Abstract
A large group of bacterial small regulatory RNAs (sRNAs) use the Hfq chaperone to mediate pairing with and regulation of mRNAs. Recent findings help to clarify how Hfq acts and highlight the role of the endonuclease RNase E and its associated proteins (the degradosome) in negative regulation by these sRNAs. sRNAs frequently uncouple transcription and translation by blocking ribosome access to the mRNA, allowing other proteins access to the mRNA. As more examples of sRNA-mediated regulation are studied, more variations on how Hfq, RNase E, and other proteins collaborate to bring about sRNA-based regulation are being found.
Keywords: Bacterial Genetics, Escherichia coli, RNA Turnover, RNA-binding Proteins, Translation Control, Hfq, sRNA
Post-transcriptional Regulation by Small Noncoding RNAs in Bacteria
The idea that RNAs could function as regulators of gene expression has been around since the earliest studies of gene regulation. In their seminal paper entitled “Genetic Regulatory Mechanisms in the Synthesis of Proteins”, Jacob and Monod originally hypothesized, “The specific 'repressor' (RNA?), acting with a given operator, is synthesized by a regulator gene” (1). Although the repressor in the case of the lac operon turned out to be the Lac repressor protein, the later discovery of small RNA (sRNA)3 regulators confirmed their original hypothesis.
Currently, examples of this form of gene regulation are widespread among organisms. Here, we will focus on pairing sRNAs in bacteria and, specifically, those that are often termed trans-encoding sRNAs. These RNAs are expressed from the DNA in trans, i.e. the sRNA genes are far from the genes encoding their mRNA target(s) and have limited complementarity with their target mRNAs. These bacterial sRNAs typically range in length from ∼50 to 300 nucleotides. Many of these sRNAs are highly expressed when cells are undergoing some type of stress (for instance, oxidative stress, sugar phosphate accumulation, or nutrient starvation). The sRNAs base pair with their mRNA targets, leading to a variety of outcomes. Base pairing can lead to stabilization and/or translational activation of an mRNA target. Usually, activation occurs by base pairing within the 5′-UTR, changing folding of the 5′-UTR to allow entry of the ribosome and translation to occur (reviewed in Refs. 2 and 3). Another mode of action by sRNAs ultimately leads to translational repression and/or degradation of an mRNA target. In the majority of characterized cases, an sRNA base pairs at or around the ribosome-binding site (RBS) of an mRNA target. This leads to inhibition of translational initiation and, in most cases, the subsequent destabilization of the target. Negative regulation can also occur in other ways, as discussed below. Degradation of the mRNA target reinforces the translational repression and makes it irreversible.
In many bacteria, an RNA chaperone, Hfq, is required for efficient base pairing between an sRNA and its target mRNA (reviewed in Ref. 4). In this minireview, we will focus on recent advances in understanding sRNA-mediated negative gene regulation in Escherichia coli and Salmonella enterica. More specifically, we will focus on the variety of pathways that lead to sRNA-mediated degradation of specific mRNA targets and how Hfq engages the various proteins associated with these pathways.
Hfq and Its Role in sRNA-mediated Regulation
Originally identified in E. coli as a host factor required for the efficient replication of the RNA bacteriophage Qβ (5), Hfq is now understood to function as an RNA chaperone within bacterial cells. Hfq has been characterized as a member of the conserved RNA-binding Lsm (like-Sm)/Sm-like protein family found in eukaryotes, bacteria, and archaea. In eukaryotes, the Sm and Lsm proteins have been implicated in many functions, including roles in mRNA splicing, RNA decapping, and RNA stabilization (reviewed in Refs. 6 and 7). This protein family is characterized as forming a ring-like multimeric complex that binds RNA. The eukaryotic Sm proteins generally form heteromultimeric rings, whereas the bacterial Hfq proteins characterized thus far are homomultimeric.
In recent years, Hfq has gained much attention after it was shown to play a critical role in sRNA-mediated gene regulation. It is required for efficient stabilization and annealing of sRNAs to their mRNA targets (reviewed in Refs. 7 and 8). Homologs have been found in >50% of sequenced bacteria (9). Although the mechanism by which Hfq facilitates these interactions remains unclear, crystal structures in complex with RNA molecules and in vitro analyses have shed some light on its multiple RNA-binding surfaces. The Staphylococcus aureus Hfq crystal structure in complex with a uridine-rich aptamer revealed that RNA winds around the central cavity of the ring structure on one face of Hfq termed the proximal face (Fig. 1A) (10). A crystal structure of E. coli Hfq in complex with a poly(A) aptamer demonstrated that the face opposite to that where poly(U) binds, termed the distal face, preferentially binds adenine-rich sequences (11).
FIGURE 1.
