Abstract
Tel1 is the budding yeast ortholog of the mammalian tumor suppressor and DNA damage response (DDR) kinase ATM. However, tel1-Δ cells, unlike ATM-deficient cells, do not exhibit sensitivity to DNA-damaging agents, but do display shortened (but stably maintained) telomere lengths. Neither the extent to which Tel1p functions in the DDR nor the mechanism by which Tel1 contributes to telomere metabolism is well understood. To address the first question, we present the results from a comprehensive genome-wide screen for genetic interactions with tel1-Δ that cause sensitivity to methyl methanesulfonate (MMS) and/or ionizing radiation, along with follow-up characterizations of the 13 interactions yielded by this screen. Surprisingly, many of the tel1-Δ interactions that confer DNA damage sensitivity also exacerbate the short telomere phenotype, suggesting a connection between these two phenomena. Restoration of normal telomere length in the tel1-Δ xxx-Δ mutants results in only minor suppression of the DNA damage sensitivity, demonstrating that the sensitivity of these mutants must also involve mechanisms independent of telomere length. In support of a model for increased replication stress in the tel1-Δ xxx-Δ mutants, we show that depletion of dNTP pools through pretreatment with hydroxyurea renders tel1-Δ cells (but not wild type) MMS-sensitive, demonstrating that, under certain conditions, Tel1p does indeed play a critical role in the DDR.
Keywords: DNA damage, telomeres, Saccharomyces cerevisiae
THE ATM tumor suppressor kinase is a major signaling component of the DNA damage response (DDR) pathway, and patients with homozygous ATM mutations are afflicted with the cancer-prone disorder ataxia telangiectasia (AT) (Savitsky et al. 1995; Shiloh 2003). ATM-deficient cell lines are sensitive to DNA damage, exhibit pronounced checkpoint and double-strand break (DSB) repair defects (Painter and Young 1980; Kastan et al. 1992; Kuhne et al. 2004), and exhibit significantly reduced phosphorylation levels of DDR targets (Canman et al. 1998). Cells from AT patients exhibit accelerated telomere shortening (Metcalfe et al. 1996), and ATM is thought to play a role in telomere length regulation through interactions with telomere binding proteins (Wu et al. 2007).
The Saccharomyces cerevisiae ortholog for mammalian ATM is TEL1 (Greenwell et al. 1995; Morrow et al. 1995; Mallory and Petes 2000). Tel1p is recruited to DSBs via an interaction with the Mre11-Rad50-Xrs2 (MRX; MRN in mammals) DNA-binding complex (Nakada et al. 2003), and Tel1 both facilitates efficient end resection through an unknown mechanism and participates in phosphorylation of downstream DDR substrates (Mantiero et al. 2007). Following DSB resection, the related kinase Mec1 [ATR in mammals (Cimprich et al. 1996)] recognizes RPA-coated, single-strand DNA (ssDNA) at ssDNA–double-strand DNA (dsDNA) junctions via an interaction with Ddc2, and the DNA damage checkpoint is activated (Paciotti et al. 2000). The distinct sensing of double-strand and single-strand damaged DNA structures by Tel1p and Mec1p bears a striking resemblance to the different roles of their ATM and ATR counterparts in mammalian cells (Zou and Elledge 2003; Lee and Paull 2007). However, while the loss of MEC1 results in severe sensitivity to DNA-damaging agents (Weinert et al. 1994), Tel1p is not functionally required for checkpoint activation in response to intrachromosomal DSBs, and the loss of TEL1 does not significantly sensitize cells to DNA-damaging agents (Greenwell et al. 1995; Morrow et al. 1995). Despite this, a mec1tel1 double mutant is more sensitive to DNA damage than the mec1 single mutant. These results demonstrate that, although MEC1 plays the predominant role at intrachromosomal DSBs, TEL1 does play some role in response to DNA damage in a mec1 background (Morrow et al. 1995).
While Mec1p appears to be the primary responder to DNA damage (with Tel1p functioning in a back-up role), the respective roles of Mec1p and Tel1p are reversed at telomeres. In S. cerevisiae, the telomerase enzyme preferentially associates with short telomeres for elongation through an interaction with Cdc13, and this preferential association is dependent on TEL1 and the MRX complex (Sabourin et al. 2007). MRX recruits Tel1p to DNA ends (Fukunaga et al. 2011), at which Tel1p phosphorylates one or more substrates to facilitate telomerase recruitment by Cdc13 via an as-yet poorly understood mechanism (Gao et al. 2010; Martina et al. 2012). tel1 mutant cells exhibit a decreased frequency of telomere elongation events and decreased telomerase processivity at telomeres (Arneric and Lingner 2007; Chang et al. 2007) that leads to progressive telomere shortening (Greenwell et al. 1995; Mallory and Petes 2000). Telomeres in tel1 cells are shortened but are stably maintained; this depends on MEC1 (Ritchie et al. 1999). Telomere erosion in a mec1tel1 mutant leads to aneuploidy, senescence, and cell death (Craven et al. 2002; Vernon et al. 2008; McCulley and Petes 2010). Despite the requirement for MEC1 in telomere homeostasis in the absence of TEL1, Mec1p is not detected at telomeres in wild-type or tel1-Δ cells, and the specific role that Mec1p plays in facilitating telomere maintenance in the absence of TEL1 is not yet understood (McGee et al. 2010).
For Tel1p’s role in both the DDR and telomere metabolism, significant questions remain. While the kinase was once thought to be functionally redundant with Mec1p in the DDR, recent studies have identified distinct Mec1-independent roles for Tel1p in checkpoint signaling (Mantiero et al. 2007), replication fork stability (Doksani et al. 2009), and the suppression of genome rearrangements (Lee et al. 2008). None of the mechanisms underlying these roles are well understood. At telomeres, the straightforward model consisting of Tel1p phosphorylation of Cdc13 leading to a conformational change that allows for recruitment of the Est1 subunit of telomerase has recently given way to a model of more complex interactions potentially involving multiple kinases, rates of telomere end resection, and other, possibly novel intermediates (Gao et al. 2010; Martina et al. 2012; Wu et al. 2013). Moreover, the mechanism(s) by which MRX and Tel1 are targeted to short telomeres is poorly understood but likely involves constituents of the shelterin complex (Marcand et al. 1997; Teixeira et al. 2004).
Despite recent characterizations of Mec1-independent roles for Tel1p in the DDR, these roles are apparently either nonessential, infrequently utilized, or redundant with other pathways as the fact remains that the loss of TEL1 alone does not confer sensitivity to DNA-damaging agents. To comprehensively characterize the contexts by which Tel1p fits into the DNA damage response, we performed genome-wide screens for TEL1 genetic interactions that cause sensitivity to two different genotoxic agents, methyl methanesulfonate (MMS) and ionizing radiation (IR). From these screens, we have identified a diverse set of mutant backgrounds for which TEL1 is required for survival upon exposure to DNA damage. We report that, despite the diversity of tel1-Δ interactions identified here, most share an additional common phenotype of an exacerbated telomere defect.
Materials and Methods
Media and growth conditions
Yeast-extract-peptone-dextrose (YEPD) and dropout media have been previously described (Paulovich et al. 1998). MMS and hydroxyurea (HU) were purchased from Sigma. YEPD and synthetic plates containing MMS were freshly prepared ~15 hr prior to use.
Yeast strains and plasmids
S. cerevisiae strains used in this study are listed in Table 1. Strain BY4741 and the haploid yeast knockout collection were purchased from Open Biosystems. Plasmid p4339 and strain Y7092 were gifts from Charles Boone and Brenda Andrews. Strain SLY60 was a gift from Sang Eun Lee; strain UCC3508 and plasmid pRS313-Y′ were a gift from Daniel Gottschling, and plasmid pVL1107 was a gift from Vicki Lundblad. All gene disruptions were achieved by homologous recombination at their chromosomal loci by standard polymerase chain reaction (PCR)-based methods (Brachmann et al. 1998). Briefly, a deletion cassette with a 0.5-kb region flanking the target ORF was amplified by PCR from the corresponding xxx::kanMX strain of the deletion array (Open Biosystems) and transformed into the target strain for gene knockout. The primers used in the gene disruptions were designed using 20- to 23-bp sequences that are 0.5 kb upstream and downstream of the target gene. A list of primer sequences for all knockouts used in this study is available upon request from the authors.
Table 1. S. cerevisiae strains.
