Abstract
Silencing the mutant huntingtin gene (muHTT) is a direct and simple therapeutic strategy for the treatment of Huntington disease (HD) in principle. However, targeting the HD mutation presents challenges because it is an expansion of a common genetic element (a CAG tract) that is found throughout the genome. Moreover, the HTT protein is important for neuronal health throughout life, and silencing strategies that also reduce the wild-type HTT allele may not be well tolerated during the long-term treatment of HD. Several HTT silencing strategies are in development that target genetic sites in HTT that are outside of the CAG expansion, including HD mutation-linked single-nucleotide polymorphisms and the HTT promoter. Preclinical testing of these genetic therapies has required the development of a new mouse model of HD that carries these human-specific genetic targets. To generate a fully humanized mouse model of HD, we have cross-bred BACHD and YAC18 on the Hdh−/− background. The resulting line, Hu97/18, is the first murine model of HD that fully genetically recapitulates human HD having two human HTT genes, no mouse Hdh genes and heterozygosity of the HD mutation. We find that Hu97/18 mice display many of the behavioral changes associated with HD including motor, psychiatric and cognitive deficits, as well as canonical neuropathological abnormalities. This mouse line will be useful for gaining additional insights into the disease mechanisms of HD as well as for testing genetic therapies targeting human HTT.
INTRODUCTION
Huntington disease (HD) is a dominantly inherited neurodegenerative disorder characterized by loss of voluntary motor control, psychiatric disturbance and cognitive decline that usually leads to death 15–20 years after symptom onset (1). The identification of the monogenic cause of HD led to the development of an accurate genetic test for the disorder 25 years ago (2). Despite this predictive ability to identify mutation carriers decades before disease onset, there is currently no effective therapy for HD that can delay onset or slow disease progression. Development of small molecule therapies for HD has been hindered by difficulties identifying and validating tractable drug targets within the disorder's complex pathogenesis. However, the mutant copy of the HTT gene (muHTT) provides an upstream and universal therapeutic target for the treatment of HD.
There is significant evidence that reducing the expression of muHTT will result in amelioration of HD. Inactivation of the muHTT transgene in symptomatic conditional HD transgenic mice leads to not only halting of disease progression, but reversal of neuropathological and behavioral HD-like phenotypes (3). Even in late-stage animals where significant, irreversible neuron loss has already occurred, inactivation of muHTT prevents further neuron loss, allows recovery of dopamine- and cyclic AMP-regulated phosphoprotein (DARPP-32) expression in the remaining neurons and results in full motor recovery (4). This provides evidence that muHTT silencing could provide both preventative and potentially restorative therapeutic benefit. Several studies have demonstrated the efficacy of non-selective or species-selective HTT silencing in mouse models of HD (5–11). Recently, it has even been shown that therapeutic benefit can persist well beyond the period of HTT silencing (12), indicating that transient relief from muHTT-induced toxicity can allow partial functional recovery and provide lasting benefit. However, the silencing reagents used in these studies would induce non-selective silencing of both mutant and wild-type (wt) HTT in humans, which may result in loss of wtHTT function-associated pathology.
The HTT protein is required for embryonic development and loss of one copy of Hdh (the mouse HTT homolog) throughout development results in behavioral and morphological abnormalities (13,14). The postnatal HTT requirements are less well understood. The HTT protein is neuroprotective and plays roles in several vital cellular processes including axonal and vesicle transport, transcriptional regulation and control of synaptic activity (15). While total loss of HTT in the adult forebrain leads to progressive neurodegeneration in mice (16), several short-term studies have demonstrated the relative safety of partial HTT reduction in the brains of adult mice and non-human primates (8,12,17,18). However, these studies have not adequately evaluated the threshold of non-pathological postnatal HTT reduction. The non-human primate studies achieved a maximal 45% reduction in the HTT protein, a value that is likely to be exceeded in treatment of HD. Greater HTT reduction was achieved in the transgenic HD mouse model studies; however, as these models express both the normal complement of Hdh and transgenic human HTT, they have more total HTT to begin with and would therefore be more resistant to HTT reduction. For example, a 75% reduction in the total HTT protein in a transgenic mouse model that initially has twice as much the HTT protein as a wt mouse would still result in ∼50% of endogenous HTT levels remaining. Moreover, while the amount of HTT reduction in genetic models is homogeneous in all cells, HTT reduction following treatment with silencing reagents is heterogeneous and calculated by tissue averages. Following treatment, HTT reduction may vary from 0 to 100% from cell to cell, further confounding the interpretation of postnatal HTT reduction safety data. Additionally, while these studies were conducted over a matter of months, therapeutic HTT silencing in humans would likely span decades. The safety of long-term partial wtHTT reduction in the brain has not yet been investigated. Considering that HD pathogenesis includes the aspects of both gain and loss of HTT function and that symptom onset usually occurs in mid-life, despite the lifelong presence of the mutation, it is likely that the effects of wtHTT silencing could not be fully evaluated over the course of 6 months, which has been the longest post-HTT-silencing interval tested in animals. For these reasons, allele-specific silencing strategies that selectively lower mutant but not wtHTT are preferable and more likely to be well tolerated.
The mutation that causes HD is the expansion of a polyglutamine-encoding CAG tract in exon 1 of HTT to >36 repeats (19). As CAG repeats are a normally occurring genetic element throughout the genome (20), the development of ‘selective’ silencing strategies has required identification of target sites other than the CAG tract in HTT. Over 2000 single-nucleotide polymorphisms (SNPs) have been identified in the HTT region and ∼200 of these have minor allele frequencies greater than 1% (http://www.ncbi.nlm.nih.gov/projects/SNP/), providing a wealth of potential silencing targets. Using direct sequencing and SNP genotyping, we have previously identified haplotypes in the HTT region that are strongly associated with CAG expansion (21). These haplotypes contain numerous SNPs that, when heterozygous in an individual, can be used to selectively target the CAG-expanded HTT allele for knockdown. Combinatorial analysis demonstrated that >85% of HD patients would have the correct genotypes to receive allele-specific treatment using a therapeutic panel of silencing reagents targeting as few as three of these SNPs (21,22). Several SNP-targeted antisense oligonucleotide (ASO) drugs have been identified that selectively silence muHTT in HD patient fibroblasts (23). Preclinical testing of these ASOs must be performed in an animal model with the same SNP genotypes as the prospective patient population.
Sequence analysis at 91 SNPs of the human HTT transgene in multiple mouse models of HD revealed that BACHD mice (24), which express full-length human muHTT with 97Q, share 85 target SNP alleles with the majority of Caucasian HD patients. Of these, 27 SNPs occur in the pre-mRNA and are heterozygous in greater than 30% of the sequenced HD population, making them viable targets for human therapy. In contrast, YAC18 mice (25), which express full-length human wtHTT with 18Q, do not contain the target SNP alleles at these 27 therapeutically relevant SNPs in their human transgenes (23). Thus, when co-expressed the BACHD and YAC18, transgenes are heterozygous at both the HD mutation and 27 potential allele-specific silencing target SNP sites. To generate a suitable mouse model for preclinical testing of human HTT SNP-targeted silencing reagents, we have cross-bred BACHD and YAC18 on the Hdh−/− background. This fully humanized HD model, Hu97/18, lacks the mouse homolog, but has one human muHTT gene and one human wtHTT gene, making it the first mouse model that is genetically equivalent to human HD patients. The HD-like phenotypes of the BACHD mouse, which include motor and cognitive deficits as well as neuropathology (24), are expected to be penetrant on the Hu97/18 strain. Therefore, we have characterized these mice using endpoints previously validated in the BACHD model.
RESULTS
Genetics
The YAC and BACHD transgenic mice were previously genotyped at 91 SNPs in the HTT gene region using the ‘96SNP-assay’ (23). The YAC mice were determined to have haplotype ‘C’ and the BACHD mice are haplotype ‘A4’ according to the haplotype definitions established previously (21). In the Caucasian HD population, the majority of expanded CAG HTT alleles have A haplotypes, while the most common single haplogroup for non-expanded CAG HTT alleles is C (21). Thus, when co-expressed with the CAG expansion on the BAC transgene, these two HTT alleles recapitulate the genetics of the most common combination of alleles in the Caucasian HD population. There are 39 SNP positions in the HTT region that were found to be different between the YAC and BACHD transgenes (Table 1, adapted from 23; Supplementary Material, Table S1). Of these, 27 were found to be within the HTT transcript and individually heterozygous in greater than 30% of the sequenced HD population. These therapeutically relevant SNPs are highlighted in grey in Table 1. The transgenic mouse lines were homozygous for all other SNPs in the 96SNP assay.
Table 1.
