Abstract
Nematodes compose an abundant and diverse invertebrate phylum with members inhabiting nearly every ecological niche. Panagrellus redivivus (the “microworm”) is a free-living nematode frequently used to understand the evolution of developmental and behavioral processes given its phylogenetic distance to Caenorhabditis elegans. Here we report the de novo sequencing of the genome, transcriptome, and small RNAs of P. redivivus. Using a combination of automated gene finders and RNA-seq data, we predict 24,249 genes and 32,676 transcripts. Small RNA analysis revealed 248 microRNA (miRNA) hairpins, of which 63 had orthologs in other species. Fourteen miRNA clusters containing 42 miRNA precursors were found. The RNA interference, dauer development, and programmed cell death pathways are largely conserved. Analysis of protein family domain abundance revealed that P. redivivus has experienced a striking expansion of BTB domain-containing proteins and an unprecedented expansion of the cullin scaffold family of proteins involved in multi-subunit ubiquitin ligases, suggesting proteolytic plasticity and/or tighter regulation of protein turnover. The eukaryotic release factor protein family has also been dramatically expanded and suggests an ongoing evolutionary arms race with viruses and transposons. The P. redivivus genome provides a resource to advance our understanding of nematode evolution and biology and to further elucidate the genomic architecture leading to free-living lineages, taking advantage of the many fascinating features of this worm revealed by comparative studies.
Keywords: Panagrellus redivivus, microRNA, BTB domain, F-box, eRF1, cullin
NEMATODES are highly prolific organisms that originated during the Precambrian or Cambrian explosion over 500 million years ago and have subsequently evolved exquisite adaptations, allowing them to inhabit nearly all ecological niches (Malakhov and Hope 1994; Ayala and Rzhetsky 1998; Blaxter et al. 1998; Rodriguez-Trelles et al. 2002). Most nematodes are adapted to “free-living” lifestyles (i.e., nonparasitic and not associated with plants or animals, or only transiently associated as in phoresy) in terrestrial, freshwater, and marine environments, while others have parasitic lifestyles (Malakhov and Hope 1994). Free-living nematodes such as Caenorhabditis elegans have proven to be invaluable models for elucidating developmental and behavioral processes, leading to major discoveries including the genetic pathways underlying programmed cell death and the discovery of microRNA (miRNAs) and RNA interference, among others (Ambros and Horvitz 1984; Yuan et al. 1993; Fire et al. 1998). There is a huge repertoire of culturable free-living species for comparative studies, potentially making it difficult to decide which to prioritize for sequencing (Blaxter et al. 1998).
The free-living nematode Panagrellus redivivus has been used as a model system since the days of Linnaeus and is an established free-living comparative taxon to C. elegans (Sternberg and Horvitz 1981, 1982; Srinivasan et al. 2008). Fascinating differences in cell lineages and in behavior have been observed between the two (Sternberg and Horvitz 1981, 1982; Sulston et al. 1983; Srinivasan et al. 2008). For example, P. redivivus exhibits several distinguishing morphological characteristics: it is a gonochoristic (male–female) species requiring both sexes for reproduction and is ovoviviparous eggs hatch in utero and the young larvae are subsequently released through the vulva (Figure 1B). The larvae undergo four molts post hatch. Males have 9 chromosomes while females have 10 (Hechler 1970; Sternberg and Horvitz 1981). P. redivivus adults average 2 mm in size, twice as long as C. elegans adults (Hechler 1970; Sternberg and Horvitz 1982).
Figure 1.
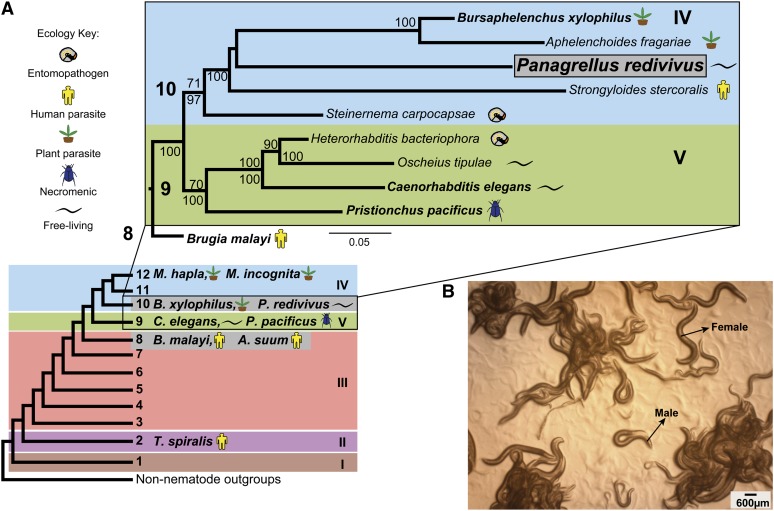
Phylogenetic classification of the nematode phylum and the position of the nematode P. redivivus. (A) A schematic representation of the division of the phylum Nematoda into clades, with the 12-clade designation after Holterman et al. (2006) and the 5-clade designation after Blaxter et al. (1998) in Roman numerals. Blaxter clades are encompassed in colored boxes, and nematode ecologies are represented by colored icons. The diagram zooms into a maximum-likelihood (ML) tree of representative taxa in clades 9 and 10, based on small subunit ribosomal DNA sequences. ML bootstrap support values ≥70 are shown above nodes while Bayesian posterior probabilities ≥70, concordant with the ML analysis, are shown below supported nodes. The closest sequenced nematode neighbor to P. redivivus in clade 10 is the migratory endoparasitic nematode B. xylophilus. The scale bar shows the amount of nucleotide changes per site that have occurred across taxa. (B) P. redivivus is a gonochoristic species comprising males and females. It is larger in size than C. elegans and lays young ones instead of eggs.