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| Y7092 | MATα can1Δ::STE2pr-Sp_his5 lyp1Δ his3Δ1 leu2delta0 ura3Δ0 met15Δ0 | Tong and Boone (2006) |
| UCC3508 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 trp1-Δ1/TRP1 leu2-Δ1/leu2-Δ1 adh4::URA3-TEL/adh4::URA3-TEL DIA5-1/DIA5-1 ppr1::HIS3/ppr1::LYS2 TLC1/tlc1:LEU2 | Singer et al. (1998) |
| SLY60 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO | Lee et al. (2008) |
| yBP1020-22 | MATα can1Δ::STE2pr-Sp_his5 lyp1Δ his3Δ1 leu2delta0 ura3Δ0 met15Δ0 tel1::natMX | This study |
| yBP1406-08 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 | This study |
| yBP1416-18 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1:natMX | This study |
| yBP1423 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX | This study |
| yBP1490-91 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 pop2::kanMX | This study |
| yBP1502-04 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 sap30::kanMX | This study |
| yBP1505-07 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX rad17::kanMX | This study |
| yBP1508-10 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX ddc1::kanMX | This study |
| yBP1511-13 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX nup60::kanMX | This study |
| yBP1517-19 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX nup133::kanMX | This study |
| yBP1520-21 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX lsm7::kanMX | This study |
| yBP1524-26 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX sap30::kanMX | This study |
| yBP1527-29 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX hda3::kanMX | This study |
| yBP1550-52 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 rad17::kanMX | This study |
| yBP1553-55 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 nup60::kanMX | This study |
| yBP1558-60 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX rad17::kanMX | This study |
| yBP1564-66 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX nup60::kanMX | This study |
| yBP1576-78 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX sap30::kanMX | This study |
| yBP1585-87 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX hda3::kanMX | This study |
| yBP1608-10 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 nup133::kanMX | This study |
| yBP1611-13 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX nup133::kanMX | This study |
| yBP1622-23 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX pop2::kanMX | This study |
| yBP1630-32 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 rad26::kanMX | This study |
| yBP1633-35 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX rad26::kanMX | This study |
| yBP1636-38 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX rad26::kanMX | This study |
| yBP1669-71 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 ccr4::kanMX | This study |
| yBP1672-74 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX ccr4::kanMX | This study |
| yBP1681-83 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 lsm7::kanMX | This study |
| yBP1684-86 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX lsm7::kanMX | This study |
| yBP1714-16 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 hda3::kanMX | This study |
| yBP1717-19 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 ddc1::kanMX | This study |
| yBP1720-22 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX ddc1::kanMX | This study |
| yBP1738-40 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX pop2::kanMX | This study |
| yBP1787-89 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 fyv4::kanMX | This study |
| yBP1790-92 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX fyv4::kanMX | This study |
| yBP1793-95 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 yku80::kanMX | This study |
| yBP1796-98 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX yku80::kanMX | This study |
| yBP1799-1801 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 rad27::kanMX | This study |
| yBP1802-04 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX rad27::kanMX | This study |
| yBP1805-07 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 rad24::kanMX | This study |
| yBP1808-10 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxt3::URA3 tel1::natMX rad24::kanMX | This study |
| yBP1838-40 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX ccr4::kanMX | This study |
| yBP1841-43 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO yku80::kanMX | This study |
| yBP1844-46 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX yku80::kanMX | This study |
| yBP1847-49 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO rad27::kanMX | This study |
| yBP1850-52 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX rad27::kanMX | This study |
| yBP1859-61 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO rad24::kanMX | This study |
| yBP1862-64 | MATΔ3′:intron:ura3Δ5′ hoΔ hmlΔ:ADE1 hmrΔ:ADE1 ade1-100 leu2-3,112 lys5 trp1:hisG ura3Δ3′:intron:HOcs ade3:GAL:HO tel1::natMX rad24::kanMX | This study |
tel1-Δ double-deletion library construction and screening
The synthetic genetic array (SGA) approach was used to construct a tel1-Δ double-deletion library following the protocol described in Tong and Boone (2006). Library replication was performed using floating-pin manual replicators (VP Scientific). For the IR screen, the library was pin-replicated onto fresh YEPD plates and exposed to gamma irradiation using a Mark II 137Cs irradiator (JL Shepherd & Associates) operated at varying dose rates. Plates were analyzed by manual inspection at 24 and 36 hr following IR. For the MMS screen, the library was pin-replicated onto plates containing 0.01% and 0.03% MMS, grown for 2 days at 30°, and analyzed by visual inspection.
MMS/IR spot and colony assays
For serial-dilution spot assays, log-phase cells were serially diluted in PBS and spotted onto YEPD or YEPD + MMS plates using a pin replicator. A subset of the plates was immediately irradiated using the conditions described above. Plates were incubated at 30° and analyzed by visual inspection at 24 and 36 hr.
For colony-based survival assays, three independent transformants were analyzed for each mutant, along with wild-type and tel1-Δ controls. Log-phase cells (~5 × 107 cells) were sonicated and counted using a Beckman-Dickinson Coulter counter. Cells were serially diluted in PBS and plated onto YEPD or YEPD + MMS plates. For analyzing radiation sensitivity, cells were spread on YEPD plates and the plates were subsequently irradiated as described above. Viability was determined by plating serial dilutions of cultures onto YEPD and scoring the number of colony-forming units (CFU) after 3–4 days at 30°. Viability was calculated as CFU/total cells. For experiments utilizing a low dose rate (0.9 Gy/min), cells were irradiated in 5-ml liquid cultures over a 7.5-hr period prior to plating on YEPD to assess colony-based survival. In experiments in which a HU pretreatment was used, cells were incubated in liquid YEPD media +/− HU (Sigma) at the indicated times. Following the incubation period, cells were washed twice with PBS, counted by Coulter counter, serially diluted, and plated onto YEPD or YEPD + MMS plates.
Gross chromosomal rearrangement and translocation assays
For the measurement of gross chromosomal rearrangement (GCR) frequencies, log-phase cells grown at 30° in YEPD were harvested, sonicated, and counted using a Coulter counter. Cells (1 × 108) were resuspended in 20 ml YEPD and YEPD + 0.003% MMS and grown at 30° overnight. At 15 hr, cells were washed in 5% Na2SO3, sonicated, and counted using a Coulter counter. Cells (1 × 109) were plated onto C-Arg-Ser + canavanine + 5-fluororotic acid (FOA) to measure GCR events, and serial dilutions were plated onto YEPD to measure cell viability. GCR plates were incubated for 4–5 days at 30°. Viability was calculated as CFU/total cells, and MMS-induced GCR frequencies were normalized to GCR frequencies from untreated cells.
The HO-inducible translocation assay was performed according to Lee et al. (2008). Briefly, log-phase cells were sonicated, cell number was determined using a Coulter counter (Beckman Dickinson), and serial dilutions were plated on C-Ura dropout plates containing galactose to induce HO expression. Strain growth and translocations in the absence of HO-induced DSBs were measured on synthetic complete media and C-Ura plates containing glucose.
Southern blotting
Southern blotting for telomere lengths was carried out using a previously described DNA probe targeting telomeric Y′ regions (Singer et al. 1998). DIG-labeled probe synthesis was carried out by PCR using the Roche DIG Probe Synthesis Kit following the manufacturer’s instructions. Genomic DNA was prepared using a Yeastar genomic DNA kit (Zymo Research). Genomic DNA preparations were digested overnight with XhoI (Invitrogen) and separated on 1% gels. Separated DNA molecules were transferred onto nylon membranes via blot sandwich overnight in 20× SSC buffer. DNA molecules were crosslinked onto the membrane using a UV crosslinker (Fisher Scientific) at 60 mJ/cm2, and the membrane was incubated with the Y′ telomeric DIG-labeled probe overnight. Antibody detection of the DIG probe was performed using the DIG luminescent detection kit (Roche), and blots were imaged on a ChemiDoc XRS system (Bio-Rad).
Results
Synthetic genetic array screen for interactions with tel1-Δ in response to MMS and IR
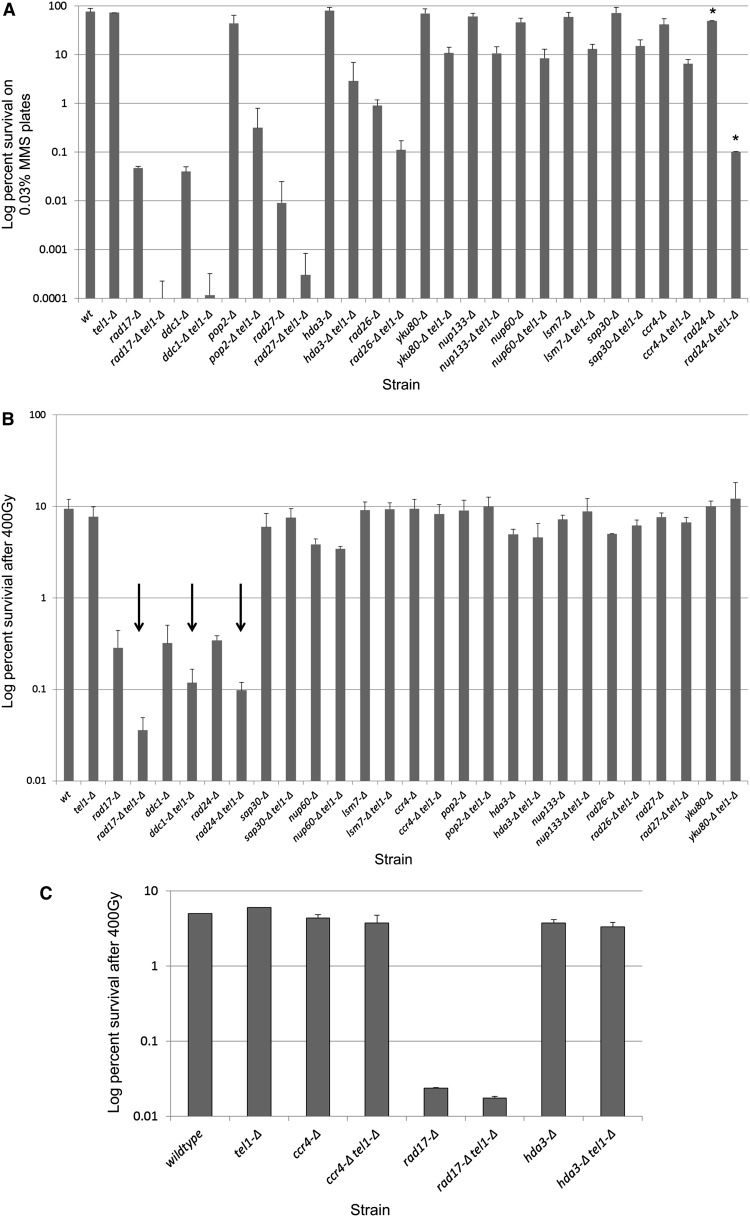
To better understand the extent of Tel1p’s role in the DDR, we sought to characterize mutant backgrounds in which TEL1 is required for survival in response to MMS and/or ionizing radiation. To achieve this, we constructed a genome-wide double-deletion library by mating a MATα tel1-Δ strain to the MATa haploid deletion library (Winzeler et al. 1999) using the SGA procedure developed by Tong et al. (2001) and Tong and Boone (2006). The tel1-Δ xxx-Δ double-deletion library was screened for survival on YEPD plates containing either 0.01% or 0.03% MMS. Plates were examined after 24 and 48 hr by visual inspection for double mutants that exhibited MMS sensitivity. Double-mutant strains exhibiting sensitivity were subsequently spotted in 10-fold serial dilutions along with the parental single-mutant strains on YEPD + MMS to confirm the interaction. As an additional verification step, we remade each single and double mutant by PCR-mediated transformation in a new BY4741 parental haploid strain. These new double-deletion mutants were then retested by serial-dilution spot assay on MMS plates and scored by visual inspection. Interactions passing this second criterion were then subjected to colony survival analyses to quantify the degree of interaction with tel1-Δ on MMS plates. After validation, 13 gene deletions showed enhanced sensitivity to MMS when paired with tel1-Δ (Figure 1A). These genes include multiple subunits of the 9-1-1 checkpoint clamp (RAD17, DDC1; ~400-fold) as well as the 9-1-1 clamp loader RAD24 (deletion of the third subunit mec3-Δ grows poorly in BY4741 and could not be evaluated in the SGA screen) and members of the CCR4-NOT deadenylase complex (CCR4 and POP2; 6- to 130-fold). Additional interactions exhibiting >10-fold increases in MMS sensitivity were between TEL1 and the base excision repair endonuclease RAD27 (~30-fold) and the histone deacetylase (HDAC) subunit HDA3 (~30-fold). Additional genes exhibiting <10-fold interactions with tel1-Δ consisted of two nucleoporins (NUP60 and NUP133), the nonhomologous end-joining (NHEJ) repair factor YKU80, a second HDAC subunit (SAP30), the RAD26 ATPase, and a member of the Sm-like mRNA decay family (LSM7). We note that, in the initial and confirmative screens, an additional tel1-Δ interaction with the uncharacterized FYV4 gene exhibited a growth defect with tel1-Δ as well as a >10-fold increase in MMS sensitivity. However, the FYV4 ORF is located ~200 bp upstream of the transcription start site of the essential mediator subunit MED6. Transforming the fyv4-Δ tel1-Δ strain with a plasmid containing the MED6 gene and its promoter completely abolished the growth defect and MMS sensitivity of this strain (data not shown), leading us to conclude that the fyv4-Δ gene replacement exerts an off-target effect on the essential MED6 gene. Due to these complications, the FYV4/MED6 candidate was removed from further consideration in this study.
