Abstract
The amplification of DNA fragments, cloned between user-defined 5′ and 3′ end sequences, is a prerequisite step in the use of many current applications including massively parallel sequencing (MPS). Here we describe an improved method, called homopolymer tail-mediated ligation PCR (HTML-PCR), that requires very little starting template, minimal hands-on effort, is cost-effective, and is suited for use in high-throughput and robotic methodologies. HTML-PCR starts with the addition of homopolymer tails of controlled lengths to the 3′ termini of a double-stranded genomic template. The homopolymer tails enable the annealing-assisted ligation of a hybrid oligonucleotide to the template's recessed 5′ ends. The hybrid oligonucleotide has a user-defined sequence at its 5′ end. This primer, together with a second primer composed of a longer region complementary to the homopolymer tail and fused to a second 5′ user-defined sequence, are used in a PCR reaction to generate the final product. The user-defined sequences can be varied to enable compatibility with a wide variety of downstream applications. We demonstrate our new method by constructing MPS libraries starting from nanogram and sub-nanogram quantities of Vibrio cholerae and Streptococcus pneumoniae genomic DNA.
Keywords: molecular cloning, annealing-assisted ligation, DNA capture, massively-parallel sequencing
Cloning DNA fragments as molecular libraries has become a core method used in many research, forensic and clinical settings. Common approaches for molecular library construction involve the ligation of double-stranded adapters of defined sequence to template DNA ends followed by PCR amplification (1, 2, 3). Due in part to the poor efficiency of the adapter ligation reaction, these techniques require large quantities of starting template DNA. Moreover, they are prone to the formation of adapter-dimers, an inhibitory side reaction that necessitates the purification of the DNA products of interest by gel electrophoresis and extraction. This requirement is a particular hindrance when such protocols are adapted for high-throughput robotic 96-well and 384-well plate based methods, thereby limiting the number of different libraries that are created at one time.
More recently, in vitro transposition has been used to facilitate library construction (4). This technology, referred to as Nextera, is marketed as a kit to create Illumina sequencing libraries. Sample DNA is first subjected to a transposition reaction in which the transposase inserts transposon end/Illumina sequence chimeric double-stranded DNA molecules into the sample. Two such transposition events, separated in the sample by roughly 50–500 nucleotides then serve as the template in a PCR reaction that creates the final library. Compared with adapter ligation protocols, Nextera has several major advantages. Its workflow is much faster; it is far less labor-intensive; and it is better suited to high-throughput methods. Nextera also requires significantly less starting template. The major disadvantages of this method are: (i) Nextera is expensive, (ii) kits are no longer marketed to construct libraries either for other sequencing platforms or for non-sequencing applications and since the technology is proprietary, homemade kits are not possible, and (iii) in order to obtain the correct number of transposition events separated by the appropriate distances, the ratio of transposase complexes to sample DNA is critical. For this reason, two different kits are available, one that uses 50 ng of template and another that uses 1 ng. Thus, the user must have accurate knowledge of sample concentration; however, for very dilute samples such knowledge may be inaccurate or lacking. A final disadvantage of the Nextera approach is, a requirement that the template DNA be at least 300 nucleotides in length (preferably longer) and hence, Nextera is not recommended for use in applications such as ChIP-seq (Nextera DNA Sample Preparation Guide; October 2011).
Here we developed a new method, HTML-PCR, which is unencumbered by the deficiencies associated with many other procedures. Compared with Nextera, it is more cost-effective, uses generally available reagents, and can be used to generate libraries for applications in addition to Illumina sequencing. Furthermore, the same HTML-PCR protocol functions with template DNA concentrations that can vary by up to five orders of magnitude and with template molecules of no minimum length. Compared with adapter ligation protocols, HTML-PCR is more efficient and streamlined, and is less labor intensive. Adapters are not used, which avoids the problem of adapter-dimers and the need for gel purification in those applications where specific size ranges are not required. Thus, HTML-PCR is more compatible with high-throughput and robotic methods. In addition, the method uses an extremely efficient ligation reaction that is facilitated by annealing of an oligo-nucleotide to a homopolymer tail. As a result, HTML-PCR can be used to clone miniscule amounts of DNA that are below the amount of starting material needed for adapter ligation protocols.