Important protein players in sRNA-mediated gene repression. A, faces of Hfq for RNA interactions, illustrating the proximal face of Hfq (orange) bound to sRNA (green), the distal face of Hfq (purple) bound to an mRNA target (blue), and the rim of Hfq (red), shown to have interactions with some sRNAs. The currently understood function of each face in sRNA-mediated regulation is summarized at the bottom of each panel. B, organization of the RNA degradosome in E. coli. The endonuclease RNase E consists of two domains. The N-terminal domain (residues 1–529) resembles RNase G and contains the catalytic site of the enzyme. The C-terminal domain (residues 530–1061) serves as a scaffold domain with which the other components of the degradosome associate to form an active complex. These components and the residues at which they have been shown to interact with RNase E are shown. Interactions of Hfq with RNase E regions closer to the C terminus have also been suggested. See text for references.
In vitro studies have confirmed that mutants in the proximal face disrupt binding of uridine-rich sequences (12). The proximal face may be the site for initial sRNA interactions. One basis for this interaction may be the Rho-independent terminators with polyuridine tails characteristic of these sRNAs. It has been demonstrated in vitro that Hfq has a strong affinity for stem-loops followed by a run of uridines (13–15). However, internal binding sites for Hfq have also been identified in many sRNAs, including OxyS (16), DsrA (17), RyhB (18), and RybB (19). A recent study on SgrS (15) further characterized this internal binding motif; in addition to the Rho-independent terminator, SgrS also required an internal U-rich sequence flanked by a stem-loop for stable binding to Hfq and subsequent sRNA-mediated regulation. The study found that this motif is present in a large portion of the currently characterized sRNAs, suggesting more extensive interactions between Hfq and sRNAs. Recent tests with mutant Hfq proteins indicated that these interactions are abrogated or weakened upon mutation of proximal site residues; these same mutations lead to reduced sRNA accumulation in vivo, consistent with loss of tight binding to the sRNAs (20).
Studies on the distal face of Hfq have further characterized it as a site for poly(A) binding and, more specifically, for mRNA interactions (12, 21). The distal face has a strong affinity for mRNAs containing an ARN-binding motif, in which R is purine and N is any base (11). The requirement for an ARN motif for Hfq binding and subsequent sRNA-mediated gene regulation was first reported with the DsrA mRNA target rpoS (22). Later studies confirmed the importance of this motif and its role in sRNA regulation (23–25). In addition, mutations in the distal face binding residues disrupt positive regulation and some negative regulation (20).
Recently, a third site for interaction has been characterized, termed the lateral surface or the rim of the Hfq ring. This surface harbors a positively conserved patch that has been implicated in sRNA binding. It has been suggested that this third interaction site may play an important role in the stepwise association with sRNAs that eventually leads to mRNA binding and dissociation from Hfq (21),4 and this model is supported by in vivo studies of regulation in rim mutants (20).
As noted above, the mechanism by which Hfq facilitates sRNA-mRNA interactions remains unclear. One current hypothesis for the role of Hfq in the cell is to increase the local concentration of the RNAs and thereby increase the likelihood of base pairing. This requires that a given Hfq hexamer bind the appropriate pair of sRNA and mRNA.
Alternatively (or in addition), Hfq may facilitate base pairing by restructuring the RNAs to take on more favorable conformations for interaction (7, 27). Both (or yet other models) could apply in different cases. Regardless, further experiments will be required to elucidate the mechanism by which Hfq facilitates these RNA-RNA interactions.
Although Hfq is an abundant protein, recent studies suggest that there is competition for binding to Hfq (28–30). Each Hfq hexamer appears to bind one sRNA and one mRNA at a time (31).
Introduction to the Degradosome and Its Role in sRNA Regulation
Studies of proteins that facilitate the role of Hfq in sRNA-dependent negative regulation have focused on RNase E and its role in degradation of target mRNAs (32–35). The N terminus of RNase E encodes an essential endonuclease. RNase E cuts RNAs at single-stranded regions and is important for both mRNA degradation and processing of some structured RNAs.
The C terminus of RNase E acts as a scaffold for the binding of several proteins, including the RNA helicase RhlB, the 3′–5′-exoribonuclease polynucleotide phosphorylase (PNPase), and the glycolytic enzyme enolase (36). The resulting complex of proteins is referred to as the degradosome. RNase E itself is active as a tetramer, and thus, it can be imagined that different subunits of the tetramer may interact with different auxiliary proteins. After endonucleolytic cleavage, PNPase helps to degrade RNAs further by its exonucleolytic activity, and the RhlB helicase is believed to help PNPase deal with secondary structures in RNA targets (reviewed in Ref. 37). Thus, these three proteins provide a machine for the stepwise degradation of RNAs. The role of enolase in this complex is not understood. In addition, the C-terminal scaffold domain has two arginine-rich domains involved in RNA binding (termed ARRBD and AR2) (Fig. 1B) (reviewed in Ref. 38).
RNase E was first shown by Massé et al. (39) to be important for the coupled degradation of RyhB and its target sodB mRNA; it is also necessary for the rapid degradation of RyhB that occurs in the absence of Hfq. Either heat inactivation of a temperature-sensitive RNase E or replacement of rne with an allele that expresses RNase E lacking the C-terminal scaffold domain (Fig. 1B) inhibits degradation of sodB (39). Subsequent studies have shown that RNase E catalyzes degradation of other target mRNAs after sRNA pairing and that the C-terminal scaffold domain of RNase E is required for degradation of these mRNAs (35, 40–42). In addition, Hfq has been found to interact with the scaffold domain of RNase E (Fig. 1B; discussed below) (35).
