Background: Class IIa HDACs shuttle in a signal-regulated manner between the nucleus and cytosol.
Results: Nuclear calcium signaling is required for the nuclear export of a subset of class IIa HDAC.
Conclusion: Transcriptional regulation by nuclear calcium signaling involves changes in the subcellular localization of specific HDACs.
Significance: Through regulating HDAC nucleo-cytoplasmic shuttling nuclear calcium signaling may cause genome-wide alterations in chromatin structure in response to synaptic activity.
Keywords: Calcium Signaling, Gene Regulation, Histone Deacetylase, Neurobiology, Nuclear Translocation, Signal Transduction
Abstract
In neurons, dynamic changes in the subcellular localization of histone deacetylases (HDACs) are thought to contribute to signal-regulated gene expression. Here we show that in mouse hippocampal neurons, synaptic activity-dependent nucleo-cytoplasmic shuttling is a common feature of all members of class IIa HDACs, which distinguishes them from other classes of HDACs. Nuclear calcium, a key regulator in neuronal gene expression, is required for the nuclear export of a subset of class IIa HDACs. We found that inhibition of nuclear calcium signaling using CaMBP4 or increasing the nuclear calcium buffering capacity by means of expression of a nuclear targeted version of parvalbumin (PV.NLS-mC) led to a build-up of HDAC4 and HDAC5 in the cell nucleus, which in the case of PV.NLS-mC can be reversed by nuclear calcium transients triggered by bursts of action potential firing. A similar nuclear accumulation of HDAC4 and HDAC5 was observed in vivo in the mouse hippocampus following stereotaxic delivery of recombinant adeno-associated viruses expressing either CaMBP4 or PV.NLS-mC. The modulation of HDAC4 activity either by RNA interference-mediated reduction of HDAC4 protein levels or by expression of a constitutively nuclear localized mutant of HDAC4 leads to changes in the mRNA levels of several nuclear calcium-regulated genes with known functions in acquired neuroprotection (atf3, serpinb2), memory consolidation (homer1, arc), and the development of chronic pain (ptgs2, c1qc). These results identify nuclear calcium as a regulator of nuclear export of HDAC4 and HDAC5. The reduction of nuclear localized HDACs represents a novel transcription-promoting pathway stimulated by nuclear calcium.
Introduction
Neurons rely on a variety of intracellular signal transduction mechanisms to adjust their functions in response to environmental stimuli. Synaptic activity-induced calcium transients play a crucial role in this process (1, 2) by activating transcription-regulating signaling pathways both in the cytosol and in the nucleus (3). Nuclear calcium has emerged as a key regulator of neuronal gene expression and is required for several neuroadaptations including neuronal survival, memory consolidation, and the development of chronic inflammatory pain (4–12). The transcription factor CREB represents the principal target of nuclear calcium signaling that mediates the induction of a large pool of genes (3, 6, 7, 13). In addition, via the activation of CREB3-binding protein (CBP) and MeCP2, as well as through increasing the expression of the de novo DNA methyltransferase, Dnmt3a2, nuclear calcium evokes more global genomic responses by altering chromatin structures through histone acetylation and DNA methylation (11, 14–19). Epigenetic gene regulatory mechanisms also involve histone deacetylases (HDACs) that interact with DNA-binding proteins (20–24) and confer transcription repressing activity by catalyzing the removal of acetyl residues from acetylated lysine located in the N-terminal tails of histones (25, 26). HDACs are divided into four different classes depending on their domain structure. Eleven members of the classical HDAC family (class I: HDAC1, 2, 3, 8; class IIa: HDAC4, 5, 7, 9; class IIb: HDAC6, 10 and class IV: HDAC11) and seven sirtuins (class III) have been identified (27). Class I, III, and IV HDACs are present in many tissues, and their expression level and subcellular localization are cell type-specific (28, 29). In contrast, the expression of class II HDACs is more restricted and, in case of class IIa HDACs, primarily in the brain, heart, and skeletal muscle (30, 31). Moreover, class IIa HDACs can shuttle in a signal-regulated manner between the cytosol and the nucleus (32, 33). Here we established a function for nuclear calcium in the regulation of nuclear export of a subset of class IIa HDACs in mouse hippocampal neurons. Our results indicate that, in addition to causing increases in histone acetylation and DNA methylation (11, 14–19), the stimulation of HDAC nuclear export by nuclear calcium represents an additional pathway that links synaptic activity to genome-wide changes in chromatin structure facilitating gene transcription.
EXPERIMENTAL PROCEDURES
Expression Constructs
All HDAC constructs were C-terminal epitope-tagged. FLAG-tagged human HDAC4, HDAC4 3SA, and HDAC5 have been described before (33, 34). HA-tagged mouse YFP-HDAC7 construct has also been described previously (35). Human HDAC11 (36), rat HDAC3, and human HDAC9 were cloned into an expression vector containing an HA tag. CaMBP4 (7, 37) was FLAG-tagged and fused to the fluorescent protein mCherry. The FLAG tag has been removed for the co-expression with FLAG-tagged HDAC constructs. Parvalbumin-NLS (38) was also expressed as a mCherry fusion protein.
Short hairpin RNAs (shRNAs) targeting HDAC4 or HDAC5 were cloned into an expression vector containing the U6 promoter driving shRNA expression and the CaMKII promoter driving mCherry expression. The sequence used for the shRNA against HDAC4 was GCGGCAGATACTCATTGCAGA (39). For the shRNA against HDAC5 two sequences were tested: shHDAC5A, CGAAAGGATGGCACTGTTATT; shHDAC5B, CATGGGATTCTGCTTCTTCAA. As a control a nontargeting shRNA (shSCR (10)) was used.
Hippocampal Neuronal Cultures and Treatments
Hippocampal neurons from newborn C57BL/6 mice were isolated and cultured as described before (8, 40). DNA transfection was done after a culturing period of 8 days in vitro (DIV) using Lipofectamine 2000 (Invitrogen) as described (41). Experiments were done on DIV10. Cells were treated with 50 μm bicuculline (Sigma) or 1 μm tetrodotoxin (TTX; Tocris Bioscience) for 2 h. Control cells were treated with vehicle (0.1% dimethyl sulfoxide; Merck).
Immunocytochemistry
Following transfection and/or treatments, cells were fixed on DIV10 for 20 min at room temperature with 4% paraformaldehyde, 4% sucrose in phosphate-buffered saline (PBS), pH 7.4. Antibodies were diluted in GDB (0.1% gelatin, 0.3% Triton X-100, 15 mm Na2HPO4, 400 mm NaCl), and cells were incubated for 2 h (4 h for staining of endogenous HDAC4) in primary antibodies and 45 min in secondary antibodies. Hoechst staining (1:6000) was used for visualization of nuclei. Coverslips were mounted with Mowiol 4-88 (Calbiochem).
Immunohistochemistry
Animals were deeply anesthetized with Nembutal, pre-perfused transcardially with PBS, and perfused with neutral phosphate-buffered 10% formalin (Sigma-Aldrich). Brains were removed and postfixed overnight in the same fixative solution. For cryoprotection, brains were incubated for 2 days in 30% sucrose in 0.1 m phosphate buffer containing 0.04% thimerosal (Sigma). 40-μm-thick frozen sections, cut at −20 °C, were collected in PBS containing 0.04% thimerosal. Sections were blocked in 1% BSA, 5% normal goat serum, 0.1% Triton X-100 in PBS for 1 h at room temperature, incubated with primary antibody diluted in 1% BSA, 1% normal goat serum, 0.1% Triton X-100 at 4 °C overnight. Sections were rinsed twice with PBS containing 0.1% Triton X-100 and incubated with the secondary antibody in the same solution as the primary antibody. Sections were incubated in Hoechst 33258 (1:5000) for 5 min, rinsed twice with distilled water, and then mounted on glass slides.
Antibodies
Mouse monoclonal anti-FLAG-M2 (1:1000; Sigma), rabbit polyclonal anti-HA (1:200; Santa Cruz Biotechnology), rabbit monoclonal anti-HDAC4 (1:100; Cell Signaling), rabbit polyclonal anti-HDAC5 (1:50; Cell Signaling), Dylight 488 goat anti-mouse and anti-rabbit and Cy3 goat anti-rabbit (1:400–1:500; Dianova).
Microelectrode Arrays Recordings
MEA recordings were done as described (42).
Stereotaxic Delivery of Recombinant Adeno-associated Viruses (rAAVs)
rAAVs were delivered by stereotaxic injection into the right ventral hippocampus of 2-month-old male C57BL/6 mice. Mice were randomly grouped and anesthetized with a mixture of medetomidin, midazolam, and fentanyl. A total volume of 1.5 μl containing 1–2 ×109 genomic virus particles was injected over a period of 20 min at the following coordinates relative to Bregma: anteroposterior, −3.4 mm; mediolateral, −3.25 mm; dorsoventral, −3.3, −3.5, and −3.7 mm from the skull surface. After stereotaxic injection, mice were allowed to recover from anesthesia by subcutaneous application of a mixture with antipamezol, flumazenil, and naloxone.
RNA Extraction and cDNA Synthesis
Total RNA was isolated at DIV10 from hippocampal primary neuron cultures with RNeasy Mini kit (Qiagen), including an optional DNase I treatment at room temperature for 15 min according to the manufacturer's instructions (Qiagen). 1.2 μg of extracted RNA was reverse transcribed into first strand cDNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems).