Summary of known heterozygosities in Hu97/18 mice and human HD patients
| SNP | Transcribed? | YAC | BAC | Human HD | % HD population |
|---|---|---|---|---|---|
| rs1263309 | No | C | T | T | 49.1 |
| rs2798296 | No | A | G | G | 51.7 |
| rs2857936 | No | T | C | C | 44.9 |
| rs762855 | No | G | A | A | 53.8 |
| rs3856973 | Yes | A | G | G | 49.6 |
| rs2285086 | Yes | G | A | A | 49.6 |
| rs7659144 | Yes | G | C | C | 41.5 |
| rs16843804 | Yes | T | C | C | 38.5 |
| rs2024115 | Yes | G | A | A | 47.9 |
| rs7691627 | Yes | A | G | G | 48.7 |
| rs4690072 | Yes | G | T | T | 49.1 |
| rs6446723 | Yes | C | T | T | 50.0 |
| rs363064 | Yes | T | C | C | 35.5 |
| rs4690073 | Yes | A | G | G | 48.7 |
| rs34315806 | Yes | T | C | C | 37.6 |
| rs363099 | Yes | T | C | C | 37.6 |
| rs363096 | Yes | C | T | T | 39.7 |
| rs2298967 | Yes | C | T | T | 37.6 |
| rs2298969 | Yes | G | A | A | 52.1 |
| rs6844859 | Yes | C | T | T | 48.7 |
| rs2798232 | Yes | C | T | C | 0.0 |
| rs7685686 | Yes | G | A | A | 48.7 |
| rs363088 | Yes | T | A | A | 37.6 |
| rs362331 | Yes | C | T | T | 48.7 |
| rs916171 | Yes | G | C | C | 48.7 |
| rs2857790 | Yes | C | A | C | 0.0 |
| rs362275 | Yes | T | C | C | 38.0 |
| rs362273 | Yes | G | A | A | 38.9 |
| rs3121419 | Yes | T | C | C | 38.0 |
| rs362272 | Yes | A | G | G | 38.0 |
| rs362271 | Yes | A | G | G | 38.5 |
| rs3775061 | Yes | G | A | A | 38.0 |
| rs362306 | Yes | A | G | G | 38.0 |
| rs362296 | No | A | C | C | 42.3 |
| rs3121417 | No | A | G | G | 41.5 |
| rs3129322 | No | C | T | T | 41.5 |
| rs108850 | No | G | C | C | 48.7 |
| rs1006798 | No | G | A | A | 41.0 |
| rs3095074 | No | A | G | G | 39.3 |
Therapeutically relevant SNPs, those that are in the HTT transcript and heterozygous in greater than 30% of the sequenced HD population, are highlighted in bold. Adapted from Carroll et al. (23).
HTT levels
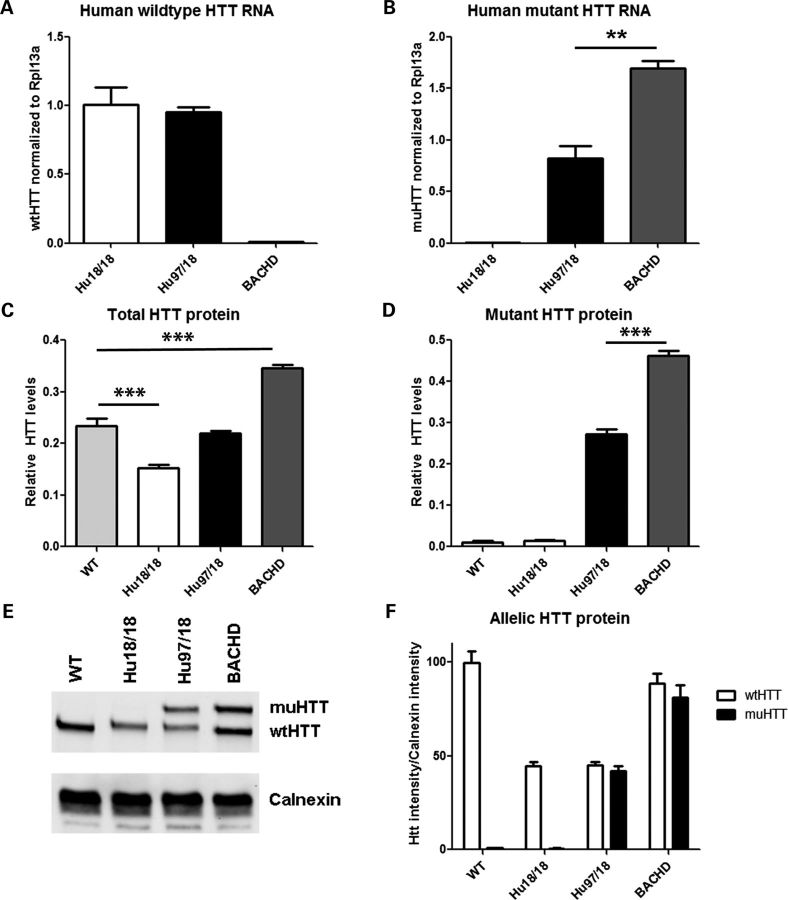
We interbred BACHD, YAC18 and Hdh+/− mice to generate BACHD+;YAC18++; Hhd−/− (Hu97/18) mice or YAC18++; Hdh−/− (Hu18/18) mice. We then evaluated the levels of HTT RNA and protein in the brains of the newly generated humanized mice and compared them with BACHD and wt mice. RNA levels were compared in four mice per genotype using Taqman SNP quantitative reverse transcription polymerase chain reaction (qRT–PCR) with probes selective for either human wt or human muHTT (Fig. 1A and B). As expected human wtHTT RNA was similar in Hu18/18 and Hu97/18 mice. However, human muHTT RNA was nearly two-fold higher in BACHD than Hu97/18 mice, despite arising from the same transgene. HTT protein levels were compared in four mice per genotype by fluorescence resonance energy transfer (FRET) using antibody pairs recognizing total HTT or expanded CAG HTT (Fig. 1C and D). We found that compared with wt mice, BACHD mice have more total HTT protein and Hu18/18 mice have less, while the level of the total HTT protein in the brains of Hu97/18 mice is similar to that of wt mice. Mirroring the RNA, the muHTT protein level in Hu97/18 mice is ∼40% less than that in BACHD mice. We next compared the level of wt and muHTT protein in three mice per genotype using allelic separation quantitative immunoblotting and found equivalent levels of the two proteins in the Hu97/18 brains (Fig. 1E and F). This assay also confirms the down-regulation of BACHD transgene-derived muHTT protein in the context of the Hu97/18 brain seen in the qRT–PCR and FRET assays. This reduction is presumably related to the replacement of mouse wtHTT with human wtHTT, though loss of mouse HTT does not change the expression level of the YAC18 transgene (Supplementary Material, Fig. S1). Data shown are from the right cortex of 2-month-old animals, though similar relative HTT levels were observed in pooled striata or in cortices from 12-month-old animals (data not shown).
Figure 1.
HTT levels in Hu97/18 mice. HTT RNA (A and B) and protein (C–F) were evaluated in cortical lysates from 2-month-old wt, Hu18/18, Hu97/18 and BACHD mice (n = 4 per group). (A) Quantitative reverse transcription polymerase chain reaction (qRT–PCR) detection of human wtHTT RNA, which is similar in Hu18/18 and Hu97/18 mice, but absent from BACHD mice. (B) qRT–PCR detection of human muHTT RNA, which is absent from Hu18/18 mice, and higher in BACHD than Hu97/18 mice. (C) FRET detection of relative levels of the total HTT protein, which is less than wt in Hu18/18 mice, similar to wt in Hu97/18 mice, and higher than wt in BACHD mice. (D) FRET detection of relative levels of muHTT protein, which is absent in wt and Hu18/18 mice and higher in BACHD mice than Hu97/18 mice. (E) Representative allelic separation quantitative immunoblot of wt and muHTT protein (MAB2166) showing that there are similar levels of human wt and muHTT protein in Hu97/18 mice. (F) Quantitation of three animals per group and two replicates per animal. **P < 0.01 and ***P < 0.001.
Body weight
Hu97/18 and Hu18/18 littermates were weighed at 2-month intervals from 2 to 12 months of age. While Hu97/18 mice display a small increase in body weight when compared with Hu18/18 mice, both genotypes display increased body weight when compared with historical body weight data from FVB mice (Fig. 2). It has previously been reported that body weight is correlated to levels of full-length HTT protein (26, 27). However, as Hu18/18 mice have less full-length HTT than FVB mice and Hu97/18 mice have a similar amount while both genotypes are heavier, it is likely that metabolic changes depend not only on total HTT levels, but on levels and composition of human HTT, indicating that there may be some functional differences between the HTT proteins of the two species. Supporting this is the finding that BACHD mice, which have roughly the same amount of human HTT as Hu97/18 mice plus mouse wtHTT, weigh about the same as Hu97/18 mice, while YAC18 mice, which have the same amount of human HTT as Hu18/18 mice plus mouse wtHTT, weigh less than Hu18/18 mice (Supplementary Material, Fig. S2). The body weight gain in humanized mice is also greater in females than in males, indicating some gender component to metabolic dysfunction.
