Abstract
Aims/hypothesis
Although obesity is associated with endoplasmic reticulum (ER) stress and activation of the unfolded protein response (UPR) in adipose tissue, it is not known how UPR signalling affects adipogenesis. To test whether signalling through protein kinase RNA-like ER kinase/eukaryotic initiation factor 2 alpha (PERK/eIF2α) or inositol-requiring enzyme 1 alpha/X-box binding protein 1(IRE1α/XBP1) is required for adipogenesis, we studied the role of UPR signalling in adipocyte differentiation in vitro and in vivo in mice.
Methods
The role of UPR signalling in adipogenesis was investigated using 3T3-L1 cells and primary mouse embryonic fibroblasts (MEFs) by activation or inhibition of PERK-mediated phosphorylation of the eIF2α- and IRE1α-mediated splicing of Xbp1 mRNA. Body weight change, fat mass composition and adipocyte number and size were measured in wild-type and genetically engineered mice fed a control or high-fat diet (HFD).
Results
ER stress repressed adipocyte differentiation in 3T3-L1 cells. Impaired eIF2α phosphorylation enhanced adipocyte differentiation in MEFs, as well as in mice. In contrast, increased eIF2α phosphorylation reduced adipocyte differentiation in 3T3-L1 cells. Forced production of CCAAT/enhancer binding protein (C/EBP) homologous protein (CHOP), a downstream target of eIF2α phosphorylation, inhibited adipogenesis in 3T3-L1 cells. Mice with deletion of Chop (also known as Ddit3) (Chop−/−) gained more fat mass than wild-type mice on HFD. In addition, Chop deletion in genetically obese Leprdb/db mice increased body fat mass without altering adipocyte size. In contrast to the eIF2α–CHOP pathway, activation or deletion of Ire1a (also known as Ern1) did not alter adipocyte differentiation in 3T3-L1 cells.
Conclusions/interpretation
These results demonstrate that eIF2α–CHOP suppresses adipogenesis and limits expansion of fat mass in vivo in mice, rendering this pathway a potential therapeutic target.
Keywords: Adipocyte, C/EBP homologous protein (CHOP), eIF2α phosphorylation, Endoplasmic reticulum, Unfolded protein response (UPR)
Introduction
The prevalence of obesity is increasing worldwide, and will be one of the most serious public health concerns in the future [1]. Obesity is frequently accompanied by insulin resistance and type 2 diabetes [2] and is characterised by increased fat mass caused by an increased adipocyte number (hyperplasia) and/or size (hypertrophy) [3]. When the demand for fat storage exceeds capacity, adipocyte number increases through proliferation and/or differentiation from pre-adipocytes [4]. Adipogenesis is mediated by a well-programmed sequence of transcriptional events beginning with the induction of two transcription factors in the CCAAT/enhancer binding protein (C/EBP) family: C/EBPβ and C/EBPδ [5, 6]. Subsequently, these proteins activate transcription of the peroxisome proliferator-activated receptor gamma (PPARγ) and C/EBPα, two central adipogenic regulators that positively control each other and cooperate to orchestrate expression of the full adipogenic programme, including induction of additional transcription factors, suppression of growth-associated genes and stimulation of insulin-dependent glucose transport [7].
When endoplasmic reticulum (ER) homeostasis is disrupted, unfolded proteins accumulate in the ER lumen, which subsequently activates the unfolded protein response (UPR) through dissociation of binding immunoglobulin protein/78 kDa glucose-regulated protein (BiP/GRP78) from protein kinase RNA-like ER kinase (PERK), inositol-requiring enzyme 1 alpha (IRE1α) and activating transcription factor 6 alpha (ATF6α) [8, 9]. Activated PERK phosphorylates eIF2α at Ser51 leading to rapid and transient attenuation of protein synthesis [10, 11]. Paradoxically, under these conditions translation of Atf4 mRNA is selectively enhanced, which induces transcription of Chop (also known as Ddit3) [12]. Activated IRE1α elicits an endoribonuclease function that initiates unconventional splicing of Xbp1 mRNA [13] to produce a novel translation product X-box binding protein 1 (XBP1), which induces expression of genes encoding functions including ER protein chaperones, lipid biosynthetic enzymes and ER-associated protein degradation (ERAD) [14, 15]. Upon ER stress, ATF6α traffics to the Golgi apparatus where it is cleaved by the processing enzymes S1P and S2P to liberate a fragment that migrates into the nucleus to induce genes encoding ER protein chaperones and ERAD functions [16, 17].
There have been some studies suggesting that ER stress might be important for adipogenesis [18–20]. However, it is not well understood how ER stress and subsequent activation of the UPR affects adipogenesis and weight gain. Interestingly, mice deleted in p58IPK (also known as Dnajc3), a co-chaperone DNAJ family member for BiP/GRP78 in the ER, as well as mice heterozygous for Grp78 (also known as Hspa5), gain less fat mass compared with wild-type mice [21, 22]. Since p58IPK deficiency and Grp78 heterozygosity decrease ER chaperone activity and increase ER stress, the reduced fat mass in these mice might result from activation of UPR signalling pathways. In addition, we reported previously that high-fat diet (HFD)-fed mice with a heterozygous mutation at the phosphorylation site in Eif2a (Ser51Ala, S/A) became significantly more obese than HFD-fed wild-type mice [23]. This prompted us to investigate the role of eIF2α phosphorylation and IRE1α signalling in adipocyte differentiation using 3T3-L1 cells, mouse embryonic fibroblasts (MEFs) and genetically engineered mice.
Methods
Animals
Animal use was in compliance with the Institute of Laboratory Animal Research Guide for the Care and Use of Laboratory Animals and approved by the University Committee on Use and Care of Animals at the University of Michigan. Chop−/− [24] and Eif2aS/A [23] mice were previously described. The genetic background of Chop−/− and Eif2aS/A mice is C57BL/6J. Leprdb/+ mice (C57BKS.Cg-m+/+ Leprdb, JAX mice) were bred with Chop−/− mice to generate heterozygous F1 mice that were intercrossed to obtain Chop−/−Leprdb/db and Chop+/+Leprdb//db+ mice. For HFD studies, 10- to 14-week-old male mice were provided with free access to HFD (catalogue D12451; Research Diets, New Brunswick, NJ, USA) or regular chow (catalogue D12450; Research Diets) for the indicated times. Body weights were measured between 15:00 and 17:00 hours. Food intake was analysed by measurement of food mass each day for 4 days and intake calculated (kJ/day) using a conversion rate of 19.79 kJ/g. For all studies, age-matched siblings were used as controls.
Cell culture and adipocyte differentiation
3T3-L1 pre-adipocytes were maintained in DMEM supplemented with 10% calf serum [25]. Two days after confluency, cell differentiation into adipocytes was induced with DMEM containing 1 μg/ml insulin (Invitrogen, Carlsbad, CA, USA), 0.5 mmol/l 3-isobutyl-1-methylxanthine (Sigma, St Louis, MO, USA) and 1 μmol/l dexamethasone (Sigma) for 4 days, and then with DMEM supplemented with 10% FBS and 1 μg/ml insulin only for another 3 days. After induction, cells were fed every other day with DMEM containing 10% FBS. Next, Oil red O (Sigma) working solution was added to formalin-fixed cells and incubated for 10 min at room temperature. Images were taken using an Olympus microscope system (Olympus, Center Valley, PA, USA). For quantification, absorbance was measured at 500 nm using spectrophotometer (SpectraMAX plus, Molecular Devices, Sunnyvale, CA, USA).
