FimZ in regulating the expression of type I fimbriae in Salmonella enterica serovar typhimurium. J. Bacteriol. 191:3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Forero M, Yakovenko O, Sokurenko EV, Thomas WE, Vogel V. 2006. Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS Biol. 4:e298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chattopadhyay S, Tchesnokova V, McVeigh A, Kisiela DI, Dori K, Navarro A, Sokurenko EV, Savarino SJ. 2012. Adaptive evolution of class 5 fimbrial genes in enterotoxigenic Escherichia coli and its functional consequences. J. Biol. Chem. 287:6150–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Swofford D. 2000. PAUP*: phylogenetic analysis using parsimony and other methods. Sinauer Associates, Sunderland, MA [Google Scholar]
- 59. Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 60. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 61. Suzuki Y, Gojobori T. 2003. Analysis of coding sequences, p 283–311 In Salemi M, Vandamme A-M, The phylogenetic handbook—a practical approach to DNA and protein phylogeny, 1st ed. Cambridge University Press, Cambridge, MA [Google Scholar]
- 62. Guharoy M, Chakrabarti P. 2005. Conservation and relative importance of residues across protein-protein interfaces. Proc. Natl. Acad. Sci. U. S. A. 102:15447–15452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of fim and sfm operons on S. enterica serovar Typhimurium LT2 and E. coli K-12 chromosomes. The positions of fim and sfm operons (red) are shown relative to the positions of the origins of replication (ORI) and housekeeping genes (adk, hisD. thrA, fumC, gyrB, and aroC) in the genome sequences of S. Typhimurium strain LT2 (GenBank accession no. NC_003197) and E. coli K-12 strain MG1655 (GenBank accession no. U00096). Download
Maximum-likelihood DNA phylograms of concatenated 3- and 6-locus MLST and fimA sequences of E. coli and S. enterica. (A) The MLST and fimA trees were constructed based on sequences from 44 E. coli strains for which fully assembled genomes are publicly available. The colored boxes mark strain clades with identical sequence types. Cross-connecting lines indicate the same fimA allele carried by different MLST type strains (black boxes). The E. coli clade of B2 strains is boxed (gray dashed line). The red asterisk indicates the presence of sfmA, and the blue asterisk indicates that the sfm operon is not complete. The scale at the bottom of the phylograms indicates phylogenetic distance and corresponds to a one-nucleotide difference. (B) Maximum-likelihood DNA phylograms of concatenated 3- and 6-locus MLST and fimA sequences of S. enterica. The MLST and fimA trees were constructed based on sequences of 24 S. enterica subsp. I strains for which fully assembled genomes are publicly available. The colored boxes mark strain clades with identical sequence types. The scale at the bottom of the phylograms indicates phylogenetic distance and corresponds to a one-nucleotide difference. Download
Z score plot by SiScan test of recombination for 10 sets of randomly chosen triplet sequences of E. coli (A) and Salmonella (B). A particular set (1 through 10) for each species includes identical randomly generated sequence numbers (see Materials and Methods for details). In each plot, the black dotted line represents the significant threshold of P = 0.05 (for Z score = 1.96). For each triplet sequences, that MaxChi statistic P value is provided. Sets 1, 2, 5, 7, and 8 of E. coli show significant recombination signals via both MaxChi and SiScan statistics. Download
BLAST analysis of fimA in different enterobacteria.
List of Salmonella enterica strains used in this study.
List of Escherichia coli strains used in this study.