Abstract
Nuclear magnetic resonance assays allow for measurement of a wide range of metabolic phenotypes. We report here the results of a GWAS on 8,330 Finnish individuals genotyped and imputed at 7.7 million SNPs for a range of 216 serum metabolic phenotypes assessed by NMR of serum samples. We identified significant associations (P < 2.31 × 10−10) at 31 loci, including 11 for which there have not been previous reports of associations to a metabolic trait or disorder. Analyses of Finnish twin pairs suggested that the metabolic measures reported here show higher heritability than comparable conventional metabolic phenotypes. In accordance with our expectations, SNPs at the 31 loci associated with individual metabolites account for a greater proportion of the genetic component of trait variance (up to 40%) than is typically observed for conventional serum metabolic phenotypes. The identification of such associations may provide substantial insight into cardiometabolic disorders.
Circulating metabolites have key roles in numerous biological pathways and consequently contribute to risk for many diseases, particularly disorders of the metabolic and cardiovascular systems1,2. Such metabolites have long been used for clinical risk assessment, diagnosis, prognosis and evaluation of treatment efficacy. Genome-wide association studies (GWAS) have discovered numerous genomic regions associated with clinically relevant metabolites, with recent large-scale meta-analyses having identified, in total, over 100 loci associated with serum concentrations of individual metabolites, such as glucose, insulin, lipids and uric acid3–5. Nevertheless, our understanding of the genetic basis and pathophysiological impact of variations in metabolite levels remains far from complete, and recent studies suggest the importance of investigating metabolite phenotypes beyond those used in traditional genetic studies. For example, in a recent longitudinal study, amino acids identified by metabolite profiling were shown to be associated with the risk for developing type 2 diabetes (T2D) in cohorts of apparently healthy individuals6.
Until recently, the search for metabolic risk variants had focused on only a few metabolites at a time, but recent technological developments in NMR and mass spectrometry have made possible the quantification of over 100 metabolites in a single analytical procedure, allowing both broader and deeper molecular profiling of large cohorts7. Previous genome-wide studies of high-throughput blood metabolites have identified over 40 loci associated with such measures8–10. We present here the results of the largest genetic investigation of metabolomic phenotypes reported to date, including 8,330 unrelated individuals and 561 twin pairs sampled from the Finnish population who were analyzed for both genome-wide SNP genotypes and for 216 phenotypic measures obtained from an NMR-based metabolomic screen of fasting serum samples. We identify 31 loci associated with one or more phenotypes at a genome-wide significance level, including 6 newly identified loci for serum amino acids, 1 for citrate and 4 for serum lipoprotein and lipid metabolites. Using the twin samples, we estimate the heritability of each of the 216 metabolic measures and determine the proportions of overall and genetic variance that can be explained by the significantly associated loci.
RESULTS
We present genome-wide association results for 117 directly measured metabolites and 99 variables derived from these measures (Supplementary Table 1). The 117 metabolites consist of 80 lipoproteins, 15 lipids and 22 low-molecular-weight metabolites (with the latter including pyruvic acid, amino acids and other small molecules participating in glycolysis, the citric acid cycle or the urea cycle). The derived variables (mainly ratios of directly measured metabolites) were selected on the basis of their prior utility in characterizing metabolic function in either normal or disease states. For example, an increase in the ratio of branched-chain amino acids (valine, leucine and isoleucine) to aromatic amino acids (phenylalanine and tyrosine) in the serum, termed Fischer’s ratio, is characteristic of liver fibrosis and is hypothesized to contribute to hepatic encephalopathy11. The rationale for the specific ratios that we analyzed is presented (Supplementary Table 2). The ratios selected in this study involve metabolites implicated in lipolysis, proteolysis, ketogenesis and glycolysis, as well as reagents and products of enzymatic reactions.
Heritability of metabolic measures
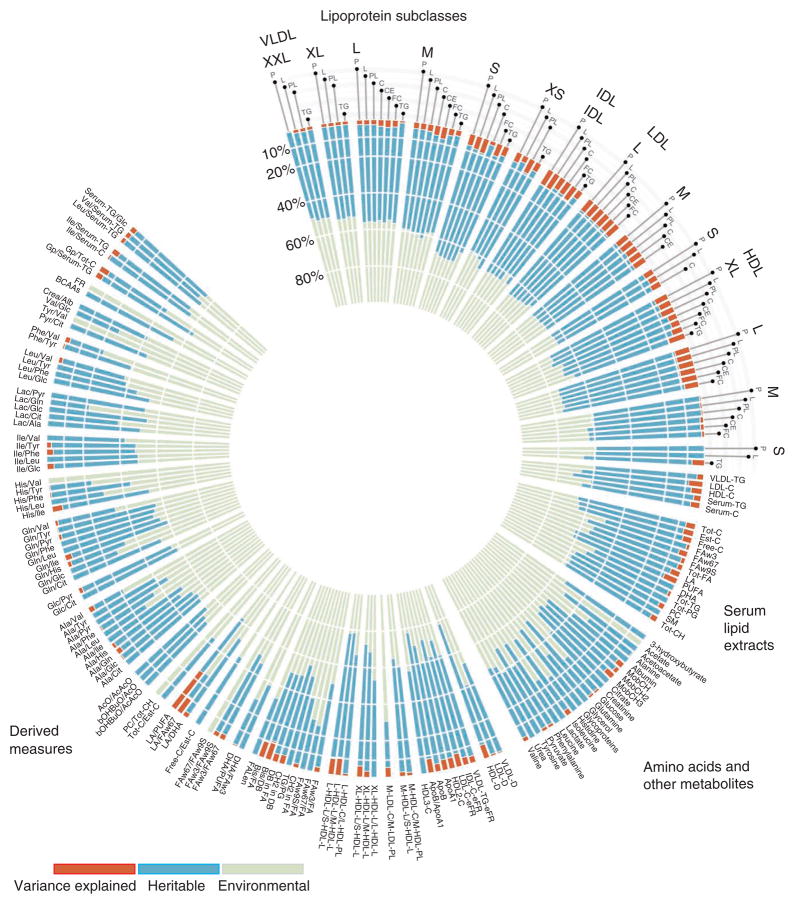
To assess the heritability of each measure, we estimated intrapair metabolite correlations for 221 monozygotic and 340 dizygotic twin sets, aged 22–25, from the Finnish Twin Cohort. For amino acids and other small-molecule metabolites, the heritability estimates ranged between 0.23–0.55. Heritability estimates were higher for both lipids (range of 0.48–0.62) and lipoproteins (range of 0.50–0.76) (Fig. 1 and Supplementary Table 3).
Figure 1.
The heritability estimates and proportion of variance explained for all traits. Heritability estimates are presented for lipids, small molecules, ratios used in this study and lipoproteins. Labels in the lipoprotein subclasses describe the properties measured in each subfraction (P, concentration of particles; L, total lipids; PL, phospholipids; C, total cholesterol; CE, cholesterol esters; FC, free cholesterol; TG, triglycerides). Abbreviations are explained in detail in Supplementary Table 1.