Generation of stable 3T3-L1 cell lines
The pBabe vector encoding Fv2E-PERK was obtained from D. Ron (University of Cambridge) [26]. For Chop, Ire1a (also known as Ern1) and Ire1aK907A constructs, cDNAs were amplified by PCR and cloned into pLVX-tight-puro (Clonetech, Mountain View, CA, USA). Tet-inducible 3T3-L1 cells, described previously [25], were transduced with viruses, followed by puromycin (2 μg/ml) selection for stable inducible expression of the transgene.
Measurement of body composition
Body composition was analysed in the conscious state with a Minispec LF90 II (Bruker Optics, Billerica, MA, USA), a nuclear magnetic resonance-based whole-body composition analyser at the mouse phenotyping core at the University of Michigan.
MEF generation
Primary MEFs were generated as described previously [27]. For differentiation, only primary MEFs at passage two were used. Differentiation was induced as described for 3T3-L1 cells except that 10 μg/ml of insulin and rosiglitazone (50 nmol/l; Cayman Chemical, Ann Arbor, MI, USA) was added for the first 4 days.
RNA extraction and real-time PCR analysis
Total RNA was extracted from cells using RNAeasy (Qiagen, Valencia, CA, USA). The relative amounts of mRNAs were calculated from the comparative threshold cycle (Ct) values relative to β-actin. Real-time primer sequences are shown in electronic supplementary material (ESM) Table 1. Data was analysed using the 2−ΔΔCt method.
Immunohistochemistry and cell size measurement
Epididymal fat pads were obtained from mice at indicated times, then fixed in formalin and paraffin embedding. Five-micrometre sections were deparaffinised and stained with hematoxylin and eosin. For each mouse two or three representative images of each slide were obtained and cell sizes were measured using ImageJ 1.43u software (http://rsb.info.nih.gov/ij).
Serum analysis
Serum levels of NEFA and total cholesterol were measured using HR Series NEFA-HR kit and Cholesterol E (Wako, Richmond, VA, USA) according to the manufacturer’s instructions. Triacylglycerol levels were measured using Infinity Triglycerides Reagent (Thermo, Middletown, VA, USA) according to the manufacturer’s instructions. Serum samples were collected after a 4–6 h fast.
Insulin tolerance tests
Mice were fasted for 6 h, followed by i.p. injection of insulin (0.75 U/kg body weight). Blood samples were collected via the tail vein and blood glucose levels were measured using an OneTouch Ultra glucometer (LifeScan, Milpitas, CA, USA).
Western blotting
Cell lysates were obtained in cell lysis buffer (50 mmol/l Tris HCl [pH 7.4], 150 mmol/l NaCl, 1% [vol./vol.] Triton X-100, 0.1% [wt/vol.] SDS, 1% [wt/vol.] sodium deoxycholate and protease inhibitors) (Roche Diagnostics, Indianapolis, IN, USA) and total protein concentration in each sample was measured using the Lowry protein assay kit (Biorad, Hercules, CA, USA). Primary antibodies were as follows: p-eIF2α (Invitrogen), eIF2α (Invitrogen), p-IRE1α (Novus, Littleton, CO, USA), IRE1α (Cell Signaling, Danvers, MA, USA), KDEL which detects GRP78 and GRP94 (Abcam, Cambridge, MA, USA), ATF4 (a kind gift from M. Kilberg, University of Florida), CHOP (Santa Cruz Biotechnology, Santa Cruz, CA, USA), PPARγ (Cell Signaling), C/EBPα (Santa Cruz Biotechnology), C/EBPβ (Santa Cruz Biotechnology), C/EBPδ (Santa Cruz Biotechnology) and tubulin (Sigma). Chemiluminescence detection was performed using ECL Western Blot detection reagents (GE Healthcare, Pittsburgh, PA, USA). Membranes were exposed to imaging film and developed using a Kodak X-OMAT processor (Kodak, Rochester, NY, USA).
Statistical analysis
All data are presented as means±SEM. The difference between groups was evaluated using Student’s t test; p<0.05 was considered significant.
Results
The UPR is activated during adipogenesis
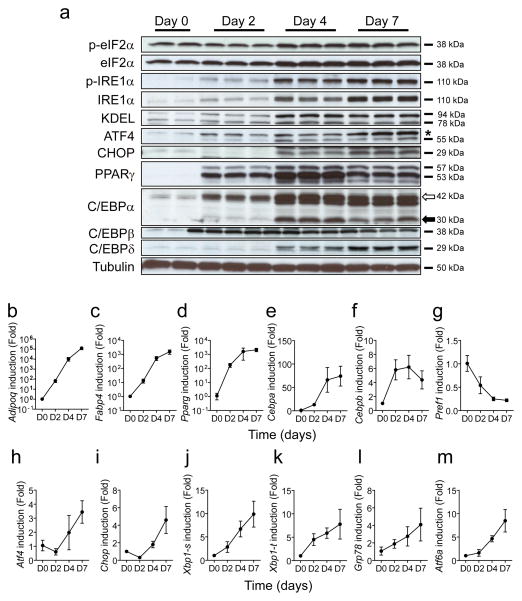
During differentiation of 3T3-L1 cells, the production of proteins encoded by the adipocyte-specific genes Pparg, Cebpa, Cebpb and Cebpd was increased (Fig. 1a). In parallel, mRNA expression of Adipoq, Fabp4, Cebpa and Cebpb was increased coinciding with the progression of adipogenesis (Fig. 1b–f). In contrast, the expression of the pre-adipocyte-specific gene Pref1 (also known as Delk1) decreased (Fig. 1g), indicating that this cell line was well differentiated. Next, we investigated the levels of UPR markers during adipogenesis (Fig. 1h–m). Interestingly, the ratio of phosphorylated eIF2α to total eIF2α was reduced at day 2 but restored at days 4 and 7 (Fig. 1a, ESM Fig. 1a). Consistent with this pattern, the production of CHOP, a transcription factor induced by eIF2α phosphorylation, decreased at day 2 and subsequently increased between days 4 and 7 (Fig. 1a,i). In addition, the level of both phosphorylated and unphosphorylated IRE1α increased during adipocyte differentiation (Fig. 1a, ESM Fig. 1b), where the levels of spliced Xbp1 (Xbp1-s), as well as total Xbp1 (Xbp1-t), mRNAs were upregulated during the later period of adipocyte differentiation (Fig. 1j,k).
Fig. 1.