Figure 1.
Quantitative survival analysis for tel1 interactions in MMS and IR via colony-forming assay. (A) Quantitative survival analysis in MMS. Log-phase cultures for three independent transformants of each single and double mutant were serially diluted in PBS and spread onto YEPD or YEPD + 0.03% MMS plates (asterisks indicate that screening was done in 0.015% MMS due to extreme MMS sensitivity). Viable cells were determined by the number of CFU after 3 days at 30°. (B) Quantitative survival analysis in IR. Log-phase cultures for three independent transformants of each single and double mutant were serially diluted in PBS and spread onto YEPD plates and irradiated at 400 Gy at 8 Gy/min. Viable cells were determined by the number of CFU after 3 days at 30°. Arrows indicate interactions identified in the genome-wide screen. Error bars represent the standard deviation of values from three independent transformants. (C) Quantitative survival analysis using continuous low-dose-rate IR. Log-phase cultures for two independent transformants of each single and double mutant were diluted in YEPD in 15-ml tubes and irradiated with 400 Gy delivered at a continuous dose rate of 0.9 Gy/min over 7.5 hr. Following delivery of IR, cells were counted, serially diluted, and plated for colony survival analysis. Error bars show the range of values for two independent transformants.
As our MMS screen revealed a diverse set of interactions that cause enhanced MMS sensitivity with tel1-Δ, we asked whether a different set of mutants would interact with tel1-Δ in response to a different DNA-damaging agent, γ-irradiation. To test for genetic interactions with tel1-Δ in ionizing radiation, the tel1-Δ xxx-Δ double-deletion library was plated onto YEPD and exposed to either 200 or 400 Gy of ionizing radiation. In contrast to the MMS screen, only the 9-1-1 checkpoint genes rad17-Δ, ddc1-Δ, and rad24-Δ exhibited interactions with tel1-Δ in response to IR, and these interactions were minor (<10-fold) in comparison to the 9-1-1-Δ tel1-Δ interactions in MMS (>100-fold) (Figure 1B). To confirm that the tel1-Δ xxx-Δ interactions identified in the MMS sensitivity screen were indeed not also sensitive to IR, we tested each of the 13 MMS-sensitive tel1-Δ xxx-Δ strains for IR sensitivity. Consistent with the screen results, only the 9-1-1-Δ tel1-Δ double mutants exhibited enhanced IR sensitivity (Figure 1B).
As MMS is often referred to as a “radiomimetic” agent, the finding that many of the MMS interactions were not recapitulated using IR was unexpected. One possible explanation for this is that the 400 Gy of IR was delivered as a pulse over a short period of time (8 Gy/min), while for MMS treatment cells were grown continuously in 0.03% MMS. [DNA damage phenotypes can differ significantly when the agent is delivered as a pulse or chronic treatment (Murakami-Sekimata et al. 2010).] To test this hypothesis, three of the tel1-Δ xxx-Δ double mutants identified in our screen (ccr4-Δ tel1-Δ, hda3-Δ tel1-Δ, and rad17-Δ tel1-Δ) were examined for sensitivity to the same 400-Gy cumulative dose of IR (as in Figure 1B), but this time delivered chronically over a period of 7.5 hr (0.9 Gy/min). As seen in Figure 1C, the total IR sensitivity for wild-type and single-mutant strains was increased somewhat in the chronic exposure relative to the pulse of 400 Gy (Figure 1B); however, no additional (i.e., aside from 9-1-1) interactions with tel1-Δ were observed, and the rad17-Δ tel1-Δ interaction was reduced. From this, we conclude that, unlike the MMS case, tel1-Δ interactions in IR are limited to mutations in the 9-1-1 pathway.
Loss of telomerase is associated with a progressive increase in MMS sensitivity
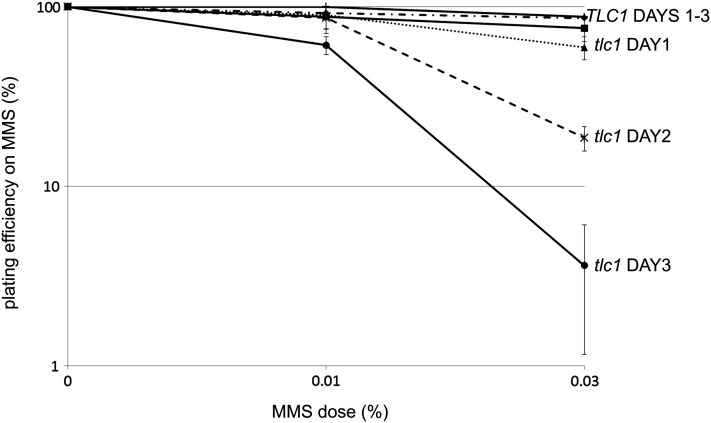
Mammalian cells with shortened telomeres exhibit increased sensitivity to DNA-damaging agents via an as-yet-unknown mechanism (Goytisolo et al. 2000; Wong et al. 2000; Gonzalez-Suarez et al. 2003; Nakamura et al. 2005; Agarwal et al. 2008; Soler et al. 2009; Drissi et al. 2011; Woo et al. 2012). Based on this precedent, we hypothesized that resistance to DNA-damaging agents in yeast would also be tightly linked to telomere length and that yeast cells would become more sensitive to DNA damage in a progressive manner as telomeres shorten. To evaluate this possibility, we employed a heterozygous diploid tlc1 strain, which, upon sporulation into haploid progeny, exhibits progressive telomere shortening that leads to eventual replicative senescence (Singer and Gottschling 1994). After inducing sporulation, we subcultured TLC1 and tlc1 haploid progeny over a series of days, and each day we removed an aliquot of cells for testing of survival on YEPD or YEPD + MMS plates. In the absence of MMS, tlc1 strains exhibited progressive telomere shortening over the 3-day period, while telomere lengths in the TLC1 strains remained unchanged over the same period (data not shown). When tested for viability on plates containing either 0.01% or 0.03% MMS, TLC1 strains showed minimal MMS sensitivity that was unchanged over the course of the experiment (Figure 2). In contrast, the tlc1 mutant strains exhibited a progressive and dose-dependent increase in MMS sensitivity that was most pronounced by day 3 on 0.03% MMS plates (survival rates in MMS were normalized to rates on YEPD alone to correct for MMS-independent loss of viability). As the half-life of telomerase RNA is a few hours (Chapon et al. 1997) and the MMS sensitivity manifests after days, we conclude that MMS sensitivity is telomere length-dependent in tlc1 cells, rather than due to TLC1 loss alone. These results raised the interesting possibility that the DNA damage sensitivity exhibited by the identified tel1-Δ xxx-Δ interactions results from an exacerbation of the well-known telomere length defect caused by loss of TEL1.
Figure 2.
Telomerase-null cells exhibit a progressive increase in MMS sensitivity. TLC1 and tlc1 haploid spores from freshly dissected tetrads were subcultured in YEPD over multiple days. Each day, an aliquot was removed and assayed for MMS sensitivity by colony-forming assay. Error bars represent the standard deviation of values from three independent spores.