Because of the high heritability estimated for many of the lipoprotein subclasses, we also compared the heritability of composite lipid phenotypes derived from NMR measures with that determined using conventional lipid measures (enzymatically measured lipid levels), using data from a subset of the twins (n = 256) for whom data from both types of assays were available. The NMR-based and enzymatic measures gave similar heritability estimates, except for triglycerides, for which estimated heritability was 0.68 for the NMR-based measure and 0.55 for the enzymatic measure (Supplementary Note and Supplementary Tables 4 and 5).
Genome-wide association analysis identifies 31 metabolite loci
We used stochastic imputation methods to augment the directly genotyped SNPs in the various cohorts (see Table 1 for a complete list of the cohorts) to generate a marker set for association analyses consisting of 7.7 million SNPs. The genotype imputation panel that we employed incorporated phased haplotypes from the 1000 Genomes Project12, HapMap 3 (ref. 13) and the Finnish extension to HapMap 3 (ref. 14). Using an additive genetic model, we tested for univariate associations between these 7.7 million genetic markers and 216 metabolic measures. To correct for multiple testing, genome-wide statistical significance was set to P < 2.31 × 10−10 (standard univariate genome-wide significance threshold of 5 × 10−8 / 216 phenotypes tested). Genome-wide inflation factors ranged from 0.99 to 1.06 (all are listed in Supplementary Table 6). Thirty-one loci were significantly associated with at least one metabolic measure (Tables 2 and 3); all SNPs that reached the genome-wide level of significance are presented in Supplementary Table 7). The associations for these SNPs are shown in 2-Mb windows surrounding the SNP with the lowest P value to provide a graphical view of the associated regions (Supplementary Fig. 1), and quantile-quantile plots were created for the lead traits in Tables 2 and 3 and Supplementary Fig. 2) to graphically present the overall inflation of the test statistics. For each cohort, box plots of raw phenotype values for each genotype class of the SNP with the lowest P value are provided (Supplementary Fig. 3) to show the effects of the SNPs per genotype class per cohort. Four of the 11 newly associated loci presented here (1-159807481, rs17610395, 17-7083575 and rs6917603; indicated in Tables 2 and 3) would not have been identified using only the HapMap 2 imputation panel instead of the expanded panel that we employed for our association testing (see Online Methods).
Table 1.
Sample demographics
| Study | n | Mean age (years ± s.d.) | Female (%) | BMI ± s.d. | |
|---|---|---|---|---|---|
| NFBC1966 | Northern Finland Birth Cohort 1966 | 4,703 | 31.0 ± 0.0 | 51 | 24.6 ± 4.1 |
| YF | The Cardiovascular Risk in Young Finns Study | 1,904 | 37.7 ± 5.0 | 54 | 26.0 ± 4.7 |
| HBCS | Helsinki Birth Cohort Study | 708 | 61.3 ± 2.9 | 60 | 27.1 ± 4.1 |
| GenMets | Health2000 GenMets Study | 572 | 55.8 ± 7.3 | 57 | 27.2 ± 4.5 |
| DILGOM | The Dietary, Lifestyle, and Genetic determinants of Obesity and Metabolic Syndrome | 443 | 50.1 ± 13.5 | 56 | 26.5 ± 4.6 |
| Twins | FinnTwin12 (FT12) and FinnTwin16 (FT16) cohort studies | 1,269 | 23.9 ± 2.1 | 55 | 23.1 ± 3.7 |
BMI, body mass index; n, number of individuals with both genotype and metabolic phenotypes included in this study.
Table 2.
Fourteen loci associated with small-molecule phenotypes with genome-wide significance in meta-analysis of five Finnish cohorts
| Newly identified loci | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr. | Lead trait | Position | Coded allele | Other allele | P value | CAF | Effect size | s.e.m. | eQTL | Candidate gene | Genotyped |
| rs2160387 | 2 | Ala/Val | 65074414 | C | T | 2.62 × 10−22 | 0.41 | −0.17 | 0.02 | – | SLC1A4 | rs10211524 |
| rs1440581 | 4 | Fischer’s ratio | 89445446 | C | T | 1.96 × 10−16 | 0.47 | 0.13 | 0.02 | – | PPM1K | rs6841731 |
| rs2545801 | 5 | Phe | 176773945 | T | C | 8.70 × 10−11 | 0.73 | −0.12 | 0.02 | – | F12 | rs2731672 |
| rs2297644 | 10 | Gln/His | 99349729 | C | T | 1.23 × 10−12 | 0.20 | 0.15 | 0.02 | – | DHDPSL | rs11817730 |
| rs4788815 | 16 | Phe/Tyr | 70192312 | T | A | 1.54 × 10−17 | 0.64 | 0.15 | 0.02 | – | TAT | rs4788547 |
| 17-7083575 (rs117616209)a | 17 | Fischer’s ratio | 7083575 | T | C | 2.64 × 10−14 | 0.02 | −0.51 | 0.07 | – | SLC2A4 | –b |
| rs807669 | 22 | Citrate | 17534522 | C | T | 3.30 × 10−16 | 0.62 | −0.14 | 0.02 | CLTCL1 | SLC25A1 | rs712964 |
| Known loci | ||||||||||||
|
| ||||||||||||
| SNP | Chr. | Lead trait | Position | Coded allele | Other allele | P value | CAF | Effect size | s.e.m. | eQTL | Candidate gene, trait | |
|
| ||||||||||||
| rs1260326 | 2 | Ala/Gln | 27584444 | C | T | 2.59 × 10−18 | 0.65 | −0.15 | 0.02 | – | GCKR, TG31 | |
| rs560887 | 2 | Glc | 169471394 | G | A | 2.19 × 10−17 | 0.69 | 0.15 | 0.02 | – | G6PC2, Glc20 | |
| rs4241816 | 4 | His/Val | 187389321 | T | A | 5.58 × 10−13 | 0.57 | 0.12 | 0.02 | – | KLKB1, bradykinin8 | |
| rs6900341 | 6 | Ala/Tyr | 111901102 | G | T | 3.68 × 10−15 | 0.37 | 0.13 | 0.02 | – | SLC16A10, Ile, Tyr8 | |
| rs10830963 | 11 | Glc | 92348358 | G | C | 3.19 × 10−11 | 0.28 | 0.14 | 0.02 | – | MTNR1B, Glc21 | |
| rs2638315 | 12 | Gln/Glc | 55151323 | C | G | 2.43 × 10−35 | 0.17 | −0.29 | 0.02 | SPRYD4 | GLS2, Gln8 | |
Chr., chromosome; position, SNP position in NCBI human genome build 36; CAF, coded allele frequency; candidate gene, potential candidate gene in the region; candidate gene, trait, gene reported for the known locus and lead trait with which the gene was associated; genotyped, genome-wide significant SNP genotyped in at least three study cohorts in the region of association; Ala, alanine; Glc, glucose; Gln, glutamine; His, histidine; Ile, isoleucine; Phe, phenylalanine; TG, total triglyceride content; Tyr, tyrosine; Val, valine.