Real Time Quantitative PCR
Quantitative reverse transcriptase PCR (QRT-PCR) was done on an ABI7300 thermal cycler using Universal QRT-PCR master mix with TaqMan Gene Expression Assays for the indicated genes (Applied Biosystems). Expression of target genes was normalized against the expression of Gusb as endogenous control gene. Data were derived from at least five independent experiments.
Data Analysis
Stained cells were screened using a Leica DM IRBE inverted fluorescence microscope. Numbers of cells with a nuclear or cytoplasmic staining were counted. Cells with a strong fluorescent signal of the HDAC of interest in the nucleus were scored under the category “nuclear” (i.e. see Fig. 1, HDAC5, TTX). These cells were scored as “N.” Cells were counted as “cytoplasmic” when there was cytoplasmic HDAC localization featuring a clear border between a missing nuclear staining and a high cytoplasmic signal (Fig. 1, HDAC4, untreated and TTX). These cells were scored as “C.” Cells that showed an equal distribution of the HDAC signal between the compartments were scored in both categories (i.e. Fig. 1, HDAC3, untreated, bic, TTX). These cells were scored as “E.” Images of representative cells were taken using a Leica TCS SP2 confocal microscope with the resolution of 1024 × 1024 pixels and processed in ImageJ and Adobe Photoshop.
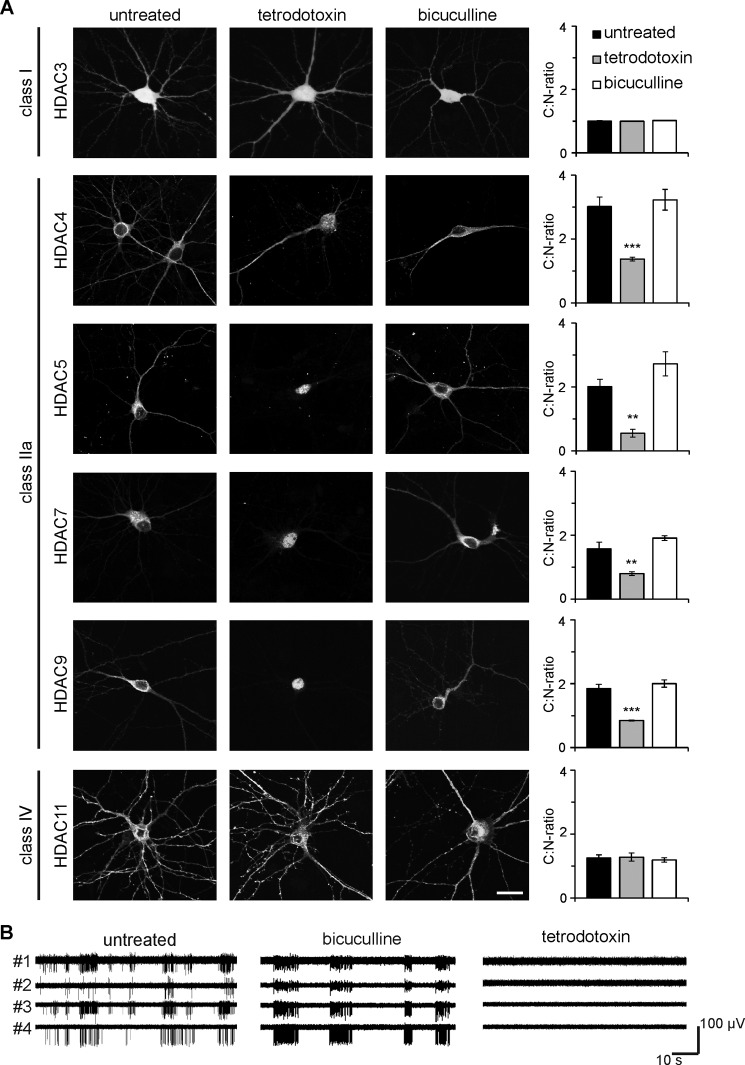
FIGURE 1.
Nucleo-cytoplasmic shuttling of class IIa HDACs is dependent on neuronal activity. A, representative micrographs of cultured hippocampal neurons transfected with epitope-tagged HDAC constructs (HDAC3-HA, HDAC4-FLAG, HDAC5-FLAG, HDAC7-HA, HDAC9-HA, and HDAC11-HA) and either left untreated or treated with TTX or bicuculline as indicated. Scale bar is 20 μm. Graphs show the ratio between cytoplasmic and nuclear localization. 400–1000 cells were analyzed for each tested HDAC and experimental condition from a minimum of three independent preparations. Statistically significant differences are indicated with asterisks (***, p < 0.001; **, p < 0.01, one-way ANOVA, Dunnett's post hoc test). Error bars, S.E. B, typical examples of MEA recordings obtained from untreated cultured hippocampal neurons (left), and neurons after exposure to bicuculline (50 μm, center), or TTX (1 μm, right). Simultaneous recordings from four separate electrodes are shown in each panel.
The ratio between the number of cells showing a cytoplasmic localization of a specific HDAC and cells with a nuclear localization was calculated (C:N ratio (C+E)/(N+E)). The resulting number indicates whether the protein is located predominantly in the cytoplasm (>1) or the nucleus (<1) or whether it is equally distributed (≈1). Quantitative fluorescent intensity analysis has been done by selecting the nuclei in the Hoechst channel by threshold application and manual identification of somata. The average fluorescent intensity of both areas was measured, and the relative intensity of the nucleus compared with the cytoplasm was calculated. Data are presented as means ± S.E. Statistical analysis has been done using one-way analysis of variance (ANOVA) with Dunnett's post hoc test. Unless otherwise stated, a two-sample unpaired Student's t test was performed for the comparison of two conditions. Results were considered to be statistically significant with a significance level of p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***).
RESULTS
We first characterized synaptic activity-dependent nucleo-cytoplasmic shuttling of different HDAC classes and transfected cultured mouse hippocampal neurons with expression vectors for HDAC3, HDAC4, HDAC5, HDAC7, HDAC9, and HDAC11 (Fig. 1A). The cultures were subsequently either silenced using TTX, a sodium channel blocker, or exposed to bicuculline, a GABAA receptor antagonist that blocks inhibitory inputs and triggers action potential (AP) bursting (13). The effects of TTX and bicuculline on the hippocampal network firing patterns were confirmed using MEA (Fig. 1B). MEA recordings revealed spontaneous firing activity of single spikes and short trains of activity under basal conditions, which switched to recurrent synchronous bursting after the addition of bicuculline to the media. Action potential firing was completely blocked by the application of TTX (Fig. 1B).
As described in detail under “Experimental Procedures,” transfected hippocampal neurons were divided into three groups based on the localization of the exogenously expressed HDAC (nuclear, cytoplasmic, equal distribution nucleus versus cytoplasm). We calculated the cytoplasm-to-nucleus-ratio (C:N ratio) to assess the subcellular localization of HDACs and detect changes in response to treatments. In untreated controls, HDAC3, a class I HDAC, was located in both the cytoplasm and the nucleus (Fig. 1A). The C:N ratio calculated for HDAC3 was 1.0 ± 0.01, as it was equally distributed between the cytoplasm and nucleus. The C:N ratio for HDAC11, the class IV HDAC, was 1.25 ± 0.1 indicating a moderately higher expression of this protein in the cytoplasm compared with the nucleus. The subcellular localization of HDAC3 and HDAC11 was largely invariant with the treatments indicating that HDAC class I and IV subcellular distribution is not regulated by neuronal activity (Fig. 1A).
We next analyzed class IIa HDACs (i.e. HDAC4, -5, -7, and -9), all of which were located predominantly in the cytoplasm in untreated hippocampal neurons (Fig. 1A). Upon TTX treatment, all class IIa HDACs underwent a shift toward a nuclear localization, whereas stimulating AP bursting with bicuculline did not lead to observable changes in their subcellular distribution (Fig. 1A). The C:N ratio for HDAC4 decreased significantly from 3.02 ± 0.29 to 1.37 ± 0.06 (p < 0.001, one-way ANOVA, Dunnett's post hoc test, F = 17.29) after TTX treatment and remained virtually unaltered after bicuculline treatment (3.23 ± 0.32, p = 0.79). Similar results were obtained for HDAC5, -7, and -9. The C:N ratio for HDAC5 in controls was 2.01 ± 0.23, indicating a predominantly cytoplasmic localization, but decreased after TTX treatment (0.55 ± 0.12; p = 0.004, F = 17.5), reflecting redistribution toward the nucleus, and moderately increased upon bicuculline stimulation (2.72 ± 0.38, p = 0.14). Compared with HDAC4 and HDAC5, the synaptic activity-induced subcellular redistributions of HDAC7 and HDAC9 were less dramatic. Nevertheless, we detected a decrease of the C:N ratio following TTX treatment and a moderate increase following bicuculline treatment for both HDAC7 (control, 1.58 ± 0.2; TTX, 0.8 ± 0.06, p = 0.004; bicuculline, 1.91 ± 0.07, p = 0.18; F = 19.28) and HDAC9 (control, 1.85 ± 0.13; TTX, 0.85 ± 0.01, p < 0.001; bicuculline, 2.0 ± 0.12, p = 0.49; F = 37.13). These results indicate that neuronal activity is required for nuclear export of all class IIa HDACs and that silencing neuronal activity with TTX leads to their accumulation in the nucleus.