Many of the tel1-Δ MMS interactions exhibit shortened telomeres
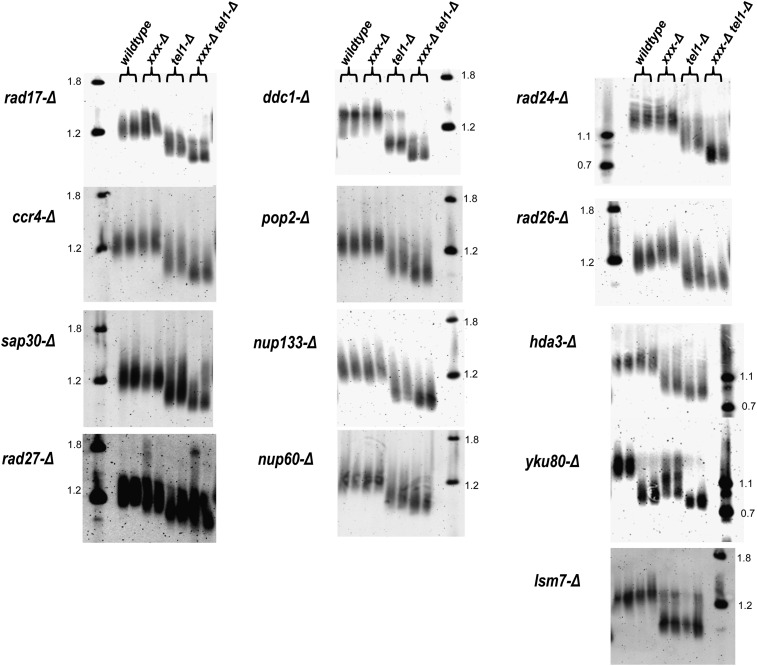
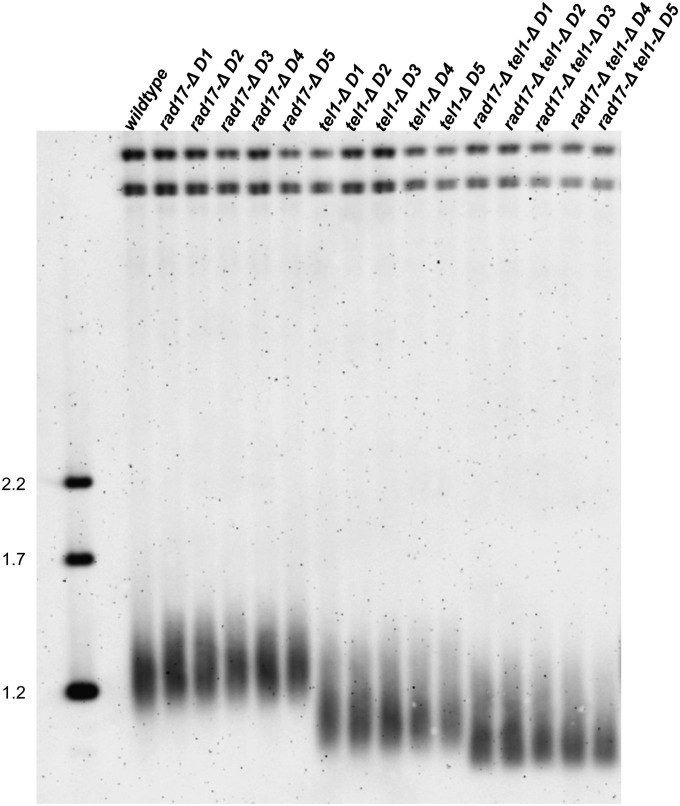
As cellular sensitivity to MMS increases progressively with telomere shortening (Figure 2), we hypothesized that some or all of the interactions identified in the tel1-Δ screen exacerbate the tel1-mediated telomere length defect and that this may be a cause for DNA damage sensitivity in these cells. Thus we asked whether any of the tel1-Δ xxx-Δ double mutants exhibited telomere lengths that were significantly shorter than either corresponding single mutant. To answer this question, we isolated genomic DNA from single and double mutants for each of the 13 tel1-Δ xxx-Δ interactions and analyzed XhoI fragments by Southern blotting with a Y′ subtelomeric probe (Singer et al. 1998). As expected (Lustig and Petes 1986; Greenwell et al. 1995; Morrow et al. 1995), the tel1-Δ single mutant exhibited shorter telomere lengths relative to a wild-type strain (Figure 3). Additionally, a number of the other single mutants exhibited shorter telomere lengths relative to the wild type, including yku80-Δ, rad27-Δ, and sap30-Δ, with the yku80-Δ mutant being the only single mutant exhibiting a shorter telomere length than tel1-Δ (Figure 3). Notably, the 9-1-1 mutants ddc1-Δ and rad17-Δ were shown in a previous study to exhibit a minor telomere defect (Longhese et al. 2000). However, we did not observe discernible shortening of these mutants relative to the wild type (Figures 3 and 4); this may reflect differences in the strain background used in these studies. Notably, the 9-1-1-Δ tel1-Δ double mutants (rad24-Δ tel1-Δ, rad17-Δ tel1, and ddc1-Δ tel1-Δ) exhibited very short telomeres relative to tel1-Δ, and a second class consisting of sap30-Δ tel1-Δ, ccr4-Δ tel1-Δ, pop2-Δ tel1-Δ, hda3-Δ tel1-Δ, nup133-Δ tel1-Δ, nup60-Δ tel1-Δ, rad27-Δ tel1-Δ, and yku80-Δ tel1-Δ also exhibited shorter telomeres relative to tel1-Δ. The rad26-Δ tel1-Δ and lsm7-Δ tel1-Δ double mutants exhibited telomere lengths that were identical to tel1-Δ. Our finding that 11 of the 13 tel1-Δ xxx-Δ interactions exhibited decreased telomere lengths relative to tel1-Δ is unexpected, since many of identified genes play no known role in telomere metabolism. To exclude the possibility that the tel1-Δ xxx-Δ short telomere phenotype was not merely an artifact due to a previously characterized phenotypic lag for tel1 telomeres [~150 generations (Lustig and Petes 1986)], we examined telomere lengths for a selection of single and double mutants over additional subculturing for a period of 5 days. During the repeated subculturing, we did not observe any further changes in telomere length by Southern blot (Figure 4). From these data, we conclude that the majority of tel1-Δ interactions identified in the MMS sensitivity screen also confer shorter telomeres, suggesting a possible connection between the two phenotypes.
Figure 3.
Telomere lengths for tel1-Δ MMS-sensitive interactions. For each strain, XhoI-digested DNA was analyzed by Southern blot using a probe complementary to the Y′ subtelomere. Each xxx-Δ mutant is listed to the left of each corresponding blot, and duplicates representing independent transformants for each strain are loaded side by side (duplicates are indicated by the brackets above). DNA ladders (in kb) are indicated in the far left or right lane of each blot.
Figure 4.
Repeated subculturing does not alter telomere lengths. Wild-type, tel1-Δ, rad17-Δ, and rad17-Δ tel1-Δ cultures were diluted in fresh YEPD media and grown overnight. Genomic DNA was harvested the following day, and a portion of the cells was diluted in fresh medium and cultured overnight. The process was repeated over a period of 5 days (D1–D5). XhoI-digested DNA was analyzed by gel electrophoresis and Southern blotting with a probe recognizing subtelomeric Y′ sequence. Molecular weight markers are indicated on the left (in kb).