Expression of ER stress genes is upregulated during adipogenesis in 3T3-L1 cells. (a) Protein expression profiles for adipocyte-related genes and UPR-related genes. Cell lysates were collected at the indicated times after the start of adipocyte differentiation and analysed by western blotting analysis. The white and black arrows indicate larger and smaller isoforms of C/EBPα (* indicates a non-specific band). p-eIF2α and p-IRE1α indicate phosphorylated forms of eIF2α and IRE1α, respectively. KDEL antibody can detect GRP78 and GRP94 proteins (b–m) Gene expression profiles for adipocyte-related and UPR-related genes. Total RNA was isolated from 3T3-L1 cells at the indicated times during adipogenesis and mRNA levels were measured by quantitative real-time RT-PCR. Xbp1-s and Xbp1-t indicate spliced and total Xbp1, respectively. D0, D2, D4 and D7 indicate day 0, day 2, day 4 and day 7 after initiation of adipogenesis. Data are presented as means±SEM of three independent experiments with triplicates
ER stress represses adipocyte differentiation
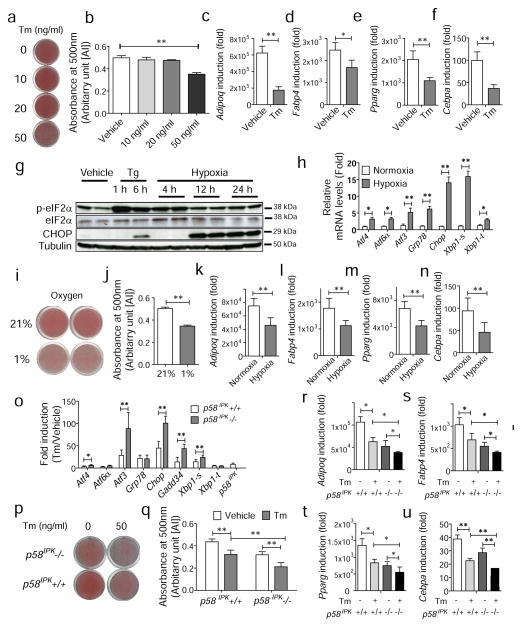
Next, we asked whether exogenous ER stress affects adipogenesis. Induction of ER stress by treatment with a low dose of tunicamycin (Tm), an inhibitor of N-linked glycosylation that is not cytotoxic [28], significantly inhibited adipogenesis quantified by Oil red O staining (Fig. 2a,b) and expression of mature adipocyte marker genes (Fig. 2c–f). We also investigated the effects of a physiologically relevant inducer of ER stress, hypoxia [29], on adipogenesis. As early as 4 h under hypoxic conditions, eIF2α was phosphorylated and subsequently, after 12 h of hypoxia, its downstream target, CHOP, was upregulated as were other UPR-induced genes (Fig. 2g,h). When hypoxia was introduced during adipogenesis, adipocyte differentiation was significantly attenuated, with reduced expression of mature adipocyte marker genes (Fig. 2in). Since mice deficient in p58IPK, which assists chaperone-mediated protein maturation in the ER [30, 31], display reduced adipose mass [22], we also investigated the effect of p58IPK deletion on adipocyte differentiation. We observed that UPR-related genes were upregulated to a significantly greater degree in response to ER stress in p58IPK−/− compared with p58IPK+/+ pre-adipocytes (Fig. 2o), indicating that p58IPK-deficient MEFs are more susceptible to ER stress. Consistent with the in vivo observation, adipogenesis in p58IPK−/− pre-adipocytes was reduced compared with p58IPK+/+ pre-adipocytes in the presence or absence of ER stress (Fig. 2p,q), with reduced expression of mature adipocyte marker genes (Fig. 2r–u).
Fig. 2.
ER stress suppresses adipogenesis in 3T3-L1 cells. (a, b) Suppression of adipocyte differentiation by tunicamycin (Tm) treatment. (a) Confluent 3T3-L1 cells were cultured in differentiation media in the presence of Tm at the indicated concentrations. On day 10, cells were fixed and stained with Oil red O. Representative images of Oil red O staining are shown. (b) Quantification of Oil red O staining in (a). **p<0.01 vs vehicle (c–f) Gene expression profiles for markers of mature adipocytes. Total RNA was isolated from 3T3-L1 cells at the indicated times during adipogenesis in the absence or presence of Tm (50 ng/ml) and mRNA levels were measured by quantitative real-time RT-PCR. (g–n) Suppression of adipogenesis by hypoxia. (g) Induction of eIF2α phosphorylation and CHOP production. Cell lysates were collected at the indicated times after hypoxic treatment and analysed by western blotting. Tg indicates thapsigargin at 300 nmol/l. (h) Gene expression profiles after hypoxic treatment for 24 h. Total RNA was isolated from 3T3-L1 cells at the indicated times during adipogenesis and mRNA levels were measured by quantitative real-time RT-PCR. White bar, normoxic conditions; grey bar, hypoxia. (i) Oil red O staining of 3T3-L1 cells after adipogenesis under normoxic (21%) or hypoxic (1%) conditions. (j) Quantification of Oil red O staining in (i). (k–n) Gene expression profiles for markers of mature adipocytes under normoxic or hypoxic conditions. (o–u) Suppression of adipogenesis by p58IPK deletion. (o) Gene expression profiles in p58IPK+/+ (white bars) or p58IPK−/− cells (grey bars) in the presence of Tm (1 μg/ml). Total RNA was isolated from p58IPK+/+ and p58IPK−/− pre-adipocytes at 24 h after treatment with Tm and mRNA levels were measured by quantitative real-time RT-PCR. Data were plotted as fold induction relative to the vehicle-treated sample. (p) Oil red O staining of p58IPK+/+ and p58IPK−/− MEFs after differentiation in the absence or presence of Tm (50 ng/ml). (q) Quantification of Oil red O staining in (p). White bars, vehicle; grey bars, Tm. (r–u) Gene expression profiles for markers of mature adipocytes in p58IPK+/+ and p58IPK−/− MEFs in the absence or presence of Tm (50 ng/ml). Data are presented as means±SEM of three independent experiments with triplicates. *p<0.05, **p<0.01. AU, arbitrary units
Phosphorylation of eIF2α represses adipogenesis
The experiments above suggest that UPR activation appears to decrease adipogenesis. Since all three UPR subpathways are activated under conditions of ER stress, it is difficult to attribute an inhibitory effect to any specific UPR subpathway. Therefore, we investigated the effect of mutations that singly inactivate each of the UPR subpathways. First, we generated stable 3T3-L1 cell lines producing Fv2E-PERK (3T3-L1-Fv2EPERK), which forms dimers in the presence of the drug AP20187, to preemptively phosphorylate eIF2α without an ER stress signal [26] (ESM Fig. 2). After AP20187 treatment, eIF2α phosphorylation was dramatically increased at 1 h then slightly declined up to 24 h (Fig. 3a). ATF4 induction was observed at 2 h and CHOP was also upregulated at 2–4 h after AP20187 treatment (Fig. 3a). Addition of AP20187 induced eIF2α phosphorylation to a level comparable with that observed in MEFs treated with thapsigargin (ESM Fig. 3) and significantly reduced adipogenesis in the 3T3-L1-Fv2EPERK stable cell line compared with vehicle-treated cells as detected by reduced lipid accumulation (Fig. 3b,c) and expression of mature adipocyte marker genes (Fig. 3d–g).