Nuclear Calcium Signaling in HDAC Nucleo-cytoplasmic Shuttling
Nuclear calcium is a signaling end point in synapse-to-nucleus communication and an important mediator of synaptic activity-driven gene transcription (3, 7, 13). To investigate a possible role of nuclear calcium in nucleo-cytoplasmic shuttling of HDACs, we used the nuclear calcium/calmodulin signaling inhibitor CaMBP4. This protein consists of four copies of the M13 peptide derived from myosin light chain kinase; it is expressed in the cell nucleus where it binds to and inactivates the nuclear calcium/calmodulin complex thereby preventing the activation of its downstream targets (37). CaMBP4 has been characterized in great detail and was used in many studies to determine the role of nuclear calcium in neuronal gene expression important for acquired neuroprotection, memory formation, and chronic inflammatory pain (4–8, 10–12). Because CaMBP4 is nuclear localized and fused to mCherry for detection (CaMBP4-mC); we used an expression vector for mCherry-NLS as a control (mC.NLS). We selected the class I HDAC3 as control for these experiments because its subcellular localization is not influenced by neuronal activity (Fig. 1). Indeed, we found that CaMBP4 expression had no effect on the subcellular localization of HDAC3 (Fig. 2); the C:N ratio for HDAC3 was close to 1 in both mC.NLS- and CaMBP4-expressing cells (mC.NLS, 0.98 ± 0.01; CaMBP4, 1.01 ± 0.05).
FIGURE 2.
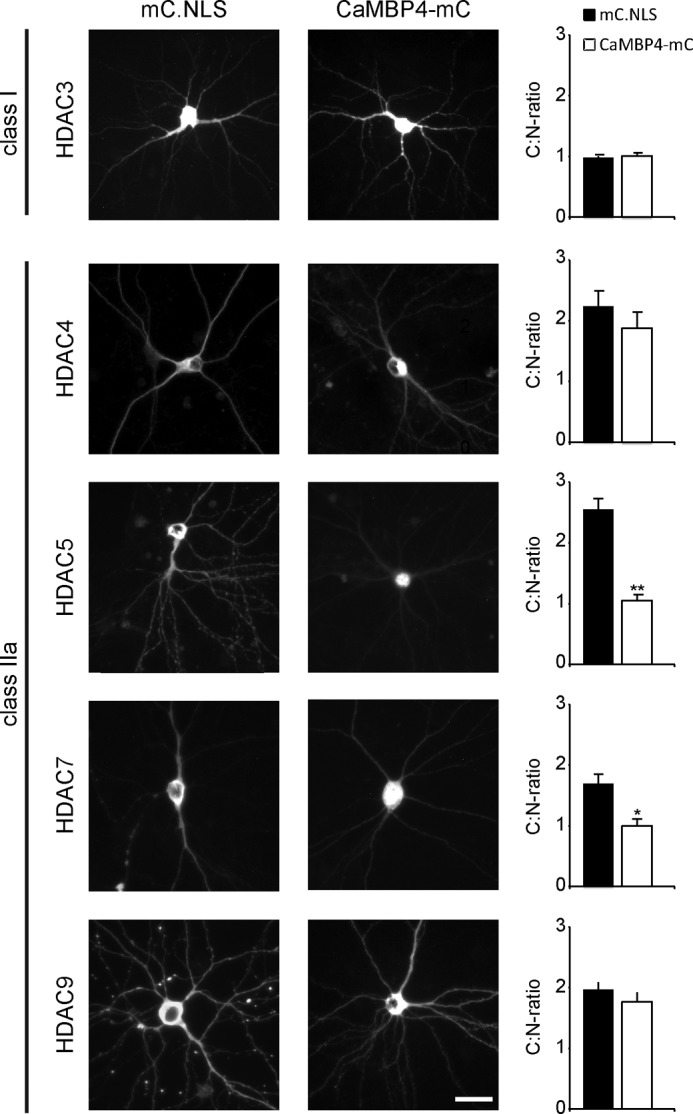
Nuclear calcium differentially regulates the shuttling of class IIa HDACs. Representative images show hippocampal neurons transfected with HDAC constructs with expression vector for CaMBP4-mCherry (CaMBP4-mC) or mCherry-NLS (mC.NLS) as indicated. HDACs subcellular localization was analyzed only in CaMBP4- or mC.NLS-expressing cells, selected by mCherry expression. Scale bar is 20 μm. Graphs show the ratio between cytoplasmic and nuclear localization. 140–700 cells were analyzed for each condition from a minimum of three independent experiments. Statistically significant differences are indicated with asterisks (**, p < 0.01; *, p < 0.05, two-sided Student's t test). Error bars, S.E.
We next determined the effects of CaMBP4 expression on the overexpressed forms of class IIa HDACs as well as on their endogenous counterpart (see below). We found that CaMBP4 did not significantly affect the subcellular localization of the overexpressed forms of HDAC4 and -9 (Fig. 2: HDAC4 mC.NLS, 2.24 ± 0.24; CaMBP4, 1.88 ± 0.25; HDAC9 mC.NLS, 1.97 ± 0.11, CaMBP4, 1.76 ± 0.14). In contrast, the C:N ratio of the overexpressed versions of HDAC5 and also that of HDAC7 decreased from 2.55 ± 0.18 to 1.05 ± 0.07 (p = 0.001, Student's t test) and from 1.69 ± 0.13 to 0.99 ± 0.11 (p = 0.04, Student's t test), respectively, upon blockage of nuclear calcium signaling with CaMBP4 (Fig. 2).
Because neuronal activity drives nuclear export of HDACs (see Fig. 1), we next investigated whether increasing neuronal activity via bicuculline treatment would be sufficient to counteract the nuclear accumulation of HDACs observed following blockade of nuclear calcium signaling. To interfere with nuclear calcium, we used CaMBP4 and, in addition, the calcium-binding protein parvalbumin targeted to the cell nucleus by means of fusion to a nuclear localization signal (38). Overexpression of parvalbumin.NLS (which is fused to mCherry; PV.NLS-mC) increases the nuclear calcium buffering capacity, which in nonneuronal cell types has been reported to prevent the activation of nuclear calcium-dependent downstream targets (43, 44). Because HDAC4 and HDAC5 are expressed in the mammalian brain at far higher levels than HDAC7 and HDAC9 (45) we focused our studies on these two class IIa HDACs. We co-transfected cultured mouse hippocampal neurons with expression vectors for HDAC4 or HDAC5 constructs alongside expression vectors for either CaMBP4 or PV.NLS-mC; AP bursting was induced with bicuculline treatment (Fig. 3). Because CaMBP4 and PV.NLS-mC were fused to mCherry, we used an expression vector for mCherry as a control. Similar to the results obtained with CaMBP4 (see Fig. 2), PV.NLS-mC had no effect on the subcellular localization of overexpressed HDAC4 (Fig. 3, B and D). The C:N ratio for HDAC4 was between 3 and 4 in all experimental conditions, indicating a predominantly cytoplasmic localization. In contrast, the localization of overexpressed HDAC5 was sensitive to nuclear calcium blockade. Similar to the results shown in Fig. 2, expression of CaMBP4 led to a nuclear shift of HDAC5 localization in untreated hippocampal neurons (Fig. 3, A and B; mCherry, 3.41 ± 0.55; CaMBP4, 2.18 ± 0.21). A similar result was also observed with PV.NLS-mC expression (Fig. 3, C and D; mC.NLS, 2.66 ± 0.41; PV.NLS-mC, 1.71 ± 0.34).
FIGURE 3.