Multiple Pathways for Negative Regulation
Through bioinformatics and experimental approaches, many Hfq-binding sRNAs were identified in E. coli and S. enterica serovar Typhimurium. One particularly powerful approach has been to isolate Hfq from cells and analyze both the bound sRNAs and mRNAs that are presumably targets for these sRNAs (43–45). Although computational approaches to match sRNAs with their targets are available, these programs still miss many targets and return many false positives. Target mRNAs have been identified for many of these sRNAs by two general experimental approaches. One is to monitor the change in abundance or the turnover of the target mRNA; this approach has been used to identify targets in microarray experiments following short-term overexpression of the sRNA or after expression of the sRNA in response to the natural inducing signal (46, 47). The second approach has been to create a translational fusion of GFP or lacZ to a gene of interest and to monitor the effect of deleting or overexpressing a set of sRNAs on this translational fusion (see, for instance, Refs. 48 and 49). Both of these approaches have identified mRNA targets that are negatively regulated by sRNAs. Characterization of the mechanism by which the sRNA leads to the down-regulation of these target mRNAs has revealed that there are multiple pathways for sRNAs to cause degradation of their target mRNAs.
Pairing of the sRNA with a target mRNA can result in translational inhibition by blocking the RBS or ribosome-loading site (Fig. 2B). Even binding of an sRNA early within the first few codons of an mRNA can block translation (50). For several sRNAs and their mRNA targets, translational inhibition in the absence of mRNA degradation is sufficient for the negative regulation of gene expression, and cleavage of the mRNA by RNase E is a subsequent step. For example, pairing of the SgrS sRNA with ptsG mRNA is sufficient to inhibit translation even if degradation of the ptsG mRNA is blocked (in an rne mutant). However, in a wild-type strain, pairing subsequently leads to degradation of the mRNA (51, 52). In a similar fashion, RyhB pairing with the sodB mRNA leads to inhibition of the translation of SodB whether or not the mRNA is degraded (52).
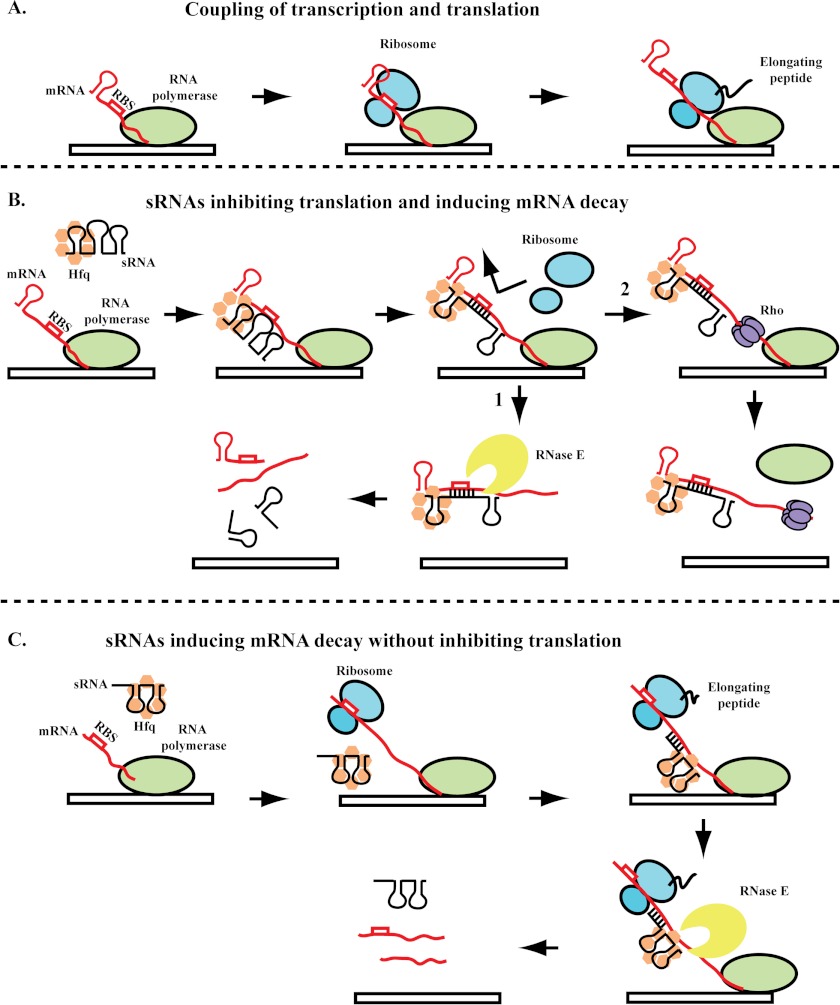
FIGURE 2.