Figure 2.
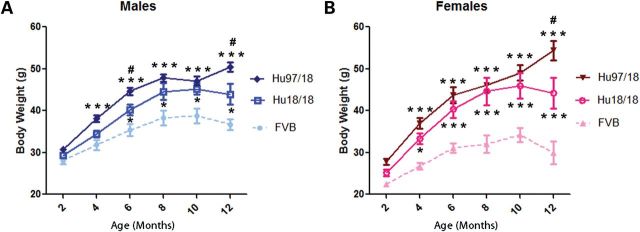
Body weight is increased in both Hu18/18 and Hu97/18 mice. Body weight was assessed at 2 month intervals in (A) male and (B) female Hu97/18 and Hu18/18 mice and compared with historical FVB body weight data (unpublished). While Hu97/18 mice are slightly heavier than Hu18/18 mice, both humanized genotypes are significantly heavier than wt mice. Different from FVB: *P < 0.05 and ***P < 0.001; different from Hu18/18: #P < 0.05.
Motor performance
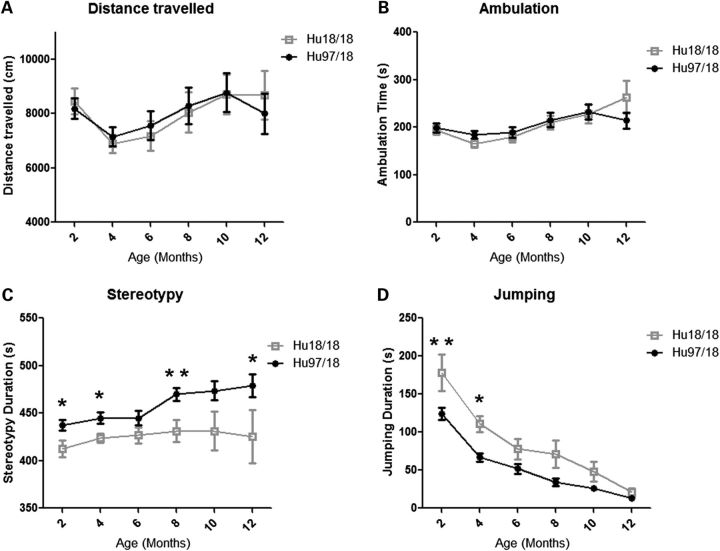
To evaluate motor performance, Hu97/18 and Hu18/18 littermates were assessed longitudinally at 2-month intervals from 2 to 12 months of age using an accelerating rotarod, climbing and spontaneous activity assays. Two-month-old Hu97/18 mice display a motor learning deficit as evidenced by increased falls and decreased latency to fall during rotarod training [Fig. 3A; genotype P < 0.01, trial P < 0.001, two-way analysis of variance (ANOVA)]. After training at this age, they perform similar to Hu18/18 mice, indicating that this difference is truly the result of a learning deficit rather than a motor deficit. In longitudinal rotarod testing, both Hu97/18 and Hu18/18 mice display declining performance with age (Fig. 3B; age P < 0.001, two-way ANOVA). For males, there is a significant difference between the two genotypes (genotype P < 0.01, two-way ANOVA), indicating greater impairment in Hu97/18 than Hu18/18 mice, though no single time point shows a significant difference by post hoc analysis. There is no genotypic difference in performance for females. In both humanized genotypes, we found a significant linear relationship between body weight and rotarod performance (Fig. 3C; P < 0.0001, r2 = .55 for Hu18/18 and 0.58 for Hu97/18, regression), and both genotypes are overweight (Fig. 2), an effect which is exaggerated in females, indicating that poor rotarod performance may be due in part to increased body weight. However, there is a scatter around the line for both genotypes, indicating that other factors besides body weight likely contribute to declining motor performance in these animals. This includes the HD mutation for Hu97/18 mice. In Hu18/8 mice, the declining rotarod performance may also be related to the reduction in overall HTT levels compared with wt mice (Fig. 1C) or the failure of human HTT to fully compensate for the loss of mouse HTT. During a spontaneous climbing test, 2-month-old Hu97/18 mice trend toward an increased latency to begin climbing (P = 0.08, Student's t-test) and decreased number of climbing events (P = 0.07, Student’s t-test) and spend significantly less time climbing (P = 0.04, Student’s t-test) than Hu18/18 mice (Fig. 4). However, after this time point, climbing in both genotypes of mice decreased dramatically to a level that could not be meaningfully evaluated (data not shown). We also assessed motor performance longitudinally by spontaneous activity. During a 30 min trial, there was no difference in distance traveled or ambulation time between Hu97/18 and Hu18/18 mice, indicating similar activity during exploration (Fig. 5A and B). However, compared with Hu18/18 littermates, Hu97/18 mice did show increased stereotypy, or repetitive movement, and decreased jumping (Fig. 5C and D; genotype P < 0.001, two-way ANOVA). Taken together, these data indicate that Hu97/18 mice have significant motor deficits, though the difference in performance from the control humanized line is measurable but modest.
Figure 3.
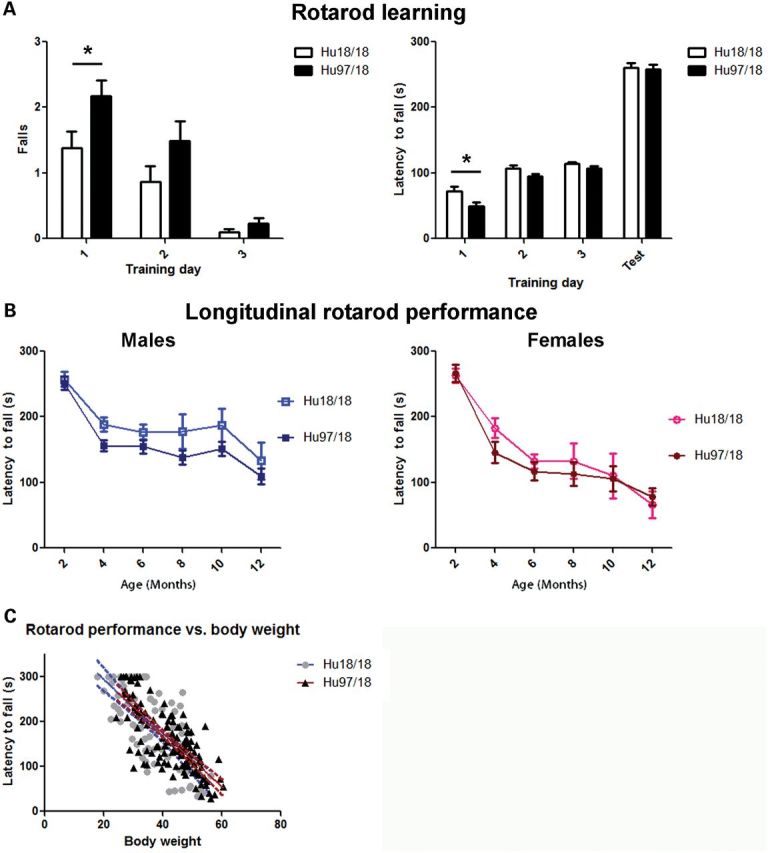
Hu97/18 mice display a motor learning deficit. (A) Two-month-old mice were trained on a fixed speed rotarod over 3 consecutive days and tested in an accelerating program on the fourth day. The number of falls during training and latency to the first fall during training and testing were scored. On day 1 of training, Hu97/18 mice fell from the rotarod sooner and more frequently. However, during testing on the fourth day, there was no genotypic difference indicating that the difference in training performance is the result of a motor learning deficit. (B) Mice were tested on an accelerating rotarod at 2 month intervals from 2 to 12 months of age. Performance declines with age for both genotypes (age P < 0.001). For males, there is a genotypic difference in performance with Hu18/18 mice scoring slightly better (genotype P < 0.01), but for females, there is no effect of genotype. (C) Regression analysis of rotarod performance and body weight. Linear relationship P < 0.0001 both genotypes, r2 = 0.55 for Hu18/18 and 0.58 for Hu97/18, indicating that rotarod performance is partially dependent on body weight. *P < 0.05.
Figure 4.
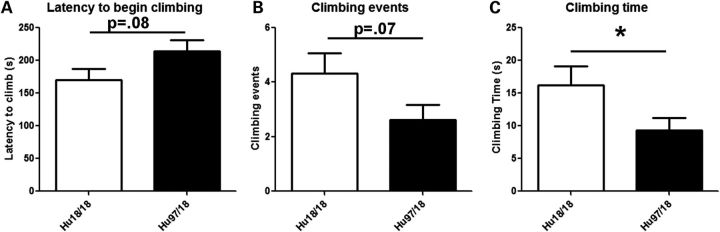
Hu97/18 mice display a climbing deficit. Climbing was assessed at 2 months of age by placing mice in a wire mesh container and recording activity for 5 min. (A) Hu97/18 mice display a trend toward increased latency to begin climbing and (B) decreased number of climbing events, and (C) a significant decrease in total time spent climbing. *P < 0.05.