Fig. 3.
eIF2α phosphorylation represses adipogenesis. (a) Protein expression profile in a stable 3T3-L1 cell line that expresses chimeric Fv2E-PERK. Cell lysates were collected at the indicated times after AP20187 treatment (5 nmol/l) for western blot analysis (* indicates a non-specific band). (b) Oil red O staining. Adipogenesis was stimulated in 3T3-L1 cells expressing Fv2E-PERK in the absence (vehicle) or presence of AP20187. On day 10, cells were fixed and stained with Oil red O. Representative images from indicated cell lines are shown. Scale bar indicates 100 μm. (c) Quantification of Oil red O staining in (b). (d–g) Gene expression profiles for markers of mature adipocytes in Fv2E-PERK 3T3-L1 cells in the absence or presence of AP20187 (5 nmol/l). (h) Oil red O staining of Eif2aS/S and Eif2aA/A MEFs after adipogenesis. Differentiation was induced in primary MEFs from wild-type (Eif2aS/S) or homozygous mutant (Eif2aA/A) for 14 days. Cells were then fixed and stained with Oil red O. Representative images are shown and scale bar indicates 100 μm. (i) Quantification of Oil red O staining in (h). (j–n) Gene expression profiles during adipocyte differentiation of primary MEFs. Total RNA was isolated from cells at the indicated times during adipogenesis, and mRNA levels were measured by quantitative real-time RT-PCR. Circles, Eif2aS/S; squares, Eif2aA/A. D0, D2, D4, D7, D10 indicate day 0, day 2, day 4, day 7 and day 10 after initiation of adipocyte differentiation. Data are presented as means±SEM of three independent experiments with triplicates. **p<0.01, *p<0.05 vs Eif2aA/A. AU, arbitrary units
To analyse the effect of impaired eIF2α phosphorylation on adipogenesis, primary MEFs were isolated from mouse embryos with wild-type eIF2α alleles (Eif2aS/S) or with two mutant eIF2α alleles that had alanine substitutions at Ser51 to prevent phosphorylation (Eif2aA/A) [11]. When induced for adipogenesis, there was significantly more lipid accumulation in the Eif2aA/A MEFs compared with the Eif2aS/S MEFs (Fig. 3h,i). Similar results were obtained in two additional independent preparations of Eif2aS/S and Eif2aA/A primary MEFs. The lipid accumulation correlated with significantly reduced induction of Chop in the Eif2aA/A mutant MEFs during the entire period of adipocyte differentiation compared with wild-type Eif2aS/S MEFs (Fig. 3j). In contrast, expression of the main adipogenic regulators Pparg and Cebpa in Eif2aA/A MEFs was significantly increased at day 10 (Fig. 3k–n). These results suggest that eIF2α phosphorylation and subsequent activation of downstream signalling pathways represses adipocyte differentiation.
Eif2aS/A mice fed HFD become more obese than wild-type mice through an increase in adipocyte number
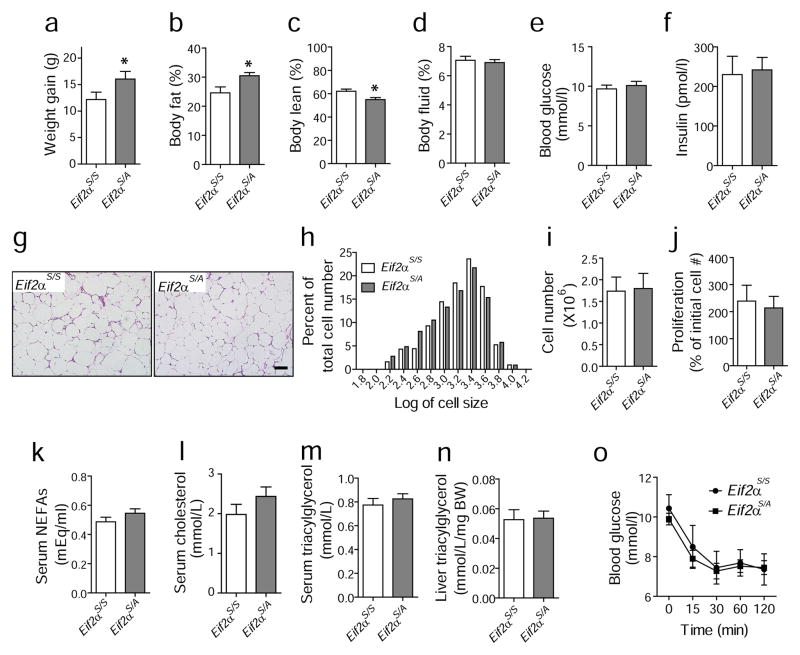
We then investigated the effect of eIF2α phosphorylation in vivo using heterozygous Eif2aS/A and wild-type mice (Eif2aS/S) since homozygous Eif2aA/A is perinatal lethal. The Eif2aS/A mice gained more body weight than Eif2aS/S mice fed an HFD [23]. Although there were no differences in food intake (ESM Fig. 4a), the Eif2aS/A mice displayed increased body fat and less lean body mass compared with Eif2aS/S mice (Fig. 4a–f), indicating that most of the weight gain was due to increased fat mass. The size distribution of adipocytes was not significantly different between the genotypes (Fig. 4g,h). In addition, there was no significant difference in the number of cells or their rate of proliferation from the stromal vascular fraction (SVF) of epididymal fat tissues between Eif2aS/A and Eif2aS/S mice (Fig. 4i,j). These results suggest that the increased fat mass in the HFD-fed Eif2aS/A mice results from increased adipocyte number. Although obesity can cause hyperlipidaemia, the levels of serum NEFA, total cholesterol, adiponectin and triacylglycerol, as well as liver triacylglycerol and insulin sensitivity, were not significantly different between the genotypes fed an HFD (Fig. 4k–o; ESM Fig. 4b).
Fig. 4.