Artificial elongation of telomeres in tel1-Δ 9-1-1-Δ mutants partially suppresses MMS sensitivity
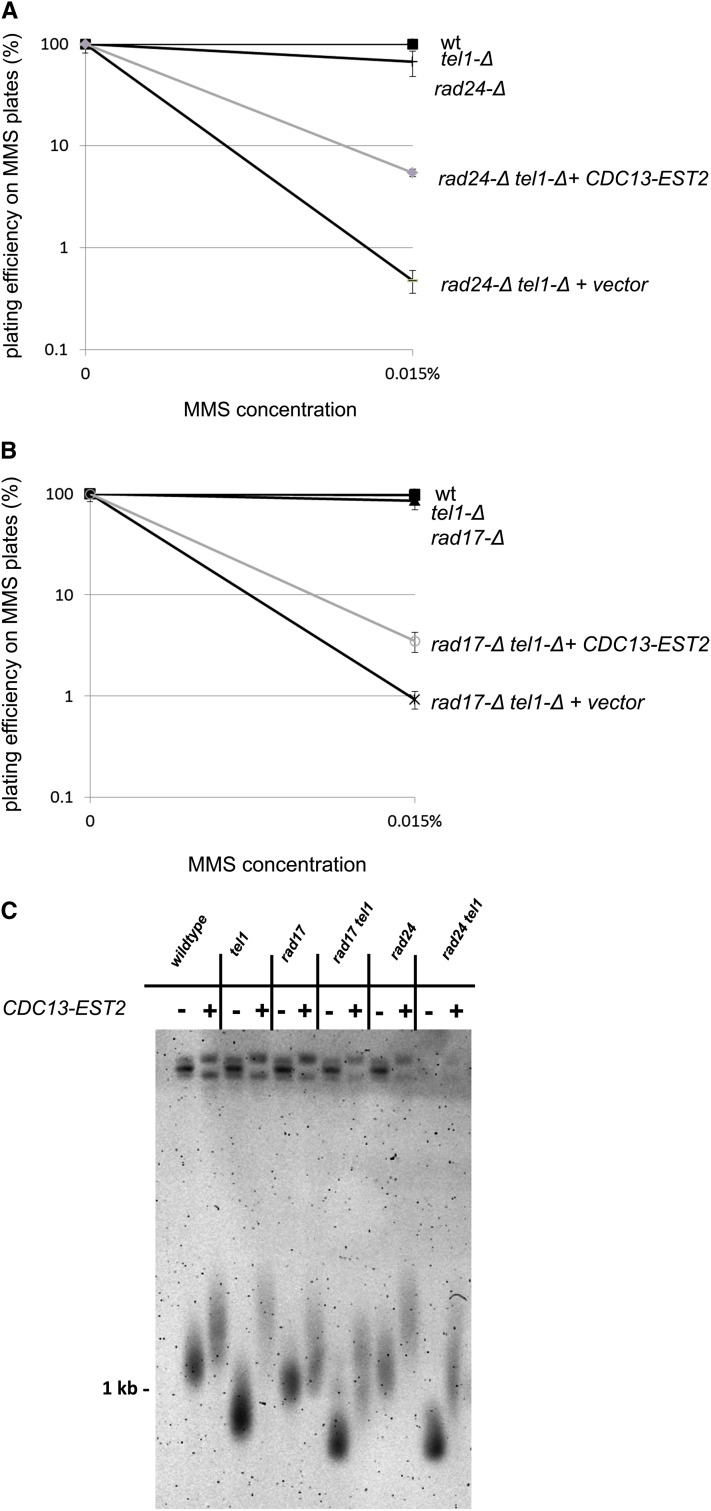
As telomere shortening was shown to be causative for MMS sensitivity in the tlc1 case (Figure 2), we next hypothesized that the exacerbated telomere defect exhibited by the majority of tel1-Δ xxx-Δ strains (Figure 3) may be causative for enhanced MMS sensitivity. Thus, reversal of the telomere length defect would also reduce the MMS sensitivity of these mutants. To test this, we transformed each of the tel1-Δ xxx-Δ single and double mutants with a plasmid expressing a fusion of the Cdc13 capping protein to the Est2 subunit of telomerase (Evans and Lundblad 1999). This fusion has been previously shown to alleviate the short telomere phenotype in a tel1 mutant (Tsukamoto et al. 2001). A panel of tel1-Δ xxx-Δ mutant strains with and without the CDC13-EST2 plasmid was screened for sensitivity by spotting cells on MMS plates (supporting information, Figure S1). Of the tested tel1-Δ xxx-Δ interactions, the rad24-Δ tel1-Δ strain exhibited a visible increase in survival on MMS plates when transformed with the CDC13-EST2 fusion plasmid (and not the vector). None of the other tel1-Δ xxx-Δ interactions exhibited any change in MMS sensitivity upon transformation with CDC13-EST2. We confirmed the suppression of MMS sensitivity in rad24-Δ tel1-Δ as well as a second 9-1-1 component (rad17-Δ tel1-Δ) by a quantitative colony-forming assay (Figure 5, A and B), and again the fusion plasmid conferred discernible (but not total) resistance to MMS (11-fold for rad24-Δ tel1-Δ and 4-fold for rad17-Δ tel1-Δ vs. the vector). Telomeres in these strains were significantly elongated to wild-type levels by addition of the CDC13-EST2 fusion and were hyper-elongated in wild-type and tel1-Δ strains (Figure 5C). (CDC13-EST2 was able to elongate telomeres to an identical degree in other non-9-1-1-related tel1-Δ xxx-Δ interactions (not shown) despite having no effect on MMS resistance.) While expression of CDC13-EST2 suppresses strong (>100-fold) MMS sensitivity in 9-1-1-Δ tel1-Δ interactions by ~10-fold (Figure 5), the fact that this suppression is not total and that CDC13-EST2 expression does not affect the MMS sensitivity of the other tel1-Δ xxx-Δ interactions suggests that there are additional telomere-length-independent defects that contribute to the MMS sensitivity of tel1-Δ xxx-Δ interactions.
Figure 5.
Suppression of MMS sensitivity in rad24-Δ tel1-Δ and rad17-Δ tel1-Δ by a CDC13-EST2 fusion plasmid. (A and B) Strains of the indicated genotype were transformed either with an empty vector or with a CDC13-EST2 fusion plasmid (pVL1107) and screened for MMS sensitivity by colony-forming assay on MMS plates. Error bars represent the standard deviation of values from three independent transformants. (C) Telomere lengths for tel1-Δ interactions with or without the CDC13-EST2 fusion plasmid. Cells with the CDC13-EST2 fusion plasmid or empty vector were propagated on –Leu media and diluted in fresh rich medium overnight. Genomic DNA was harvested the following day and analyzed by electrophoresis and Southern blotting. The blot was probed with sequence complementary to a region in the Y′ subtelomeric element.
tel1-Δ xxx-Δ interactions do not affect the frequency of NHEJ-mediated translocations
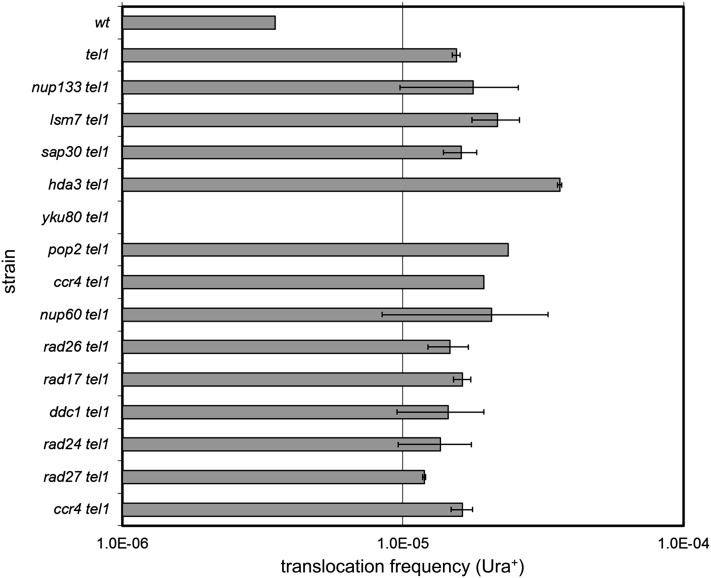
Lee et al. (2008) previously described an 11-fold increase in the frequency of DSB-induced, NHEJ-mediated translocations for a tel1-Δ mutant, reflecting a role for TEL1 in preventing deleterious chromosomal fusions through an as-yet-undefined mechanism. That a tel1-Δ strain is not sensitive to DNA-damaging agents despite this defect suggests that the occurrence of these events even in the presence of DNA-damaging agents is a rarity. Thus, one possibility is that the tel1-Δ xxx-Δ interactions identified here may increase cellular dependence on TEL1 to prevent deleterious chromosomal fusions. We tested this possibility by determining whether the tel1-Δ xxx-Δ double mutants experience enhanced frequencies (compared to tel1) of chromosomal translocations. We cloned each of the 13 single mutants and tel1-Δ xxx-Δ double mutants into a strain background harboring the translocation assay construct (Lee et al. 2008) that employs two GAL-inducible HO cuts on chromosomes V and VII, where each breakpoint contains a nonfunctional fragment of the URA3 gene. Translocations are measured by the reconstitution of a functional URA3 allele, which is dependent on Ku70/80-mediated NHEJ (Lee et al. 2008). We measured the frequency of translocations after the induction of GAL-HO for the panel of tel1-Δ xxx-Δ interaction strains (Figure 6). While we were able to reproduce the Ku-dependent increase in Ura+ translocations for tel1-Δ, none of the other double mutants exhibited frequencies that differed from tel1-Δ. From this we conclude that an increased frequency of DSB-induced chromosomal translocations is unlikely to be the cause of the MMS sensitivity exhibited by tel1-Δ xxx-Δ interactions. This is supported by the fact that the tel1-Δ xxx-Δ MMS interactions were also largely insensitive to IR (Figure 1), which directly induces DSBs [whether or not MMS produces DSBs is a current source of controversy (Lundin et al. 2005)].
Figure 6.
HO-induced translocation frequency for tel1-Δ interactions. NHEJ-mediated translocation frequency for tel1-Δ double mutants following GAL-HO induction of DSBs on chromosome V and chromosome VII. Frequencies are measured as the fraction of colonies that survive on −Ura plates. Error bars indicate the standard deviation of values from three independent transformants.
GCRs in tel1-Δ xxx-Δ strains
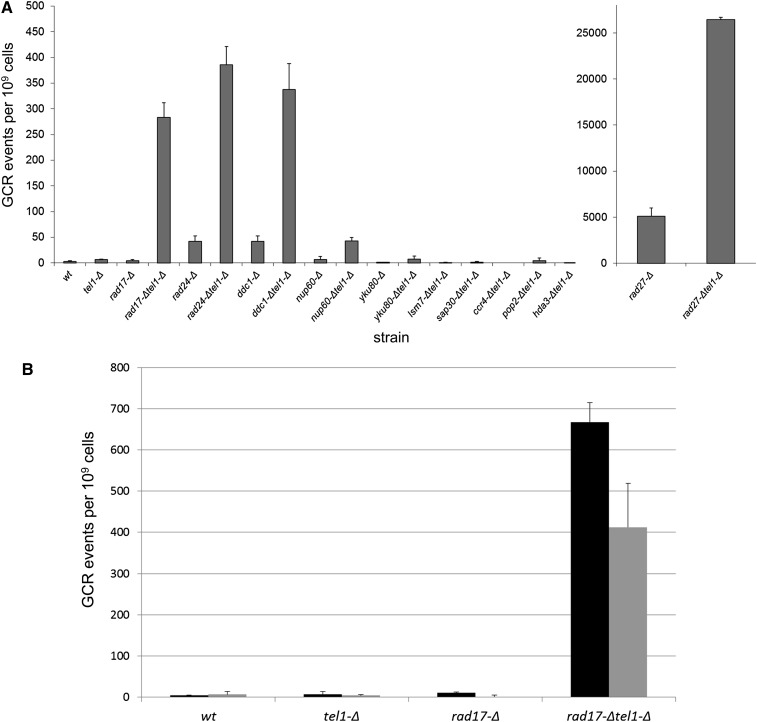
Kolodner and colleagues have previously shown that one double mutant identified in this screen (rad24tel1) causes an increased frequency of spontaneous chromosome breakage and rearrangement involving the left arm of chromosome V (the GCR arrangement assay) (Myung and Kolodner 2002). As MMS has also been shown to induce higher GCR frequencies (Myung and Kolodner 2003; Stellwagen et al. 2003), we asked whether the MMS sensitivity exhibited by the tel1-Δ xxx-Δ double mutants may reflect an increased frequency of MMS-induced genome rearrangements. To do this, we grew single- and double-mutant strains in the presence of 0.003% MMS for 15 hr to induce GCR events, which were detected by selecting for the loss of two nearby markers (CAN1 and URA3) on the left arm of chromosome V, as previously described (Chen and Kolodner 1999). The 0.003% MMS exposure resulted in an ~10-fold induction of GCR events for wild-type cells. For the tel1-Δ xxx-Δ double mutants, members of the 9-1-1 complex showed an ~300-fold induction of MMS-induced GCR events when combined with tel1-Δ (Figure 7A). The rad27-Δ tel1-Δ mutant and nup60-Δ tel1-Δ each showed a minor ~5-fold increase in MMS-induced GCR. None of the other double mutants exhibited an increased GCR frequency (Figure 7A). From these data, we conclude that a subset of tel1-Δ xxx-Δ interactions (rad17-Δ tel1-Δ, ddc1-Δ tel1-Δ, rad24-Δ tel1-Δ, rad27-Δ tel1-Δ, and nup60-Δ tel1-Δ) exhibit increased genome instability as measured by the GCR assay.
