Abstract
Cells respond to DNA damage during S phase by slowing chromosome replication. Recent results have shed light on the mechanism by which this ‘intra-S phase’ checkpoint is implemented.
Damage to chromosomes is potentially catastrophic, and cells are never more vulnerable than when in the process of replicating their DNA. Lesions caused by DNA-modifying agents such as MMS (the alkylating agent methyl methanesulphonate) must be repaired before the DNA is copied, to prevent the replication machinery pairing an incorrect base with the modified one. Other agents, such as hydroxyurea, have the potential to damage chromosomes by interfering directly with the progress of replication forks, leading to incomplete replication and subsequent chromosome breakage. Cells respond to such problems encountered during chromosome replication by slowing progression through S phase, as a consequence of activating a RAD53 and MEC1–mediated checkpoint [1]. Two recent publications give the first picture of how this ‘intra-S checkpoint’ is enacted. These studies reveal a two-pronged cellular response, with existing replication forks being stabilised, whilst unfired replication origins are prevented from initiating new replication forks. Since DNA damaging drugs are used in cancer chemotherapy, understanding the cellular response to them may lead to more effective treatment regimes.
Initial work on the intra-S checkpoint showed that when yeast cells were treated with MMS, progress through S phase was retarded [1]. That this retardation was due to a cellular checkpoint, rather than simply non-specific inhibition of the replication machinery, was demonstrated by examining cells deficient in mec1 and rad53, two classic checkpoint genes. Mutants for either gene appeared to complete replication of MMS-damaged DNA at almost normal speeds, as assessed by flow cytometry. In an undisturbed S phase, replication origins are activated according to a predetermined programme, with some initiating replication early and others late in S phase. Two subsequent papers showed that when exposed to MMS, checkpoint-proficient yeast cells initiate replication from early origins but then pause the temporal programme and do not activate late replication origins. rad53 and mec1 mutants were found to be incapable of this late origin repression; when treated with MMS they still proceed with initiation at late origins [2, 3]. This observation was consistent with work on mammalian cells showing that DNA alkylation can lead to a specific inhibition of initiation from other origins [4, 5]. But is inhibition of late origin firing sufficient to explain the Rad53/Mec1-mediated delay in S phase progression, or is the rate of movement of existing replication forks also inhibited?
Checkpoint-independent slowing of replication forks on damaged DNA
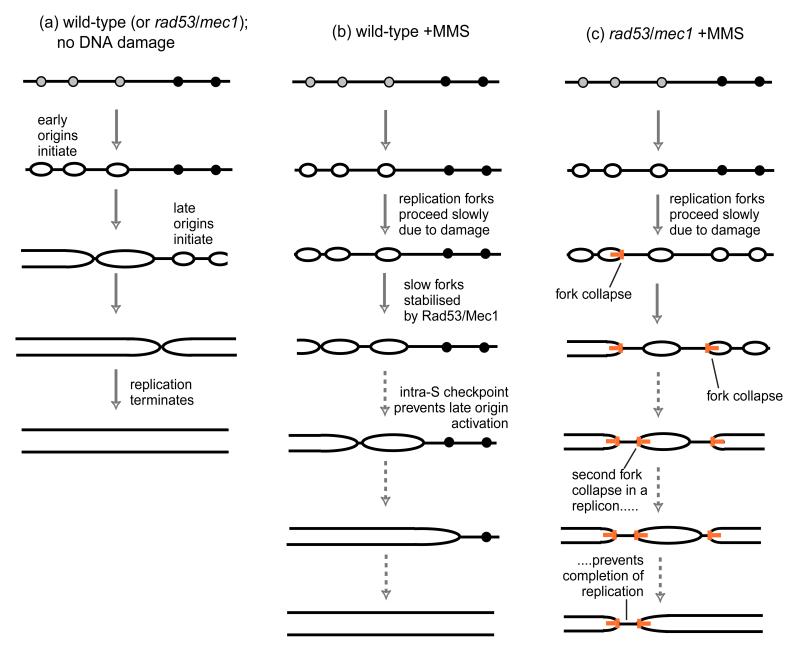
Tercero and Diffley [6] have addressed this question by using a modification of the classic Meselson and Stahl density substitution technique [7] to monitor the replication timing of different chromosome segments in yeast. Tercero and Diffley found that in the presence of MMS, replication forks indeed proceed only extremely slowly (at about 300 bp/min compared to approximately 3000 bp/min in an undisturbed S phase; Fig. 1A,B). However this fork slowing did not require the Rad53/Mec1 checkpoint—forks in rad53 and mec1 mutant cells moved just as slowly in the presence of MMS (Fig. 1C). This result implies that the failure of rad53/mec1 checkpoint mutants to slow S phase progression in MMS does not arise from faster fork progression, but is instead entirely due to their inability to repress late origins. There was, however, a crucial change in the behaviour of the forks in the absence of Rad53 or Mec1. Although forks replicating MMS-damaged DNA in wild-type cells progressed very slowly, they did make steady progress, copying the DNA at about the same overall rate over 4 hours. But in rad53 and mec1 mutants exposed to MMS, an increased proportion of replication forks appeared to stall irreversibly or collapse completely: origin-proximal sequences were efficiently replicated, but sequences further away were replicated in only a proportion of cells.