Historically, free-living nematodes have served as useful models for understanding basic biology such as organ development and signal transduction (Sternberg and Horvitz 1981, 1982; Srinivasan et al. 2008). In addition to comparative developmental studies, P. redivivus has been used in aquatic and soil toxicity studies, revealing interesting insights into the effects of pollutants and toxins on reproduction, movement, and feeding (Ager et al. 1984; Debus and Niemann 1994; Hempel et al. 1995; Boyd and Williams 2003; Niu et al. 2010). Small metabolites isolated from several fungal species have been successfully tested for their nematocidal activity using P. redivivus (Li et al. 2005; Huang et al. 2009; da Cruz et al. 2011). P. redivivus has been used to isolate male and female sex pheromones (Choe et al. 2012). It has also been used as a model for studying infection using human bacterial pathogens (Laws et al. 2005). Hence, P. redivivus has been used as a model system extensively in many diverse fields of biology in addition to being a free-living comparative taxon with C. elegans, making it a standout among free-living nematode sequencing candidates.
A molecular phylogenetic approach based on small subunit ribosomal DNA suggests the presence of 12 monophyletic clades in Nematoda (Figure 1A) (Holterman et al. 2006; van Megan et al. 2009). According to this phylogeny, P. redivivus belongs to clade 10, whereas C. elegans belongs to clade 9 (Figure 1). Sequencing efforts have focused primarily on the crown clades of Chromadoria with >13 sequenced genomes. All of the sequenced free-living nematode genomes currently available are restricted to clade 9 and are within the Caenorhabditis genus (Dillman et al. 2012). Other than the caenorhabditids, nematode sequencing efforts have prioritized either plant or animal parasites—including some of the most devastating agricultural and human pathogens such as plant parasites within Meloidogyne and the human parasites Brugia malayi and Trichinella spiralis (Ghedin et al. 2007; Opperman et al. 2008; Mitreva et al. 2011), which cause elephantiasis and trichinosis, respectively. P. redivivus represents the first noncaenorhabditid free-living nematode to be sequenced. Although little is known about its natural ecology, published literature suggests that P. redivivus has been isolated from a variety of environments, including felt beer hall mats, insect frass, slime from tree wounds, rotting fruit, insects, and wheat paste (Ferris 2009; Félix and Duveau 2012). These are acidic and nutrient-rich environments and have considerable overlap with the nutrient-rich natural habitats of C. elegans, which has also been isolated from rotting/decaying matter, especially rotting fruit (Kiontke and Sudhaus 2006; Félix and Duveau 2012). Given this ecological overlap, it is interesting to consider the architecture of free-living nematode genomes and how they might adapt to their respective niches. The phylogenetic position of P. redivivus and its ecological overlap with C. elegans make it an excellent species for studying the evolution of development, behavior, and adaptation (Figure 1A) (Blaxter et al. 1998; Holterman et al. 2006).
Here we describe the de novo assembly and characterization of a draft genome, transcriptome, and the complement of small RNAs of P. redivivus.
Materials and Methods
Strain culturing and maintenance of P. redivivus
For genomic and transcriptomic analysis, we used the P. redivivus strain PS2298/MT8872 (Sternberg and Horvitz 1981) originally obtained from D. J. Hooper (Rothamsted Experimental Station, Harpenden, Hertsfordshire, England). This strain was raised at 20° using standard methods.