This locus would not have been identified using the HapMap 2 imputation reference panel.
Imputed SNPs that were validated by sequencing.
Table 3.
Genome-wide significant loci identified for the lipid metabolism phenotypes in the meta-analysis of five Finnish cohorts
| New loci
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr. | Lead trait | Position | Coded allele | Other allele | P value | CAF | Effect size | s.e.m. | eQTL | Candidate gene | Genotyped |
| 1-159807481 (rs67418890)a | 1 | XL-HDL-CE | 159807481 | A | T | 1.21 × 10−10 | 0.11 | 0.19 | 0.03 | FCGR2B, FCGR2A | FCGR2B, FCGR2A | rs6427609 |
| 4-73541429 (rs115136538) | 4 | Albumin | 73541429 | C | T | 4.84 × 10−18 | 0.02 | −0.51 | 0.06 | – | ALB | rs16850360 |
| rs6917603a | 6 | XXL-VLDL-P | 30125050 | C | T | 2.81 × 10−29 | 0.20 | −0.24 | 0.02 | – | PPP1R11 | rs6917603 |
| rs17610395a | 11 | LA/PUFA | 68318904 | T | C | 7.57 × 10−12 | 0.16 | 0.17 | 0.02 | – | CPT1A | –b |
| Known loci | ||||||||||||
|
| ||||||||||||
| SNP | Chr. | Lead trait | Position | Coded allele | Other allele | P value | CAF | Effect size | s.e.m. | eQTL | Candidate gene, trait | |
|
| ||||||||||||
| 1-55889093 (rs72669744) | 1 | L-LDL-FC | 55889093 | T | C | 1.10 × 10−19 | 0.02 | −0.59 | 0.06 | – | PCSK9, LDL31 | |
| rs1168029 | 1 | MobCH | 62741990 | A | G | 2.66 × 10−13 | 0.70 | 0.13 | 0.02 | – | ANGPTL3, TG31 | |
| rs13247874 | 7 | VLDL-D | 72648378 | T | C | 8.43 × 10−14 | 0.20 | −0.16 | 0.02 | – | MLXIPL, TG31 | |
| 8-19956650 (rs115849089) | 8 | M-VLDL-PL | 19956650 | A | G | 4.26 × 10−15 | 0.10 | −0.22 | 0.03 | – | LPL, TG31 | |
| rs2575876 | 9 | Tot-C/Est-C | 106705560 | A | G | 1.63 × 10−11 | 0.19 | −0.14 | 0.02 | – | ABCA1, HDL31 | |
| rs174547 | 11 | LA/PUFA | 61327359 | C | T | 8.02 × 10−262 | 0.42 | 0.57 | 0.02 | – | FADS1-FADS2-FADS3, PC10 | |
| rs651821 | 11 | Val/serum TG | 116167789 | T | C | 7.98 × 10−20 | 0.91 | 0.27 | 0.03 | – | APOA1-APOC3-APOA4- APOA5, LDL33 | |
| rs35853021 | 15 | XL-HDL-TG | 56467935 | T | G | 7.11 × 10−76 | 0.39 | 0.31 | 0.02 | – | LIPC, HDL31 | |
| rs11075253 | 16 | LA/PUFA | 15056147 | A | C | 4.98 × 10−15 | 0.28 | −0.14 | 0.02 | – | PDXC1, C20:2 and C20:3 fatty acid metabolism8 | |
| rs3764261 | 16 | HDL-C | 55550825 | A | C | 1.23 × 10−36 | 0.28 | 0.22 | 0.02 | – | CETP, HDL32 | |
| rs7228085* | 18 | XL-HDL-TG | 45414812 | G | A | 6.70 × 10−11 | 0.46 | 0.11 | 0.02 | – | LIPG, HDL31 | |
| rs55791371 | 19 | M-LDL-C/M-LDL-PL | 11049153 | C | A | 8.21 × 10−17 | 0.10 | −0.26 | 0.03 | – | LDLR, LDL31 | |
| rs7412 | 19 | L-LDL-FC | 50103919 | T | C | 2.52 × 10−58 | 0.04 | −0.75 | 0.05 | – | APOE-APOC1-APOC2, LDL31 | |
| rs6065904 | 20 | L-HDL-L/M-HDL-L | 43968058 | A | G | 2.29 × 10−31 | 0.24 | −0.22 | 0.02 | PLTP | PLTP, TG32 | |
Chr., chromosome; position, SNP position in NCBI human genome build 36; CAF, coded allele frequency; candidate gene, potential candidate gene in the region; candidate gene, trait: gene reported for the known locus and lead trait the gene was associated with; genotyped, genome-wide significant SNP genotyped in at least three study cohorts in the region of association; L-HDL-L, lipid content in L-HDL; M-HDL-L, lipid content in medium HDL; FC, free cholesterol; LA, linoleic acid; PUFA, polyunsaturated fatty acids; C, cholesterol; PL, phospholipid; MobCH, double-bond protons of mobile lipids; Tot-C, total cholesterol; Est-C, esterified cholesterol; Val, valine; serum TG, serum total triglyceride content; D, particle diameter; CE, cholesterol ester; P, particle concentration.
These loci would not have been identified using the HapMap 2 imputation reference panel.
Imputed SNPs that were validated by sequencing.
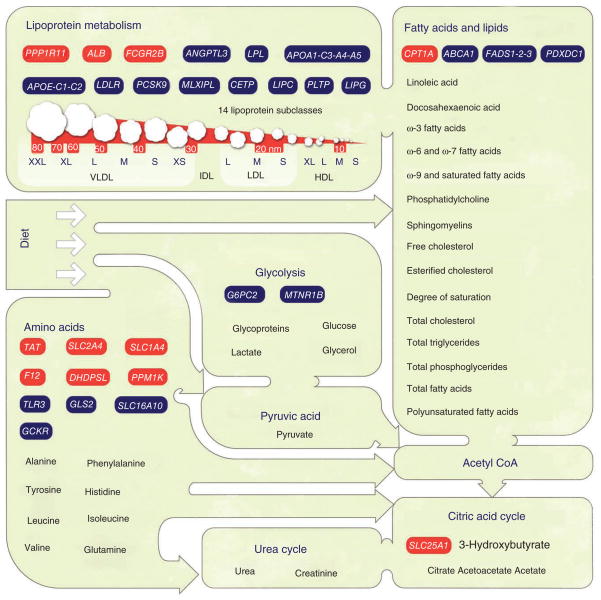
The 31 loci for which we detected association to NMR-based measures are shown in relation to the relevant metabolic pathways (Fig. 2). Multiple loci had not previously been observed to be associated with metabolic measures, either in the National Human Genome Research Institute (NHGRI) catalog of published GWAS15 or in previous blood metabolomics genome-wide screens8–10. Thirteen of these loci, seven of which were newly identified here, were associated with amino acids and other small molecules (Table 2). The remaining 18 loci were associated with NMR-based measures of lipid metabolites (Table 3). Fourteen of these loci, three of which were newly identified here, were most strongly associated with lipoprotein measures, and four loci, including one newly identified locus, showed the strongest associations to other lipid-related measures. Below, we categorize the loci reaching genome-wide significance by phenotype and present potential candidate genes from the associated regions.