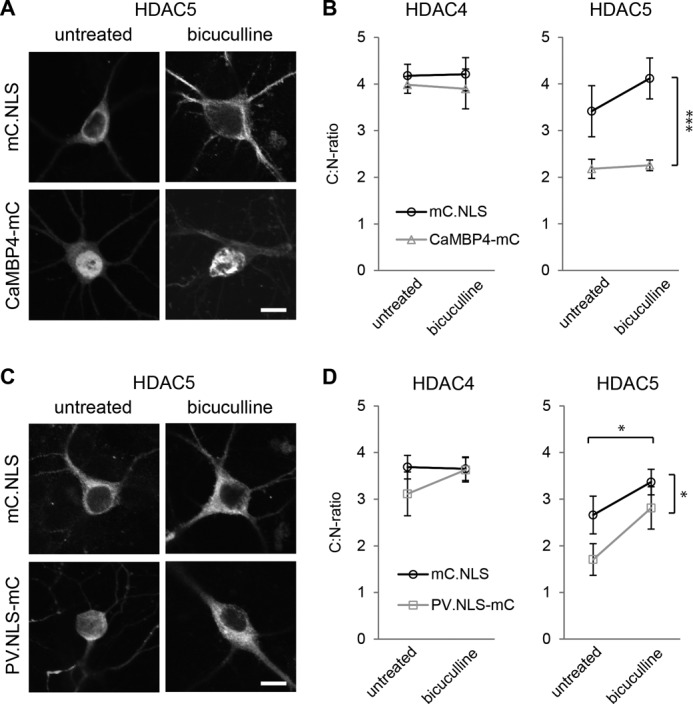
Increasing neuronal activity can overcome parvalbumin-dependent but not CaMBP4-dependent block of HDAC5 nuclear export. A, representative images of hippocampal neurons transfected with HDAC5 and with mCherry (mC.NLS) or CaMBP4-mCherry (CaMBP4-mC) as indicated. Neurons were either left untreated or treated with bicuculline. Scale bar is 10 μm. B, quantitative analysis of the subcellular localization of HDAC4 or HDAC5 in hippocampal neurons. Neurons were transfected with HDAC4 or HDAC5 and with mCherry or CaMBP4 and treated with bicuculline or not as indicated. Graphs show the ratio between cytoplasmic and nuclear localization. In total, 140–400 cells were analyzed for each condition from a minimum of five independent experiments. C, representative images of hippocampal neurons transfected with HDAC5 and with mCherry (mC.NLS) or parvalbumin-NLS-mCherry (PV.NLS-mC) as indicated. Neurons were either left untreated or treated with bicuculline. Scale bar is 10 μm. D, quantitative analysis of the subcellular localization of HDAC4 or HDAC5 in hippocampal neurons. Neurons were transfected with HDAC4 or HDAC5 and with mCherry or PV.NLS-mC and treated with bicuculline or not as indicated. Graphs show the ratio between cytoplasmic and nuclear localization. In total, 140–200 cells were analyzed for each condition from a minimum of five independent experiments. Statistically significant differences are indicated with asterisks (***, p < 0.001; *, p < 0.05, two-way ANOVA). Error bars, S.E.
Whereas both CaMBP4 and PV.NLS-mC attenuated the nuclear export of overexpressed HDAC5 in unstimulated hippocampal neurons, only CaMBP4 expression was sufficient to maintain the nuclear accumulation of HDAC5 after bicuculline stimulation (Fig. 3, A and B). Bicuculline caused an increase in the C:N ratio of HDAC5 from 3.41 ± 0.55 to 4.12 ± 0.44 in mCherry-expressing cells whereas virtually no increase in the C:N ratios (i.e. without bicuculline treatment, 2.18 ± 0.21; with bicuculline treatment, 2.26 ± 0.11) was observed in CaMBP4-expressing neurons (Fig. 3, A and B; two-way ANOVA: mCherry versus CaMBP4, p < 0.001, F = 29.86; untreated versus bicuculline, p = 0.28, F = 1.21). In contrast, in PV.NLS-mC-expressing neurons, bicuculline treatment increased the C:N ratio of HDAC5 from 1.71 ± 0.34 to 2.81 ± 0.45 (Fig. 3, C and D; two-way ANOVA: mCherry versus PV, p = 0.034, F = 5.29; untreated versus bicuculline, p = 0.023, F = 6.14), indicating that despite increased nuclear calcium buffering, synaptic activity can initiate the redistribution of HDAC5 from the nucleus to the cytosol. This result is expected because bicuculline-induced AP bursting gives rise to very robust nuclear calcium transients (13, 46) that will saturate the nuclear calcium buffers allowing HDAC nuclear export to be induced. The results obtained with CaMBP4 are different from those using PV.NLS because CaMBP4 does not buffer free calcium but instead blocks in a competitive manner the nuclear calcium/calmodulin complex irrespective of the amplitude and duration of the nuclear calcium transients.
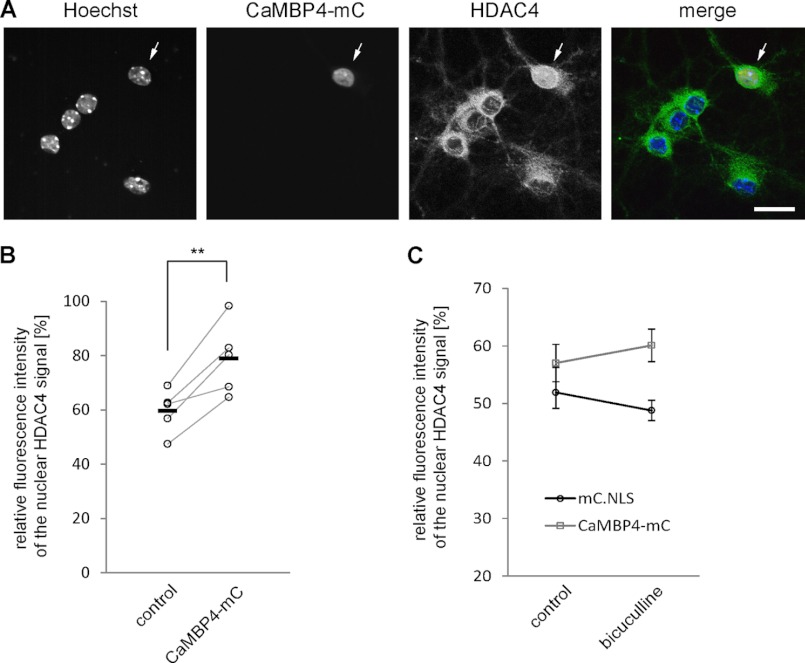
We next analyzed the shuttling behavior of endogenous HDACs in cultured hippocampal neurons. We were able to only investigate endogenous HDAC4 due to a lack of antibodies against HDAC5 that were suitable for immunocytochemistry (although we did succeed in using HDAC5 antibodies on brain sections; see Fig. 6). Similar to the results obtained using the overexpressed HDAC4 (see Fig. 1), the endogenous HDAC4 was located predominantly in the cytoplasm under control conditions as well as after bicuculline stimulation and accumulated in the nucleus after TTX treatment (Fig. 4). We next investigated the role of nuclear calcium in the shuttling of endogenous HDAC4. Although we were unable to detect a significant change in the subcellular distribution of endogenous HDAC4 in PV.NLS-expressing neurons (data not shown), the expression of CaMBP4 led to an increase in the nuclear localization of HDAC4 compared with their surrounding nontransfected control cells, which persisted even after bicuculline stimulation (Fig. 5, A and B). These results indicate that the localization of the endogenous HDAC4 is controlled by nuclear calcium signaling, even though we failed to detect an effect of CaMBP4 expression on the subcellular localization of the overexpressed form of HDAC4 (see Fig. 2). The reason for this apparent discrepancy remains unexplained but is most likely the result of gross differences in the expression levels of the endogenous compared with the overexpressed protein.
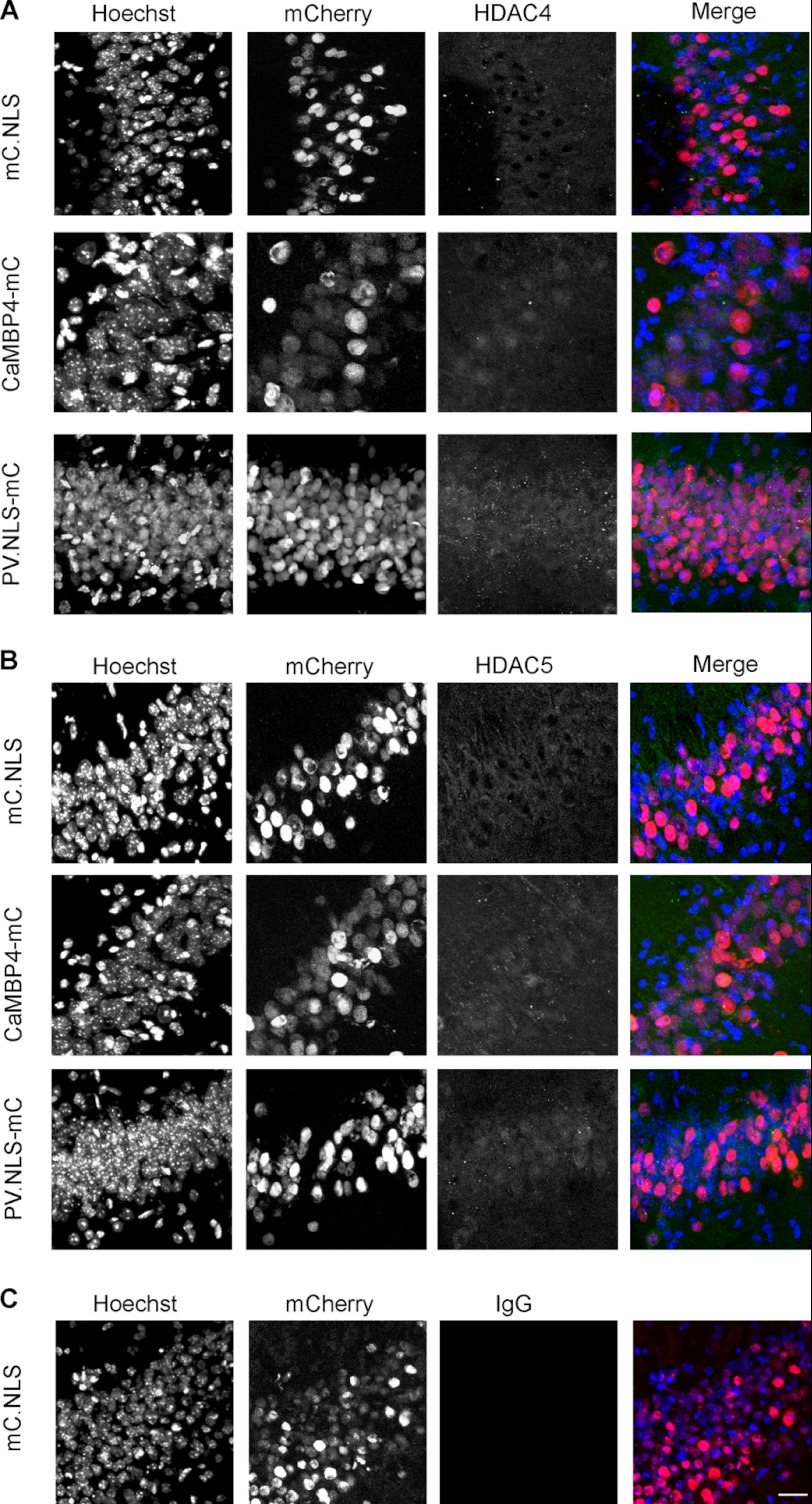
FIGURE 6.