Materials and methods
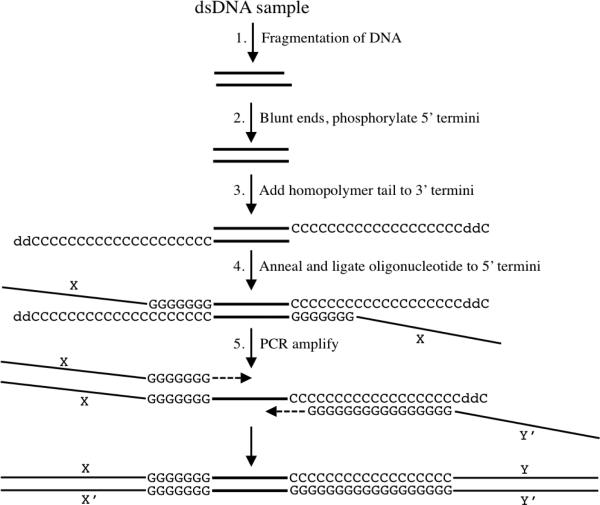
Figure 1 illustrates the application of HTML-PCR for capturing and amplifying double-stranded DNA. Sample DNA is first fragmented into a size range that is appropriate for the downstream application. The ends of the DNA are blunted and 5′ ends are phosphorylated to allow for later ligation. A homopolymer tail (e.g., poly(dC)) of controlled length is added to the 3′ termini using terminal deoxynucleotidyl transferase (TdT) and a mixture of deoxynucleotide triphosphate (e.g., dCTP) and chain-terminating dideoxynucleotide triphosphate (e.g., ddCTP). For oligo(dC) tailing, an average tail length of 20 was achieved by adjusting the ratio of dCTP to ddCTP to 19:1 (Supplementary Figure S1) (5).
Figure 1. Schematic diagram of homopolymer tail-mediated ligation PCR (HTML-PCR).
Details of each numbered step are provided in the text. The sample double-stranded DNA sequence is indicated by the thick double lines and is not to scale. For simplicity only a single double-stranded DNA molecule is depicted and only one strand is shown for the PCR step. X and Y represent user-defined oligonucleotide sequences and X′ and Y′ are their respective complementary sequences. C, 2’-deoxycytosine monophosphate; G, 2’-deoxyguanosine monophosphate; ddC, 2′,3′-dideoxycytosine monophosphate.
After oligo(dC) tail addition, a chimeric oligonucleotide with a defined sequence X at its 5′ end and 4–7 complementary deoxyguanosines (7 is optimal [Supplementary Figure S2]) at its 3′ end is annealed to the homopolymer tail and joined to the 5′ end of the opposing strand using T4 DNA ligase. Due to the stable nature of the seven dC:dG base pairs, this ligation event is extremely efficient. The DNA is next amplified by PCR using the same oligonucleotide used for ligation as the forward primer, and a chimeric reverse primer composed of Y’ at its 5′ end and 16 complementary deoxyguanosines at its 3′ end that are used to prime DNA synthesis from the oligo(dC) tail.
The reverse primer can anneal to and prime from anywhere along the homopolymer tail. In the absence of a chain terminator, the tail length generated can exceed hundreds of nucleotides (Supplementary Figure S1). By using ddCTP in the tailing reaction, the contribution of poly(dC) to the final product is effectively limited. Although either titration of TdT or reduction of reaction time could also be used to limit tail length, we found that the use of chain terminators in the context of excess enzymatic activity yielded the most precise and reproducible results (Supplementary Figure S1 and data not shown). Here HTML-PCR was used exclusively to generate Illumina sequencing libraries. The complete details of each enzymatic reaction together with oligonucleotide and sequencing primer sequences are provided in the Supplementary Materials and Methods section. Some bioinformatic methods of analysis were previously described (6) and additional detail is provided in the Supplementary Materials and Methods section.