Figure 7.
GCR frequency in 0.003% MMS. (A) GCR frequency for tel1-Δ interactions. Strains were grown in YEPD + 0.003% MMS for 15 hr and subsequently plated onto C-Arg-Ser + canavanine + 5-FOA to simultaneously select for the loss of CAN1 and URA3 markers on the left end of chromosome V. Error bars represent standard deviations from two independent cultures per strain, each plated two times. The rad27-Δ mutants are plotted separately due to scale. (B) GCR frequency in 0.003% MMS with or without the CDC13-EST2 fusion plasmid. The indicated strains containing either an empty vector (black bars) or the CDC13-EST2 fusion (gray bars) were grown in YEPD + 0.003% MMS for 15 hr and subsequently plated onto C-Arg-Ser + canavanine + 5-FOA to select for simultaneous loss of CAN1 and URA3 markers. Error bars represent standard deviations from two independent cultures per strain, each plated two times.
As restoration of telomere lengths through addition of the CDC13-EST2 fusion plasmid restored a proportion of MMS resistance to the 9-1-1-Δ tel1-Δ mutant strains, we asked whether the CDC13-EST2 fusion would also suppress the increased MMS-induced GCR frequency of a 9-1-1-Δ tel1-Δ mutant as well. We tested a rad17-Δ tel1-Δ mutant along with the corresponding single mutants for the induction of GCR events with or without the fusion construct. As can be seen in Figure 7B, the rad17-Δ tel1-Δ double-mutant strain harboring the fusion plasmid had a reduced GCR frequency relative to the same strain carrying an empty vector. Consistent with a partial reduction in MMS sensitivity, the CDC13-EST2 fusion did not completely abolish MMS-induced gross chromosomal rearrangements in the rad17-Δ tel1-Δ strain. From these data, we conclude that a proportion of the MMS sensitivity exhibited by 9-1-1-Δ tel1-Δ strains is due to MMS-induced genomic instability that is caused by telomere shortening. However, much of the increased GCR in 9-1-1-Δ tel1-Δ is unexplained by telomere length effects; thus additional mechanisms (i.e., aside from altered telomere length) contribute to the sensitivity of tel1-Δ xxx-Δ interactions.
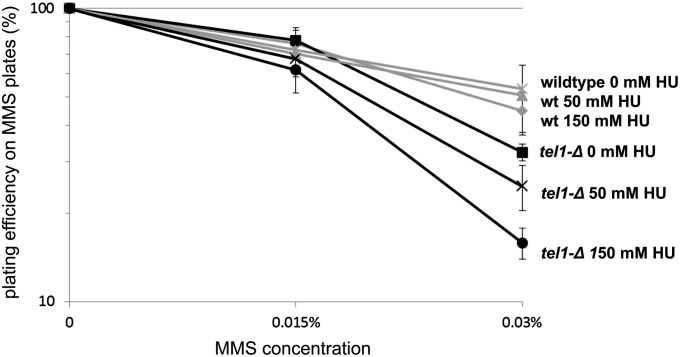
A tel1-Δ strain is rendered sensitive to MMS by predepletion of nucleotide pools
Prior studies have implicated both the 9-1-1 complex and the CCR4-NOT complex as key regulators of ribonucleotide reductase, and mutants in these pathways exhibit depleted nucleotide pools and are sensitive to replication stress (Zhao et al. 2001; Westmoreland et al. 2004; Mulder et al. 2005; Traven et al. 2005; Woolstencroft et al. 2006). Moreover, a ccr4-Δ tel1-Δ strain has been previously shown to exhibit enhanced sensitivity to the ribonucleotide reductase inhibitor hydroxyurea (Woolstencroft et al. 2006). Thus we hypothesized that a decrease in dNTP pools in 9-1-1-Δ tel1-Δ and ccr4-Δ/pop2-Δ tel1-Δ may contribute to the MMS sensitivity exhibited by these strains. From this, we predicted that depletion of nucleotide pools (e.g., by pretreating cells with hydroxyurea) in tel1-Δ cells should phenocopy deletion of CCR4 in a tel1-Δ background, thus sensitizing tel1-Δ cells to MMS. To test this prediction, wild-type and tel1-Δ cells were cultured in rich medium with 0, 50, or 150 mM HU for a period of 4 hr, after which the HU was removed, and cells were plated onto MMS plates to assess viability. As expected, the MMS sensitivity of a wild-type strain does not change, regardless of whether the cells have been pretreated with HU (Figure 8). In contrast, while a tel1-Δ strain is insensitive to the HU pretreatment alone, when HU pretreatment is followed by plating on MMS plates, tel1-Δ cells exhibit enhanced MMS sensitivity in a dose-dependent manner, with the greatest MMS sensitivity observed in 150 mM HU (Figure 8). From this we conclude that depletion of nucleotide pools renders tel1-Δ sensitive to the DNA-damaging agent MMS, consistent with a model in which increased replication stress contributes to the MMS sensitivity exhibited by the 9-1-1-Δ tel1-Δ and ccr4-Δ tel1-Δ/pop2-Δ tel1-Δ mutants (and possibly other tel1-Δ xxx-Δ double mutants isolated in the screen; see Discussion).
Figure 8.
MMS sensitivity following pretreatment with HU. The indicated strains were grown in the presence of the indicated dose of HU for 4 hr to deplete nucleotide pools. Cells were then washed two times and plated onto MMS plates to assay MMS sensitivity by colony-forming assay. The error bars indicate the standard deviation of values from three independent cultures.
Discussion
Categorizing the tel1-Δ interactions
While a tel1-Δ mutant exhibits interactions with a diverse set of 13 mutants, we found that these interactions fell into three phenotypic classes based upon our follow-up characterizations (Table 2). The first class is composed of mutants in the 9-1-1 complex (rad17-Δ and ddc1-Δ) and the 9-1-1 clamp loader (rad24-Δ); these tel1-Δ interactions conferred a rather large (>100-fold) increase in MMS sensitivity (Figure 1A), cross-sensitivity to IR (Figure 1, B and C), a pronounced telomere defect (Figure 3), and a synergistic increase in GCR events (Figure 7). For this class, the DNA damage sensitivity and the increase in GCR frequencies were partially suppressed by elongating telomeres using the CDC13-EST2 fusion construct (Figures 5 and 7B). The second class of interactions comprises ccr4-Δ, pop2-Δ, sap30-Δ, hda3-Δ, yku80-Δ, rad27-Δ, nup133-Δ, and nup60-Δ (Table 2); these exhibited a somewhat milder interaction with tel1-Δ in MMS and no cross-sensitive interactions to IR, but exhibited a discernible telomere length defect with tel1-Δ (Figure 3). The third class of mutants, rad26-Δ and lsm7-Δ, showed similar characteristics to class 2, but did not exhibit any discernible telomere length defect (Figure 3). There is likely some overlap between these classes in the mechanism that causes their interactions with tel1-Δ (discussed below).
Table 2. Tabulation of phenotypes for tel1-Δ interactions identified in the MMS screen.
| Strain | MMS interaction | IR interaction | GCR | Short telomere | CDC13-EST2 rescue | Description | |
|---|---|---|---|---|---|---|---|
| Class 1 | rad24-Δtel1-Δ | ++ | + | ++ | ++ | +/− | 9-1-1 complex |
| rad17-Δtel1-Δ | ++ | + | ++ | ++ | +/− | 9-1-1 complex | |
| ddc1-Δtel1-Δ | ++ | + | ++ | ++ | +/− | 9-1-1 complex | |
| Class 2 | nup60-Δtel1-Δ | + | — | + | + | — | Nucleoporin |
| rad27-Δtel1-Δ | + | — | + | + | — | Flap endonuclease | |
| sap30-Δtel1-Δ | + | — | — | + | — | Deacetylase | |
| pop2-Δtel1-Δ | + | — | — | + | — | Deadenylase | |
| ccr4-Δtel1-Δ | + | — | — | + | — | Deadenylase | |
| hda3-Δtel1-Δ | + | — | — | + | — | deacetylase | |
| yku80-Δtel1-Δ | + | — | — | + | — | NHEJ | |
| nup133-Δtel1-Δ | + | — | — | + | — | Nucleoporin | |
| Class 3 | lsm7-Δtel1-Δ | + | — | — | — | — | mRNA decap |
| rad26-Δtel1-Δ | + | — | — | — | — | TCR |
For each of the tel1-Δ xxx-Δ genetic interactions identified in the MMS screen a “+” indicates whether a double mutant exhibited a positive result in each of the assays tested (e.g., increased GCR frequency, shorter telomere, etc.); a “++” indicates a more severe phenotype; and a “+/−” indicates partial suppression. TCR, transcription-coupled repair.