Mice with mutation in eIF2α (Eif2aS/A) display increased body weight compared with wild-type Eif2aS/S mice without differences in adipocyte size or serum lipid levels. (a–f) Weight gain, body fat mass, body lean mass, fluid mass, blood glucose and insulin levels. Mice were fed an HFD for 13 weeks and metabolic variables were measured using an NMR-based analyser. Eif2aS/A (n=12) and Eif2aS/S (n=7). (g, h) No difference in adipocyte size between genotypes. After HFD for 13 weeks, epididymal fat tissues were obtained for histochemistry and representative images of epididymal fat pads are shown in (g). Scale bar indicates 100 μm. (h) Histogram shows the distribution of adipocyte size. The x-axis represents the logarithm of cell size in pixels, while the y-axis shows the percentage of cells having a given size for each group of mice. White bars, Eif2aS/S; grey bars, Eif2aS/A. (i) The number of cells from the SVF of adipose tissues from Eif2aS/A (n=4) and Eif2aS/S (n=4) mice at age of 6–8 weeks on a regular diet. (j) Proliferation rate of cells from SVF. Cells from SVF were plated in replicate and the number of cells was counted after 4 days of culture using an automated cell counter. Each suspension was counted twice. The y-axis represents the percentage of the initial cell number. Plasma levels of non-esterified NEFA (k), cholesterol (l) and triacylglycerol (m) after 13 weeks of HFD. Sera from Eif2aS/A (n=11) and Eif2aS/S (n=9) mice were collected and measured for NEFA and cholesterol as indicated in methods. (n) Hepatic triacylglycerol levels. Liver samples were collected for analysis after 13 weeks of HFD. (o) Insulin tolerance tests (ITTs). After 13 weeks of HFD, blood glucose levels were measured at the indicated times after insulin injection (0.75 U/kg body weight). Eif2aS/A (n=12, squares) and Eif2aS/S (n=9, circles). *p <0.05. BW, body weight
CHOP production inhibits adipogenesis
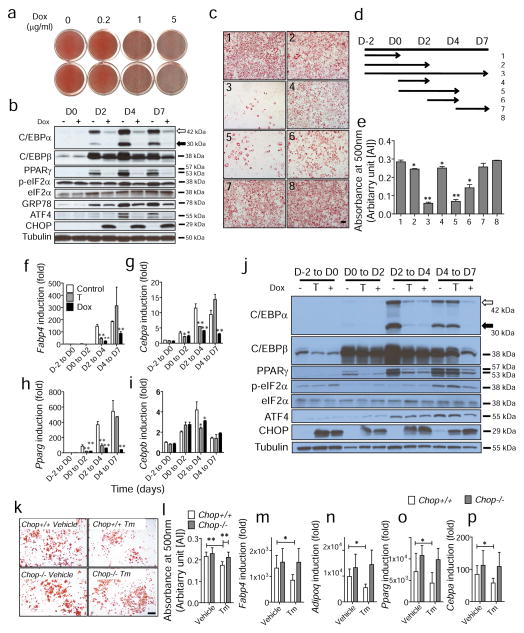
Since CHOP is induced through eIF2α phosphorylation and inhibits adipogenesis by interfering with other C/EBP family members [32], we examined the effect of CHOP on adipogenesis more intensively using an inducible expression system. First, conditional 3T3-L1 cell lines were generated that express CHOP under the control of a tetracycline-inducible promoter (ESM Fig. 5). It is notable that CHOP production itself is not apoptotic unless there is a stress signal [33]. When CHOP production was induced during adipocyte differentiation, adipogenesis was significantly inhibited in a doxycycline-dose-dependent manner (Fig. 5a). The level of C/EBPβ was slightly attenuated at days 2 and 4 after doxycycline treatment, and substantially reduced by day 7 (Fig. 5b). Upon doxycycline treatment, C/EBPα and PPARγ were barely detected during adipogenesis, suggesting that adipogenesis was significantly impaired by overproduction of CHOP (Fig. 5b). Next, we identified the most critical time point for CHOP to exert its inhibitory effect on adipogenesis. Whereas doxycycline treatment during the entire period of adipogenesis almost completely blocked differentiation (Fig. 5c-3, d,e), doxycycline treatment during the earlier period (Fig. 5c-1,c-2,d,e) or the later period (Fig. 5c-7,d,e) had negligible effects on adipogenesis. However, induction of CHOP by doxycycline treatment from day 2 to day 4 significantly repressed adipogenesis (Fig. 5c-6,d,e) and doxycycline treatment from day 0 to day 4 further repressed adipocyte differentiation in the CHOP-inducible 3T3-L1 cell line (Fig. 5c-5,d,e), indicating that CHOP production during day 2 to day 4 is critical for its inhibitory effect. Consistent with this observation, transient expression of Chop from day 2 to day 4 during adipogenesis significantly attenuated induction of Cebpa and Pparg mRNAs (Fig. 5f–i) and their related proteins (Fig. 5j). However, treatment with doxycycline from day 4 to day 7 did not alter expression of these genes. These results indicate that CHOP inhibits adipogenesis through suppression of C/EBPα and PPARγ production and the most critical time period is around day 2 when the production of CHOP is reduced during adipogenesis (Fig. 1a). To study the requirement for CHOP in suppressing adipogenesis in response to ER stress, we studied adipogenesis in Chop+/+ and Chop−/− MEFs in the absence or presence of ER stress induced by Tm treatment (50 ng/ml). Whereas Tm treatment significantly reduced adipogenesis in Chop+/+ MEFs, adipogenesis was not significantly reduced in Chop−/− MEFs, indicating that CHOP is essential for ER stress-mediated suppression of adipogenesis (Fig. 5k–p). However, there was no significant difference either in cAMP levels or in insulin sensitivity, during adipogenesis, between Chop+/+ and Chop−/− MEFs (ESM Fig. 6).
Fig. 5.
CHOP production inhibits adipogenesis in 3T3-L1 cells. (a) Inhibition of adipogenesis by CHOP. 3T3-L1 cells producing CHOP under the control of tetracycline-inducible promoter (Tet-CHOP) were differentiated in the presence of doxycycline (Dox) at various concentrations. On day 10, cells were fixed and stained with Oil red O. (b) Protein production profile during adipogenesis in the absence (−) or presence (+) of doxycycline. Cell lysates were collected at indicated time points and subjected to western blotting. D0, D2, D4 and D7 indicate day 0, day 2, day 4 and day 7 after initiation of adipocyte differentiation. The white and black arrows indicate larger and smaller isoforms of C/EBPα, respectively. (c–e) Identification of critical time period for inhibitory effect of CHOP on adipogenesis. Adipocyte differentiation was stimulated in Tet-CHOP cells in the presence of doxycycline for various time intervals as indicated (d). On day 10, cells were stained with Oil red O and representative images from each condition are shown (c). Staining was quantified and presented as means ± SEM of three independent experiments with triplicates (e). Scale bar indicates 100 μm. (f–j) Effect of transient CHOP overproduction on (f–i) gene expression and (j) protein production during adipogenesis. Tet-CHOP cells were differentiated in the absence (white bars) or presence of doxycycline during the entire differentiation period (black bars) or transient doxycycline treatment (grey bars) as indicated. D–2 indicates 2 days before the adipocyte differentiation; D0, D2, D4 and D7 indicate day 0, day 2, day 4 and day 7 after initiation of adipogenesis. (f–i) At the days indicated, total RNAs were isolated for analysis by real-time RT-PCR. Data are presented as means±SEM of three independent experiments with triplicates. *p<0.05 and **p<0.01 vs control. (j) Protein production was analysed by western blot under the same conditions as (f–i). (k) Oil red O staining. Adipogenesis was stimulated in primary Chop+/+ and Chop−/− MEFs in the absence (vehicle) or presence of Tm (50 ng/ml). On day 10, cells were fixed and stained with Oil red O. Representative images from indicated MEF lines are shown. Scale bar indicates 100 μm. (l) Quantification of Oil red O staining in (k). White bars, Chop+/+; grey bars, Chop−/−. (m–p) Gene expression profiles for markers of mature adipocytes in Chop+/+ (white bars) and Chop−/− MEFs (grey bars) in the absence or presence of Tm (50 ng/ml). *p<0.05 and **p<0.01. AU, arbitrary units
Fig. 7.
