Abstract
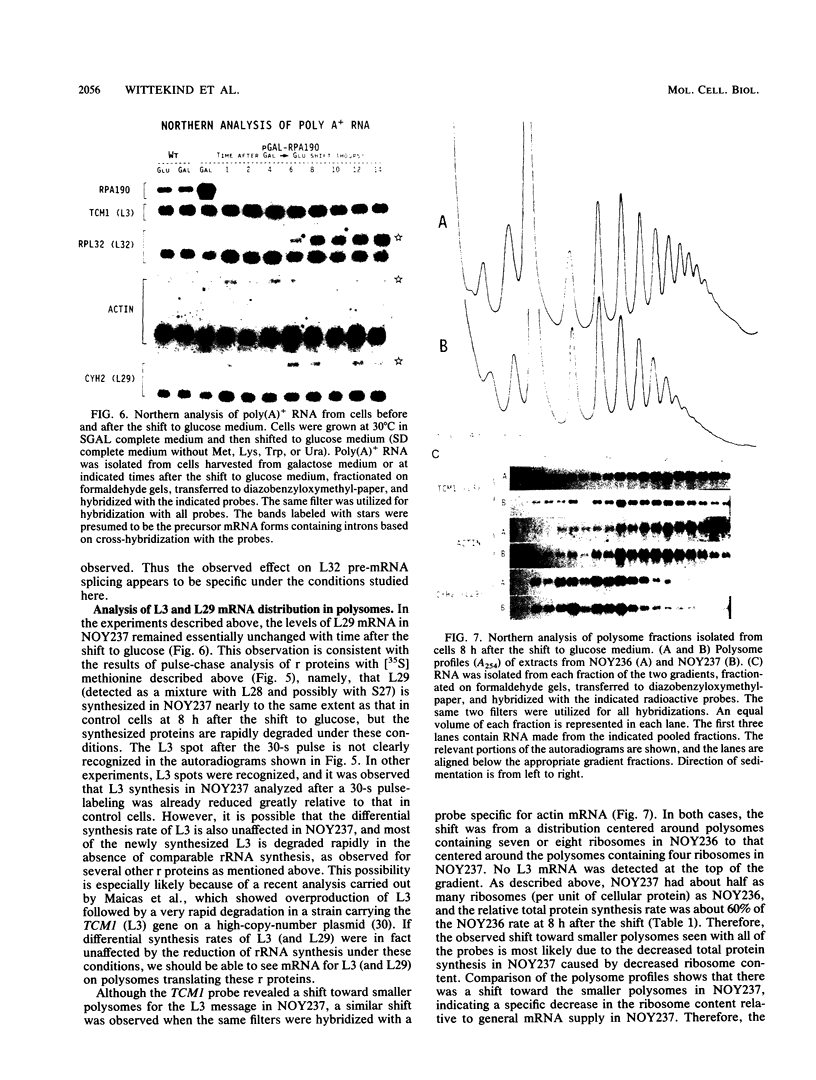
The synthesis of ribosomal proteins (r proteins) under the conditions of greatly reduced RNA synthesis were studied by using a strain of the yeast Saccharomyces cerevisiae in which the production of the largest subunit (RPA190) of RNA polymerase I was controlled by the galactose promoter. Although growth on galactose medium was normal, the strain was unable to sustain growth when shifted to glucose medium. This growth defect was shown to be due to a preferential decrease in RNA synthesis caused by deprivation of RNA polymerase I. Under these conditions, the accumulation of r proteins decreased to match the rRNA synthesis rate. When proteins were pulse-labeled for short periods, no or only a weak decrease was observed in the differential synthesis rate of several r proteins (L5, L39, L29 and/or L28, L27 and/or S21) relative to those of control cells synthesizing RPA190 from the normal promoter. Degradation of these r proteins synthesized in excess was observed during subsequent chase periods. Analysis of the amounts of mRNAs for L3 and L29 and their locations in polysomes also suggested that the synthesis of these proteins relative to other cellular proteins were comparable to those observed in control cells. However, Northern analysis of several r-protein mRNAs revealed that the unspliced precursor mRNA for r-protein L32 accumulated when rRNA synthesis rates were decreased. This result supports the feedback regulation model in which excess L32 protein inhibits the splicing of its own precursor mRNA, as proposed by previous workers (M. D. Dabeva, M. A. Post-Beittenmiller, and J. R. Warner, Proc. Natl. Acad. Sci. USA 83:5854-5857, 1986).