Mechanisms of sRNA-mediated regulation. A, the coupling of transcription by RNA polymerase (green) and translation by the ribosome (blue) in the absence of sRNA regulation. Growing evidence suggests the possibility of interactions between RNA polymerase and the ribosome to ensure coupling (see text). B, sRNAs can block ribosome entry by pairing with sequences at or near the RBS of the mRNA. Pairing of the sRNA with the target mRNA is facilitated by Hfq (orange). As a result of blocking ribosome binding, the sRNA can decouple transcription and translation. Arrow 1, the unprotected mRNA may then be subject to cleavage by RNase E (yellow). Arrow 2, in addition to access to RNase E, the untranslated RNA can also provide a binding site for Rho (purple), leading to transcription termination. C, alternatively, sRNAs may induce cleavage of the mRNA by recruiting RNase E, causing mRNA cleavage without inhibiting translation. Recruitment of RNase E may be through interactions with Hfq, the RNAs, or both.
Is translational inhibition by an sRNA and the resulting absence of ribosomes on the mRNA sufficient to lead to degradation of the mRNA by RNase E? Normally, transcription and translation are tightly coupled in E. coli, and thus, most mRNAs may be covered with ribosomes soon after they emerge from RNA polymerase (Fig. 2A) (53, 54). There is a variety of evidence suggesting that blocking translation generally increases mRNA degradation, presumably because RNase E is able to bind to sites in the mRNA that would otherwise be protected by translating ribosomes (Fig. 2B) (reviewed in Ref. 2). However, there is also evidence that inhibition of translation is not always sufficient, suggesting that pairing has a second important role in recruiting RNase E. For ptsG mRNA, translational inhibition of the ptsG mRNA by addition of the translational inhibitor kasugamycin does not lead to destabilization of this mRNA unless SgrS is expressed (52). This suggests that pairing of SgrS with the ptsG mRNA leads to active recruitment of RNase E for degradation. Similarly, for RyhB regulation of sodB, a specific RNase E site far from the site of pairing was found to be necessary for sodB degradation, and blocking translation by inserting a stop codon early in the ORF was not sufficient to recruit RNase E; pairing by RyhB significantly improved RNase E-dependent cleavage (55).
Another consequence of sRNA-dependent decoupling of translation from transcription is exemplified by the regulation of the chiPQ mRNA by ChiX. The sRNA ChiX pairs with a region of the chiPQ mRNA that encompasses the RBS of chiP (56). Pairing of ChiX with the chiPQ mRNA leads to termination of the transcription of the mRNA by decoupling transcription and translation and therefore allowing entry of Rho onto the mRNA (Fig. 2B) (57). As a result, the downstream gene is not transcribed. This mechanism of negative regulation by decoupling of transcription and translation may be particularly important for polycistronic mRNAs.
Negative regulation of gene expression by an sRNA does not always result from translational inhibition. In S. enterica serovar Typhimurium, the MicC sRNA binds far downstream of the start codon of ompD, which encodes an outer membrane protein. Binding of the sRNA does not disrupt translation of the ompD mRNA by ribosomes but leads to mRNA degradation (Fig. 2C) (41). The mechanism by which MicC stimulates degradation of the ompD mRNA has been recently elucidated. The cleavage of mRNAs by RNase E is stimulated by a monophosphate at the 5′-end of the sRNA. Bandyra et al. (58) have shown that, after pairing with the ompD mRNA, the 5′-monophosphate on MicC stimulates cleavage of the ompD mRNA. Stimulated cleavage requires pairing between the sRNA and mRNA. This type of regulation presumably allows RNase E to recognize target mRNAs that would otherwise lack an ideal RNase E-binding site. Regulation of lpxR by the MicF sRNA is also partially dependent on stimulation of RNase E cleavage, in this case, by changes in the folding of the lpxR mRNA upon MicF binding (59).
Interaction of RNase E with Hfq May Be Important for RNase E Recruitment
The regulatory examples discussed above all imply recruitment of RNase E as a result of sRNA-mRNA pairing. Thus, it might not be surprising if RNase E and Hfq interact directly. In fact, several co-immunoprecipitation studies have found that Hfq interacts with RNase E and other components of the degradosome, although whether this interaction is direct or via RNA (or both) remains unclear (33, 35, 60–62).
By co-immunoprecipitation experiments with crude extracts or purified Hfq and RNase E, Hfq was found to bind to a peptide composed of residues 711–844 of RNase E, and this interaction still occurred after treatment with RNases (35). These results suggest that the interaction between Hfq and RNase E is direct and not indirect through RNA. Consistent with an important site in this region, deletion of residues 701–845 of RNase E caused a defect in regulation of the ptsG mRNA by SgrS (35). This deletion removes the region of RNase E containing the RhlB-binding site and an arginine-rich domain (AR2) involved in RNA binding but leaves the other arginine-rich binding domain (ARRBD) intact. Consistent with the idea that RhlB and Hfq occupy similar sites on RNase E, independent of RNA binding, overexpression of RhlB resulted in a decrease in the amount of Hfq bound to RNase E, and Hfq was pulled down with an RNase E derivative containing only the first 750 amino acids, lacking the AR2 domain but containing the ARRBD (35). These experiments do not rule out the possibility that either the ARRBD or AR2 domain is sufficient to bind Hfq through RNA.