Nuclear calcium controls HDAC4 and -5 nucleo-cytoplasmic shuttling in vivo. A, immunohistochemistry of HDAC4 in the CA1 region of the hippocampus of adult mice stereotaxically injected with rAAV-mCherry-NLS (mC.NLS), rAAV-CAMBP4-mCherry (CaMBP4-mC) or with rAAV-parvalbumin.NLS-mCherry (PV.NLS-mC) using an anti-HDAC4 antibody, Hoechst to visualize the nuclei, and mCherry fluorescence to detect virus expression. B, immunohistochemistry of HDAC5 in the CA1 region of the hippocampus of adult mice stereotaxically injected with rAAV-mCherry-NLS (mC.NLS), rAAV-CAMBP4-mCherry (CaMBP4-mC), or with rAAV-parvalbumin.NLS-mCherry (PV.NLS-mC) using an anti-HDAC5 antibody, Hoechst to visualize the nuclei, and mCherry fluorescence to detect virus expression. C, immunohistochemistry of the CA1 region of the hippocampus of adult mice stereotaxically injected with rAAV-mCherry.NLS (mC.NLS), using a normal rabbit IgG antibody to test the specificity of the immunostainings shown in A and B, Hoechst to visualize the nuclei, and mCherry fluorescence to detect virus expression. Scale bar is 20 μm.
FIGURE 4.
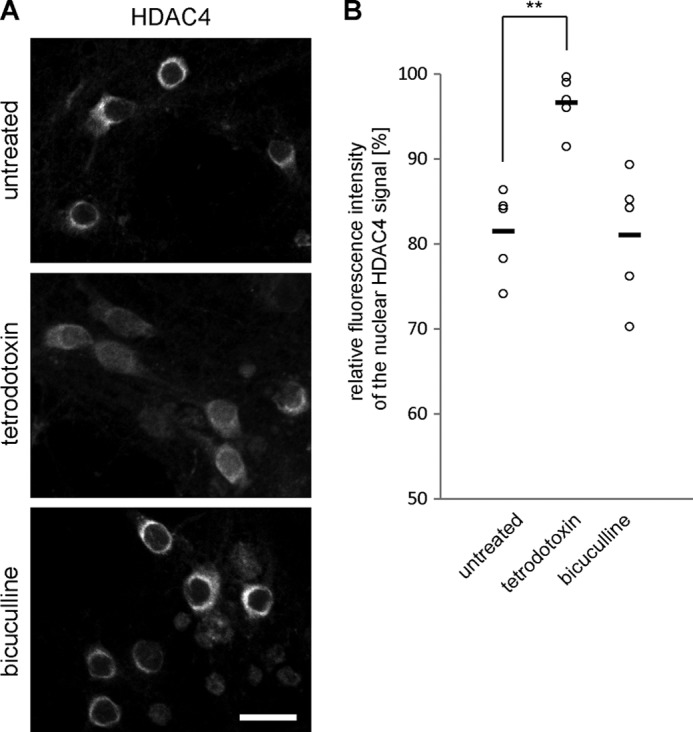
Nucleo-cytoplasmic shuttling of endogenous HDAC4 is dependent on neuronal activity. A, representative images of hippocampal neurons left untreated or treated with tetrodotoxin or bicuculline and immunostained for endogenous HDAC4. Scale bar is 20 μm. B, quantitative measurements of the relative fluorescent intensity of HDAC4 signal in the nucleus compared with the signal of the cytosol. Each circle represents an independent experiment. Horizontal bars show mean values (control, 81.5 ± 2.3%; bicuculline, 81.0 ± 3.4%, p = 0.99; TTX, 96.6 ± 1.4% p = 0.002; one-way ANOVA, Dunnett's post hoc test, F = 12.34). Statistically significant difference is indicated with asterisks (**).
FIGURE 5.
Shuttling of endogenous HDAC4 is responsive to nuclear calcium signaling. A, representative images showing the subcellular localization of endogenous HDAC4 in hippocampal neuron transfected with CaMBP4-mCherry (arrow) and nontransfected surrounding control cells. Scale bar is 20 μm. B, quantitative measurements of the relative fluorescent intensity of HDAC4 signal in the nucleus compared with the signal of the cytosol in untransfected (control) and CaMBP4-expressing cells. Each circle represents an independent experiment. Horizontal bars show mean values (control, 59.6 ± 3.6%; CaMBP4, 78.9 ± 5.9% p = 0.007 paired Student's t test). Statistically significant difference is indicated with asterisks (**). C, quantitative measurements of the relative fluorescent intensity of HDAC4 signal in the nucleus compared with the signal of the cytosol in mC.NLS- and CaMBP4-mC-expressing cells. Neurons were left untreated or treated with bicuculline and immunostained for endogenous HDAC4. The averaged results of three independent experiments are shown. Error bars, S.E.
We next investigated the role of nuclear calcium signaling on HDAC shuttling in vivo. rAAVs containing the expression cassettes for mCherry-NLS, CaMBP4-mCherry, or PV.NLS-mC were stereotaxically delivered into the ventral mouse hippocampus of 2-month-old C57BL/6 male mice; infected neurons could readily be identified by mCherry fluorescence (Fig. 6). Immunohistochemical analysis of the subcellular localization of endogenous HDAC4 and HDAC5 in brain slices derived from the injected mice revealed that, very similar to the results obtained with cultured neurons (see Fig. 1), the nuclei of hippocampal neurons of the CA1 region showed a predominant cytoplasmic localization of HDAC4 and HDAC5 under control conditions (i.e. expression of mCherry.NLS; Fig. 6, A and B). These observations are in agreement with previous studies on the subcellular localization of HDACs in the brain (39, 47). However, in the presence of either CaMBP4 or PV.NLS both HDAC4 and HDAC5 showed a clear redistribution from the cytosol toward the nucleus (Fig. 6, A and B). These results indicate that nucleo-cytoplasmic shuttling of HDAC4 and HDAC5 in vivo is regulated by nuclear calcium and that either blockade of nuclear calcium signaling or increasing the nuclear calcium buffering capacity leads to a nuclear accumulation of HDAC4 and HDAC5.
HDAC4 Regulates Expression of Genes Involved in Various Forms of Neuroadaptations
Given the known role of HDACs in the modulation of gene transcription, we next analyzed the effects of HDACs on the expression of genes that are known targets of nuclear calcium signaling (Fig. 7 (7, 12)). DNA sequences encoding shRNAs designed to target the mouse HDAC4 (rAAV-shHDAC4) or HDAC5 (rAAV-shHDAC5) mRNA were inserted downstream of the U6 promoter of an rAAV vector. Immunoblot analysis revealed that the rAAVs reduced protein levels of their respective targets leaving unaltered the expression of other HDAC family members (Fig. 7A). QRT-PCR revealed that compared with uninfected neurons or neurons infected with an rAAV-shSCR (scrambled control), infections with rAAV-shHDAC4 altered the mRNA expression levels of several nuclear-calcium target genes that have functions in different forms of neuroadaptations (7, 8, 12, 48, 49) including neuronal survival (serpinb2, atf3), chronic pain (ptgs2, c1qc), and synaptic plasticity and memory (homer1, arc) (Fig. 7B). Infection of hippocampal neurons with rAAV-shHDAC5 had very little effect on the expression levels of the genes analyzed (Fig. 7B).
FIGURE 7.