Figure 2.
Overall summary of basic metabolism, key constituents of the NMR-measurable serum metabolome and associated genetic loci. The green shapes represent various dietary ingredients and systemic metabolic measures reflecting human body functions. A key selection of metabolites quantified in each metabolic category is given. Red rectangles represent the newly identified genetic loci found in this work, and blue rectangles indicate loci that were previously identified. The genes are categorized by the lead trait associations given in Tables 2 and 3. In particular, ALB and FCGR2B were associated with serum cholesterol, PPP1R11 with very high VLDL measures, CPT1A with the LA/PUFA ratio, SLC25A1 with citrate, F12 with phenylalanine, TAT with tyrosine, DHDPSL with glutamine, SLC1A4 with valine and PPM1K and SLC2A4 with Fischer’s ratio.
Associations with amino acids and other small molecules
For the metabolites involved in processes such as glycolysis, the citric acid cycle and amino acid metabolism, we identified, in total, seven new and six previously described loci (Table 2). Five of these associations (to valine, phenylalanine, tyrosine, isoleucine and leucine or their composite measure, Fischer’s ratio) involve amino acids previously shown to be associated with the risk for T2D6. In addition, we identified one new locus associated with serum citrate levels and one new locus associated with serum glutamine levels.
The strongest associations at chromosome 2p14 (rs2160387) were with the ratio of alanine to valine (P = 2.6 × 10−22) and with circulating valine levels (P = 8.4 × 10−11). The associated SNP is located in the first intron of SLC1A4 (encoding solute carrier family 1 member 4), a neutral amino acid transporter. A marker (rs1440581) at 4q22 was associated with both Fischer’s ratio (P = 2.0 × 10−16) and valine (P = 6.4 × 10−14), and a SNP (rs2545801) at 5q35 was associated with phenylalanine (P = 8.7 × 10−11). The rs4788815 SNP at 16q22 was associated with both tyrosine levels (P = 1.2 × 10−10) and the ratio of phenylalanine to tyrosine (P = 1.5 × 10−17). This marker is located 25 kb upstream of TAT (encoding tyrosine aminotransferase), an enzyme that catalyzes the conversion of tyrosine to hydroxyphenylpyruvate. Mutations in TAT have been shown to cause type 2 tyrosinemia (MIM 276600), whose symptoms include keratitis, painful palmoplantar hyperkeratosis, intellectual disability and elevated serum tyrosine levels16. Two additional markers at this locus (rs34042070 and rs3213423) were independently associated with the ratio of glycoproteins and total cholesterol and remained significantly associated (P = 2.4 × 10−14 and 3.2 × 10−12, respectively) after conditioning on rs4788815. These associations with the ratio of glycoproteins to total cholesterol may reflect a known association with total cholesterol at this locus5. At 17p13, a SNP located 45 kb from SLC2A4, encoding a facilitated glucose transporter, was associated with Fischer’s ratio (17-7083575; rs117616209; P = 2.6 × 10−14).
Two additional loci were associated with amino acid levels, small molecules or measures derived from these metabolite levels. The rs807669 SNP near SLC25A1 was associated with serum citrate concentration (P = 3.3 × 10−16), and rs2297644 in the first intron of DHDPSL (encoding DHDPS-like protein isoform 1) was associated with the ratio of glutamine to histidine (P = 1.2 × 10−12), as well as with serum glutamine levels (P = 4.7 × 10−11).
Six of the 13 loci determined by this study to be associated with small molecules have previously been reported. At chromosome 6q21, rs6900341 showed association with the ratio of alanine to tyrosine (P = 3.7 × 10−15), and, in a previous study, the rs7760535 SNP in the same locus associated with the ratio of isoleucine to tyrosine8. rs6900341 is located within an intron of REV3L (encoding the REV3-like catalytic subunit of DNA polymerase ζ) and is 250 kb from a possible functional candidate gene, SLC16A10 (encoding solute carrier family 16 member 10), which transports aromatic amino acids across the plasma membrane. In addition, in line with previous findings8, we found that rs2638315 in the 3′ UTR of GLS2 (encoding mitochondrial phosphate-activated glutaminase) was associated with serum glutamine levels (P = 8.6 × 10−28), as well as with the ratio of glutamine to glucose (P = 2.4 × 10−35). Moreover, we extend a previous finding of an association on chromosome 4 between a SNP in KLKB1 and bradykinin. In our study, the rs4241816 SNP in the same locus was associated with both serum histidine levels (P = 2.2 × 10−11) and the ratio of histidine to valine (P = 5.6 × 10−13). The decarboxylation of histidine by the HDC enzyme leads to the formation of histamine, a central proinflammatory mediator. A possible functional candidate gene, TLR3 (encoding Toll-like receptor 3), lies 145 kb from the associated SNP. TLR3 encodes a receptor that regulates the release of histamine-rich granules from mast cells17,18. A previous study provided further support for the potential involvement of TLR3, having shown that bradykinin regulates histamine release from mast cells through a mechanism that remains unclear19.
The SNPs in G6PC2 and MTNR1B that showed associations with serum glucose in previous GWAS20,21 were also associated with NMR-measured glucose levels in this study. Previous studies have shown association between SNPs in GCKR (encoding glucokinase regulator) and a variety of metabolic traits, including triglycerides5, glucose3 and the glucose to mannose ratio8. In our study, a SNP in GCKR (rs1260326) was associated with the ratio of alanine to glutamine and other amino acids measures, in addition to measures of very-low-density lipoprotein (VLDL) and total triglycerides.
Associations with NMR-based measures of lipid metabolism
Previous investigations have identified nearly 100 genetic loci associated with measures of serum lipid concentrations that are typically used in clinical practice5. These measures (total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL) and triglycerides), however, are heterogeneous aggregates that reflect multiple biological processes. We assessed here association to a much wider range of lipoprotein and lipid measures (n = 95) hypothesized to quantify more homogenous phenotypes. We identified a total of 18 such associations, 4 of which have not previously been reported (Table 3). These four loci include one associated with a cholesterol measure, one associated with a VLDL measure, one associated with the ratio of linoleic acid to other polyunsaturated fatty acids (LA/PUFA) and one associated with serum albumin levels as well as lipoprotein measures.
The dosage of the C allele at the 4-73541429 SNP (rs115136538) was associated with decreased concentration of serum albumin (P = 4.8 × 10−18) and with elevated concentration of 37 other metabolites, including apoB-containing lipoprotein particles, several cholesterol- related measures and sphingomyelin (Supplementary Table 7). This SNP was also positively associated with the levels of enzymatically measured total cholesterol (P = 1.5 × 10−19) and LDL cholesterol (P = 2.64 × 10−13). These observations are in contrast to the mostly positive correlations observed in the metabolic phenotype data between albumin and apoB-containing lipoprotein measures (range for correlations of −0.03 to 0.5; mean = 0.31; median = 0.37). Rare variants in the albumin gene have previously been associated with analbuminemia, hypercholesterolemia and hyperlipidemia22–24; however, to our knowledge, this is the first report of a common genetic variant at ALB being associated with any lipoprotein or lipid measure or albumin levels.