Ikeda et al. (35) also noted that Hfq is likely to interact with RNase E at multiple sites, even if the site they identified is critical for regulation. These additional interactions may help to explain contrasting results obtained by Worrall et al. (62), who demonstrated in vitro that purified Hfq bound to a C-terminal fragment of RNase E from residues 628 to 843 but that this interaction did not occur unless RNA was present. This region does contain the two arginine-rich RNA-binding domains (Fig. 1B), and it is possible that the indirect binding of RNase E to Hfq by RNA can occur at lower Hfq concentrations than the direct binding reported by Ikeda et al. (35). It is certainly possible that both protein-protein interactions and RNA-mediated interactions take place in vivo. Future experiments should clarify this situation, but the data all point to an association of Hfq and RNase E that plays a role in negative regulation.
PNPase May Block RNase E Recruitment of Hfq-bound sRNAs Prior to mRNA Pairing
As discussed above, Hfq interacts directly or indirectly with the C-terminal scaffold domain of RNase E. In addition, degradation of the sRNA and target mRNA after pairing has been shown to require the C-terminal scaffold domain (35, 40–42). Together, these results suggest that Hfq may deliver the sRNA and target mRNA to the degradosome after pairing. If this is the case, what prevents Hfq from delivering the sRNA to the degradosome before the sRNA pairs with the mRNA?
For some sRNAs, PNPase blocks recruitment of Hfq-bound sRNAs by RNase E (42, 63). PNPase is a 3′–5′-exoribonuclease that catalyzes the decay of RNAs via a nucleophilic attack of the phosphodiester backbone with an inorganic phosphate. PNPase can also catalyze the reverse reaction, adding ribonucleotides to RNA, under conditions of high nucleotide diphosphate and low phosphate concentrations. PNPase can associate with the C-terminal scaffold domain of RNase E and with Hfq (32).
Two recent studies have shown that PNPase can protect sRNAs from premature degradation by RNase E (42, 63). In both studies, deletion of pnp resulted in a decreased stability of several sRNAs. Deletion of pnp also resulted in the abrogation of sRNA-dependent regulation, presumably because the sRNAs were unable to accumulate (42). In the absence of PNPase, these sRNAs were degraded by RNase E prior to pairing with target mRNAs. Premature degradation of sRNAs in the absence of PNPase required recruitment via the C-terminal domain of RNase E because deletion of this domain increased the stability of sRNAs in a pnp mutant (42). Presumably, it is Hfq-bound sRNAs that are recruited to the C-terminal scaffold domain of RNase E in the absence of pnp.
It is not yet clear whether PNPase protects only a subset of Hfq-dependent sRNAs from premature degradation or protects them only under some conditions. PNPase also contributes to degradation of sRNAs, as expected for an exonuclease, although these data reflect studies done in stationary phase, whereas the studies in which PNPase protected sRNAs were done during exponential growth. Viegas et al. (64) demonstrated that PNPase is important for degradation of two sRNAs, the Hfq-dependent MicA sRNA and the Hfq-independent SraL sRNA in S. enterica serovar Typhimurium, in stationary phase. It was also found that PNPase is critical for the degradation of MicA and RybB sRNAs, again during stationary phase, because deletion of pnp increased their stability in E. coli (65). It appears that PNPase degrades sRNAs that are not bound to Hfq because the effect of a pnp deletion in stabilizing MicA sRNA was much greater in an hfq mutant (63). RyhB and SgrS accumulated to higher levels, and their stability was modestly higher in an hfq pnp double mutant compared with the stability in an hfq or a pnp single mutant, particularly in stationary phase (63). This result is different from that found in exponential phase in our studies (42). A possible explanation for the increased degradation of these sRNAs in the hfq− pnp+ strain is that PNPase acts as a polymerase, extending the 3′-tail of the sRNA and thus providing a foothold for 3′–5′-exoribonucleases, including PNPase itself; this polymerase activity is significantly increased in the absence of Hfq (32). PNPase may also act by regulating access of the sRNA to RNase E, known to degrade sRNAs in the absence of Hfq (39), protecting them when they are bound to Hfq and accelerating their recognition and/or degradation by RNase E in the absence of Hfq.