Figure 5.
Hu97/18 mice display similar activity but increased stereotypy and decreased jumping during spontaneous activity. Spontaneous activity was assessed by infrared beam breaks at 2-month intervals from 2 to 12 months of age, during a 30 min exploration. Hu97/18 and Hu18/18 mice display similar (A) total distance traveled and (B) time spent during ambulation. Compared with Hu18/18 mice, Hu97/18 mice display (C) increased stereotypy and (D) decreased jumping. *P < 0.05 and **P < 0.01.
Psychiatric-like behavior
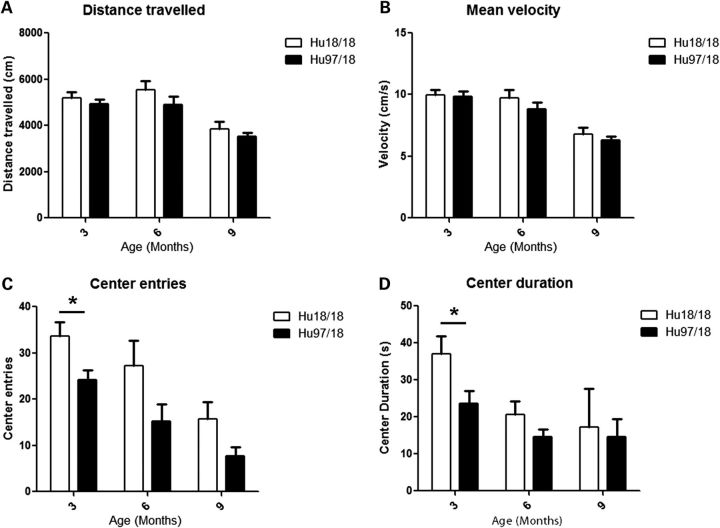
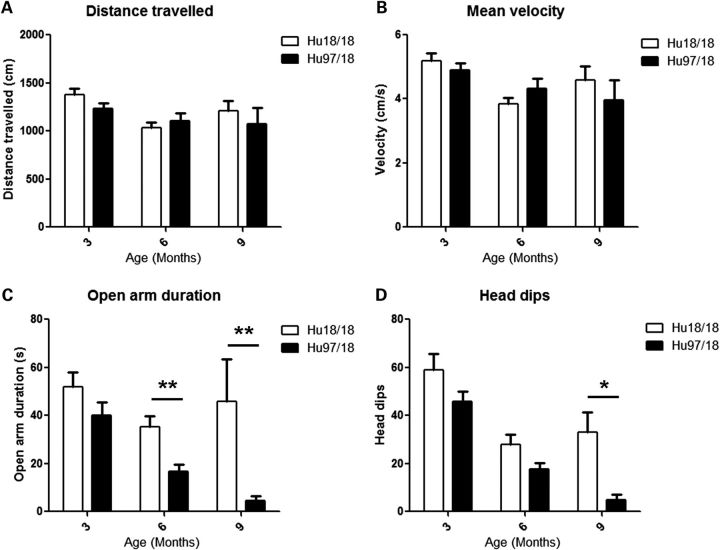
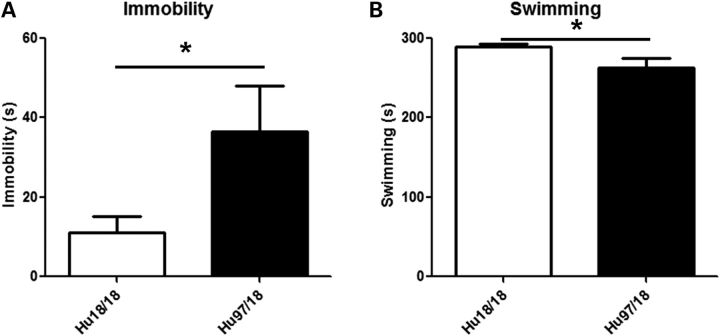
To evaluate anxiety, we assessed open field and elevated plus maze exploration in Hu97/18 and Hu18/18 mice aged 3, 6 or 9 months. Mice were tested in each paradigm only once and multiple cohorts were used to provide data at different ages. During open-field exploration, both genotypes had similar distance traveled and mean velocity (Fig. 6A and B), indicating a similar exploratory activity. However, Hu97/18 mice were more anxious than their Hu18/18 littermates, entering the center of the field less frequently and spending less total time in the center of the field (Fig. 6C and D; genotype P < 0.05, two-way ANOVA). During elevated plus maze exploration, both genotypes again displayed similar exploratory activity (Fig. 7A and B), but Hu97/18 mice spent less time in the open arms and less frequently dipped their head off the edges of the open arms (Fig. 7C and D; genotype P < 0.001, two-way ANOVA), indicating increased anxiety. Depressive behavior was evaluated in 12-month-old Hu97/18 and Hu18/18 mice using the Porsolt forced swim test. When compared with Hu18/18 mice, Hu97/18 mice spent more time in immobile and less time in swimming (Fig. 8; P = 0.04, Student’s t-test), indicating increased depressive-like behavior.
Figure 6.
Hu97/18 mice display increased anxiety during open-field exploration. Mice aged 3, 6 or 9 months were allowed to explore an open field under bright lighting for 10 min. Exploration was recorded by a ceiling-mounted video camera and animals were tracked using Ethovision XT 7 software. Both genotypes had similar (A) total distance traveled and (B) mean velocity during the exploration period. Compared with Hu18/18 mice, Hu97/18 mice (B) less frequently entered into the center portion of the field and (D)spent less total time in the center of the field, though these differences only reached significance by post hoc at 3 months of age. *P < 0.05.
Figure 7.
Hu97/18 mice display increased anxiety during elevated plus maze exploration. Mice aged 3, 6 or 9 months were allowed to freely explore an elevated plus maze under normal lighting during 5 min. Exploration was recorded by a ceiling-mounted video camera and animals were tracked using Ethovision XT 7 software. Both genotypes had similar (A) total distance traveled and (B) mean velocity during the exploration period. Compared with Hu18/18 mice, Hu97/18 mice (C) spent less time in the open arms and (D) less frequently dipped their heads off the edges of the open arms, though these differences only reached significance by post hoc at 6 and 9 months of age. *P < 0.05 and **P < 0.01.
Figure 8.
Hu97/18 mice display depressive-like behavior. Twelve-month-old mice were placed in cylinders filled with room temperature water and recorded for 6 min with a video camera. During the final 5 min, compared with Hu18/18 mice, Hu97/18 mice (A) spent more time immobile and (B) less time swimming. *P < 0.05.