The cholesterol ester content of extra large HDL (XL-HDL-CE) particles was associated (P = 1.2 × 10−10) with a single SNP (1-159807481; rs67418890). In a subset of the samples (n = 585) with leukocyte expression data and genome-wide genotype data available, we identified a cis expression quantitative trait locus (eQTL) for this SNP in the FCGR2A and FCGR2B genes that encode the α and β subunits of the Fc fragment of the IgG low-affinity II receptor, respectively (linear regression P = 3.9 × 10−10 and 1.3 × 10−20, respectively; Online Methods and Supplementary Table 8). There were several SNPs that acted as eQTLs in the FCGR2B locus, including our lead SNP (rs67418890). All of the best associated SNPs with eQTL activity are in tight correlation with each other (r > 0.87), suggesting that they represent the same possibly functional variant in this shared haplotype. FCGR2A and FCGR2B encode the components of the CD32 cell surface receptor, an IgG-mediated B-cell coreceptor that represses antibody production in the presence of IgG; however, CD32 also has a role in cardiovascular disease, with FCGR2B having been shown to modify the size of atherosclerotic plaques in mice25,26. Our findings suggest a role for CD32 in the cholesterol balance in peripheral tissue and potentially in reverse cholesterol transport.
A class I major histocompatibility (MHC) locus HLA-A also harbored metabolite signals, with the top SNP, rs6917603, being strongly associated with the concentration of chylomicrons and extremely large VLDL particles (XXL-VLDL-P) (P = 2.8 × 10−29). The associated variant is upstream of PPP1R11, an inhibitor of PP1, a highly conserved serine/threonine phosphatase with a central role in glycogen metabolism and blood glucose levels27. The association of rs6917603 with VLDL was also independent of the two other SNPs in the human leukocyte antigen (HLA) locus that have been shown to be associated with triglycerides, total cholesterol and LDL cholesterol5.
A SNP at 11q13.2 was found to be associated with the LA/PUFA ratio (rs17610395; P = 7.6 × 10−12). The associated SNP is within 6 Mb of the FADS gene cluster (encoding fatty acid desaturase), a known lipoprotein locus, and it is likely to be independent of the previously identified SNP in FADS (rs174547). In our study, the association remained significant after conditioning on the earlier marker (P = 7.6 × 10−12) (Supplementary Note)9,28,29. rs17610395 is a nonsynonymous SNP (encoding a p.Ala275Thr amino acid change) located in CPT1A, a gene encoding carnitine palmitoyltransferase IA, a liver-expressed enzyme involved in long-chain fatty acid oxidation30. Rare mutations in CPT1A cause CPT IA deficiency in an autosomal recessive metabolic disorder of long-chain fatty acid oxidation (MIM 255120).
Fourteen loci that were reported in previous GWAS to be associated with lipid measures were also associated with NMR-based lipid measures in this study. FADS1 was originally associated with a range of glycero-phosphatidylcholines10. The authors in the previous study also found that a SNP in FADS1 was associated with several lipid and phosphatidylcholine measures, whereas we showed association with measures of fatty acid saturation and with omega-3 fatty acids and several omega-3 fatty acid ratios. PDXDC1 (encoding pyridoxal-dependent decarboxylase domain–containing protein 1) was recently shown to associate with the ratio of eicosatrienoylglycerophospholipids8, indicating a role for it in the metabolism of 20:2 and 20:3 fatty acids. In our study, the same locus was associated with the LA/PUFA ratio, with linoleic acid being one of the metabolites involved in eicosanoid synthesis. Four loci previously reported to be associated with serum triglycerides (ANGPTL3, MLXIPL, LPL and PLTP)31,32 were associated with VLDL measures in our study. The PLTP locus was also associated with a large number of HDL measures. Four loci have previously been associated with total LDL cholesterol (LDL-C) (PCSK9, the APOA1 region, LDLR and the APOE region)31,33. In our study, the APOA1 region showed association with a large number of VLDL and triglyceride measures, whereas the other three loci showed association with LDL and cholesterol measures. Finally, associations to four loci previously determined to be involved in total HDL cholesterol (HDL-C) levels (ABCA1, LIPC, CETP and LIPG) are replicated in our data31,32.
Proportion of variance explained
For each phenotype, we estimated the proportion of variance explained by the 33 significantly associated SNPs in an independent sample of 436 individuals from the Finnish twin cohort (Supplementary Table 3). For the direct metabolite measures, the proportion of variance explained by the SNPs ranged between 0.2–9.1% for HDL subclasses, 5.0–8.0% for LDL subclasses, 0.5–8.2% for VLDL subclasses, 4.8–9.5% for intermediate-density lipoprotein (IDL) subclasses and up to 7.6% for the other lipids and molecules. When comparing the proportion of variance explained in the lipoprotein subclasses and the composite measures for the corresponding lipids, associated SNPs explained much more of the variance for the larger size lipoprotein subclasses than for the corresponding composite measures (Fig. 1 and Supplementary Table 3).
For derived measures, the SNPs explained up to 25% (18–32%, 95% confidence interval (CI)) of the total variance in the LA/ PUFA ratio, corresponding to 40% of the heritability. This particularly high proportion of explained variance is driven by the association in the FADS1-FADS2-FADS3 locus, where each risk allele was accompanied by an increase of 0.57 s.d. in the fatty acid ratio in a common variant (coded allele frequency (CAF) = 42%). This strong association shows that a few variants in specific metabolite measures can explain a high proportion of variation, as has been shown previously for the FADS gene cluster9,10.
DISCUSSION
In this study, we report the heritability of metabolite measures assayed by NMR and identify 11 new genetic associations for these measures through genome-wide analysis of five population-based cohorts comprising >8,000 individuals. We further show that, compared to the associations typically observed for complex traits in comparably sized samples, the associations identified in this study explain a greater proportion of trait variance.
The results support several conclusions. First, almost 40% of the metabolites assayed in this study showed an estimated heritability of >0.6, a figure higher on average than that typically reported for clinically used measures (heritability estimates of plasma lipids have been reported to range from 0.39 to 0.62 for total cholesterol (TC), 0.39 to 0.83 for HDL-C, 0.24 to 0.50 for LDL-C, 0.20 to 0.55 for triglycerides and 0.07 to 0.28 for fasting plasma glucose)34,35. This observation supports the hypothesis that the more detailed metabolite measures obtained by NMR are more reflective of underlying biology than are the composite measures used in clinical practice. This idea is in line with the heritability estimates of composite measures being similar between enzymatic- and NMR-based measures of HDL-C and LDL-C. For triglycerides, the higher heritability estimate for NMR-based measures may reflect its smaller variance compared to enzymatic triglyceride measures. Smaller variance for the NMR-based measure has also been shown previously28.