This was not the first evidence supporting a role of pnp in protecting RNAs from degradation. In a genome-wide expression profiling experiment, deletion of pnp was shown to result in a significant increase in the expression of >500 mRNAs, as might be expected if PNPase was important for degradation of these RNAs (66); some of these increases may also reflect failure of sRNA-based negative regulation. However, deletion of pnp also led to a substantial decrease in the expression of 140 genes, suggesting the possibility that PNPase also protects many mRNAs from degradation, directly or indirectly (via failure of sRNA positive regulation, for instance) (66). Our studies of mRNA targets of sRNA regulation also support a decrease in the steady-state level of some mRNAs in pnp mutants (42). Consistent with this idea of PNPase being able to protect RNAs from degradation, Briani et al. (67) showed that overexpression of the ribosomal protein S1 results in stabilization of mRNAs and that this stabilization requires PNPase. This is intriguing because S1 binds RNA with a similar sequence specificity as Hfq and clearly carries out RNA chaperoning activities independent of the ribosome (reviewed in Ref. 68). One could imagine that PNPase regulates the accessibility to RNase E of RNAs bound to both of these proteins. Although many of the details of how PNPase affects sRNA trafficking remain unexplored, these studies all suggest that Hfq, PNPase, and RNase E interact with each other in ways that have significant effects on the fate of both sRNAs and their mRNA targets and that reflect previously unsuspected roles for PNPase (42, 66, 67).
Conclusions, Comparisons, and Future Directions
sRNA-dependent regulation is now well established in bacterial cells. The list of Hfq-dependent sRNAs in E. coli and S. enterica serovar Typhimurium is large, and each sRNA has multiple mRNA targets. Although these sRNA-mRNA pairs share dependence on some common machinery (primarily Hfq) for regulation to occur, it is becoming increasingly clear that there are many variations on how regulation actually occurs. These variations are helping us to understand more about how translational regulation in general can occur.
One important insight is the impact of the usual coupling of transcription and translation and how uncoupling this affects the ability of sRNAs to act (Fig. 2). The uncoupling of transcription and translation that underlies the ability of both RNase E and Rho to access the mRNA makes clear the important role that competition between sRNAs and ribosomes plays in translational regulation, and thus, ribosomes and their ability to load on a given mRNA must be considered as important components of the sRNA-dependent regulatory network. The growing understanding of ribosome occupancy on different mRNAs (see, for instance, Ref. 69) will help in unraveling these issues. Other protein participants in sRNA trafficking seem likely to be discovered in the near future. The complex interactions of the translation apparatus, the mRNA degradation apparatus, and the sRNAs themselves are likely to take longer to unravel.
To what extent do findings for bacterial sRNAs mirror our understanding of the eukaryotic microRNAs (miRNAs) and siRNAs (reviewed in Ref. 70)? In their mature form, miRNAs function as 21–23-nucleotide RNAs that can bind their target through imperfect complementarity, usually in the 3′-UTR, leading to gene silencing. For silencing to take effect, the mature miRNA must be presented to its targets in the context of a set of proteins called the RNA-induced silencing complex (RISC). The first job of RISC is to take the pre-miRNA exported from the nucleus and process it to a mature single-stranded miRNA. This task presumably is not essential for bacterial sRNAs that are frequently, if not always, active in their unprocessed form.
At the core of RISC are a glycine-tryptophan repeat-containing protein of 182 kDa (GW182) and the Argonaute protein (Ago), which includes domains for nucleic acid binding and a PIWI domain that has ribonuclease activity. RISC loaded with an miRNA can then bind to an mRNA target through RNA-RNA interactions. Presumably, RISC is important to stabilize and present the very short miRNA, a role that Hfq also plays with its longer sRNA substrates. RISC is also directly involved in the outcome of pairing/recruitment. If perfect complementarity exists between the miRNA and mRNA target, then annealing can induce endonuclease activity by Ago, and subsequent degradation of the target can occur. In the case of imperfect complementarity to a target mRNA, miRNA-mediated translational repression and/or subsequent deadenylation and decapping will occur, leading to mRNA decay (reviewed in Ref. 26). Ago proteins both present the sRNA to the mRNA and serve as the platform to recruit factors for RNA processing. Hfq and RNase E may serve in similar capacities, with Hfq certainly promoting pairing. We understand less about how Hfq presents the paired RNAs to RNase E and how the RNase E scaffold acts to recruit other players.
As found with bacterial sRNA-mediated repression, multiple pathways of miRNA-mediated translational repression have been characterized, blocking initiation in multiple ways or, in some cases, blocking translation beyond initiation (reviewed in Ref. 70). For eukaryotic mRNAs, there is certainly no direct coupling of transcription and translation, but the coupling of maturation of mRNA and translation is achieved by the direct communication of the poly(A) tail with the initiation of translation.
It seems clear that prokaryotes and eukaryotes have evolved different machineries to process and present regulatory RNAs, but these sRNAs facilitate similar outcomes, well regulated control of translation, using machinery that is sensitive to the state of the mRNA and its ability to be translated and that can be tuned in multiple ways to fit the physiological requirements. Therefore, future lessons learned in eukaryotes may also hold true for prokaryotes and vice versa: what is true for an elephant may actually be true for E. coli.
Acknowledgments
We thank Gisela Storz, Aurelia Battesti, Hyun-Jung Lee, and Nadim Majdalani for comments on the manuscript.
The preparation of this review was supported in whole by the National Institutes of Health Intramural Research Program of the National Cancer Institute Center for Cancer Research.