Cognitive performance
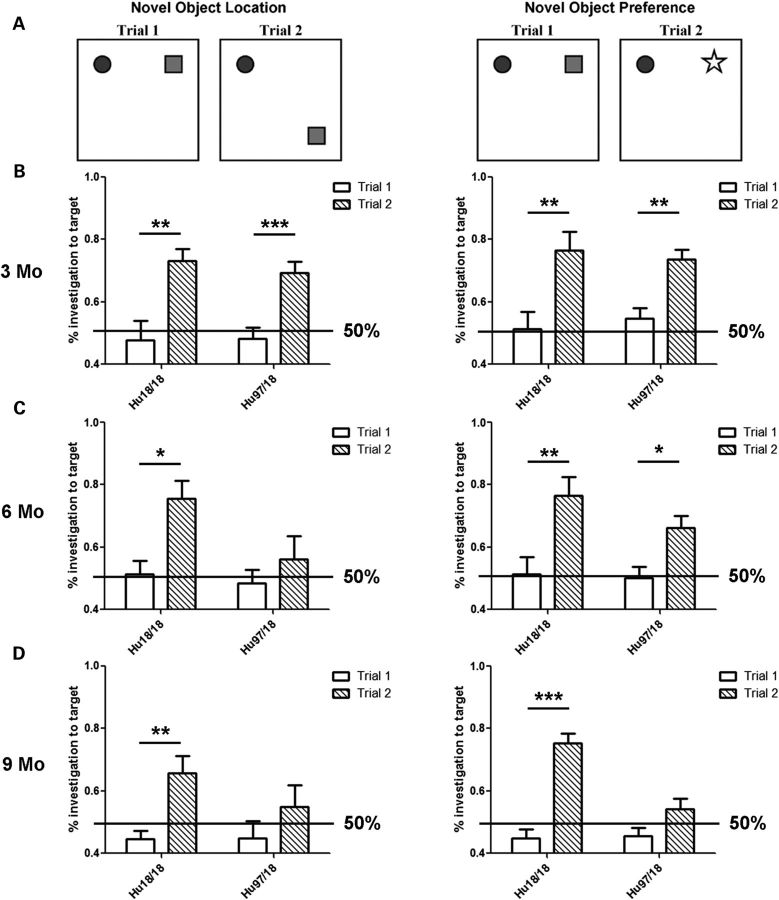
Aside from the rotarod training task in which Hu97/18 mice display a motor learning deficit (Fig. 3A), we also evaluated spatial learning by preference for a known object in a novel location and object recognition by preference for an unknown object (Fig. 9) at 3, 6 or 9 months of age. Mice were tested only once and multiple cohorts were used to evaluate performance at different ages. In the spatial learning task, mice are presented with two unknown objects in trial 1. If there is no basal preference for one object over the other, investigation of the target object, the one on the right, should be at the level of chance (50%). In trial 2, the mice are presented with the same two objects, but the right object has been moved to a novel location. Mice with adequate spatial learning will prefer to investigate the target object, the one in the new location, resulting in a significant increase in the percentage of investigations to the target object between trials 1 and 2. Mice with impaired spatial learning will investigate both objects at the level of chance resulting in no difference in investigations to the target object between trials 1 and 2. In the object preference task, the mice are again presented with the same two objects in their original locations in trial 1. As this is the third exposure to this object pair, and they are both known, there should be no preference for one object over the other resulting in ∼50% of the investigations to the right object. In trial 2, the right object is replaced with a completely novel object. Mice with adequate object recognition will prefer to investigate the novel object resulting in greater than 50% investigations and a significant increase in target object investigations between trials 1 and 2. Mice with impaired object recognition will investigate the known and novel objects equally, resulting in no difference in target object investigations between trials 1 and 2. At 3 months of age, both genotypes of mice display normal spatial learning and object recognition showing preference to investigate both a known object in a novel location and an unknown object (Fig. 9B). At 6 months of age, Hu18/18 mice still display normal learning in both assays, while Hu97/18 mice display a spatial learning deficit (Fig. 9C). By 9 months of age, Hu18/18 mice still display normal learning, but Hu97/18 mice display deficits in both spatial learning and object recognition (Fig. 9D), indicating a progressive cognitive deficit.
Figure 9.
Hu97/18 mice display progressive cognitive deficits. Mice aged 3, 6 or 9 months were evaluated for spatial learning and object recognition using the novel object location and novel object preference learning assays. (A) Diagram of the two tests. In the object location test, the mice are presented with two unknown objects in trial 1, and two known objects (the same objects), one in a novel location, in trial 2. In the object preference test, mice are presented with two known objects (the same from object location testing) in trial 1, and one known and one novel object in trial 2. (B) At 3 months of age, both genotypes display normal spatial learning and object recognition, i.e. they prefer to investigate the target object, either the one in the novel location or the novel object, in trial 2. (C) At 6 months of age, Hu18/18 mice display normal learning. Hu97/18 mice display impaired spatial learning (they do not prefer the target object in trial 2), but normal object recognition. (D) At 9 months of age, Hu18/18 mice display normal learning, while Hu97/18 mice have deficits in both tasks indicating impaired spatial learning and object recognition. Mo, month; *P < 0.05, **P < 0.01 and ***P < 0.001.
Neuropathology
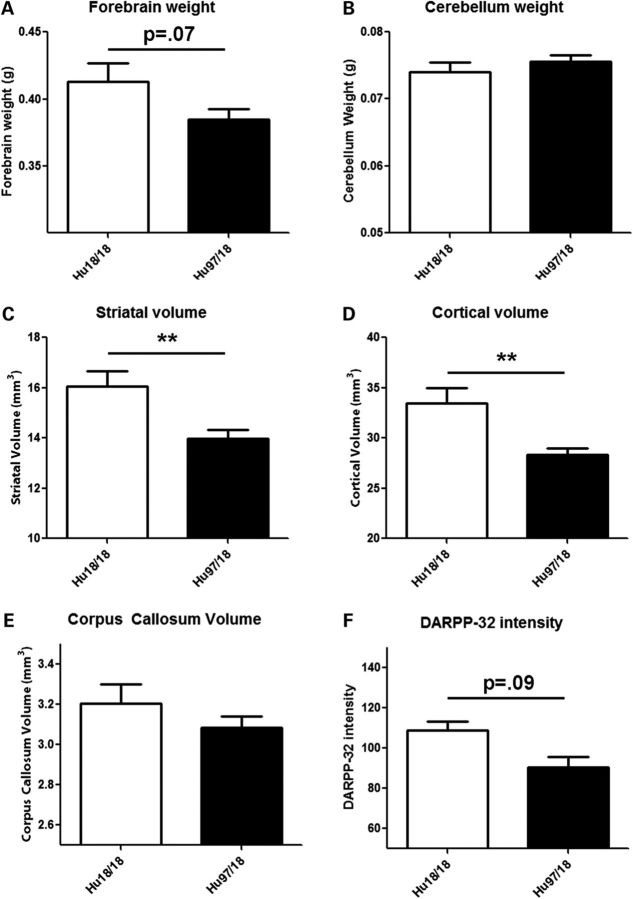
To evaluate neuropathological changes in Hu97/18 mice, we assessed forebrain and cerebellum weight, striatal and cortical volume, corpus callosum volume and DARPP-32 staining intensity in the brains of 12-month-old mice. Compared with Hu18/18 mice, Hu97/18 mice have a trend toward reduced forebrain weight (P = 0.07, Student's t-test) with no change in cerebellum weight (Fig. 10A and B). Hu97/18 mice show striking reductions in both striatal volume (12.9%, P < 0.01, Student's t-test) and cortical volume (15.3%, P < 0.01, Student's t-test; Fig. 10C and D). Additionally, there is a trend toward reduced corpus callosum volume and a 16.8% reduction in DARPP-32 staining intensity that did not reach significance (P = 0.09, Student's t-test; Fig. 10E and F). These data are consistent with human HD in which forebrain atrophy, striatal volume loss, cortical shrinkage and white matter loss are observed, but the cerebellum is relatively spared (28,29).
Figure 10.
Hu97/18 mice display neuropathological abnormalities consistent with HD. The brains of 12-month-old mice were evaluated for physical changes seen in HD. (A) Hu97/18 trend toward reduced forebrain weight (P = 0.07), (B) but have no change in cerebellum weight. Stereological analysis shows that compared with Hu18/18 mice, Hu97/18 mice have significantly decreased (C) striatal and (D) cortical volume and (E) a trend toward reduced corpus callosum volume. (F) There is also a trend toward reduced DARPP-32 staining intensity in the Hu97/18 brain. **P < 0.01.
DISCUSSION
In an effort to generate a mouse model suitable for preclinical testing of therapies specifically targeting human HTT, we have generated the first fully humanized transgenic mouse model of HD. The Hu97/18 mouse lacks both endogenous mouse Hdh alleles but expresses two copies of the full-length human HTT gene. One transgene copy is CAG-expanded (97Q) while the other is wt (18Q) to model the dominant effects of the mutation in HD patients. The CAG-expanded transgene is derived from the BACHD mouse, while the wt transgene originates from the YAC18 mouse. This means that in addition to the difference in the CAG-tract length, Hu97/18 mice are also heterozygous at 27 therapeutically relevant SNP sites in the human HTT gene that are potential allele-specific silencing targets. The Hu97/18 mouse therefore allows efficacy testing of candidate silencing reagents in an appropriate genetic background. In parallel to developing the Hu97/18 mouse line, we have generated the humanized control mouse line Hu18/18, which also lacks both Hdh alleles but expresses only human wtHTT from the YAC18 transgene.
We predicted that Hu97/18 mice would have much of the same HD-like phenotypes as BACHD mice due to the shared CAG-expanded BACHD transgene. Though the expression of additional wtHTT from the YAC18 transgene could reduce phenotypic severity in BACHD mice, we predicted that deletion of both copies of the wt Hdh gene would nullify any benefit. We previously found that the addition of the YAC18 transgene to YAC128 mice only slightly ameliorates striatal neuropathology and has no effect on motor deficits (30). In contrast, the deletion of both copies of the Hdh gene increased the severity of HD-like phenotypes in YAC128 mice (31). Thus, we have characterized the newly generated humanized mouse models with endpoints previously validated in the BACHD mouse.
We find that Hu97/18 and BACHD mice have similar phenotypes (Table 2). Both lines display increased body weight. Interestingly, Hu18/18 mice, which have less total HTT than wt mice, also display increased body weight (Figs 1 and 2), indicating that weight gain in HD mouse models is not only correlated with the amount of full-length HTT protein (26) but may be more dependent on the level and composition of full-length human HTT. Human HTT is able to rescue the embryonic lethality of loss of mouse HTT (25), indicating the appropriate developmental expression of exogenous human HTT in mice and the functional overlap of the two proteins during development. This led to the long-standing assumption that human HTT could fully compensate for the loss of mouse HTT. However, the metabolic changes evidenced by increased body weight in Hu18/18 control mice indicate that the functional overlap of the two proteins is not complete. Although Hdh is 86% identical to human HTT at the DNA level, there are several key genetic differences cis- to the CAG tract, in the gene promoter, and throughout the gene loci that could alter function in ways such that deficits are not observed until adulthood (32). The humanized mouse lines will be important tools for evaluating the functional differences in human and mouse HTT proteins and give valuable insight to the interpretation of data generated in transgenic versus knock-in mouse models of HD.