Second, we have identified newly associated loci that may help to characterize the biochemical pathways by which serum amino acids influence risk for T2D. Five newly identified loci were associated with metabolic measures of tyrosine, phenylalanine and valine, three of the five amino acids recently shown to be associated with the risk for developing T2D6.
Third, by employing NMR-based metabolite measures for GWAS, we were able to identify new loci associated with lipid metabolism using a much smaller sample size than has been required to identify most such loci for composite lipid traits. There are several potential explanations for why these signals have not previously been detected by large GWAS consortia. For example, the loci containing FCGR2B and PPP1R11 showed association patterns in the lipoprotein spectrum that likely confounded composite measures, and the CPT1A locus was associated with a specific metabolic measure (the LA/PUFA ratio) that is not usually considered in the analyses of lipid phenotypes. These observations suggest that further enhancing the range of measures of lipid metabolism used in genetic association analyses will likely provide a more complete picture of the loci modifying lipids and lipoproteins.
The results reported here represent only the first step in the genetic dissection of high-resolution metabolic phenotypes. The metabolome contains many more types of molecules and particles than those measured in the 216 phenotypes assessed here. The phenotypes assessed here were assayed in a high-throughput and cost-effective manner in order to attain the large sample numbers required for genome-wide association analyses. Measurements using mass spectrometry methods broaden the range of metabolites beyond those identifiable using NMR methods, and both approaches are required to obtain a more comprehensive view of the metabolome9. NMR, which is less expensive and more automatable than mass spectrometry, is therefore more suitable for large-scale genetic investigations. It can also extract information from a range of lipoprotein particles not assayable using mass spectrometry. On the other hand, mass spectrometry provides analytical opportunities that are outside the reach of NMR spectroscopy. Mass spectrometry is several fold more sensitive and can be used as a discovery tool to identify new compounds as well as assay a wider spectrum of known molecules.
The use in this study of the most comprehensive available imputation panel increased the opportunities for identifying new loci, as evidenced by the fact that 4 of the 11 newly associated loci reported here would not have been detected with the commonly used HapMap 2 panel. On the other hand, imputation on this scale can also enhance the chances of identifying false positive loci. For this reason, we applied particularly stringent quality control filters to prevent false positives caused by imputation errors. For example, we restricted our search to only variants that had a minor allele frequency (MAF) of >0.01, good imputation quality and similar effect sizes in all cohorts. Although 9 of the 11 newly identified loci showed the strongest association with an imputed marker, all but two loci also showed association to directly genotyped SNPs in three or more cohorts. The two remaining loci showed high accuracy of imputation in sequencing undertaken to validate the imputed genotypes. Thus, we used highly conservative procedures to diminish the risk of false positive associations, and these strict filters may have abolished some true positive signals.
In conclusion, the study highlights the value of enhancing the specificity of metabolic phenotyping for genetic association analyses. The availability of 216 metabolite measures by NMR provides a substantial enhancement over the measures obtained by classical clinical chemistry methods that are typically available in large population cohorts. Using these phenotypes with the higher-resolution genetic analysis now possible using the 1000 Genomes Project variant catalog, we identified several new loci associated with serum metabolites, some of which may be important in risk for T2D.
URLs
1000 Genomes Project imputation reference panel, http://mathgen.stats.ox.ac.uk/impute/data_download_1000G_pilot_plus_hapmap3.html.
ONLINE METHODS
We performed a GWAS for metabolites and metabolite ratios in five cohorts from Finland totaling 8,330 individuals (cohorts are described in Table 1 and the Supplementary Note). Written informed consent was obtained from all participants. Studies were approved by the following ethical committees: Ethical Committee of Oulu University Faculty of Medicine for NFBC 1966, Ethics Committee of the National Public Health Institute for Health2000 and HBCS, Helsinki University Hospital Coordinating Ethical Committee for DILGOM and Twins and Ethics Committee of the Hospital District of Southwest Finland for YF. The genomic positions indicated throughout this study are based on NCBI human genome build 36. Some of the markers from the 1000 Genomes imputation reference set highlighted in this manuscript have since been assigned rs numbers (1-55889093, rs72669744; 1-159807481, rs67418890; 4-73541429, rs115136538; 8-19956650, rs115849089; and 17-7083575, rs117616209).
Genotyping and imputation
All cohorts were genotyped using commercially available Illumina genotyping arrays. NFBC1966 was genotyped using the HumanHap CNV 370k array. DILGOM and GenMets were genotyped with the Illumina HumanHap 610k array. YF and HBCS were genotyped using a custom-generated HumanHap 670k array that largely overlapped with the HumanHap 610k array but had additional copy-number probes. Quality control analysis was performed independently for each study before imputation. Poor quality markers (for which genotyping failed in >5% of samples) and poor quality DNA samples (for which genotyping failed at >5% of markers) were removed from further analysis. In addition, individuals with excessive genome-wide heterozygosity (indicating sample contamination) or gender discrepancies as well, as closely related individuals, were removed from the data. Imputation was performed on the cleaned data using IMPUTE36. Imputation included a 1000 Genomes imputation reference and a HapMap 3 imputation reference, which included an additional Finnish imputation reference in HapMap 3 depth. The reference used included HapMap3 and 1000 Genomes in NCBI build 36, where the HapMap 3 files were from release 2 (February 2009) and the 1000 Genomes files were from the low-coverage pilot genotypes released in March 2010 (see URLs). The benefit of the additional Finnish reference set has previously been discussed in detail14. The regional plots of the associated loci and genotyping status or imputation reference are presented (Supplementary Fig. 1). The SNPs reported in Tables 2 and 3, other than those directly genotyped, have imputation quality of >0.7. The quantile-quantile plots for phenotypes that show the strongest association with newly identified loci are presented (Supplementary Fig. 2), as are the box plots of the associations (Supplementary Fig. 3).
To assess the accuracy of imputation, we compared imputed genotypes for 316 markers showing genome-wide significance with directly genotyped Cardiometabochip SNPs available for the DILGOM study sample (Supplementary Fig. 4). The concordance between genotyped SNPs on the Cardiometabochip and imputed genotypes in DILGOM was high (94% of the SNPs had r2 > 0.8), indicating imputation was accurate for the reported SNPs. Imputation accuracy was also relatively similar across the whole region of MAFs ranging from 1% to 50% (Supplementary Fig. 4).
We tested the effect of using the comprehensive imputation reference panel described above by analyzing the same 31 loci reported in this study using the HapMap 2 release 22 Utah residents of Northern and Western European ancestry (CEU) imputation reference panel with the lead trait for each locus reported in Tables 2 and 3. Individual cohorts were imputed with the HapMap 2 reference panel using MACH37 and analyzed using ProbABEL38, and the results from individual cohorts were combined with an inverse variance meta-analysis using GWAMA39.