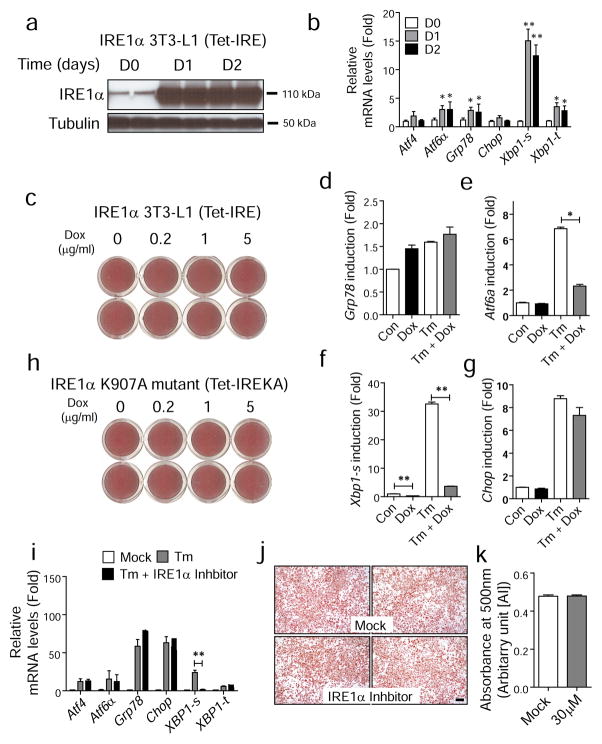
IRE1α signalling is not required for adipogenesis in vitro or in vivo. (a, b) Generation of 3T3-L1 cells that produce wild-type IRE1α under the control of a tetracycline-inducible promoter (Tet-IRE). Total protein lysates and total RNA were collected at indicated time points after doxycycline (5 μg/ml) treatment and analysed by western blot (a) and quantitative real-time PCR (b). White bars, day 0 (D0); grey bars, day 1 (D1); black bars, day 2 (D2). **p<0.01, *p<0.05 vs D0. (c) IRE1α overproduction does not affect adipogenesis. Differentiation was induced in Tet-IRE cells in the presence of doxycycline (Dox) at various doses. On day 10, cells were stained with Oil red O. Representative images from each condition are shown. (d–g) Gene expression profile for stable 3T3-L1 cells expressing dominant-negative mutant Ire1aK907A under the control of a tetracycline-inducible promoter (Tet-IREKA) in the absence or presence of Tm-induced ER stress. Data are presented as means±SEM of three independent experiments with triplicates. (h) Images of Oil red O staining for 3T3-L1 cells expressing mutant Ire1aK907A under the same conditions as (d–g). (i) Quantitative real-time RT-PCR to verify IRE1α inhibition. 3T3-L1 cells were treated with Tm (2 μg/ml) in absence (grey bars) or presence of IRE1α inhibitor (30 μmol/l, black bars) for 16 h for analysis of gene expression. ‘Mock’ indicates DMSO treatment (white bars). (j) Oil red O staining. Adipogenesis was stimulated in 3T3-L1 cells in the absence (vehicle) or presence of IRE1α inhibitor (30 μmol/l). On day 10, cells were fixed and stained with Oil red O. Representative images are shown. Scale bar indicates 100 μm. (k) Quantification of Oil red O staining is shown in right panel. Data are presented as means±SEM. **p<0.01, *p<0.05. AU, arbitrary units
Fig. 6.
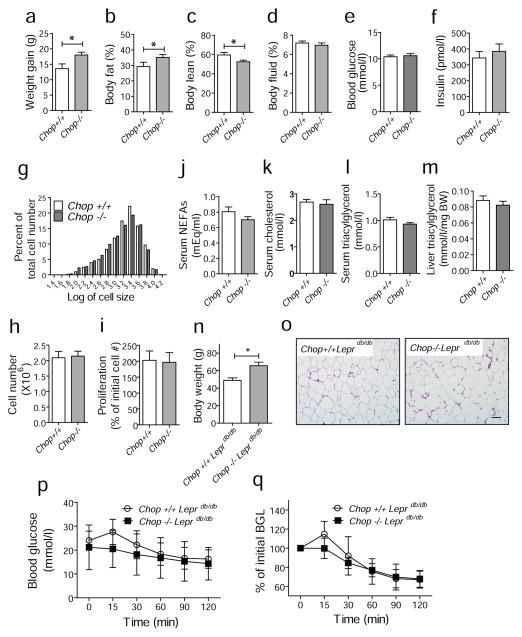
Chop deletion increases body fat mass in mice fed a HFD or in genetically obese Leprdb/db mice. (a–f) Weight gain, body fat mass, body lean mass, fluid mass, blood glucose and insulin levels. Chop+/+ (n=10) and Chop−/− (n=8) mice were fed an HFD for 13 weeks and weight gain, body fat mass, body lean mass and fluid mass were measured using an NMR-based analyser. (g) Histogram showing the distribution of adipocyte size in mice fed an HFD for 13 weeks. The x-axis represents the logarithm of cell size in pixels, while the y-axis shows the percentage of cells having a given size for each group of mice. White bars, Chop+/+; black bars, Chop−/−. (h) The number of cells from the SVF of adipose tissues from Chop+/+ (n=4) and Chop−/− (n=4) mice at the age of 6–8 weeks on regular diet. (i) Proliferation rate of cells from SVF. Cells from SVF were plated in replicate and the number of cells was counted after 4 days using an automated cell counter. Each suspension was counted twice. The y-axis represents the percentage of the initial cell number. Serum levels of NEFA (j), cholesterol (k) and triacylglycerol (l) from mice fed an HFD for 13 weeks. Sera from Chop+/+ (n=10) and Chop−/− (n=8) mice were collected and analysed. (m) Hepatic triacylglycerol levels. Liver samples were collected after 13 weeks of HFD for analysis. (n) Body weights of Chop+/+Leprdb/db (n=5) and Chop−/−Leprdb/db (n=5) mice at 6 months of age. (o) Representative images of epididymal fat pads from Chop+/+Leprdb/db and Chop−/−Leprdb/db mice at 6 months of age. Scale bar indicates 100 μm. (p, q) ITTs. Chop+/+Leprdb/db (n=5, circles) and Chop−/− Leprdb/db (n=5, squares) mice at 6 months of age were challenged with insulin (0.75 U/kg body weight) and blood glucose levels were measured. Data are presented as means±SEM. **p<0.01, *p<0.05. BW, body weight
Chop deletion increases obesity in vivo
Next, we investigated the effect of Chop deletion in mice. After being fed with an HFD, Chop−/− mice gained more body weight, had a higher percentage of body fat and a lower percentage of lean mass compared with Chop+/+ mice (Fig. 6a–f). Analysis of the distribution of adipocyte sizes demonstrated no significant difference between genotypes of HFD-fed mice (Fig. 6g, ESM Fig. 7a). In addition, there was no significant difference in number of cells, or their proliferation rates, from the SVF of epididymal fat tissues between Chop+/+ and Chop−/− mice (Fig 6h,i). These results suggest that the increased fat mass in the HFD-fed Chop−/− mice results from increased adipocyte number. In addition, there was no significant difference in the levels of serum NEFA, cholesterol, adiponectin or triacylglycerol, as well as liver triacylglycerol (Fig. 6j–m; ESM Fig. 7b) between the genotypes. Similar results were obtained from one-year old mice, where the body weight and fat mass was significantly greater in Chop−/− compared with wild-type mice, without significant differences in the size distribution of adipocytes (ESM Fig. 8).
We also investigated the effect of Chop deletion in the leptin receptor-deficient mouse, Leprdb/db. Since Leprdb/db mice lose appetite control due to deficiency of the leptin receptor, these mice show severe obesity without an HFD. Chop−/− mice were crossed with Leprdb/db mice to generate Chop−/−Leprdb/db and Chop+/+Leprdb/db mice. Strikingly, Chop deletion increased body weight in the Leprdb/db mice (Fig. 6n), without a significant difference in adipocyte size (Fig. 6o). However, there was no difference in insulin sensitivity between the genotypes despite the increased body weight in Chop−/−Leprdb/db mice (Fig. 6p,q).