A replication defect underlies sensitivity to MMS in multiple classes of tel1-Δ interactions
While the tel1-Δ interactions composing classes 1 and 2 (Table 2) exhibit shortened telomeres relative to the corresponding single mutants, only in the class 1 case is MMS sensitivity suppressed by telomere elongation (Figure 5), and even in this class of double mutants the suppression is modest. While it is formally possible that telomere elongation due to the expression of the CDC13-EST2 fusion creates a structure that is somehow physiologically different from a natural telomere and thus is not a good substitute, a more straightforward model is that only a minor proportion of MMS sensitivity is directly caused by telomere shortening in class 1 mutants, while the majority of MMS sensitivity in these and other tel1-Δ xxx-Δ interactions reflects an underlying replication defect that manifests a dual-pronged effect on telomere metabolism and MMS resistance.
Our data (and other studies) support a model in which increased replication stress, combined with a tel1-Δ-mediated defect in replication fork stability, causes both MMS sensitivity and telomere shortening in tel1-Δ xxx-Δ interactions. First, aside from a modest effect in the class 1 mutants, none of the identified in tel1-Δ xxx-Δ interactions exhibit cross-sensitivity to ionizing radiation, regardless of whether the IR was administered as a pulse (Figure 1B) or as chronic treatment (Figure 1C). Unlike MMS treatment, IR does not induce detectable replication fork stalling (Merrick et al. 2004), so while there is a minor IR interaction in tel1-Δ 9-1-1-Δ cells (Figure 1, B and C) (likely through an additive defect in Mec1/Tel1 DSB sensing), replication fork stalling/collapse is the likely major lethal lesion in tel1-Δ xxx-Δ interactions. Additionally, recent studies have uncovered a TEL1-dependent role in the preservation of fork stability through the prevention of fork reversion and degradation into abnormal cruciform structures (Doksani et al. 2009). Consistent with this, Kaochar et al. (2010) showed that tel1-Δ exhibits an increased frequency of dicentric chromosomes due to the fusion of inverted repeats likely due to fork reversion. As the reason why tel1-Δ cells exhibit short telomeres is poorly understood, it is formally possible that a failure to preserve fork stability in telomeric regions in tel1-Δ cells is causative for the short telomere phenotype [telomeres are enriched for replication pause sites such as G-quadruplex structures (Ivessa et al. 2002; Bochman et al. 2012)].
Many of the tel1-Δ interactions identified in the MMS screen fit a model for increased replication stress. Members of class 1 (9-1-1 components) (Table 2) are required for the MEC1-dependent degradation of the ribonucleotide reductase inhibitor Sml1 following MMS treatment (Zhao et al. 2001; Chabes et al. 2003); the resultant increase in dNTP production following this process is thought to facilitate DNA synthesis at stalled forks to prevent fork collapse (Fasullo et al. 2010). In addition, members of class 2 (CCR4 and POP2, members of the CCR4-NOT deadenylation complex) are known regulators of ribonucleotide reductase, and mutants in ccr4-Δ and pop2-Δ are sensitive to replication inhibitors such as HU (Westmoreland et al. 2004; Mulder et al. 2005; Traven et al. 2005; Woolstencroft et al. 2006). As telomere shortening has recently been shown to occur upon dNTP depletion (Gupta et al. 2013), it is likely that the short telomeres in CCR4-NOT and 9-1-1 mutants are at least partially due to this mechanism. Consistent with a model for increased replication stress in tel1-Δ xxx-Δ cells, depleting nucleotide pools by pretreatment with HU (effectively phenocopying the loss of 9-1-1 or CCR4/POP2) sensitizes tel1-Δ cells to MMS in a dose-dependent manner, whereas a wild-type strain is unaffected by the HU pretreatment (Figure 8).
Other mutants composing class 2 are also linked to preventing replication stress via counteracting fork regression [RAD27 (Kang et al. 2010)] or stabilizing sites of active transcription [NUP60/NUP133 (Palancade et al. 2007; Bermejo et al. 2011)]. Additionally, the mutants composing class 3 (Table 2) are linked to increased replication stress due to defects in histone regulation [LSM7 (Herrero and Moreno 2011; Tkach et al. 2012)] or through defective targeting of transcription-coupled repair [RAD26 (Kapitzky et al. 2010; Malik et al. 2010)].
Progressive telomere shortening is a cause for MMS sensitivity
Recently, numerous studies have described a connection between short telomeres and enhanced sensitivity to DNA-damaging agents across a variety of organisms (Wong et al. 2000; Lin et al. 2009; Soler et al. 2009; Drissi et al. 2011); the reason for this relationship is poorly understood. Here, we show that, in yeast, cellular sensitivity to MMS progressively increases as telomeres shorten (Figure 2), suggesting that the progressive loss of telomere protection renders cells sensitive to MMS. In concordance with this, a proportion of MMS sensitivity and genome instability can be suppressed in 9-1-1-Δ tel1-Δ mutants by alleviating the short telomere phenotype in these cells (Figures 5 and 7B).
There are multiple possible mechanisms for how short telomeres cause MMS sensitivity. Loss of telomeric protection can render telomeres as targets for the DDR, and the loss of telomerase activity is associated with a gradual increase in constitutive Rad53 phosphorylation (Grandin et al. 2005); accordingly, in telomerase-deficient cells telomeres are enriched for DDR proteins while nontelomeric DSBs exhibit reduced binding of DDR factors (Lin et al. 2009). Thus, the recruitment of DDR factors to short telomeres may interfere with the ability of the cell to cope with MMS-induced stress elsewhere in the genome. Alternatively, de-protected telomeres themselves may be problematic in the presence of MMS due to the potential for lethal chromosomal fusions with DSBs resulting from MMS-induced collapsed forks. Supporting this, a subset of GCR events can be suppressed by elongating telomeres in 9-1-1-Δ tel1-Δ (Figure 7B), and a previous study has shown that a 9-1-1-Δ tel1-Δ double mutant exhibits an increased frequency of spontaneous telomere–telomere fusions that can also be suppressed by elongating telomeres (Mieczkowski et al. 2003).
For the other identified interactions (class 2, Table 2), despite a lack of MMS suppression by CDC13-EST2, the telomere defect in these cells may still be a cause of MMS sensitivity. For example, an increase in ssDNA at telomeres would create a structure that is more susceptible to MMS-induced lesions [fork-blocking lesions occur predominantly in ssDNA in MMS (Shrivastav et al. 2010)]. Accordingly, a rad27 mutant is associated with abnormally large regions of ssDNA in telomeres (Parenteau and Wellinger 1999). As DNA damage in telomeres has recently been shown to be uniquely irreparable (Fumagalli et al. 2012), it is likely that telomeres exhibiting abnormal structures are both more susceptible to MMS-induced damage and less able to survive it.
Implications of the tel1-Δ screen for mammalian cells
From this study, we show that tel1-Δ cells are rendered sensitive to MMS by increased replication stress or exacerbation of the short telomere phenotype. Thus, targeting these mechanisms may be an effective strategy for killing tumor cells that have lost ATM activity. Intriguingly, a recent study found that the specific combination of an ATM (TEL1 ortholog) inhibitor drug combined with a telomerase inhibitor rendered tumor cells extremely sensitive to the chemotherapy agent etoposide (Tamakawa et al. 2010). Furthermore, based on the replication stress model described above, targeting ATM for inhibition, combined with agents such as MMS, would be expected to confer a synergistic effect in cells experiencing oncogene-induced replication stress.
Acknowledgments
We thank Brenda Andrews, Charles Boone, Daniel Gottschling, Vicki Lundblad, and Sang Eun Lee for strains and plasmids used in this study and Mary Ellard-Ivey for access to equipment. We thank members of the Brewer/Raghuraman and Gottschling labs and Wenyi Feng and Lindsey Williams for helpful discussions related to this study. We thank our anonymous reviewers and the GENETICS Editor for helpful comments on the manuscript. B.D.P. was supported by the Fred Hutchinson Cancer Research Center Dual Mentor Program and a U.S. Department of Defense Breast Cancer Research Program predoctoral fellowship. This work was supported by National Institutes of Health grant R01 CA 129604.