Results and discussion
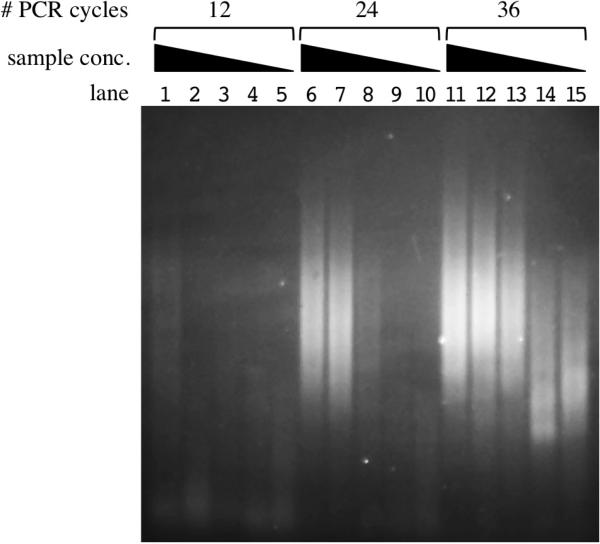
We first tested the utility of HTML-PCR by using the method, in conjunction with MPS, to determine the sequence of a previously unsequenced bacterial strain, V. cholerae E7947. After fragmenting the genomic DNA by high intensity sonication, DNA concentrations over a range of four orders of magnitude (100–0.01 ng) were individually blunted, 5′ end phosphorylated and treated with TdT in the presence of a 19:1 ratio of dCTP:ddCTP to generate 3′ oligo(dC) tails averaging 20 nucleotides in length. The tailed substrate was then ligated to the chimeric oligonucleotide olj623, which has seven dG nucleotides at its 3′ end and a sequence required for Illumina sequencing at its 5′ end. Finally, the products of this reaction were amplified by PCR using primers olj623 and a barcode-containing primer that contains sixteen dG nucleotides at its 3′ end and a second sequence required for Illumina sequencing at its 5′ end. The reactions with 1–100 ng of input template yielded a range of products from approximately 150–1000 bp (Figure 2, lanes 1, 6–8 and 11–13). Twelve cycles of PCR were sufficient only for the highest amount (100 ng) of input genomic DNA (Figure 2, lane 1), while 24 cycles was sufficient for input amounts down to 1 ng (Figure 2, lanes 6–8). For the lowest input amounts (0.1 and 0.01 ng), visible products were only observed in the 36 cycle samples (Figure 2, lanes 14 and 15): However, the size range of the resulting products was distinctly lower than in the other lanes. MPS revealed that the products in lanes 14 and 15 were a mixture of bona fide V. cholerae sequences and unintended sequences derived from primers and contaminating human (possibly investigator) DNA (see below).
Figure 2. Sensitivity of HTML-PCR using Vibrio cholerae genomic DNA.
HTML-PCR products generated using a range of starting sample amounts as visualized by 2% agarose gel electrophoresis. The following amounts of fragmented V. cholerae E7946 gDNA were used as input for the homopolymer tail addition step of HTML-PCR: 100 ng for lanes 1, 6 and 11; 10 ng for lanes 2, 7 and 12; 1 ng for lanes 3, 8 and 13; 0.1 ng for lanes 4, 9 and 14; and 0.01 ng for lanes 5, 10 and 15. Lanes 1, 6, 7, 8, 11, 12 and 13 show products of a successful reaction, namely DNA smears ranging from approximately 140–600 base pairs that represent V. cholerae library fragments (see text). The lower molecular weight DNA smears in lanes 14 and 15 represent a mixture of V. cholerae library fragments plus artifactual products (see text).