Figure 1. Replication defects caused by DNA damage in cells mutant for the intra-S phase checkpoint.
Grey circles represent early and black circles late origins. (a) progress of normal chromosome replication (b) replication of chromosomes damaged by MMS. Replication forks much more slowly than normal but are stable and not subject to collapse. Repression of late origin activation depends directly or indirectly on Rad53 and Mec1. (c) replication of MMS-damaged chromosomes in rad53 or mec1 mutants. Replication forks proceed as slowly as in wild-type cells subject to DNA damage. Late origins are activated, and replication forks are unstable, with about 40% stalling irreversibly or collapsing (orange T shapes), so that some DNA regions cannot be replicated. Dotted arrows indicate that the S phase period is extended even more than illustrated here: S phase is complete within 1 hour under normal circumstances, but lasts more than 3 hours in the presence of MMS.
Checkpoint genes prevent accumulation of abnormal DNA structures at stalled forks
An accompanying paper by Lopes et al [8] sheds light on the nature of the irreversible fork stalling in the checkpoint mutants. This study examined forks stalled by hydroxyurea (HU), a drug that blocks ribonucleotide reductase and hence reduces the availability of dNTP precursors needed for DNA synthesis. Lopes and co-workers used a 2-dimensional agarose gel technique that allows examination of the replication intermediates present in any DNA fragment of interest [9]. In wild-type cells treated with HU, most replication forks arrested close to the replication origin. When HU was subsequently removed, these forks then resumed replication and moved away from the origin region. In rad53 mutant cells treated with HU, fewer replication forks were apparent, but instead replication intermediates were seen in positions corresponding to X and small Y-shaped molecules. Although the exact pathway by which they arise is not known, such X-shaped and small Y structures are believed to result when stalled replication forks collapse and undergo abnormal DNA processing events, such as may occur during abortive recombinational repair [10]. Subsequent removal of HU in rad53 mutants did not result in the resumption of replication and consequent disappearance of these intermediates. The abnormal structures instead appear to be unrepairable, consistent with the previous observation that rad53 mutant cells are unable to recover from a hydroxyurea block.
Co-ordination of replication fork traffic
The effects of MMS and hydroxyurea on checkpoint-mutant cells suggest that a Rad53/Mec1- dependent pathway (preventing fork collapse) is enacted by both treatments. However it seems unlikely that collapse of a single replication fork is a lethal event, because the DNA lying between two origins can potentially be replicated by a fork from either origin. If one fork collapses, the fork from the neighbouring origin should simply carry on replicating the intervening DNA until it eventually encounters the collapsed fork. DNA replication will only fail if both forks converging from adjacent origins collapse before they meet (see Fig. 1C). From the observed rates of replication fork collapse in rad53 or mec1 mutant cells in MMS, Tercero and Diffley calculate that such double fork collapse would occur in approximately 16% of replicons. Given that yeast cells have about 400 origins, some such occurrences are a virtual certainty for rad53 or mec1 cells treated with MMS, and will doom the cells as they attempt mitosis, because chromosomes with unreplicated sections cannot be properly segregated. If such increased fork collapse is the cause of the extreme sensitivity of rad53/mec1 mutants to MMS, then one prediction is that the sensitivity to MMS will be mitigated if the cells are prevented from attempting S phase. Tercero and Diffley tested this possibility directly, and were able to show that the extreme toxicity of MMS to checkpoint mutants was alleviated if cells were exposed to MMS only while in G1 or G2.
These important findings as usual raise a new set of interesting questions. One question is the immediate cause of the replication fork slowing seen when the DNA of either wild-type or rad53/mec1 mutant cells is damaged. Since it is now clear that the fork slowing does not depend on the Rad53/Mec1 checkpoint, the question arises as to whether a different (unidentified) checkpoint pathway is responsible, or whether alternatively the observed fork slowing results from a physical block to polymerase passage posed by alkylation damage. The relatively constant rate of fork collapse observed by Tercero and Diffley could be consistent with a physical impediment to fork passage posed by damage incurred at a relatively constant density within the DNA. While we favour this model, current data cannot preclude the action of an unidentified fork-slowing checkpoint.