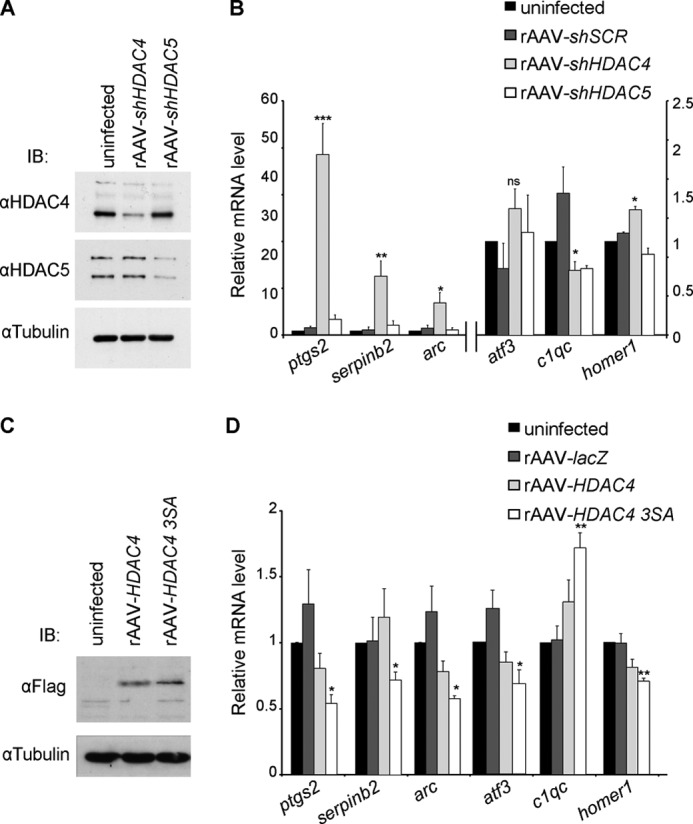
HDAC4 regulates expression of genes involved in neuroadaptations. A, immunoblot analysis of uninfected hippocampal neurons and of hippocampal neurons infected with the indicated rAAVs. Tubulin immunoblot is shown as control for protein loading. B, QRT-PCR analysis of expression of ptgs2, serpinb2, arc, atf3, c1qc, and homer1 in uninfected hippocampal neurons and in hippocampal neurons infected with rAAV-shSCR, rAAV-shHDAC4, or with rAAV-shHDAC5 (n = 5). ptgs2, arc, and serpinb2 values refer to the axis on the left. C, immunoblot analysis of uninfected hippocampal neurons and of hippocampal neurons infected with the indicated rAAVs. rAAV-HDAC4 and rAAV-HDAC4 3SA all carry a FLAG cassette which was used for detection via an anti-FLAG antibody. Tubulin immunoblot is shown as control for protein loading. D, QRT-PCR analysis of ptgs2, serpinb2, arc, atf3, c1qc, and homer1 expression in uninfected hippocampal neurons and in hippocampal neurons infected with rAAVs giving rise to the indicated proteins (n = 5). Statistically significant differences are indicated with asterisks (***, p < 0.0005; **, p < 0.05; *, p < 0.05 one-way ANOVA, Dunnett's post hoc test). Error bars, S.E.
Because blocking nuclear calcium causes a shift of HDAC class IIa localization toward the nucleus (see above), we finally tested the effects of nuclear localization of HDAC4 on the expression levels of the selected group of nuclear-calcium target genes (see above). Hippocampal neurons were infected with rAAVs containing an expression cassette for either HDAC4 (rAAV-HDAC4) or for the triple mutant (S246A/S467A/S632A) HDAC4 3SA (rAAV-HDAC4 3SA; Fig. 7C), which is constitutively nuclear as it cannot be phosphorylated and transported to the cytoplasm (34); uninfected neurons or neurons infected with an rAAV-LacZ served as controls. QRT-PCR revealed that indeed expression of HDAC4 3SA caused changes in the expression level of the nuclear-calcium target genes (Fig. 7D) in a direction opposite to the one obtained after lowering HDAC4 levels through RNAi (Fig. 7B).
DISCUSSION
In this study, we establish a link among neuronal activity, nuclear calcium signaling, and HDAC nucleo-cytoplasmic shuttling. Interfering with either neuronal activity or nuclear calcium in vitro and in vivo causes the nuclear accumulation of members of class IIa HDACs.
HDAC Nucleo-cytoplasmic Shuttling and Synaptic Activity-dependent Transcription
Neurons respond and adapt to synaptic stimuli by initiating structural and functional changes, ranging from remodeling of fine anatomical structures to alterations in gene expression (1, 50), which at the behavioral level may ultimately lead to various forms of neuroadaptations including the formation of memories. In recent years, attention has been paid not only to the characterization of signal-regulated sequence-specific DNA-binding proteins and how they control transcription of their target genes but also to epigenetic modifications. Epigenetic changes of chromatin are generally thought to be a heritable mark in most cell types (51, 52), but they are also subject to regulation by environmental signals particularly, although not exclusively, in the nervous system (53). HDACs are part of the cellular machinery that bridges external stimuli to epigenetic control of gene expression. Class IIa HDACs, all of which as shown in this study undergo synaptic activity-driven nucleo-cytoplamic shuttling, may be particularly important for the coupling of synaptic activity to genomic responses. In contrast, the subcellular localization of members of class I and class IV HDACs is largely invariant to changes in neuronal activity, indicating that these forms of HDACs may represent a continuously present (i.e. signal-independent) transcription repressing activity in the nucleus.
Our HDAC4 loss-of-function and gain-of-function experiments support that concept that class IIa HDACs take part in the regulation of transcription following synaptic activity and the generation of nuclear calcium signals. They are also in line with the recently proposed role of HDAC4 in the control of genes involved in synaptic plasticity and the formation of memories (54, 55). However, given that HDAC4 activity can modulate the expression levels of many functionally diverse nuclear calcium-regulated genes (see Fig. 7, B and D), the importance of nucleo-cytoplasmic shuttling of class IIa HDACs in the brain may not be restricted to learning and memory but also relevant for many other neuroadaptations including, besides changes in cognitive abilities, acquired neuroprotection, and the development of chronic pain.
Stimulus-induced changes in the subcellular distribution of class IIa HDACs have also been observed in other cell types. In skeletal muscle, reduced neural activity following denervation causes nuclear accumulation of HDAC4 (56). A similar redistribution of HDAC4 and HDAC7 from the cytosol to the nucleus has been observed in cerebellar granule neurons exposed to toxic stimuli (39, 57); this change in localization precedes cell death consistent with a pro-death activity of nuclear of HDAC4 and a pro-survival activity of cytosolic HDAC4 (58). Conversely, the nuclear export of class IIa HDACs following induction of AP bursting may represent a neuroprotective measure that could be part of the known survival-enhancing effects afforded by synaptic activity and nuclear calcium signaling (5–7, 46, 59, 60).
Nuclear Calcium Regulation of HDAC Nucleo-cytoplasmic Shuttling
Our study revealed that the synaptic activity-dependent nucleo-cytoplasmic shuttling is a common feature of all members of class IIa HDACs, which distinguishes them from other HDAC classes that do not undergo changes in their subcellular localization upon stimulation of neuronal activity. However, the effect of inhibiting nuclear calcium signaling or increasing nuclear calcium buffering capacity on the shuttling process varied considerably among the four different class IIa HDACs. We found that nuclear calcium is required for nuclear export of the endogenously expressed and/or overexpressed forms of HDAC4, HDAC5, and, to a lesser extent, of HDAC7; in contrast, inhibition of nuclear calcium signaling had virtually no effect on the localization of the overexpressed form of HDAC9. This indicates that nuclear calcium-dependent but also nuclear calcium-independent signaling pathways are responsible for regulating the subcellular distribution of class IIa HDACs in hippocampal neurons. This is in line with several reports in literature implicating a variety of signaling molecules in HDAC shuttling (61–63). The nuclear import and export of HDACs, which is best characterized for HDAC4, is primarily regulated by protein kinases and phosphatases (64–66). A tripartite nuclear localization signal (NLS) within the N-terminal regulatory unit of HDAC4 allows for an importin α-dependent nuclear import (34, 67), whereas a C-terminal nuclear export signal (NES) mediates the transport HDAC4 from the nucleus to the cytoplasm via CRM1-dependent mechanism (67, 68). The NES is only active after phosphorylation of HDAC4 at three serine residues (34) leading to interaction with 14-3-3 protein (68), which itself contains an NES (69). The dephosphorylation of HDAC4 in the cytoplasm, which can be catalyzed by protein phosphatase PP2A (65, 70, 71), unmasks the intrinsic nuclear localization signal (NLS) and initiate nuclear import of HDAC4. NLS, NES, and the critical regulatory serine residues are highly conserved in HDAC5, and HDAC7 and HDAC9, which can also interact with 14-3-3 proteins (34, 72, 73). Precisely how synaptic activity drives nuclear export of class IIa HDACs in hippocampal neurons remains to be investigated. In nonneuronal cells, in particular in cardiac and skeletal muscle but also in COS cells and chondrocytes, the calcium/calmodulin-dependent protein kinases (CaMK) I, II, and IV have been shown to be functionally relevant for HDAC shuttling (32, 61, 64, 70, 74–77). Pharmacological studies using hippocampal neurons also indicate an involvement of CaMKs in regulating the subcellular localization of HDAC5 and, although to a lesser extent, of HDAC4 (33). This suggests that CaMKII and CaMKIV, which are present in the cell nucleus of hippocampal neuron (19, 78), are key signaling intermediates in the nuclear calcium-dependent export of HDAC4 and HDAC5 from the nucleus. Given the presence of CaMKII in the cytosol, CaMKII may also be part of the nuclear calcium-independent HDAC nucleo-cytoplasmic shuttling. Alternatively, or in addition, the ERK-MAP kinase pathway may be involved in this process. ERK-MAP kinase signaling is activated by synaptic activity-induced, cytosolic (near-plasma membrane) calcium transients (40, 79). Once activated, ERK-MAP kinases can enter the nucleus and phosphorylate transcriptional regulators (41, 80–82). In C2C12 myoblasts, ERK-MAP kinases can associate with HDAC4 but seem to promote HDAC4 nuclear accumulation rather than stimulating its export (83).