IRE1α signalling does not alter adipogenesis
Since the expression and phosphorylation of IRE1α and Xbp1 mRNA splicing were increased during adipogenesis (Fig. 1a,b), we investigated the requirement for IRE1α in adipogenesis. For this purpose, stable 3T3-L1 cell lines (Tet-IRE) were generated which induce production of IRE1α under the control of a doxycycline-responsive promoter (Fig. 7a). Forced production of IRE1α by doxycycline treatment induced splicing of Xbp1 mRNA, as well as Atf6α and Grp78 mRNA expression (Fig. 7b). Induction of IRE1α by doxycycline treatment in Tet-IRE cells did not affect adipocyte differentiation, indicating that pre-emptive activation of IRE1α had little effect on adipogenesis (Fig. 7c).
Next, we investigated the requirement for IRE1α in adipogenesis. First, a stable 3T3-L1 cell line (Tet-IREKA) was generated with inducible expression of the dominant-negative Ire1aK907A RNase mutant (ESM Fig. 9). The Ire1α K907A mutant cannot initiate Xbp1 mRNA splicing [15] and inhibits Xbp1 splicing by wild-type IRE1α, even in the presence of ER stress (Fig. 7d–g). Overexpression of the K907A mutant during adipogenesis did not alter adipogenesis in the 3T3-L1 cells (Fig. 7h). Second, we treated 3T3-L1 cells with an IRE1α inhibitor [34] to inhibit Xbp1 mRNA splicing (Fig. 7i) during adipogenesis. Consistent with the above results, the IRE1α inhibitor did not affect adipocyte differentiation in 3T3-L1 cells (Fig. 7j,k).
Discussion
In this study, we demonstrate that eIF2α phosphorylation increases CHOP production to repress adipocyte differentiation in response to ER stress in vitro and in vivo. Our conclusion is supported by the following observations. First, activation of the UPR inhibited adipogenesis in 3T3-L1 cells. Second, pre-emptive phosphorylation of eIF2α by forced dimerisation of Fv2E-PERK inhibited adipogenesis in 3T3-L1 cells. Third, homozygous Ser51Ala mutation in eIF2α to prevent phosphorylation significantly enhanced adipogenesis in MEFs compared with wild-type MEFs. Fourth, heterozygous Ser51Ala mutation in eIF2α increased obesity and adipocyte number in HFD-fed mice. Fifth, transient induction of CHOP was sufficient to inhibit adipogenesis in 3T3-L1 cells. Sixth, Chop deletion increased obesity and adipocyte number upon HFD feeding, as well as in Leprdb/db mice. Finally, the IRE1α pathway had little effect on adipogenesis.
Several lines of evidence suggest that the UPR is activated during cellular differentiation [18, 35–39]. Consistent with these findings, during adipogenesis, we observed eIF2α phosphorylation and IRE1α activation. However, we also found that the ratio of phosphorylated eIF2α to total eIF2α was reduced at day 2, suggesting protein synthesis is increased during the early phase of adipogenesis, possibly due to increased synthesis of proteins required for differentiation. An elevated rate of protein synthesis would, in turn, increase phosphorylation of eIF2α later in differentiation and lead to the induction of downstream target molecules, including CHOP, as observed in our study.
Our results are consistent with the findings of Basseri et al that showed both eIF2α phosphorylation and the total amount of eIF2α were reduced at days 1–2, increased at days 3–7 and again reduced in the late period of 3T3-L1 differentiation [18]. In addition, the induction pattern of GRP78 and CHOP was also similar to what we observed. However, in contrast to our conclusion, they suggested that ER stress is required for adipocyte differentiation because the chemical chaperone 4-phenyl butyric acid (PBA) reduced ER stress and inhibited adipogenesis [18]. The difference might result from the source of ER stress and mechanism of UPR activation. Physiological UPR activation that occurs during differentiation might facilitate adipogenesis, whereas more severe UPR activation caused by exogenous stimuli might inhibit adipogenesis. It is also possible that PBA is doing something other than improving ER protein folding. For example, it is notable that PBA is a histone deacetylase inhibitor [40].
Our findings also show that the amount of both phosphorylated and total IRE1α increases during adipogenesis. Induction of IRE1α appeared as early as day 2 and was sustained at high levels up to day 7. Xbp1 mRNA splicing correlated with increased phosphorylation of IRE1α. However, these kinetics were quite different from those of eIF2α phosphorylation, which was reduced at day 2, suggesting that each subpathway of the UPR is regulated independently. Interestingly, either increasing IRE1α activation via IRE1α overexpression or inhibiting IRE1α via expression of a dominant-negative Ire1a RNase mutant, did not alter adipocyte differentiation. Therefore, it remains unclear why IRE1α is activated during adipogenesis. As recent studies identified a role for XBP1 and IRE1α in lipid metabolism [41–43], it is possible that IRE1α affects lipid metabolism in the differentiated mature adipocyte. Our findings conflict with those of Sha et al that suggest IRE1α activation and subsequent Xbp1 mRNA splicing is required for adipogenesis [19]. They proposed that physiological UPR activation occurs during the early period of adipogenesis and is maintained at a relatively low level in mature adipocytes. Although it is not possible to know the reason(s) for the different observations, adipogenesis is very inefficient and variable in immortalised MEFs. This may be due to variability of the differentiation state in immortalised MEFs that may change with passage number and growth conditions [44]. Our studies analysed the role of the IRE1α pathway in the well-established system of 3T3-L1 cells. We believe the role of IRE1α and XBP1 in adipogenesis deserves further investigation.
In obesity, mature adipocytes are exposed to elevated NEFA, inflammation and nutrient and oxygen deprivation [14, 29, 45, 46]. Since these factors can cause ER stress to activate the UPR, they likely influence pre-adipocytes and/or adipogenic stem cells [14]. As the demand for fat storage increases during the progression of obesity, a defect in adipose tissue expansion would result in storage of excessive NEFA in other peripheral tissues such as liver and muscle. The deposition of lipid in liver and muscle would increase insulin resistance [47–50]. Therefore, increasing the number of functional adipocytes would preserve insulin sensitivity by providing a storage depot for fatty acids. Under conditions of extreme obesity, UPR activation in pre-adipocytes would inhibit the generation of new adipocytes. This would limit the capacity to store excessive lipids inside fat depots, which in turn would exacerbate insulin resistance in peripheral tissues. In support of this notion, we found that although HFD-fed Eif2aS/A mice and HFD-fed Chop−/− mice were more obese, there was no significant increase in the serum NEFA, cholesterol or triacylglycerol, or in hepatic triacylglycerol or insulin resistance compared with HFD-fed Eif2aS/S and Chop+/+ littermates, respectively.
In conclusion, our findings show that eIF2α phosphorylation and CHOP production are reduced during the early phase of adipogenesis in order to permit adipocyte differentiation. Factors that would cause ER stress, such as inflammation, nutrient deprivation and elevated NEFA, would increase eIF2α phosphorylation and CHOP production. In this manner, eIF2α phosphorylation would exacerbate the metabolic consequences of obesity by inhibiting adipogenesis and limiting lipid storage in adipose tissue. Therefore, tight regulation of eIF2α phosphorylation is required to optimise adipogenesis, preserve metabolic homeostasis and limit lipid deposition in liver and muscle.