Samples from Figure 2, lanes 11–15 were subjected to Illumina sequencing and the resulting sequences were aligned to the complete genome sequence of a closely related V. cholerae reference strain, N16961 (7). We also used conventional adapter ligation-mediated Illumina library preparation to sequence E7946. We found that when compared with the published sequence of the N16961 reference strain, the E7946 sequence contained 92 single nucleotide polymorphisms (SNPs) and 100 deletion/insertion polymorphisms (DIPs) (Supplementary Tables S1 and S2). For the samples from Figure 2, lanes 11–13, 96.8%, 94.6% and 68.1% of the raw unfiltered sequencing reads could be mapped to the N16961 reference genome respectively. After filtering for quality, 99.7%, 99.1% and 89.5% of the respective reads were mapped to the reference sequence. Importantly, all of the SNPs and DIPs observed with the conventional Illumina library preparation were observed with the samples from Figure 2, lanes 11–13. In other words, the traditional method and HTML-PCR yielded identical results; however, while 5 μg of genomic DNA were used to prepare the traditional library, 5,000 fold less DNA (1 ng; Figure 2, lane 13) was needed for preparation by HTML-PCR. For the samples from Figure 2, lanes 14 and 15, even after the reads were filtered for quality, only 56.9% and 11.0% respectively were mapped to the N16961 genome. Still, there was sufficient data from each sample to cover greater than >99% of the E7946 genome and >90% of the SNPs were detected. Therefore, HTML-PCR was at least partially successful down to 0.01 ng of input DNA, which is 100,000–500,000-fold less than that recommended by Illumina for their adapter-based method. Although smaller amounts of starting material can be used in different adapter-based methodologies, including the standard Illumina approach, HTML-PCR is clearly effective at concentrations well below those employed with these methods.
As a point of reference, 0.01 ng is the amount of DNA present in one and one-third human diploid cells. Thus, a few contaminating human cells could provide the majority of DNA present in a 0.01 ng bacterial gDNA sample. When preparing and processing subnanogram quantities of DNA, one can go to extraordinary lengths to avoid exogenous DNA contamination. No such measures were taken during our sample preparation and our experience with DNA contamination underscores the importance of employing these procedures when minute quantities of DNA are processed.
We next compared Illumina sequencing libraries created by HTML-PCR to those created by Nextera using a strain that had been previously sequenced, namely the TIGR4 isolate of Streptococcus pneumoniae (8). In each case, 50 ng of starting gDNA template was used. The Nextera library was prepared from the kit as per the manufacturer's instructions while the HTML-PCR library was prepared as above. Following library preparation, the samples were sequenced and then subjected to bioinformatic analysis. HTML-PCR uses mechanical shearing to create the ends from which molecules are sequenced while Nextera uses transposition. Although mechanical shearing is known to be an unbiased process, transposases can select target DNA in a biased and nonrandom manner. We therefore anticipated that Nextera might show a greater bias toward and/or against particular gDNA target sequences. In an effort to compensate for this and obtain as many Nextera-generated ends as possible, 5-fold more of that sample relative to the HTML-PCR sample was loaded within a single lane of the Illumina flow-cell.
After trimming the reads for quality, in each case they were mapped to the reference genome (8). The HTML-PCR library yielded 17,995,348 filtered reads of which 99.4% mapped to the reference genome while the Nextera sample yielded 91,242,087 filtered reads of which 96.5% were mapped. We next looked for whether particular regions of the genome represented hotspots for transposition or shearing by each method. Similarly, if there were any sequences that were strongly favored in the earliest rounds of PCR, these jackpot events would also appear as hotspots. In each case, the position in the genome with the highest sequence coverage (strongest hotspot) was identified and its coverage was divided by the average coverage of the entire genome. By this measure the higher the value obtained, the greater the hot spot preference. To our surprise, Nextera did not show a significant preference for any sequence, with the highest value being 3.652, which was essentially the same as the highest value for HTML-PCR, which was 3.646. We conclude that neither method is prone to hotspot insertion or jackpot PCR biases.
As a measure of the extent of genome coverage we analyzed the number of unique 5′ ends generated by each method. Since each strand of the genome is independently sequenced, the maximum theoretical number of unique 5′ ends is twice the genome length. With Nextera, of 91,242,087 filtered reads, there were 3,192,276 unique 5′ ends represented (73.9% of the maximum) while with the 17,995,348 filtered HTML-PCR reads, 2,185,530 unique ends were represented (50.6% of the maximum). For both libraries, there were no unsequenced positions in the reference genome and both yielded the same consensus genome sequence.
We also compared the coverage distribution of sites throughout the genome in the libraries generated by the two methods. As shown in Supplementary Figure S3, the resulting plots were quite similar. We also examined coverage as a function of the GC content of specific regions throughout the genome. We observed coverage bias with both methods although it was more severe with the Nextera-generated library. For instance, regions of the genome with a 20% GC content were covered 5.52-fold lower than those with a 50% content with the Nextera library while there was only a 2.36-fold bias with the HTML-PCR library (Supplementary Figure S4). We conclude that for the two library construction methods, the quality of sequencing data obtained was similar although HTML-PCR is better suited for sequencing regions or genomes with low GC content.