In conclusion, class IIa HDACs are intracellular signal transducers and part of the process through which synaptic activity and the nuclear calcium transients associated with it are coupled to the regulation of transcription.
Acknowledgments
We thank I. Bünzli-Ehret for help with the preparation of hippocampal cultures; Drs. B. Ehrlich and M. Nathanson for parvalbumin.NLS; Dr. K. Tucker for HDAC3 and HDAC9; Dr. Hung-Ying Kao for HDAC7; and Dr. E. Seto for HDAC11.
This work was supported by the Deutsche Forschungsgemeinschaft, an ERC Advanced Grant (to H. B.), and the Sonderforschungsbereich 636 of the Deutsche Forschungsgemeinschaft.
- CREB
- cAMP response element-binding protein
- ANOVA
- analysis of variance
- AP
- action potential
- CaM
- calcium/calmodulin
- CaMBP
- CaM-binding protein
- CaMK
- calcium/calmodulin-dependent kinase
- CBP
- CREB-binding protein
- DIV
- days in vitro
- HDAC
- histone deacetylase
- mC
- mCherry
- MEA
- microelectrode-array
- NES
- nuclear export signal
- NLS
- nuclear localization signal
- QRT-PCR
- quantitative reverse transcriptase PCR
- rAAV
- recombinant adeno-associated virus
- TTX
- tetrodotoxin.
REFERENCES
- 1. Greer P. L., Greenberg M. E. (2008) From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59, 846–860 [DOI] [PubMed] [Google Scholar]
- 2. Hagenston A. M., Bading H. (2011) Calcium signaling in synapse-to-nucleus communication. Cold Spring Harb. Perspect. Biol. 3, a004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hardingham G. E., Chawla S., Johnson C. M., Bading H. (1997) Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature 385, 260–265 [DOI] [PubMed] [Google Scholar]
- 4. Limbäck-Stokin K., Korzus E., Nagaoka-Yasuda R., Mayford M. (2004) Nuclear calcium/calmodulin regulates memory consolidation. J. Neurosci. 24, 10858–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Papadia S., Stevenson P., Hardingham N. R., Bading H., Hardingham G. E. (2005) Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J. Neurosci. 25, 4279–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang S. J., Steijaert M. N., Lau D., Schütz G., Delucinge-Vivier C., Descombes P., Bading H. (2007) Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53, 549–562 [DOI] [PubMed] [Google Scholar]
- 7. Zhang S. J., Zou M., Lu L., Lau D., Ditzel D. A., Delucinge-Vivier C., Aso Y., Descombes P., Bading H. (2009) Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 5, e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang S. J., Buchthal B., Lau D., Hayer S., Dick O., Schwaninger M., Veltkamp R., Zou M., Weiss U., Bading H. (2011) A signaling cascade of nuclear calcium-CREB-ATF3 activated by synaptic NMDA receptors defines a gene repression module that protects against extrasynaptic NMDA receptor-induced neuronal cell death and ischemic brain damage. J. Neurosci. 31, 4978–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bengtson C. P., Freitag H. E., Weislogel J. M., Bading H. (2010) Nuclear calcium sensors reveal that repetition of trains of synaptic stimuli boosts nuclear calcium signaling in CA1 pyramidal neurons. Biophys. J. 99, 4066–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mauceri D., Freitag H. E., Oliveira A. M., Bengtson C. P., Bading H. (2011) Nuclear calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron 71, 117–130 [DOI] [PubMed] [Google Scholar]
- 11. Oliveira A. M., Hemstedt T. J., Bading H. (2012) Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 15, 1111–1113 [DOI] [PubMed] [Google Scholar]
- 12. Simonetti M., Hagenston A. M., Vardeh D., Freitag H. E., Mauceri D., Lu J., Satagopam V. P., Schneider R., Costigan M., Bading H., Kuner R. (2013) Nuclear calcium signaling in spinal neurons drives a genomic program required for persistent inflammatory pain. Neuron 77, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hardingham G. E., Arnold F. J., Bading H. (2001) Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat. Neurosci. 4, 261–267 [DOI] [PubMed] [Google Scholar]
- 14. Chawla S., Hardingham G. E., Quinn D. R., Bading H. (1998) CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science 281, 1505–1509 [DOI] [PubMed] [Google Scholar]
- 15. Hardingham G. E., Chawla S., Cruzalegui F. H., Bading H. (1999) Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron 22, 789–798 [DOI] [PubMed] [Google Scholar]
- 16. Hu S. C., Chrivia J., Ghosh A. (1999) Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron 22, 799–808 [DOI] [PubMed] [Google Scholar]
- 17. Tian F., Hu X. Z., Wu X., Jiang H., Pan H., Marini A. M., Lipsky R. H. (2009) Dynamic chromatin remodeling events in hippocampal neurons are associated with NMDA receptor-mediated activation of Bdnf gene promoter 1. J. Neurochem. 109, 1375–1388 [DOI] [PubMed] [Google Scholar]
- 18. Lee J. Y., Lee T. H. (2012) Effects of histone acetylation and CpG methylation on the structure of nucleosomes. Biochim. Biophys. Acta 1824, 974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buchthal B., Lau D., Weiss U., Weislogel J. M., Bading H. (2012) Nuclear calcium signaling controls methyl-CpG-binding protein 2 (MeCP2) phosphorylation on serine 421 following synaptic activity. J. Biol. Chem. 287, 30967–30974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu J., McKinsey T. A., Nicol R. L., Olson E. N. (2000) Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 4070–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dressel U., Bailey P. J., Wang S. C., Downes M., Evans R. M., Muscat G. E. (2001) A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276, 17007–17013 [DOI] [PubMed] [Google Scholar]
- 22. Chan J. K., Sun L., Yang X. J., Zhu G., Wu Z. (2003) Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278, 23515–23521 [DOI] [PubMed] [Google Scholar]
- 23. Davis F. J., Gupta M., Camoretti-Mercado B., Schwartz R. J., Gupta M. P. (2003) Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase HDAC4: implications in cardiac muscle gene regulation during hypertrophy. J. Biol. Chem. 278, 20047–20058 [DOI] [PubMed] [Google Scholar]
- 24. Cohen T. J., Barrientos T., Hartman Z. C., Garvey S. M., Cox G. A., Yao T. P. (2009) The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J. 23, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tse C., Sera T., Wolffe A. P., Hansen J. C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18, 4629–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peserico A., Simone C. (2011) Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011, 371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang X. J., Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baltan S., Bachleda A., Morrison R. S., Murphy S. P. (2011) Expression of histone deacetylases in cellular compartments of the mouse brain and the effects of ischemia. Transl. Stroke Res. 2, 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ajamian F., Suuronen T., Salminen A., Reeben M. (2003) Up-regulation of class II histone deacetylases mRNA during neural differentiation of cultured rat hippocampal progenitor cells. Neurosci. Lett. 346, 57–60 [DOI] [PubMed] [Google Scholar]
- 31. Grozinger C. M., Hassig C. A., Schreiber S. L. (1999) Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. U.S.A. 96, 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McKinsey T. A., Zhang C. L., Lu J., Olson E. N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chawla S., Vanhoutte P., Arnold F. J., Huang C. L., Bading H. (2003) Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85, 151–159 [DOI] [PubMed] [Google Scholar]
- 34. Grozinger C. M., Schreiber S. L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3--dependent cellular localization. Proc. Natl. Acad. Sci. U.S.A. 97, 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao C., Ho C. C., Reineke E., Lam M., Cheng X., Stanya K. J., Liu Y., Chakraborty S., Shih H. M., Kao H. Y. (2008) Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol. Cell. Biol. 28, 5658–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Glozak M. A., Seto E. (2009) Acetylation/deacetylation modulates the stability of DNA replication licensing factor Cdt1. J. Biol. Chem. 284, 11446–11453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J., Campos B., Jamieson G. A., Jr., Kaetzel M. A., Dedman J. R. (1995) Functional elimination of calmodulin within the nucleus by targeted expression of an inhibitor peptide. J. Biol. Chem. 270, 30245–30248 [DOI] [PubMed] [Google Scholar]
- 38. Pusl T., Wu J. J., Zimmerman T. L., Zhang L., Ehrlich B. E., Berchtold M. W., Hoek J. B., Karpen S. J., Nathanson M. H., Bennett A. M. (2002) Epidermal growth factor-mediated activation of the ETS domain transcription factor Elk-1 requires nuclear calcium. J. Biol. Chem. 277, 27517–27527 [DOI] [PubMed] [Google Scholar]
- 39. Bolger T. A., Yao T. P. (2005) Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 25, 9544–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bading H., Greenberg M. E. (1991) Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science 253, 912–914 [DOI] [PubMed] [Google Scholar]
- 41. Wiegert J. S., Bengtson C. P., Bading H. (2007) Diffusion and not active transport underlies and limits ERK1/2 synapse-to-nucleus signaling in hippocampal neurons. J. Biol. Chem. 282, 29621–29633 [DOI] [PubMed] [Google Scholar]