Table 2.
Summary of HD-like phenotypes of Hu97/18 and BACHD mice
| Phenotype | Onset (months) Hu97/18 | BACHD | References |
|---|---|---|---|
| Body weight | 4 | 3 | (33) |
| Rotarod learning | 2a | 2a | (24) |
| Rotarod | 4 | 4 | (33) |
| Climbing | 2a | 6a | (34) |
| Spontaneous activity | 2a | 1a | (41) |
| Open-field exploration | 3a | 6a | (34) |
| Elevated plus maze | 6 | 3a | (36) |
| Forced swim | 12a | 12a | (33) |
| Spatial learning | 6 | 6a | (34) |
| Object recognition | 9 | 6a | (34) |
| Forebrain weight | P = 0.07 12a | 12a | (33) |
| Striatal volume | 12a | 12a | (24) |
| Cortical volume | 12a | 12a | (24) |
| Corpus callosum | no change 12a | ND | |
| DARPP-32 staining | P = 0.09 12a | no change 7a | (34) |
ND, not determined. Age of onset (months) of HD-like behavioral and neuropathological phenotypes of full-length human BACHD-derived muHTT transgenic HD mouse models. BACHD mice are compared with WT littermates, and Hu97/18 mice are compared with Hu18/18 littermates.
aEarliest evaluated time point.
Both BACHD and Hu97/18 mice display a motor learning deficit at 2 months of age during rotarod training and progressive decline in accelerating rotarod performance with age (Fig. 3) (33). To a lesser degree, rotarod performance also declines with age in Hu18/18 mice, which is likely at least in part the result of increased body weight. Declining rotarod performance in Hu18/18 mice may also be related to the reduction in the total HTT level in these mice when compared with wt mice. However, these mice retain ∼65% normal wtHTT levels, which is well above the 50% level in Hdh+/− animals. While Hdh+/− mice do display several behavioral and neuropathological abnormalities (13), they do not have rotarod performance deficits (unpublished data). Thus, it is more likely that the motor deficits of Hu18/18 mice result from the failure of human HTT to fully compensate for the loss of mouse HTT, indicating additional potential functional differences between the two proteins. BACHD and Hu97/18 mice both display a climbing deficit (Fig. 4) (34). However, this deficit can only be evaluated at a very young age in the humanized mice because at older ages, neither genotype performs the task. Motor abnormalities were also observed in Hu97/18 mice throughout the characterization period from 2 to 12 months of age during spontaneous activity including increased stereotypy and decreased jumping (Fig. 5). Though it is difficult to accurately assess motor performance in overweight mice, there are clear deficits in the Hu97/18 line compared with the Hu18/18 line. Reducing the body weight of both humanized lines with either diet or interventions previously shown to normalize body weight in full-length human HTT transgenic models, such as 17-β-estradiol treatment (26) or the targeted hypothalamic disruption of BACHD expression (35), may allow greater differentiation of motor performance between Hu18/18 and Hu97/18 mice.
In addition to motor abnormalities, Hu97/18 mice also display behavioral and cognitive changes consistent with BACHD mice and human HD. Both BACHD and Hu97/18 mice display increased anxiety behaviors (Figs 6 and 7) (34), increased depressive-like behavior during the Porsolt forced swim test (Fig. 8) (33) and reduced spatial memory and object recognition (Fig. 9) (34). However, in some cases, these behavioral phenotypes appear slightly delayed in the Hu97/18 line. For instance, BACHD mice display increased anxiety during elevated plus maze exploration as early as 3 months of age (36), while Hu97/18 mice do not display significant anxiety in this assay until 6 months of age (Fig. 7). Additionally, by 6 months of age, BACHD mice display impaired spatial learning and object recognition (34), while Hu97/18 mice only display spatial learning deficits at this time point and do not show object recognition deficits until 9 months of age (Fig. 9).
The delay in onset of certain phenotypes in Hu97/18 mice is likely a result of the down-regulation of the BACHD transgene (Fig. 1). Though muHTT is expressed from an identical transgene and insertion in both lines, there is ∼40% less muHTT protein in the Hu97/18 brain than in the BACHD brain. The HTT gene is known to regulate HTT expression via an antisense transcript (HTT AS), which is modulated by CAG expansion (37). It is possible that there are species specific factors in HTT AS regulation that also modulate it's function. Removal of endogenous wtHTT and replacement with human wtHTT in Hu97/18 mice may alter this regulation resulting in reduced expression of BACHD-derived muHTT. This provides additional evidence of functional differences between mouse and human HTT. Though the cause of BACHD down-regulation in Hu97/18 mice remains unexplained, it actually increases the validity of the model: while BACHD mice have nearly twice as much total HTT as wt mice, Hu97/18 mice have approximately the same level of total HTT protein as wt mice and approximately equivalent levels of human wt and muHTT protein (Fig. 1).
Importantly, even with the modest delay in onset for some behavioral phenotypes, other phenotypes are apparent in very young Hu97/18 mice. Motor learning deficits, climbing deficits and abnormal behavior during spontaneous activity are observed at 2 months of age, and anxiety during open-field exploration is observed at 3 months of age. These early onset phenotypes increase the usefulness of the Hu97/18 model allowing for preclinical evaluation of early changes, which are more likely to translate to observed clinical changes considering that clinical trials are generally performed in early HD individuals for durations of 1–2 years.
In addition to behavioral abnormalities, both BACHD (24) and Hu97/18 mice have neuropathological changes that are consistent with HD. At 12 months of age, Hu97/18 mice have significantly reduced cortical and striatal volumes relative to Hu18/18 littermates, while there is no change in the cerebellum (Fig. 10). The earliest time point of these neuropathological changes in Hu97/18 mice is not yet determined.
The humanized Hu97/18 transgenic mouse precisely models the genetic condition of the human HD patient. The Hu97/18 mice are heterozygous for haplotypes in the entire HTT gene loci, not just the CAG-tract size, and are therefore the ideal model for testing genetic therapies intended for human HD patients. Hu97/18 mice recapitulate many of the behavioral changes associated with HD including motor, psychiatric and cognitive deficits as well as canonical neuropathological abnormalities. The Hu97/18 model will serve as a crucial tool for testing potential therapies, characterizing the function of the HTT protein and investigating other research questions addressing HD pathogenesis that were not possible using previous HD mouse models.
MATERIALS AND METHODS
Mice
BACHD mice were generated previously in William Yang's laboratory (24). YAC18 and Hhd+/− mice were generated previously in Michael Hayden's laboratory (13,25). The BACHD and YAC18 lines were generated and maintained on the FVB/N strain background. The Hdh+/− line was generated on a mixed 129/SV and C57Bl/6J strain background and backcrossed a minimum of 10 generations to the FVB/N so that all three preexisting lines were on the same strain background. Animals were maintained under a 12 h light:12 h dark cycle (lights on at 2300) in a clean facility and given free access to food and water. Experiments were performed with the approval of the animal care committee at the University of British Columbia.
Breeding and genotyping
BACHD or YAC18 mice were bred to Hdh+/− mice to generate BACHD+;Hdh+/− and YAC18+;Hdh+/− mice. These mice were then interbred to generate BACHD+;YAC18+;Hdh−/− mice and YAC18+:Hdh−/− mice. These mice were then interbred to generate BACHD+;YAC18++;Hdh−/− mice (Hu97/18) and YAC18++;Hdh−/− mice (Hu18/18). As the YAC transgene contains ∼4 kb more 5′ sequence of the HTT gene than the BAC transgene (33), YAC18 zygosity was determined by qPCR using primers in the region contained only in the YAC transgene (forward primer: 5′-CAGCCTGGTGACAGAGCAAG-3′, reverse primer: 5′-AGGCAAAACGGATCTCCAAA-3′). A PCR across the region containing the two loxP sites in the BAC but not in YAC transgene was used to distinguish between the two human HTT transgenes (forward primer: 5′-ATTACAGTCTCACCACGCCC-3′, reverse primer: 5′-CTTCATCAGCTTTTCCAGGG-3′) yielding product sizes of 424 bp for the YAC and 458 bp for the BAC. Hu18/18 and Hu97/18 matings were then established producing offspring of 50% each genotype. Once YAC18 and Hdh−/− homozygosity were achieved, only the loxP site spanning PCR was necessary to genotype the offspring as either Hu18/18 or Hu97/18.