Footnotes
Communicating editor: N. M. Hollingsworth
Literature Cited
- Agarwal M., Pandita S., Hunt C. R., Gupta A., Yue X., et al. , 2008. Inhibition of telomerase activity enhances hyperthermia-mediated radiosensitization. Cancer Res. 68: 3370–3378. [DOI] [PubMed] [Google Scholar]
- Arneric M., Lingner J., 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 8: 1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Capra T., Jossen R., Colosio A., Frattini C., et al. , 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M. L., Paeschke K., Zakian V. A., 2012. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13: 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., et al. , 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281: 1677–1679. [DOI] [PubMed] [Google Scholar]
- Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., et al. , 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401. [DOI] [PubMed] [Google Scholar]
- Chang M., Arneric M., Lingner J., 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21: 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C., Cech T. R., Zaug A. J., 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3: 1337–1351. [PMC free article] [PubMed] [Google Scholar]
- Chen C., Kolodner R. D., 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Cimprich K. A., Shin T. B., Keith C. T., Schreiber S. L., 1996. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. USA 93: 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R. J., Greenwell P. W., Dominska M., Petes T. D., 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y., Bermejo R., Fiorani S., Haber J. E., Foiani M., 2009. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell 137: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi R., Wu J., Hu Y., Bockhold C., Dome J. S., 2011. Telomere shortening alters the kinetics of the DNA damage response after ionizing radiation in human cells. Cancer Prev. Res. (Phila.) 4: 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. K., Lundblad V., 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120. [DOI] [PubMed] [Google Scholar]
- Fasullo M., Tsaponina O., Sun M., Chabes A., 2010. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 38: 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K., Kwon Y., Sung P., Sugimoto K., 2011. Activation of protein kinase Tel1 through recognition of protein-bound DNA ends. Mol. Cell. Biol. 31: 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Rossiello F., Clerici M., Barozzi S., Cittaro D., et al. , 2012. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 14: 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Toro T. B., Paschini M., Braunstein-Ballew B., Cervantes R. B., et al. , 2010. Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics 186: 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E., Goytisolo F. A., Flores J. M., Blasco M. A., 2003. Telomere dysfunction results in enhanced organismal sensitivity to the alkylating agent N-methyl-N-nitrosourea. Cancer Res. 63: 7047–7050. [PubMed] [Google Scholar]
- Goytisolo F. A., Samper E., Martin-Caballero J., Finnon P., Herrera E., et al. , 2000. Short telomeres result in organismal hypersensitivity to ionizing radiation in mammals. J. Exp. Med. 192: 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Bailly A., Charbonneau M., 2005. Activation of Mrc1, a mediator of the replication checkpoint, by telomere erosion. Biol. Cell 97: 799–814. [DOI] [PubMed] [Google Scholar]
- Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., et al. , 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823–829. [DOI] [PubMed] [Google Scholar]
- Gupta A., Sharma S., Reichenbach P., Marjavaara L., Nilsson A. K., et al. , 2013. Telomere length homeostasis responds to changes in intracellular dNTP pools. Genetics 193: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A. B., Moreno S., 2011. Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J. 30: 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A., 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16: 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. J., Lee C. H., Kang Y. H., Cho I. T., Nguyen T. A., et al. , 2010. Genetic and functional interactions between Mus81-Mms4 and Rad27. Nucleic Acids Res. 38: 7611–7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaochar S., Shanks L., Weinert T., 2010. Checkpoint genes and Exo1 regulate nearby inverted repeat fusions that form dicentric chromosomes in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 107: 21605–21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitzky L., Beltrao P., Berens T. J., Gassner N., Zhou C., et al. , 2010. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol. Syst. Biol. 6: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M. B., Zhan Q., el-Deiry W. S., Carrier F., Jacks T., et al. , 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71: 587–597. [DOI] [PubMed] [Google Scholar]
- Kuhne M., Riballo E., Rief N., Rothkamm K., Jeggo P. A., et al. , 2004. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 64: 500–508. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T., 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26: 7741–7748. [DOI] [PubMed] [Google Scholar]
- Lee K., Zhang Y., Lee S. E., 2008. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454: 543–546. [DOI] [PubMed] [Google Scholar]
- Lin Y. H., Chang C. C., Wong C. W., Teng S. C., 2009. Recruitment of Rad51 and Rad52 to short telomeres triggers a Mec1-mediated hypersensitivity to double-stranded DNA breaks in senescent budding yeast. PLoS ONE 4: e8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M. P., Paciotti V., Neecke H., Lucchini G., 2000. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics 155: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C., North M., Erixon K., Walters K., Jenssen D., et al. , 2005. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33: 3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A. J., Petes T. D., 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83: 1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Chaurasia P., Lahudkar S., Durairaj G., Shukla A., et al. , 2010. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 38: 1461–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory J. C., Petes T. D., 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97: 13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D., Clerici M., Lucchini G., Longhese M. P., 2007. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8: 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Gilson E., Shore D., 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990. [DOI] [PubMed] [Google Scholar]
- Martina M., Clerici M., Baldo V., Bonetti D., Lucchini G., et al. , 2012. A balance between Tel1 and Rif2 activities regulates nucleolytic processing and elongation at telomeres. Mol. Cell. Biol. 32: 1604–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley J. L., Petes T. D., 2010. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc. Natl. Acad. Sci. USA 107: 11465–11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee J. S., Phillips J. A., Chan A., Sabourin M., Paeschke K., et al. , 2010. Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat. Struct. Mol. Biol. 17: 1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick C. J., Jackson D., Diffley J. F., 2004. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 279: 20067–20075. [DOI] [PubMed] [Google Scholar]
- Metcalfe J. A., Parkhill J., Campbell L., Stacey M., Biggs P., et al. , 1996. Accelerated telomere shortening in ataxia telangiectasia. Nat. Genet. 13: 350–353. [DOI] [PubMed] [Google Scholar]
- Mieczkowski P. A., Mieczkowska J. O., Dominska M., Petes T. D., 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc. Natl. Acad. Sci. USA 100: 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow D. M., Tagle D. A., Shiloh Y., Collins F. S., Hieter P., 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82: 831–840. [DOI] [PubMed] [Google Scholar]
- Mulder K. W., Winkler G. S., Timmers H. T., 2005. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 33: 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami-Sekimata A., Huang D., Piening B. D., Bangur C., Paulovich A. G., 2010. The Saccharomyces cerevisiae RAD9, RAD17 and RAD24 genes are required for suppression of mutagenic post-replicative repair during chronic DNA damage. DNA Repair (Amst.) 9: 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Kolodner R. D., 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Kolodner R. D., 2003. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst.) 2: 243–258. [DOI] [PubMed] [Google Scholar]
- Nakada D., Matsumoto K., Sugimoto K., 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17: 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Masutomi K., Kyo S., Hashimoto M., Maida Y., et al. , 2005. Efficient inhibition of human telomerase reverse transcriptase expression by RNA interference sensitizes cancer cells to ionizing radiation and chemotherapy. Hum. Gene Ther. 16: 859–868. [DOI] [PubMed] [Google Scholar]
- Paciotti V., Clerici M., Lucchini G., Longhese M. P., 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14: 2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Painter R. B., Young B. R., 1980. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc. Natl. Acad. Sci. USA 77: 7315–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., et al. , 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell 18: 2912–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J., Wellinger R. J., 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19: 4143–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A. G., Armour C. D., Hartwell L. H., 1998. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics 150: 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K. B., Mallory J. C., Petes T. D., 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M., Tuzon C. T., Zakian V. A., 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27: 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K., Sfez S., Tagle D. A., Ziv Y., Sartiel A., et al. , 1995. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum. Mol. Genet. 4: 2025–2032. [DOI] [PubMed] [Google Scholar]
- Shiloh Y., 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3: 155–168. [DOI] [PubMed] [Google Scholar]
- Shrivastav N., Li D., Essigmann J. M., 2010. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis 31: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. S., Gottschling D. E., 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409. [DOI] [PubMed] [Google Scholar]
- Singer M. S., Kahana A., Wolf A. J., Meisinger L. L., Peterson S. E., et al. , 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler D., Pampalona J., Tusell L., Genesca A., 2009. Radiation sensitivity increases with proliferation-associated telomere dysfunction in nontransformed human epithelial cells. Aging Cell 8: 414–425. [DOI] [PubMed] [Google Scholar]
- Stellwagen A. E., Haimberger Z. W., Veatch J. R., Gottschling D. E., 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17: 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamakawa R. A., Fleisig H. B., Wong J. M., 2010. Telomerase inhibition potentiates the effects of genotoxic agents in breast and colorectal cancer cells in a cell cycle-specific manner. Cancer Res. 70: 8684–8694. [DOI] [PubMed] [Google Scholar]
- Teixeira M. T., Arneric M., Sperisen P., Lingner J., 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335. [DOI] [PubMed] [Google Scholar]
- Tkach J. M., Yimit A., Lee A. Y., Riffle M., Costanzo M., et al. , 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Boone C., 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313: 171–192. [DOI] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science (New York, N.Y.) 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Traven A., Hammet A., Tenis N., Denis C. L., Heierhorst J., 2005. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics 169: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Taggart A. K., Zakian V. A., 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11: 1328–1335. [DOI] [PubMed] [Google Scholar]
- Vernon M., Lobachev K., Petes T. D., 2008. High rates of “unselected” aneuploidy and chromosome rearrangements in tel1 mec1 haploid yeast strains. Genetics 179: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H., 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8: 652–665. [DOI] [PubMed] [Google Scholar]
- Westmoreland T. J., Marks J. R., Olson J. A., Jr, Thompson E. M., Resnick M. A., et al. , 2004. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryot. Cell 3: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science (New York, N.Y.) 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wong K. K., Chang S., Weiler S. R., Ganesan S., Chaudhuri J., et al. , 2000. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat. Genet. 26: 85–88. [DOI] [PubMed] [Google Scholar]
- Woo S. R., Park J. E., Juhn K. M., Ju Y. J., Jeong J., et al. , 2012. Cells with dysfunctional telomeres are susceptible to reactive oxygen species hydrogen peroxide via generation of multichromosomal fusions and chromosomal fragments bearing telomeres. Biochem. Biophys. Res. Commun. 417: 204–210. [DOI] [PubMed] [Google Scholar]
- Woolstencroft R. N., Beilharz T. H., Cook M. A., Preiss T., Durocher D., et al. , 2006. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J. Cell Sci. 119: 5178–5192. [DOI] [PubMed] [Google Scholar]
- Wu Y., Xiao S., Zhu X. D., 2007. MRE11–RAD50–NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 14: 832–840. [DOI] [PubMed] [Google Scholar]
- Wu Y., Dimaggio P. A., Jr, Perlman D. H., Zakian V. A., Garcia B. A., 2013. Novel phosphorylation sites in the S. cerevisiae Cdc13 protein reveal new targets for telomere length regulation. J. Proteome Res. 12: 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chabes A., Domkin V., Thelander L., Rothstein R., 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20: 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S. J., 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548. [DOI] [PubMed] [Google Scholar]