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Gritz L., Tung L., Rosbash M. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Dec;5(12):3429–3435. doi: 10.1128/mcb.5.12.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal M. G., Bowman L. H. Transcriptional and translational regulation of ribosomal protein formation during mouse myoblast differentiation. J Biol Chem. 1987 Apr 5;262(10):4868–4875. [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baim S. B., Pietras D. F., Eustice D. C., Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985 Aug;5(8):1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen G. H., Cohen L. H., Mager W. H., Klaassen A. W., Planta R. J. Isolation of cloned ribosomal protein genes from the yeast Saccharomyces carlsbergensis. Gene. 1981 Sep;14(4):279–287. doi: 10.1016/0378-1119(81)90160-8. [DOI] [PubMed] [Google Scholar]
- Bowman L. H. The synthesis of ribosomal proteins S16 and L32 is not autogenously regulated during mouse myoblast differentiation. Mol Cell Biol. 1987 Dec;7(12):4464–4471. doi: 10.1128/mcb.7.12.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabeva M. D., Post-Beittenmiller M. A., Warner J. R. Autogenous regulation of splicing of the transcript of a yeast ribosomal protein gene. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5854–5857. doi: 10.1073/pnas.83.16.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabeva M. D., Warner J. R. The yeast ribosomal protein L32 and its gene. J Biol Chem. 1987 Nov 25;262(33):16055–16059. [PubMed] [Google Scholar]
- Fallon A. M., Jinks C. S., Strycharz G. D., Nomura M. Regulation of ribosomal protein synthesis in Escherichia coli by selective mRNA inactivation. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3411–3415. doi: 10.1073/pnas.76.7.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Finley D., Bartel B., Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989 Mar 30;338(6214):394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- Fried H. M., Nam H. G., Loechel S., Teem J. Characterization of yeast strains with conditionally expressed variants of ribosomal protein genes tcm1 and cyh2. Mol Cell Biol. 1985 Jan;5(1):99–108. doi: 10.1128/mcb.5.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Sures I. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 May;77(5):2546–2550. doi: 10.1073/pnas.77.5.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenstein C., Warner J. R. Coordinate regulation of the synthesis of eukaryotic ribosomal proteins. Proc Natl Acad Sci U S A. 1976 May;73(5):1547–1551. doi: 10.1073/pnas.73.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelfarb H. J., Vassarotti A., Friesen J. D. Molecular cloning and biosynthetic regulation of cry1 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1984;195(3):500–506. doi: 10.1007/BF00341453. [DOI] [PubMed] [Google Scholar]
- Howard G. A., Traut R. R. Separation and radioautography of microgram quantities of ribosomal proteins by two-dimensional polyacrylamide gel electrophoresis. FEBS Lett. 1973 Jan 15;29(2):177–180. doi: 10.1016/0014-5793(73)80555-1. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A., Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. Study with antibodies to yeast RNA polymerases subunits. J Biol Chem. 1982 Mar 10;257(5):2613–2618. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay M. A., Jacobs-Lorena M. Selective translational regulation of ribosomal protein gene expression during early development of Drosophila melanogaster. Mol Cell Biol. 1985 Dec;5(12):3583–3592. doi: 10.1128/mcb.5.12.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H., Warner J. R. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983 Mar;3(3):457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T., Planta R. J. Analysis of the protein composition of yeast ribosomal subunits by two-dimensional polyacrylamide gel electrophoresis. Mol Biol Rep. 1974 Sep;1(7):409–415. doi: 10.1007/BF00385674. [DOI] [PubMed] [Google Scholar]