HTT RNA and protein quantitation
To evaluate HTT RNA and protein levels, 2-month-old BACHD and wt littermates and Hu18/18 and Hu97/18 littermates were euthanized with 2.5% intraperitoneal (IP) avertin. The brains were removed and placed on ice for 5 min, then microdissected into regional tissue samples that were submerged in RNAlater (Ambion) overnight at 4°C. Total protein and RNA was then isolated from right cortices using the Ambion PARIS kit according to the manufacturer's instructions. Protein concentrations were determined by DC assay (Biorad) and RNA concentrations were determined by absorbance at 260 nm.
For quantitation of human wtHTT RNA or human muHTT RNA by qRT–PCR, RNA was treated with DNase I (Invitrogen) and 1 μg of RNA was reverse-transcribed, using SuperScript III (Invitrogen) according to the manufacturer's instructions, to generate cDNA for qRT–PCR. Exon spanning Rpl13a primers were designed using Primer3 software. Sequences were confirmed with in silico PCR and tested using SYBR Green Power master mix (Applied Biosystems, USA) and the ABI Prism 7500 Sequence Detection System. Mouse Rpl13a (5′-GGAGGAGAAACGGAAGGAAAAG and 5′-CCGTAACCTCAAGATCTGCTTCTT) were purchased from integrated device technology. Human mutant and wtHTT qRT–PCR was done with TaqMan Universal PCR Master Mix (Roche) and ABI Taqman assay [C_2231945_10; Context Sequence (VIC/FAM) TGTGACCCACGCCTGCTCCCTCATC(C/T)ACTGTGTGCACTTCATCCTGGAGGC]. Gene expression was calculated using the delta–delta Ct method. An N of four animals per genotype and three replicates per animal was used.
For the quantitation of the total HTT or muHTT protein by FRET, 10 µg of the total protein from cortical brain lysates was mixed with 0.2 ng/µl terbium (FRET donor)-labeled BKP1 anti-HTT antibody (38) and 2 ng/µl of D2 (FRET acceptor)-labeled either 2166, an anti-HTT antibody (Chemicon) to detect total HTT, or MW1, an anti-expanded CAG antibody (39) to detect muHTT, in a white 384-well plate. After excitation at 340 nm, FRET was measured as the ratio of 655 nm (D2)/615 nm (TB) emission. An N of four animals per genotype and three replicates per animal was used.
For the quantitation of wt and muHTT protein by immunoblotting, 40 µg of total protein were denatured in LDS sample buffer (Invitrogen) with 100 mm dithiothreitol and heated to 70°C for 10 min. Samples were resolved on 10% low-bis acrylamide gels (23). Proteins were transferred to 0.45 µm nitrocellulose. Membranes were blocked with 5% milk in PBS, and then blotted for HTT (MAB2166, Millipore) and calnexin (Sigma C4731) loading control. Proteins were detected with IR dye 800CW goat anti-mouse (Rockland 610-131-007) and AlexaFluor 680 goat anti-rabbit (Molecular Probes A21076)-labeled secondary antibodies and the LiCor Odyssey Infrared Imaging system. Mutant and wtHTT intensities were normalized to calnexin loading control and then to the average level of wtHTT from wt mice on the same membrane. Three mice/genotype and two replicate gels were evaluated.
Behavior testing
All behavior experiments were carried out under white lighting in the animal's dark phase by an experimenter blind to genotype.
Rotarod
Two-month-old mice were trained over 3 consecutive days on the rotarod (Ugo Basille) at a fixed speed of 18 revolutions per minute (RPM). Mice received 3 × 120 s training trials per day with a 1 h inter-trial interval (ITI). Mice that fell from the rod were immediately replaced for the duration of the trial. The latency to the first fall and number of falls for each training trial were recorded. The average of the three trials for each mouse was scored. For longitudinal rotarod testing at 2-month intervals from 2 to 12 months of age, an accelerating program from 5 to 40 RPM over 300 s was used. Mice received three trials with a 1 h ITI and the latency to the first fall was recorded. The average of the three testing trials was scored.
Climbing
Mice were placed at the bottom end of a closed-top wire mesh cylinder (10 × 15 cm) on the tabletop and recorded with a video camera for 5 min. The latency to begin climbing (all four feet off the desk top) and the total time spent in climbing during the 5 min trial were scored. Mice were tested longitudinally at 2-month intervals from 2 to 12 months of age.
Spontaneous activity
Mice were placed in a 27 × 27 × 20.3 cm spontaneous activity box (Med Associates) for 30 min. Activity, ambulation, jumping, stereotypy and resting were scored by infrared beam breaks. Mice were tested longitudinally at 2-month intervals from 2 to 12 months of age.
Elevated plus maze
To assess anxiety during elevated plus maze exploration, mice aged 3, 6 or 9 months were placed in the center of an elevated plus maze with 30 × 10 cm arms, 20 cm high walls on the two closed arms and 50 cm legs. Exploration of the maze was recorded during a 5 min trial by a ceiling-mounted video camera and scored by Ethovision XT 7 animal tracking software (Noldus). Distance traveled and average velocity were assessed as a measure of exploratory activity. Entries into and time spent in the open arms, in addition to head dips off the edge of the open arms, were scored as a measure of anxiety. Each mouse was tested at a single time point and multiple cohorts were evaluated to provide data from different ages.
Porsolt forced swim test
A modified Porsolt forced swim test was performed as described previously (40). Briefly, 12-month-old mice were placed in individual cylinders (25 cm tall × 19 cm wide) filled with room temperature water (23–25°C) to a depth of 15 cm for a period of 6 min. The test sessions were recorded by a video camera placed directly above the cylinders. Time spent in swimming versus floating (immobile) in the last 5 min of the trial was scored as a measure of depressive behavior.
Open-field exploration and object learning
To assess anxiety during open-field exploration, mice aged 3, 6 or 9 months were placed in the lower left corner of a 50 × 50 cm open grey acrylic box with 20 cm tall sides in a room brightly lit by fluorescent ceiling lights. Open-field activity was recorded for 10 min by a ceiling-mounted video camera and scored by Ethovision XT 7. Distance traveled and average velocity were assessed as a measure of exploratory activity. Entries into and time spent in the center of the field were scored as a measure of anxiety. Mice were then removed from the apparatus for a 5 min ITI during which two different novel objects were placed in the upper two corners of the box, far enough from the sides so as to not impede movement around the outer edge (˜7 cm). Mice were reintroduced to the box in the lower left corner and recorded for 5 min by a ceiling-mounted video camera, during which the number of investigations of the novel objects was scored by Ethovision XT 7. Mice were then removed from the box for a 5 min ITI, and the object at the top right corner of the box was moved to the lower right corner of the box. To assess preference for a known object in a novel location, mice were reintroduced to the box and recorded for 5 min. The percentage of the investigations, or nose pokes, to the target object (the one in the new location) was computed. To assess preference for a novel object, the experiment was repeated on the subsequent day with the exception that, instead of repositioning the object in the top right corner, the object was replaced by a different, unfamiliar object in the same location. The percentage of the investigations of the target object (the unfamiliar one) was computed. Each mouse was tested at a single time point and multiple cohorts were used to evaluate longitudinal performance.
Neuropathology
Mice were injected with 2.5% IP avertin and intracardially perfused with phosphate-buffered saline (PBS) followed by ice-cold 4% paraformaldehyde (PFA) in PBS. The brains were removed and post-fixed in 4% PFA at 4°C for 24 h, and then cryopreserved in 30% sucrose. They were then frozen on dry ice, mounted with Tissue-TEK O.C.T. compound (Sakura) and cut by cryostat (Microm HM 500 M) into 25 µm free-floating coronal sections. Sections were stored in PBS with 0.08% sodium azide at 4°C until immunohistochemical processing.
Stereological volumetric assessments
A series of 25 µm coronal sections spaced 200 µm apart spanning the striatum were stained with the NeuN antibody (1:1000; Millipore) overnight at room temperature, followed by incubation with biotinylated anti-mouse antibody (1:1000; Vector Laboratories) for 2 h at room temperature. The ABC Elite kit (Vector) was used to amplify signal, which was detected with diaminobenzidine (DAB; Thermo Scientific). Striatal, cortical and corpus callosum volumes were determined by tracing the perimeter of the desired structure in serial sections using StereoInvestigator software (Microbrightfield) and volumes determined using the Cavalieri principle.
DARPP-32 staining intensity
Free-floating sections were incubated in primary mouse anti-DARPP32, #C24-6a (gift from Dr Paul Greengard and Dr Hugh Hemming; 1:20 000) overnight at room temperature. Sections incubated without primary antibody served as controls. Sections were washed and incubated with horseradish peroxidase-conjugated secondary antibody (1:500, Jackson ImmunoResearch Laboratories) for 2 h at room temperature. Staining was visualized using DAB. Sections were photographed using a Zeiss Axioplan 2microscope and Coolsnap HQ Digital CCD camera (Photometrics). The amount of DARPP32 was determined using MetaMorph software version 6.3 (Universal Imaging Corporation). Labeling was identified using constant threshold levels for all images and analyzed using ‘integrated morphometry’. Relative levels of staining were calculated as the sum of the integrated optical density for each image divided by the area of the region selected then multiplied by the sampling interval (8) and section thickness (25 µm). No staining was observed in a negative control without primary antibody.