- 42. Arnold F. J., Hofmann F., Bengtson C. P., Wittmann M., Vanhoutte P., Bading H. (2005) Microelectrode array recordings of cultured hippocampal networks reveal a simple model for transcription and protein synthesis-dependent plasticity. J. Physiol. 564, 3–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guatimosim S., Amaya M. J., Guerra M. T., Aguiar C. J., Goes A. M., Gómez-Viquez N. L., Rodrigues M. A., Gomes D. A., Martins-Cruz J., Lederer W. J., Leite M. F. (2008) Nuclear Ca2+ regulates cardiomyocyte function. Cell Calcium 44, 230–242 [DOI] [PubMed] [Google Scholar]
- 44. Rodrigues M. A., Gomes D. A., Leite M. F., Grant W., Zhang L., Lam W., Cheng Y. C., Bennett A. M., Nathanson M. H. (2007) Nucleoplasmic calcium is required for cell proliferation. J. Biol. Chem. 282, 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Broide R. S., Redwine J. M., Aftahi N., Young W., Bloom F. E., Winrow C. J. (2007) Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci. 31, 47–58 [DOI] [PubMed] [Google Scholar]
- 46. Hardingham G. E., Bading H. (2002) Coupling of extrasynaptic NMDA receptors to a CREB shut-off pathway is developmentally regulated. Biochim. Biophys. Acta 1600, 148–153 [DOI] [PubMed] [Google Scholar]
- 47. Darcy M. J., Calvin K., Cavnar K., Ouimet C. C. (2010) Regional and subcellular distribution of HDAC4 in mouse brain. J. Comp. Neurol. 518, 722–740 [DOI] [PubMed] [Google Scholar]
- 48. Plath N., Ohana O., Dammermann B., Errington M. L., Schmitz D., Gross C., Mao X., Engelsberg A., Mahlke C., Welzl H., Kobalz U., Stawrakakis A., Fernandez E., Waltereit R., Bick-Sander A., Therstappen E., Cooke S. F., Blanquet V., Wurst W., Salmen B., Bösl M. R., Lipp H. P., Grant S. G., Bliss T. V., Wolfer D. P., Kuhl D. (2006) Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52, 437–444 [DOI] [PubMed] [Google Scholar]
- 49. Jaubert P. J., Golub M. S., Lo Y. Y., Germann S. L., Dehoff M. H., Worley P. F., Kang S. H., Schwarz M. K., Seeburg P. H., Berman R. F. (2007) Complex, multimodal behavioral profile of the Homer1 knock-out mouse. Genes Brain Behav. 6, 141–154 [DOI] [PubMed] [Google Scholar]
- 50. West A. E., Greenberg M. E. (2011) Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol. 3, a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zaidi S. K., Young D. W., Montecino M., van Wijnen A. J., Stein J. L., Lian J. B., Stein G. S. (2011) Bookmarking the genome: maintenance of epigenetic information. J. Biol. Chem. 286, 18355–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carone B. R., Fauquier L., Habib N., Shea J. M., Hart C. E., Li R., Bock C., Li C., Gu H., Zamore P. D., Meissner A., Weng Z., Hofmann H. A., Friedman N., Rando O. J. (2010) Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143, 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Riccio A. (2010) Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat. Neurosci. 13, 1330–1337 [DOI] [PubMed] [Google Scholar]
- 54. Sando R., 3rd, Gounko N., Pieraut S., Liao L., Yates J., 3rd, Maximov A. (2012) HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151, 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim M. S., Akhtar M. W., Adachi M., Mahgoub M., Bassel-Duby R., Kavalali E. T., Olson E. N., Monteggia L. M. (2012) An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J. Neurosci. 32, 10879–10886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cohen T. J., Waddell D. S., Barrientos T., Lu Z., Feng G., Cox G. A., Bodine S. C., Yao T. P. (2007) The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J. Biol. Chem. 282, 33752–33759 [DOI] [PubMed] [Google Scholar]
- 57. Ma C., D'Mello S. R. (2011) Neuroprotection by histone deacetylase-7 (HDAC7) occurs by inhibition of c-jun expression through a deacetylase-independent mechanism. J. Biol. Chem. 286, 4819–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen B., Cepko C. L. (2009) HDAC4 regulates neuronal survival in normal and diseased retinas. Science 323, 256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hardingham G. E., Bading H. (2010) Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat. Rev. Neurosci. 11, 682–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee B., Butcher G. Q., Hoyt K. R., Impey S., Obrietan K. (2005) Activity-dependent neuroprotection and cAMP response element-binding protein (CREB): kinase coupling, stimulus intensity, and temporal regulation of CREB phosphorylation at serine 133. J. Neurosci. 25, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martin M., Kettmann R., Dequiedt F. (2007) Class IIa histone deacetylases: regulating the regulators. Oncogene 26, 5450–5467 [DOI] [PubMed] [Google Scholar]
- 62. Yang X. J., Grégoire S. (2005) Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25, 2873–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Verdin E., Dequiedt F., Kasler H. G. (2003) Class II histone deacetylases: versatile regulators. Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
- 64. Parra M., Verdin E. (2010) Regulatory signal transduction pathways for class IIa histone deacetylases. Curr. Opin. Pharmacol. 10, 454–460 [DOI] [PubMed] [Google Scholar]
- 65. Paroni G., Cernotta N., Dello Russo C., Gallinari P., Pallaoro M., Foti C., Talamo F., Orsatti L., Steinkühler C., Brancolini C. (2008) PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell 19, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao X., Ito A., Kane C. D., Liao T. S., Bolger T. A., Lemrow S. M., Means A. R., Yao T. P. (2001) The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276, 35042–35048 [DOI] [PubMed] [Google Scholar]
- 67. Wang A. H., Yang X. J. (2001) Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21, 5992–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McKinsey T. A., Zhang C. L., Olson E. N. (2001) Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21, 6312–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rittinger K., Budman J., Xu J., Volinia S., Cantley L. C., Smerdon S. J., Gamblin S. J., Yaffe M. B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4, 153–166 [DOI] [PubMed] [Google Scholar]
- 70. Liu Y., Randall W. R., Schneider M. F. (2005) Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J. Cell Biol. 168, 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Illi B., Dello Russo C., Colussi C., Rosati J., Pallaoro M., Spallotta F., Rotili D., Valente S., Ragone G., Martelli F., Biglioli P., Steinkuhler C., Gallinari P., Mai A., Capogrossi M. C., Gaetano C. (2008) Nitric oxide modulates chromatin folding in human endothelial cells via protein phosphatase 2A activation and class II histone deacetylases nuclear shuttling. Circ. Res. 102, 51–58 [DOI] [PubMed] [Google Scholar]
- 72. Kao H. Y., Verdel A., Tsai C. C., Simon C., Juguilon H., Khochbin S. (2001) Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276, 47496–47507 [DOI] [PubMed] [Google Scholar]
- 73. Sugo N., Oshiro H., Takemura M., Kobayashi T., Kohno Y., Uesaka N., Song W. J., Yamamoto N. (2010) Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur. J. Neurosci. 31, 1521–1532 [DOI] [PubMed] [Google Scholar]
- 74. McKinsey T. A., Zhang C. L., Olson E. N. (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. U.S.A. 97, 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Backs J., Backs T., Neef S., Kreusser M. M., Lehmann L. H., Patrick D. M., Grueter C. E., Qi X., Richardson J. A., Hill J. A., Katus H. A., Bassel-Duby R., Maier L. S., Olson E. N. (2009) The δ isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc. Natl. Acad. Sci. U.S.A. 106, 2342–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ling H., Zhang T., Pereira L., Means C. K., Cheng H., Gu Y., Dalton N. D., Peterson K. L., Chen J., Bers D., Brown J. H. (2009) Requirement for Ca2+/calmodulin-dependent kinase II in the transition from pressure overload-induced cardiac hypertrophy to heart failure in mice. J. Clin. Invest. 119, 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guan Y., Chen Q., Yang X., Haines P., Pei M., Terek R., Wei X., Zhao T., Wei L. (2012) Subcellular relocation of histone deacetylase 4 regulates growth plate chondrocyte differentiation through Ca2+/calmodulin-dependent kinase IV. Am. J. Physiol. Cell Physiol. 303, C33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Redmond L., Kashani A. H., Ghosh A. (2002) Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron 34, 999–1010 [DOI] [PubMed] [Google Scholar]
- 79. Hardingham G. E., Arnold F. J., Bading H. (2001) A calcium microdomain near NMDA receptors: on switch for ERK-dependent synapse-to-nucleus communication. Nat. Neurosci. 4, 565–566 [DOI] [PubMed] [Google Scholar]
- 80. Wiegert J. S., Bading H. (2011) Activity-dependent calcium signaling and ERK-MAP kinases in neurons: a link to structural plasticity of the nucleus and gene transcription regulation. Cell Calcium 49, 296–305 [DOI] [PubMed] [Google Scholar]
- 81. Chen R. H., Juo P. C., Curran T., Blenis J. (1996) Phosphorylation of c-Fos at the C terminus enhances its transforming activity. Oncogene 12, 1493–1502 [PubMed] [Google Scholar]
- 82. Gille H., Kortenjann M., Thomae O., Moomaw C., Slaughter C., Cobb M. H., Shaw P. E. (1995) ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 14, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhou X., Richon V. M., Rifkind R. A., Marks P. A. (2000) Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc. Natl. Acad. Sci. U.S.A. 97, 1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]