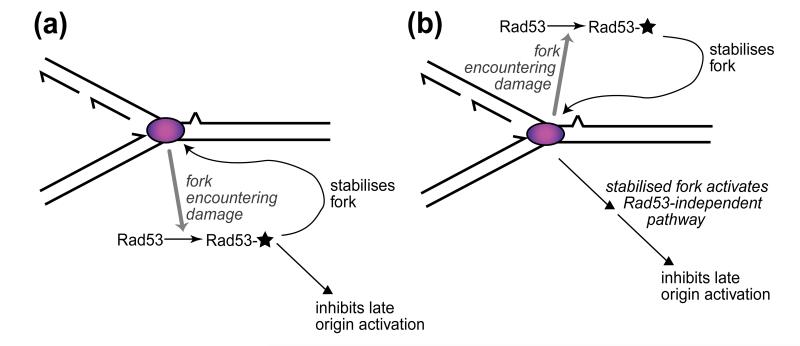
Another outstanding question is the mechanism by which the Mec1/Rad53 checkpoint pathway stabilises replication forks, and whether it is related to the mechanism by which Rad53 appears to regulate intracellular dNTP pools [11]. The target of the Mec1/Rad53 pathway in preventing collapse of stalled replication forks remains obscure. It is possible that there are targets of the intra-S checkpoint both at stalled forks (to stabilise them against breakdown) and at late origins (to prevent their inappropriate activation) (see Fig. 2A). Multiple sites of action of the checkpoint during S phase could explain the assignment of several different gene products as likely targets, including at replication forks the single-stranded DNA-binding protein RP-A [12, 13] and the polymerase α-primase complex [14], and at late origins Dbf4, the activator of the Cdc7 protein kinase (reviewed in [15]).
Figure 2. Inhibition of late origin firing by the intra-S phase checkpoint could depend either directly or indirectly on Rad53.
In either case, replication complexes (pink ovals) that are slowed or stalled due to DNA damage generate a signal that activates Rad53 (indicated by a star), which then stabilises stalled replication forks. (a) Activated Rad53 also acts to inhibit late origin firing. (b) Activated Rad53 stabilises stalled forks against breakdown, and these stabilised forks generate a separate (Rad53-independent) signal that inhibits late origins. In either case, collapsed forks are proposed to be unable to activate Rad53, so that the fork collapse that occurs in a rad53 mutant would remove the late origin inhibition, leading to late origin activation as observed.
Alternatively, the primary function of the Rad53/Mec1 checkpoint could be the stabilisation of replication forks. The effect on late-firing origins could then be caused by these stabilised forks generating a distinct signal (not directly dependent on Rad53 or Mec1) that represses late origin activation (Fig. 2B). Either of these two possible mechanisms might make physiological sense of the existence of late origins: they could be regarded as ‘backup’ initiation sites in the event that replication from early origins is unsuccessful. If cells are encountering general problems with progression of forks from early origins, it would make sense to postpone recourse to the backup late initiation sites until either the replication interference has been removed (such as occurs when MMS or HU is withdrawn from the experiment) or else the existing forks have failed irretrievably, after which they are proposed no longer to generate the signal to inhibit late origin activation.
These interpretations could potentially explain the relative sensitivity of cancer cells to chemotherapeutic DNA-damaging agents. Since they frequently have defective checkpoint pathways, cancer cells might be unable to benefit fully from the intra-S phase checkpoint: neither being able to stabilise stalled replication forks nor being able to hold late-firing origins in reserve whilst DNA damage is occurring. Failure of either or both these arms of the intra-S phase checkpoint could severely compromise the capability of cancer cells to deal with damage to their chromosomes.
Acknowledgements
We thank Conrad Nieduszynski for discussion and suggestions, and Douglas Stirling for comments on the manuscript. Our research is supported by The Royal Society and the Cancer Research Campaign.
References
- 1.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–7. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 2.Santocanale C, Diffley JFX. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–8. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 3.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–21. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 4.Zhukovskaya N, Branch P, Aquilina G, Karran P. DNA-Replication arrest and tolerance to DNA methylation damage. Carcinogenesis. 1994;15:2189–2194. doi: 10.1093/carcin/15.10.2189. [DOI] [PubMed] [Google Scholar]
- 5.Cobuzzi RJ, Burhans WC, Beerman TA. Inhibition of initiation of simian virus 40 DNA replication in infected BSC-1 cells by the DNA alkylating drug adozelesin. Journal of Biological Chemistry. 1996;271:19852–19859. doi: 10.1074/jbc.271.33.19852. [DOI] [PubMed] [Google Scholar]
- 6.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–7. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 7.Meselson M, Stahl F. The replication of DNA in Escherichia coli. Proc Natl Acad Sci USA. 1958;64:1242–1248. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–61. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 9.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–71. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 10.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 11.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–70. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brush GS, Morrow DM, Hieter P, Kelly TJ. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–80. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–96. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–72. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jares P, Donaldson A, Blow JJ. The Cdc7/Dbf4 protein kinase: target of the S phase checkpoint? EMBO Reports. 2000;1:319–22. doi: 10.1093/embo-reports/kvd076. [DOI] [PMC free article] [PubMed] [Google Scholar]