With HTML-PCR, due to the 16 dG nucleotides present at the 3′ end of one of the PCR primers used, the genomic DNA can only be amplified if it contains a stretch of complementary dC nucleotides of a similar or greater length. In most molecules, the exogenously added oligo(dC) tail provides that requirement, however, if long oligo(dC) stretches exist naturally in the genome, these sites could be amplified in a tail-independent manner. Furthermore, since amplification of endogenous sites does not depend upon the efficiency of tailing, this amplification might be very efficient resulting in the over-representation of endogenous homopolymers in the final library. For the experiments above, this theoretical objection is not applicable as nowhere within the V. cholerae or S. pneumoniae genomes are there oligo(dC) stretches that exceed 11 nucleotides in length. However, in larger, more complex genomes such as the human genome, numerous endogenous dC stretches of at least 16 nucleotides do exist (9, 10).
We therefore examined whether we could modify the specifics of the tailing and PCR reactions to prevent the amplification of endogenous homopolymer sites. While the dA:dT base pair involves only two hydrogen bonds, when the artificial base 2-amino deoxyadenosine (2-amino dA) pairs with dT, three hydrogen bonds can form (11, 12, 13, 14). We reasoned that if we created a tail composed of 2-amino dA, the added stability from pairing of this tail with an oligo(dT) primer could enable priming at PCR annealing temperatures where priming of the endogenous poly(dA) stretches would not occur.
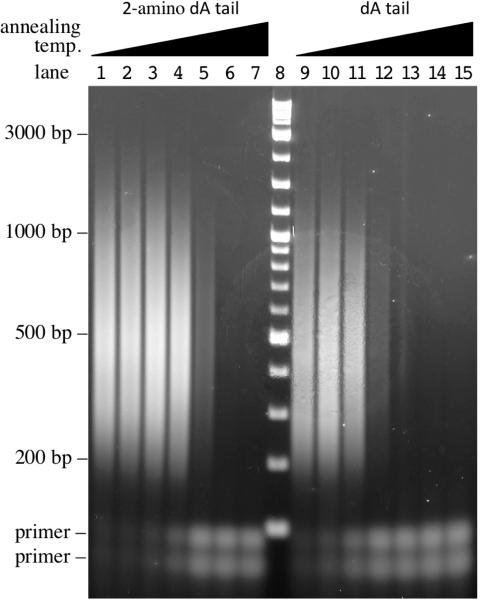
To test this hypothesis we added two different homopolymer tails to V. cholerae genomic DNA. In the first case an oligo(dA) tail with a 30 nt average length was added using TdT and a 29:1 ratio of dATP:ddATP. This tail was used as a surrogate for an endogenous dA stretch and was used to define the maximum annealing temperature at which oligo(dT) can prime from oligo(dA). The second case was identical to the first except that 2-amino dATP was substituted. Each tailed substrate was first ligated to an oligonucleotide that has seven dT nucleotides at its 3′ end and then subjected to PCR using this same oligonucleotide together with a second oligonucleotide that has 22 dT nucleotides at its 3′ end. For each tailed substrate, seven different PCR annealing temperatures were tested and the results are shown in Figure 3. The intensity of products generated with the 2-amino dA-tailed substrate at an annealing temperature of 62.4°C was very similar to that obtained with the dA-tailed substrate at 58.3°C (compare lanes 5 and 12 in Figure 3), whereas no product was formed for the dA-tailed substrate at an annealing temperature of 62.4°C (Figure 3, lane 13). Hence, the maximum allowed annealing temperature was increased by more than 4°C when 2-amino dATP was substituted for dATP in the tailing reaction. The exogenously added poly(dA) sequence is chemically equivalent to an endogenous poly(dA) sequence that might naturally occur within a genome. We therefore conclude that by using 2-amino dATP in the tailing reaction and an annealing temperature of 62.4°C during PCR, it is possible to prime from exogenous tails without priming from endogenous stretches.
Figure 3. Thermodynamic advantage of 2-amino deoxyadenosine-tailed substrates.