Statistical analysis
Data are expressed as the mean + SEM. Pairwise comparisons between genotypes at individual time points were assessed with a Student's t-test or one-way ANOVA. Longitudinal and multi-time point comparisons were assessed with two-way ANOVA for genotype and age except for object learning assays for which each time point was analyzed independently by two-way ANOVA for trial and genotype. Bonferroni post hoc analysis was used to assess genotype or trial differences for specific ages or genotypes, respectively. Differences were considered statistically significant when P < 0.05.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from ISIS Pharmaceuticals; The Canadian Institutes of Health Research; the CHDI Foundation; and postdoctoral fellowships for A.L.S. from Pfizer Ripples of Hope; The Canadian Institutes of Health Research; the Huntington Society of Canada; and The Michael Smith Foundation For Health Research.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Blair Leavitt for providing Hdh+/− mice backcrossed to the FVB/N strain, Jeremy Van Raamsdonk for providing historical YAC18 body weight data, Mahsa Amirabassi and Mark Wang for excellent animal care and Shaun Sanders for discussion and support.
Conflict of Interest statement. None declared.
References
- 1.Ross C.A., Tabrizi S.J. Huntington's disease, from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins A.K., Ho A., Hayden M.R. Lessons from predictive testing for Huntington disease: 25 years on. J. Med. Genet. 2011;48:649–650. doi: 10.1136/jmedgenet-2011-100352. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 4.Diaz-Hernandez M., Torres-Peraza J., Salvatori-Abarca A., Moran M.A., Gomez-Ramos P., Alberch J., Lucas J.J. Full motor recovery despite striatal neuron loss and formation of irreversible amyloid-like inclusions in a conditional mouse model of Huntington's disease. J. Neurosci. 2005;25:9773–9781. doi: 10.1523/JNEUROSCI.3183-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. From the Cover: RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G., et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl Acad. Sci. USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y.-L., Liu W., Wada E., Murata M., Wada K., Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci. Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., et al. Sustained effects of nonallele-specific huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 10.Machida Y., Okada T., Kurosawa M., Oyama F., Ozawa K., Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem. Biophys. Res. Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Lebron E., Denovan-Wright E.M., Nash K., Lewin A.S., Mandel R.J. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol. Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S., et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasir J., Floresco S.B., O'Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 14.Zeitlin S., Liu J.P., Chapman D.L., Papaioannou V.E., Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 15.Zuccato C., Valenza M., Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 16.Dragatsis I., Levine M.S., Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 17.McBride J.L., Pitzer M.R., Boudreau R.L., Dufour B., Hobbs T., Ojeda S.R., Davidson B.L. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease. Mol. Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grondin R., Kaytor M.D., Ai Y., Nelson P.T., Thakker D.R., Heisel J., Weatherspoon M.R., Blum J.L., Burright E.N., Zhang Z., et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135:1197–1209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 20.Butland S., Devon R., Huang Y., Mead C.-L., Meynert A., Neal S., Lee S., Wilkinson A., Yang G., Yuen M., et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warby S.C., Montpetit A., Hayden A.R., Carroll J.B., Butland S.L., Visscher H., Collins J.A., Semaka A., Hudson T.J., Hayden M.R. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfister E.L., Kennington L., Straubhaar J., Wagh S., Liu W., DiFiglia M., Landwehrmeyer B., Vonsattel J.-P., Zamore P.D., Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M., Hayden M.R. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray M., Shirasaki D.I., Cepeda C., André V.M., Wilburn B., Lu X.-H., Tao J., Yamazaki I., Li S.-H., Sun Y.E., et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodgson J.G., Agopyan N., Gutekunst C.-A., Leavitt B.R., LePiane F., Singaraja R., Smith D.J., Bissada N., McCutcheon K., Nasir J., et al. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 26.Pouladi M.A., Xie Y., Skotte N.H., Ehrnhoefer D.E., Graham R.K., Kim J.E., Bissada N., Yang X.W., Friedlander R.M., Leavitt B.R., et al. Full-length huntingtin levels modulate body weight by influencing insulin-like growth factor 1 expression. Hum. Mol. Genet. 2010:1528–1538. doi: 10.1093/hmg/ddq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Raamsdonk J.M., Gibson W.T., Pearson J., Murphy Z., Lu G., Leavitt B.R., Hayden M.R. Body weight is modulated by levels of full-length Huntingtin. Hum. Mol. Genet. 2006;15:1513–1523. doi: 10.1093/hmg/ddl072. [DOI] [PubMed] [Google Scholar]
- 28.Tabrizi S.J., Reilmann R., Roos R.A.C., Durr A., Leavitt B., Owen G., Jones R., Johnson H., Craufurd D., Hicks S.L., et al. Potential endpoints for clinical trials in premanifest and early Huntington's disease in the TRACK-HD study: analysis of 24 month observational data. Lancet Neurol. 2011:42–53. doi: 10.1016/S1474-4422(11)70263-0. [DOI] [PubMed] [Google Scholar]
- 29.Vonsattel J.P., Keller C., Cortes Ramirez E.P. Huntington's disease—neuropathology. Handb. Clin. Neurol. 2011;100:83–100. doi: 10.1016/B978-0-444-52014-2.00004-5. [DOI] [PubMed] [Google Scholar]
- 30.Van Raamsdonk J., Pearson J., Murphy Z., Hayden M., Leavitt B. Wild-type huntingtin ameliorates striatal neuronal atrophy but does not prevent other abnormalities in the YAC128 mouse model of Huntington disease. BMC Neurosci. 2006;7:80. doi: 10.1186/1471-2202-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Raamsdonk J.M., Pearson J., Rogers D.A., Bissada N., Vogl A.W., Hayden M.R., Leavitt B.R. Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2005;14:1379–1392. doi: 10.1093/hmg/ddi147. [DOI] [PubMed] [Google Scholar]
- 32.Barnes G., Duyao M., Ambrose C., McNeil S., Persichetti F., Srinidhi J., Gusella J., MacDonald M. Mouse Huntington's disease gene homolog. Somatic Cell Mol. Genet. 1994;20:87–97. doi: 10.1007/BF02290678. [DOI] [PubMed] [Google Scholar]
- 33.Pouladi M.A., Stanek L.M., Xie Y., Franciosi S., Southwell A.L., Deng Y., Butland S., Zhang W., Cheng S.H., Shihabuddin L.S., et al. Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum. Mol. Genet. 2012;21:2219–2232. doi: 10.1093/hmg/dds037. [DOI] [PubMed] [Google Scholar]
- 34.Southwell A.L., Ko J., Patterson P.H. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington's disease. J. Neurosci. 2009;29:13589–13602. doi: 10.1523/JNEUROSCI.4286-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hult S., Soylu R., Bjorklund T., Belgardt B.F., Mauer J., Bruning J.C., Kirik D., Petersen A. Mutant huntingtin causes metabolic imbalance by disruption of hypothalamic neurocircuits. Cell Metab. 2011;13:428–439. doi: 10.1016/j.cmet.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Hult S., Soylu R., Kirik D., Petersen A. Psychiatric and metabolic changes in the BACHD mouse model of Huntington disease. 2010. Society for Neuroscience Annual Meeting Abstract, 861.8.
- 37.Chung D.W., Rudnicki D.D., Yu L., Margolis R.L. A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum. Mol. Genet. 2011;20:3467–3477. doi: 10.1093/hmg/ddr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellington C.L., Ellerby L.M., Gutekunst C.-A., Rogers D., Warby S., Graham R.K., Loubser O., van Raamsdonk J., Singaraja R., Yang Y.-Z., et al. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease. J. Neurosci. 2002;22:7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko J., Ou S., Patterson P.H. New anti-huntingtin monoclonal antibodies: implications for huntingtin conformation and its binding proteins. Brain Res. Bull. 2001;56:319–329. doi: 10.1016/s0361-9230(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 40.Pouladi M.A., Graham R.K., Karasinska J.M., Xie Y., Santos R.D., Petersen A., Hayden M.R. Prevention of depressive behaviour in the YAC128 mouse model of Huntington disease by mutation at residue 586 of huntingtin. Brain. 2009;132:919–932. doi: 10.1093/brain/awp006. [DOI] [PubMed] [Google Scholar]
- 41.Menalled L., El-Khodor B.F., Patry M., Suarez-Farinas M., Orenstein S.J., Zahasky B., Leahy C., Wheeler V., Yang X.W., MacDonald M., et al. Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models. Neurobiol. Dis. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.