S. Panja and S. A. Woodson, manuscript in preparation.
- sRNA
- small RNA
- RBS
- ribosome-binding site
- PNPase
- polynucleotide phosphorylase
- ARRBD
- arginine-rich RNA-binding domain
- miRNA
- microRNA
- RISC
- RNA-induced silencing complex.
REFERENCES
- 1. Jacob F., Monod J. (1961) Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 [DOI] [PubMed] [Google Scholar]
- 2. Gottesman S. (2011) Roles of mRNA stability, translational regulation, and small RNAs in stress response regulation. in Bacterial Stress Responses (Storz G., Hengge R. eds) 2nd Ed., pp. 59–73, ASM Press, Washington, D.C. [Google Scholar]
- 3. Fröhlich K. S., Vogel J. (2009) Activation of gene expression by small RNA. Curr. Opin. Microbiol. 12, 674–682 [DOI] [PubMed] [Google Scholar]
- 4. Waters L. S., Storz G. (2009) Regulatory RNAs in bacteria. Cell 136, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carmichael G. G., Weber K., Niveleau A., Wahba A. J. (1975) The host factor required for RNA phage Qβ RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J. Biol. Chem. 250, 3607–3612 [PubMed] [Google Scholar]
- 6. Wilusz C. J., Wilusz J. (2005) Eukaryotic Lsm proteins: lessons from bacteria. Nat. Struct. Mol. Biol. 12, 1031–1036 [DOI] [PubMed] [Google Scholar]
- 7. Vogel J., Luisi B. F. (2011) Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brennan R. G., Link T. M. (2007) Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10, 125–133 [DOI] [PubMed] [Google Scholar]
- 9. Sun X., Zhulin I., Wartell R. M. (2002) Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30, 3662–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schumacher M. A., Pearson R. F., Møller T., Valentin-Hansen P., Brennan R. G. (2002) Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 21, 3546–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Link T. M., Valentin-Hansen P., Brennan R. G. (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 106, 19292–19297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mikulecky P. J., Kaw M. K., Brescia C. C., Takach J. C., Sledjeski D. D., Feig A. L. (2004) Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 11, 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Otaka H., Ishikawa H., Morita T., Aiba H. (2011) PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. U.S.A. 108, 13059–13064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sauer E., Weichenrieder O. (2011) Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. U.S.A. 108, 13065–13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishikawa H., Otaka H., Maki K., Morita T., Aiba H. (2012) The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA 18, 1062–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang A., Wassarman K. M., Ortega J., Steven A. C., Storz G. (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9, 11–22 [DOI] [PubMed] [Google Scholar]
- 17. Brescia C. C., Mikulecky P. J., Feig A. L., Sledjeski D. D. (2003) Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA 9, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geissmann T. A., Touati D. (2004) Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23, 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balbontín R., Fiorini F., Figueroa-Bossi N., Casadesús J., Bossi L. (2010) Recognition of heptameric seed sequences underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 78, 380–394 [DOI] [PubMed] [Google Scholar]
- 20. Zhang A., Schu D. J., Tjaden B. C., Storz G., Gottesman S. (2013) Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol., doi: 10.1016/j.jmb.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sauer E., Schmidt S., Weichenrieder O. (2012) Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 9396–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soper T. J., Woodson S. A. (2008) The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14, 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Salim N. N., Feig A. L. (2010) An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLOS ONE 5, e13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beisel C. L., Updegrove T. B., Janson B. J., Storz G. (2012) Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 31, 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salim N. N., Faner M. A., Philip J. A., Feig A. L. (2012) Requirements of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 40, 8021–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Höck J., Meister G. (2008) The Argonaute protein family. Genome Biol. 9, 210.211–210.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W., Wang L., Zou Y., Zhang J., Gong Q., Wu J., Shi Y. (2011) Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 25, 2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moon K., Gottesman S. (2011) Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 82, 1545–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olejniczak M. (2011) Despite similar binding to the Hfq protein, regulatory RNAs widely differ in their competition performance. Biochemistry 50, 4427–4440 [DOI] [PubMed] [Google Scholar]
- 30. Hussein R., Lim H. N. (2011) Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl. Acad. Sci. U.S.A. 108, 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Updegrove T. B., Correia J. J., Chen Y., Terry C., Wartell R. M. (2011) The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA 17, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohanty B. K., Maples V. F., Kushner S. R. (2004) The Sm-like protein Hfq regulates polyadenylation-dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54, 905–920 [DOI] [PubMed] [Google Scholar]
- 33. Morita T., Maki K., Aiba H. (2005) RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19, 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Windbichler N., von Pelchrzim F., Mayer O., Csaszar E., Schroeder R. (2008) Isolation of small RNA-binding proteins from E. coli: evidence for frequent interaction of RNAs with RNA polymerase. RNA Biol. 5, 30–40 [DOI] [PubMed] [Google Scholar]