Supplementary Material
Acknowledgments
We thank A. Kyle (University of Calgary) and J. Mitchell (University of Michigan) for assistance with manuscript preparation and the members of the Kaufman laboratory for critical input. We thank H. Mori (University of Michigan) for invaluable technical assistance and J. Patterson (MannKind Corporation) for the IRE1α inhibitor. This work used the Animal Phenotyping core of the Michigan Diabetes Research and Training Center funded by DK020572 from the National Institute of Diabetes and Digestive and Kidney Diseases. We thank ARIAD Pharmaceuticals, Inc. for providing AP20187.
Funding
This work was supported by NIH grants DK042394, DK088227, DK093074, HL052173 and HL057346 (R. J. Kaufman). Portions of this work were supported by University of Michigan CCMB Pilot Grant (J. Han).
Abbreviations
- ATF4
Activating transcription factor 4
- ATF6α
Activating transcription factor 6 alpha
- BiP/GRP78
Binding immunoglobulin protein/78 kDa glucose-regulated protein
- C/EBP
CCAAT/enhancer binding protein
- CHOP
C/EBP homologous protein
- Ct
Comparative threshold cycle
- ER
Endoplasmic reticulum
- eIF2α
Eukaryotic initiation factor 2 alpha
- ERAD
ER-associated protein degradation
- IRE1α
Inositol-requiring enzyme 1 alpha
- MEF
Mouse embryonic fibroblast
- PBA
4-Phenyl butyric acid
- PERK
Protein kinase RNA-like ER kinase
- PPAR
Peroxisome proliferator-activated receptor
- SVF
Stromal vascular fraction
- Tm
Tunicamycin
- UPR
Unfolded protein response
- XBP1
X-box binding protein 1
Footnotes
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
JH and RJK were responsible for the study concept and design and RM, BW, B. Song, SW, B. Sun, HM and RJK for acquisition of data. JH and RJK analysed and interpreted data. JH, RM, BW, B. Song, SW, B. Sun, HM and RJK drafted the manuscript. JH and RJK critically revised the manuscript for important intellectual content. RJK was responsible for study supervision. All authors approved the final version of the manuscript.
References
- 1.Barness LA, Opitz JM, Gilbert-Barness E. Obesity: genetic, molecular, and environmental aspects. Am J Med Genet A. 2007;143A:3016–3034. doi: 10.1002/ajmg.a.32035. [DOI] [PubMed] [Google Scholar]
- 2.Goossens GH. The role of adipose tissue dysfunction in the pathogenesis of obesity-related insulin resistance. Physiology & Behavior. 2008;94:206–218. doi: 10.1016/j.physbeh.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 3.de Ferranti S, Mozaffarian D. The perfect storm: obesity, adipocyte dysfunction, and metabolic consequences. Clin Chem. 2008;54:945–955. doi: 10.1373/clinchem.2007.100156. [DOI] [PubMed] [Google Scholar]
- 4.Faust I, Johnson P, Stern J, Hirsch J. Diet-induced adipocyte number increase in adult rats: a new model of obesity. Am J Physiol. 1978;235:E279–E286. doi: 10.1152/ajpendo.1978.235.3.E279. [DOI] [PubMed] [Google Scholar]
- 5.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 6.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 11.Scheuner D, Song B, McEwen E, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 12.Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 13.Calfon M, Zeng H, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 14.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum STress. Mol Cell Biol. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen J, Chen X, Hendershot L, Prywes R. ER Stress regulation of ATF6 localization by DISsociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 18.Basseri S, Lhoták Š, Sharma AM, Austin RC. The chemical chaperone 4-phenylbutyrate inhibits adipogenesis by modulating the unfolded protein response. J Lipid Res. 2009;50:2486–2501. doi: 10.1194/jlr.M900216-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sha H, He Y, Chen H, et al. The IRE1/XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe CE, Dennis RJ, Obi U, O’Rahilly S, Rochford JJ. Investigating the involvement of the ATF6alpha pathway of the unfolded protein response in adipogenesis. Int J Obes (Lond) 2012;36:1248–1251. doi: 10.1038/ijo.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye R, Jung DY, Jun JY, et al. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59:6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladiges WC, Knoblaugh SE, Morton JF, et al. Pancreatic β-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 23.Scheuner D, Mierde DV, Song B, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 24.Zinszner H, Kuroda M, Wang X, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niimi M, Tao L, Lin S-H, et al. Involvement of an alternatively spliced mitochondrial oxodicarboxylate carrier in adipogenesis in 3T3-L1 cells. J Biomed Sci. 2009;16:92–102. doi: 10.1186/1423-0127-16-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu PD, Jousse C, Marciniak SJ, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Back SH, Scheuner D, Han J, et al. Translation attenuation through eIF2 alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutkowski DT, Arnold SM, Miller CN, et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosogai N, Fukuhara A, Oshima K, et al. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes. 2007;56:901–911. doi: 10.2337/db06-0911. [DOI] [PubMed] [Google Scholar]
- 30.Rutkowski DT, Kang S-W, Goodman AG, et al. The Role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Cell Biol. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batchvarova N, Wang XZ, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153) EMBO J. 1995;14:4654–4661. doi: 10.1002/j.1460-2075.1995.tb00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volkmann K, Lucas JL, Vuga D, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286:12743–12755. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- 36.Tsang KY, Chan D, Cheslett D, et al. Surviving endoplasmic reticulum stress is coupled to altered chondrocyte differentiation and function. PLoS Biol. 2007;5:e44. doi: 10.1371/journal.pbio.0050044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugiura K, Muro Y, Futamura K, et al. The unfolded protein response is activated in differentiating epidermal keratinocytes. J Invest Dermatol. 2009;129:2126–2135. doi: 10.1038/jid.2009.51. [DOI] [PubMed] [Google Scholar]
- 38.Firtina Z, Danysh BP, Bai X, Gould DB, Kobayashi T, Duncan MK. Abnormal expression of collagen IV in lens activates unfolded protein response resulting in cataract. J Biol Chem. 2009;284:35872–35884. doi: 10.1074/jbc.M109.060384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reneker LW, Chen H, Overbeek PA. Activation of unfolded protein response in transgenic mouse lenses. Invest Ophthalmol Vis Sci. 2011;52:2100–2108. doi: 10.1167/iovs.10-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, Wang S, Malhotra J, et al. The unfolded protein response transducer IRE1 alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee A-H, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 45.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West DB, Prinz WA, Francendese AA, Greenwood MR. Adipocyte blood flow is decreased in obese Zucker rats. Am J Physiol. 1987;253:R228–R233. doi: 10.1152/ajpregu.1987.253.2.R228. [DOI] [PubMed] [Google Scholar]
- 47.Danforth E. Failure of adipocyte differentiation causes type II diabetes mellitus. Nat Genet. 2000;26:13–13. doi: 10.1038/79111. [DOI] [PubMed] [Google Scholar]
- 48.McGarry JD. Banting Lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51:7–18. doi: 10.2337/diabetes.51.1.7. [DOI] [PubMed] [Google Scholar]
- 49.Gavrilova O, Marcus-Samuels B, Graham D, et al. Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. J Clin Invest. 2000;105:271–278. doi: 10.1172/JCI7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Virtue S, Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the metabolic syndrome -- An allostatic perspective. Biochim Biophys Acta. 2010;1801:338–349. doi: 10.1016/j.bbalip.2009.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.