HTML-PCR products generated using 100 ng of template and 15 cycles of PCR as visualized by 2% agarose gel electrophoresis. Lanes 1–7 were performed using 2-amino deoxyadenosine-tailed substrate, lane 8 contains 2-Log DNA ladder (New England Biolabs, Ipswich, MA) and lanes 9–15 were performed using dA-tailed substrate. In lanes 1–7 and in lanes 8–15, the PCR annealing temperature was progressively increased as follows: 48°C, 52.1°C, 54.7°C, 58.3°C, 62.4°C, 65.7°C, or 68.4°C, respectively.
In summary, we developed a new method, HTML-PCR and used it to accurately sequence bacterial genomes, even when only 1 nanogram or less of sample DNA was used. Furthermore, by using a homopolymer tail of synthetic nucleotides, we were able to find conditions in which endogenous genomic homopolymers were ignored and only exogenously added tails were used to prime synthesis. This modification should enable the use of HTML-PCR with any genome regardless of its endogenous homopolymer content. Compared with Nextera, HTML-PCR is more versatile as it can be applied to sequencing platforms other than Illumina and to applications in addition to sequencing. It is also more cost-effective and uses reagents that are readily available from numerous sources. Finally, unlike Nextera, HTML-PCR functions with templates over a very broad range in concentration, without minimum size constraints, and even when the GC content is low. Compared with adapter ligation methods, HTML-PCR requires fewer steps, only a few inexpensive reagents and minimal hands-on time. The method does not require adapter ligation and is not prone to generating adapter-dimers or primer-dimers even when template is extremely limiting. This feature obviates the need for gel purification and size selection for many applications, thus enabling the method to be compatible with high-throughput formats and robotic assistance. In addition to research applications, HTML-PCR is also ideally suited to medical and forensic applications where the supply or integrity of sample DNA may be limited.
Supplementary Material
Acknowledgments
This work was supported by Award Numbers AI45746 (A.C.) and AI055058 (A.C.) from the National Institutes of Health. A.C. is a Howard Hughes Medical Institute investigator. This paper is subject to the NIH Public Access Policy.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/113981.
Competing Interests
The authors have a patent pending on HTML-PCR.
References
- 1.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, et al. Accurate Whole Human Genome Sequencing using Reversible Terminator Chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranade SS, Chung CB, Zon G, Boyd VL. Preparation of genome-wide DNA fragment libraries using bisulfite in polyacryl-amide gel electrophoresis slices with formamide denaturation and quality control for massively parallel sequencing by oligonucleotide ligation and detection. Anal. Biochem. 2009;390:126–135. doi: 10.1016/j.ab.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boule J-B, Rougeon F, Papanicolaou C. Terminal Deoxynucleotidyl Transferase Indiscriminately Incorporates Ribonucleotides and Deoxyribonucleotides. J. Biol. Chem. 2001;276:31388–31393. doi: 10.1074/jbc.M105272200. [DOI] [PubMed] [Google Scholar]
- 6.Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–484. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 9.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, et al. The Sequence of the Human Genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 11.Howard FB, Frazier J, Miles HT. A new polynucleotide complex stabilized by three interbase hydrogen bonds, poly-2-amino-adenylic acid + polyuridylic acid. J. Biol. Chem. 1966;241:4293–4295. [PubMed] [Google Scholar]
- 12.Rackwitz HR, Scheit KH. The Stereochemical Basis of Template Function. Eur. J. Biochem. 1976;72:191–200. doi: 10.1111/j.1432-1033.1977.tb11239.x. [DOI] [PubMed] [Google Scholar]
- 13.Scheit KH, Rackwitz HR. Synthesis and physicochemocal properties of two analogs of poly(dA): poly(2-aminopurine-9-3-D-deoxyribonucleotide) and poly 2-amino-deoxyadenylicacid. Nucleic Acids Res. 1982;10:4059–4069. doi: 10.1093/nar/10.13.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong C, Tinoco I, Jr., Chollet A. Thermodynamic studies of base pairing involving 2,6-diaminopurine. Nucleic Acids Res. 1988;16:5115–5122. doi: 10.1093/nar/16.11.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.