Serum metabolomics
All cohorts were analyzed using the same high-throughput serum NMR metabolomics platform in the same analysis laboratory as described previously40. This methodology provided information on 117 serum measures, including lipoprotein subclass distribution and lipoprotein particle concentration, low molecular weight metabolites, such as amino acids, 3-hydroxybutyrate and creatinine, and detailed molecular information on serum lipids, including free and esterified cholesterol, sphingomyelin and fatty acid saturation. Further details for the NMR spectroscopy and data analyses are provided in the Supplementary Note.
Genome-wide association analyses and meta-analysis
Data from individuals using lipid-lowering medication or pregnant individuals were removed before analysis. In all of the studies, metabolomic phenotypes were measured from fasting serum samples. Within each study, residual metabolomic concentrations or residuals of ratios were determined after regression adjustment using R software41. The selected ratios of metabolites were calculated by taking a ratio of particular metabolite measures. Outliers (≥4 s.d. from the mean) were removed from resulting ratio values before covariate adjustment, as the calculation is sensitive to very strong outliers, and strong outliers hamper the covariate correction. To calculate residuals for all metabolites and ratios, each study included as covariates age (except in NFBC66, where all individuals were examined at the age of 31 years), the first ten principal components from genetic data to correct for possible population stratification and sex. Residuals were normalized to have a mean of 0 and s.d. of 1 using inverse normal transformation. The resulting normal distributions of all 216 phenotypes were correlated against genotypes, assuming an additive genetic model using the SNPTEST program36. To combine the effect estimates from five distinct studies, we conducted a fixed-effects inverse variance meta-analysis using META36 for each phenotype. Only good-quality SNPs were included in further evaluations on the basis of the following criteria: imputation informativeness was >0.4, there was no heterogeneity in the effect sizes for the SNP between cohorts (Cochran’s Q statistic P value < 1 × 10−5), and the SNP had to have a result in all five cohorts. For SNPs successfully analyzed in each cohort, the number excluded as a result of heterogeneity in effect size ranged between 28 and 720 for different traits. The genome-wide inflation factors were calculated from each meta-analysis (Supplementary Table 4). All meta-analysis test statistics were corrected by using a genomic inflation factor for each trait. A stringent genome-wide significance level of 2.31 × 10−10 was set to correct for multiple testing of the 216 phenotypes.
P-gain values were calculated for the ratios by taking the minimum of the two P values for the individual metabolite associations divided by the P value of the ratio between both. If the P-gain value was >1, then the ratio had better power for the association than did either of the direct metabolite measures. For all ratios for which an association was presented or discussed in the manuscript, the P-gain value was >1. P-gain values are presented in Supplementary Table 7.
Cis EQTL analysis
We had whole-blood expression data available in the DILGOM cohort for 585 individuals. The expression data have been described previously42. In brief, to obtain stabilized total RNA in study III, the PAXgene Blood RNA System (PreAnalytiX) was used. Biotinylated cDNA (750 ng) was hybridized to Illumina HumanHT-12 Expression BeadChips according to the manufacturer’s protocol. The correlation of SNPs, which had significant association with metabolites, was tested for correlation with expression. All expression probes within 1 Mb of the SNP were tested for correlation with the SNP allele dosage using Spearman rank correlation in R. All eQTL associations reaching a genome-wide level of significance (P < 9 × 10−7) with a 1% genome-wide false discovery rate43 are presented (Supplementary Table 8).
Heritability estimates
Briefly, we used 561 twin pairs (221 monozygotic pairs and 340 dizygotic pairs, aged 22–25) from the Finnish population to estimate intrapair metabolite correlation for monozygotic and dizygotic pairs. For each phenotype, models estimating the hypothetical combinations of the different genetic and environmental sources of influence (including, additive genetic influences (A), shared environmental influences (C), dominance genetic influences (D) and unique environmental influences (E)) were built and tested against a saturated model, where no inference on the underlying architecture of the phenotype was assumed. The estimation of heritability is presented in detail in the Supplementary Note, as is the comparison between clinical and NMR-based lipid measures.
Proportion of variance explained
A subset of the twin cohort used for the heritability estimates also had genome-wide SNP data available. The genotyping was performed using Illumina HumanHap 670k custom arrays, and genotypes were clustered using the Illuminus algorithm44. The part of the twin cohort that was used for heritability estimation was stratified for alcohol drinking. To match the population-level alcohol consumption distribution, we randomly chose one member of the twin pair from this subset of twins. This resulted in a random population sample of 436 individuals with both metabolite data and genotypes. We assessed the proportion of variance explained in this independent population sample of 436 individuals by the 33 SNPs (31 from the GWAS and 2 from fine mapping) by creating a genetic risk score for each trait for which the significant SNP associated with a nominal genome-wide level of significance (P < 5 × 10−8).
Clinical lipid measures in population samples
The enzymatic lipoprotein measures were analyzed in the same study sample as the metabolomic phenotypes, but additional individuals (n = 2,247) were also available, who were not part of the metabolomic studies. The results from all these individuals were used to test for the association of the ALB locus with enzymatic lipoprotein measures. The sample cohorts were the same as those used in this study. Data from individuals who were pregnant or using lipid-lowering medication were removed before analysis, and lipid measures were adjusted for age, sex and the ten first principal components and transformed using inverse normal transformation. Enzymatic lipoprotein measures have been described in an earlier study5.