- Käufer N. F., Fried H. M., Schwindinger W. F., Jasin M., Warner J. R. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983 May 25;11(10):3123–3135. doi: 10.1093/nar/11.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Archer R., Zengel J. M. Transcription of the S10 ribosomal protein operon is regulated by an attenuator in the leader. Cell. 1983 May;33(1):241–248. doi: 10.1016/0092-8674(83)90353-7. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- Maicas E., Pluthero F. G., Friesen J. D. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol Cell Biol. 1988 Jan;8(1):169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Pearson N. J., Fried H. M., Warner J. R. Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell. 1982 Jun;29(2):347–355. doi: 10.1016/0092-8674(82)90151-9. [DOI] [PubMed] [Google Scholar]
- Pierandrei-Amaldi P., Beccari E., Bozzoni I., Amaldi F. Ribosomal protein production in normal and anucleolate Xenopus embryos: regulation at the posttranscriptional and translational levels. Cell. 1985 Aug;42(1):317–323. doi: 10.1016/s0092-8674(85)80127-6. [DOI] [PubMed] [Google Scholar]
- Pierandrei-Amaldi P., Bozzoni I., Cardinali B. Expression of the gene for ribosomal protein L1 in Xenopus embryos: alteration of gene dosage by microinjection. Genes Dev. 1988 Jan;2(1):23–31. doi: 10.1101/gad.2.1.23. [DOI] [PubMed] [Google Scholar]
- Schultz L. D., Friesen J. D. Nucleotide sequence of the tcml gene (ribosomal protein L3) of Saccharomyces cerevisiae. J Bacteriol. 1983 Jul;155(1):8–14. doi: 10.1128/jb.155.1.8-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel L. F., Jacobson A. Translational control of ribosomal protein synthesis during early Dictyostelium discoideum development. Mol Cell Biol. 1987 Mar;7(3):965–972. doi: 10.1128/mcb.7.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. S., Bedwell D. M., Nomura M. Regulation of alpha operon gene expression in Escherichia coli. A novel form of translational coupling. J Mol Biol. 1987 Jul 20;196(2):333–345. doi: 10.1016/0022-2836(87)90694-2. [DOI] [PubMed] [Google Scholar]
- Tsay Y. F., Thompson J. R., Rotenberg M. O., Larkin J. C., Woolford J. L., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 1988 Jun;2(6):664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- Tsurugi K., Ogata K. Preferential degradation of newly synthesized ribosomal proteins in rat liver treated with a low dose of actinomycin D. Biochem Biophys Res Commun. 1977 Apr 11;75(3):525–531. doi: 10.1016/0006-291x(77)91504-2. [DOI] [PubMed] [Google Scholar]
- Veinot-Drebot L. M., Singer R. A., Johnston G. C. Rapid initial cleavage of nascent pre-rRNA transcripts in yeast. J Mol Biol. 1988 Jan 5;199(1):107–113. doi: 10.1016/0022-2836(88)90382-8. [DOI] [PubMed] [Google Scholar]
- Warner J. R. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol. 1977 Sep 25;115(3):315–333. doi: 10.1016/0022-2836(77)90157-7. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985 Jun;5(6):1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Dodd J., Vu L., Kolb J. M., Buhler J. M., Sentenac A., Nomura M. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol. 1988 Oct;8(10):3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M., Nomura M. Deficiency in both type I and type II DNA topoisomerase activities differentially affect rRNA and ribosomal protein synthesis in Schizosaccharomyces pombe. Curr Genet. 1988 Apr;13(4):305–314. doi: 10.1007/BF00424424. [DOI] [PubMed] [Google Scholar]
- elBaradi T. T., van der Sande C. A., Mager W. H., Raué H. A., Planta R. J. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr Genet. 1986;10(10):733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]