- 35. Ikeda Y., Yagi M., Morita T., Aiba H. (2011) Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 79, 419–432 [DOI] [PubMed] [Google Scholar]
- 36. Carpousis A. J., Luisi B. F., McDowall K. J. (2009) Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog. Mol. Biol. Transl. Sci. 85, 91–135 [DOI] [PubMed] [Google Scholar]
- 37. Kushner S. R. (2007) Messenger RNA decay. in Escherichia coli and Salmonella: Cellular and Molecular Biology (Bock A., Curtiss R., III, Kaper J. B., Karp P. D., Neidhardt F. C., Nyström T., Slauch J. M., Squires C. L., Ussery D., eds) Chapter 4.6.4, ASM Press, Washington, D.C. [Google Scholar]
- 38. Carpousis A. J. (2007) The RNA degradosome of Escherichia coli: a multiprotein mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87 [DOI] [PubMed] [Google Scholar]
- 39. Massé E., Escorcia F. E., Gottesman S. (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Desnoyers G., Massé E. (2012) Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev. 26, 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pfeiffer V., Papenfort K., Lucchini S., Hinton J. C. D., Vogel J. (2009) Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 16, 840–846 [DOI] [PubMed] [Google Scholar]
- 42. De Lay N., Gottesman S. (2011) Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17, 1172–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang A., Wassarman K. M., Rosenow C., Tjaden B. C., Storz G., Gottesman S. (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 44. Sittka A., Sharma C. M., Rolle K., Vogel J. (2009) Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 6, 266–275 [DOI] [PubMed] [Google Scholar]
- 45. Chao Y., Papenfort K., Reinhardt R., Sharma C. M., Vogel J. (2012) An atlas of Hfq-bound transcripts reveals 3′-UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 31, 4005–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Massé E., Vanderpool C. K., Gottesman S. (2005) Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J. C. D., Vogel J. (2006) σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mandin P., Gottesman S. (2009) A genetic approach for finding small RNA regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol. Microbiol. 72, 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Urban J. H., Vogel J. (2009) A green fluorescent protein (GFP)-based plasmid system to study post-transcription control of gene expression in vivo. Methods Mol. Biol. 540, 301–319 [DOI] [PubMed] [Google Scholar]
- 50. Bouvier M., Sharma C. M., Mika F., Nierhaus K. H., Vogel J. (2008) Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell 32, 827–837 [DOI] [PubMed] [Google Scholar]
- 51. Maki K., Uno K., Morita T., Aiba H. (2008) RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc. Natl. Acad. Sci. U.S.A. 105, 10332–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morita T., Mochizuki Y., Aiba H. (2006) Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl. Acad. Sci. U.S.A. 103, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Burmann B. M., Schweimer K., Luo X., Wahl M. C., Stitt B. L., Gottesman M. E., Rösch P. (2010) A NusE:NusG complex links transcription and translation. Science 328, 501–504 [DOI] [PubMed] [Google Scholar]
- 54. Proshkin S., Rahmouni A. R., Mironov A., Nudler E. (2010) Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prévost K., Desnoyers G., Jacques J.-F., Lavoie F., Massé E. (2011) Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 25, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Figueroa-Bossi N., Valentini M., Malleret L., Bossi L. (2009) Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 23, 2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bossi L., Schwartz A., Guillemardet B., Boudvillain M., Figueroa-Bossi N. (2012) A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev. 26, 1864–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bandyra K. J., Said N., Pfeiffer V., Górna M. W., Vogel J., Luisi B. F. (2012) The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell 47, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Corcoran C. P., Podkaminski D., Papenfort K., Urban J. H., Hinton J. C. D., Vogel J. (2012) Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84, 428–445 [DOI] [PubMed] [Google Scholar]
- 60. Morita T., Kawamoto H., Mizota T., Inada T., Aiba H. (2004) Enolase in RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in response to metabolic stress in Escherichia coli. Mol. Microbiol. 54, 1063–1075 [DOI] [PubMed] [Google Scholar]
- 61. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 62. Worrall J. A. R., Górna M., Crump N. T., Phillips L. G., Tuck A. C., Price A. J., Bavro V. N., Luisi B. F. (2008) Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J. Mol. Biol. 382, 870–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Andrade J. M., Pobre V., Matos A. M., Arraiano C. M. (2012) The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18, 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Viegas S. C., Pfeiffer V., Sittka A., Silva I. J., Vogel J., Arraiano C. M. (2007) Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 35, 7651–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Andrade J. M., Arraiano C. M. (2008) PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14, 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mohanty B. K., Kushner S. R. (2003) Genomic analysis of Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 50, 645–658 [DOI] [PubMed] [Google Scholar]
- 67. Briani F., Curti S., Rossi F., Carzaniga T., Mauri P., Dehò G. (2008) Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli. RNA 14, 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hajnsdorf E., Boni I. V. (2012) Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie 94, 1544–1553 [DOI] [PubMed] [Google Scholar]
- 69. Li G.-W., Oh E., Weissman J. S. (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fabian M. R., Sonenberg N. (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19, 586–593 [DOI] [PubMed] [Google Scholar]