Supplementary Material
Acknowledgments
We thank all the Finnish volunteers who participated in the studies. We thank the IT Center for Science and the technology center of the Institute for Molecular Medicine Finland for providing the computational facilities required in this study. The expert technical assistance for statistical analyses provided by A. Vikman, I. Lisinen, V. Aalto and the Genotyping Facilities at the Wellcome Trust Sanger Institute are gratefully acknowledged. The study was supported through funds from The European Community’s Seventh Framework Programme (FP7/2007-2013), the BioSHaRE Consortium (261433), the Sigrid Juselius Foundation (251217 to S.R.), the Academy of Finland (137870 to P.S. and 135973 to P.W.), the Responding to Public Health Challenges Research Programme of the Academy of Finland (129269 to M.J.S., 129429 to M.A.-K., 129322 to M.P. and 139635 to V.S.), the Academy of Finland Center of Excellence in Complex Disease Genetics (213506 and 129680 to A.P., J. Kaprio, L.P., K.S. and S.R.), the Finnish Foundation for Cardiovascular Research (to M.J.S., M.A.-K., M.P., S.R. and K.H.P), the Jenny and Antti Wihuri Foundation (to A.J.K.), the Instrumentarium Science Foundation (to T.T. and P.W.), the Finnish Cultural Foundation (to T.T. and T.L.), an Aalto University School of Science and Technology researcher training scholarship (to T.T.) and the Wellcome Trust (098051 to A.P.). The Young Finns Study has been financially supported by the Academy of Finland (126925, 121584, 124282, 129378 (Salve), 117787 (Gendi) and 41071 (Skidi)), the Social Insurance Institution of Finland, the Turku University Foundation, the Yrjö Jahnsson Foundation, the Emil Aaltonen Foundation (to T.L.), the Medical Research Fund of Tampere University Hospital, the Turku University Hospital Medical Fund, the Juho Vainio Foundation, the Finnish Foundation for Cardiovascular Research (to T.L.) and the Tampere Tuberculosis Foundation (to T.L. and M.K.). The Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland (120386, 125876 and 126775 to J.E.), the Finnish Diabetes Research Society, the Novo Nordisk Foundation, the European Science Foundation (EuroSTRESS), the Wellcome Trust (89061/Z/09/Z and 089062/Z/09/Z), the Samfundet Folkhälsan and the Finska Läkaresällskapet. The FINRISK/DILGOM study was supported by the Academy of Finland (118081). Data collection for FinnTwin12 and FinnTwin16 were supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) (AA-12502, AA-09203 and AA-08315 to R.J.R. and AA-15416 to D.M.D.) and the Academy of Finland (100499, 205585, 118555 and 141054 (Skidi-Kids) to J. Kaprio). The Finnish Twin cohorts are also supported by the Novo Nordisk Foundation, the Diabetes Research Foundation, Biomedicum Helsinki and Helsinki University Central Hospital grants (all to K.H.P.). NFBC1966 received financial support from the Academy of Finland (104781, 120315, 129269, 1114194, 139900 and SALVE to M.-R.J. and Center of Excellence in Complex Disease Genetics to L.P.), University Hospital Oulu, Biocenter, University of Oulu (75617 to M.-R.J. and M.J.S.), the European Commission EURO-BLCS Framework 5 award (QLG1-CT-2000-01643 to M.-R.J.), the US National Heart, Lung, and Blood Institute (NHLBI) (5R01HL087679), the US National Institute of Mental Health (NIMH) (1RL1MH083268), European Network for Genetic and Genomic Epidemiology (ENGAGE) (HEALTH-F4-2007-201413 to L.P. and M.-R.J.), the MRC UK (G0500539, G0600705 and PrevMetSyn/Salve to M.-R.J.) and the Wellcome Trust (GR069224).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
Experiments were designed by L.P., M.P., M.A.-K., A.P. and S.R. Statistical analyses were performed by J. Kettunen, T.T., A.O.-A., E.T. and L.-P.L. Materials and/or analysis tools were contributed by J. Kettunen, T.T., A.-P.S., P.S., A.J.K., P.W., K.S., D.M.D., R.J.R., M.J.S., J.V., M.K., T.L., K.H.P., M.I.M., A.J., J.E., O.T.R., V.S., J. Kaprio, M.-R.J., N.B.F., M.A.-K., A.P. and S.R. The manuscript was written by J. Kettunen, T.T., A.J.K., M.I., N.B.F., M.A.-K., A.P. and S.R. All authors reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Schlessinger BS, Wilson FH, Jr, Milch LJ. Serum parameters as discriminators between normal and coronary groups. Circulation. 1959;19:265–268. doi: 10.1161/01.cir.19.2.265. [DOI] [PubMed] [Google Scholar]
- 2.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 3.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolz M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 8.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Illig T, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gieger C, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soeters PB, Fischer JE. Insulin, glucagon, aminoacid imbalance, and hepatic encephalopathy. Lancet. 1976;2:880–882. doi: 10.1016/s0140-6736(76)90541-9. [DOI] [PubMed] [Google Scholar]
- 12.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surakka I, et al. Founder population–specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging. Genome Res. 2010;20:1344–1351. doi: 10.1101/gr.106534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natt E, Kida K, Odievre M, Di Rocco M, Scherer G. Point mutations in the tyrosine aminotransferase gene in tyrosinemia type II. Proc Natl Acad Sci USA. 1992;89:9297–9301. doi: 10.1073/pnas.89.19.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orinska Z, et al. TLR3-induced activation of mast cells modulates CD8+ T-cell recruitment. Blood. 2005;106:978–987. doi: 10.1182/blood-2004-07-2656. [DOI] [PubMed] [Google Scholar]
- 18.Marone G, Galli SJ, Kitamura Y. Probing the roles of mast cells and basophils in natural and acquired immunity, physiology and disease. Trends Immunol. 2002;23:425–427. doi: 10.1016/s1471-4906(02)02274-3. [DOI] [PubMed] [Google Scholar]
- 19.Cross LJ, Heaney LG, Ennis M. Histamine release from human bronchoalveolar lavage mast cells by neurokinin A and bradykinin. Inflamm Res. 1997;46:306–309. doi: 10.1007/s000110050192. [DOI] [PubMed] [Google Scholar]
- 20.Bouatia-Naji N, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 21.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin Y, Vaziri ND, Willekes N, Kim CH, Joles JA. Effects of gender on hepatic HMG-CoA reductase, cholesterol 7α-hydroxylase, and LDL receptor in hereditary analbuminemia. Am J Physiol Endocrinol Metab. 2005;289:E993–E998. doi: 10.1152/ajpendo.00037.2005. [DOI] [PubMed] [Google Scholar]
- 23.Rosipal S, Debreova M, Rosipal R. A speculation about hypercholesterolemia in congenital analbuminemia. Am J Med. 2006;119:181–182. doi: 10.1016/j.amjmed.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 24.Koot BG, Houwen R, Pot DJ, Nauta J. Congenital analbuminaemia: biochemical and clinical implications. A case report and literature review. Eur J Pediatr. 2004;163:664–670. doi: 10.1007/s00431-004-1492-z. [DOI] [PubMed] [Google Scholar]
- 25.Zhao M, et al. FcγRIIB inhibits the development of atherosclerosis in low- density lipoprotein receptor–deficient mice. J Immunol. 2010;184:2253–2260. doi: 10.4049/jimmunol.0902654. [DOI] [PubMed] [Google Scholar]
- 26.Hernández-Vargas P, et al. Fcγ receptor deficiency confers protection against atherosclerosis in apolipoprotein E knockout mice. Circ Res. 2006;99:1188–1196. doi: 10.1161/01.RES.0000250556.07796.6c. [DOI] [PubMed] [Google Scholar]
- 27.Cohen PT. Protein phosphatase 1—targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 28.Chasman DI, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergstrom JD, Reitz RC. Studies on carnitine palmitoyl transferase: the similar nature of CPTi (inner form) and CPTo (outer form) Arch Biochem Biophys. 1980;204:71–79. doi: 10.1016/0003-9861(80)90008-9. [DOI] [PubMed] [Google Scholar]
- 31.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, et al. Genetic and environmental contributions to phenotypic components of metabolic syndrome: a population-based twin study. Obesity (Silver Spring) 2009;17:1581–1587. doi: 10.1038/oby.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isaacs A, et al. Heritabilities, apolipoprotein E, and effects of inbreeding on plasma lipids in a genetically isolated population: the Erasmus Rucphen Family Study. Eur J Epidemiol. 2007;22:99–105. doi: 10.1007/s10654-006-9103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 39.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soininen P, et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134:1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- 41.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2007. [Google Scholar]
- 42.Inouye M, et al. An immune response network associated with blood lipid levels. PLoS Genet. 2010;6:e1001113. doi: 10.1371/journal.pgen.1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding J, et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87:779–789. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo YY, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–2746. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.