Isolation of DNA and RNA
P. redivivus worms were grown on 5–10, 10-cm nutrient agar dishes containing Escherichia coli OP50 plates until near starvation. The worms were rinsed and collected with M9 buffer and washed multiple times to remove any E. coli. After the last wash in M9, the worms were suspended in M9 for 15–30 min. The worms were then snap-frozen in liquid nitrogen in ∼100-μl aliquots and stored at −80°. Worms were thawed and refrozen two to three times to break the cuticle before extracting either genomic DNA or bulk RNA. Genomic DNA was extracted using two rounds of proteinase K digestion followed by phenol-chloroform extraction. The genomic DNA was then treated with RNase A for digestion of any RNAs present in the sample. Bulk RNA was extracted using the Qiagen RNeasy mini kit.
Genomic and RNA-Seq library construction
A genomic library (library ID 12193) was constructed using Illumina Paired End DNA Sample Preparation Kit according to the manufacturer’s instructions. Briefly, 3 μg of genomic DNA were fragmented using nebulization. The fragments were end-repaired, 3′ adenylated, and ligated to Illumina’s paired-end adaptors. The ligation products were size-selected on an agarose gel to yield fragments of ∼350 bp. These fragments were then PCR-amplified to produce the finished library. Mate pair, a.k.a. “jumping” library (library ID 13185), was prepared using Illumina Mate Pair Library Preparation kit v2. Briefly, 7.5 μg of genomic DNA was fragmented using HydroShear device (Genomic Instrumentation Services) to generate fragments of ∼2.2 kb. Following end repair and biotinylation, the 2.2-kb fragment was gel-purified and circularized. Circular DNA was fragmented using Bioruptor NGS (Diagenode), and biotin-containing fragments were isolated using Dynabeads (Invitrogen). The fragments were end-repaired, 3′ adenylated, and ligated to NEBNext Multiplex Adaptors (NEB). The ligation products were PCR-amplified and size-selected using AMPure XP beads (Beckman Coulter) to generate the finished library of ∼450 bp in length. The RNA-Seq mixed-stage, poly(A)-selected library was created from 10 μg of total RNA using a standard unstranded protocol (Mortazavi et al. 2008, 2010). Libraries were quantified using a Qubit fluorometer (Invitrogen), and size distributions were verified using an Agilent Bioanalyzer and the High Sensitivity DNA Kit. Genomic and RNA-seq libraries were sequenced on Illumina Genome Analyzer IIx sequencer in paired-end mode with the read length of 76 nt. The jumping library was sequenced on Illumina HiSeq2000 in paired-end mode with the read length of 100 nt.
Genome assembly and annotation
The genomic libraries were built, sequenced, assembled, filtered, and repeat-masked as previously described (Mortazavi et al. 2010) using Velvet 1.2.07 and RepeatModeler 1.0.5, RepeatMasker 3.0.3, recon 1.70, and RepeatScout 1.0.5. The mixed-stage transcriptome was sequenced as previously described (Mortazavi et al. 2010) and assembled into complementary DNAs (cDNAs) using Oases 0.2.6 (Schulz et al. 2012). RNA-seq reads were submitted to the Sequence Read Archive under accession no. GSM1076725. This Whole Genome Shotgun project has been deposited at DNA Data Bank of Japan/ EMBL/GenBank under accession no. AOMH00000000. The version described in this article is the first version, AOMH01000000.
Assembled cDNAs were mapped onto the genome with blat and used as hints for gene finding using Augustus 2.6 with C. elegans settings (Stanke et al. 2008). Separately, RNA-seq reads were mapped onto the genome using TopHat 1.4 (Trapnell et al. 2009), assembled into transcripts using Cufflinks 2.02 (Trapnell et al. 2010). Candidate single nucleotide variations (SNVs) in the genome and transcriptome mapped reads were called using the samtools 0.1.13 (Li et al. 2009) pileup and varFilter options (Supporting Information, Figure S1). Candidate SNVs in the transcriptome that fell within 5 bp of exon junctions were filtered out as likely splicing artifacts.
Generation of the small RNA library
Small RNAs were isolated from mixed cultures of P. redivivus using the miRVana kit (Ambion) according to the manufacturer’s instructions. A small RNA library was then produced from the isolated RNAs using NEBNext small RNA sample prep Set 1 (New England Biolabs). The library was then size-selected on a 6% PAGE gel with the cut band corresponding to ∼90–120 bp. Library quality and size were confirmed prior to sequencing on a Bioanalyzer (Agilent).
For additional methods involving analyses of sequence information of the genome, see Supporting Information.
Results and Discussion
P. redivivus genomic assembly and transcript annotation
We sequenced 34 million, 350-bp fragments and 52 million, 2200-bp fragments of genomic P. redivivus DNA using paired-end 75-bp reads and 100-bp reads, respectively, and assembled them as described in Materials and Methods. After filtering out E. coli genomic DNA, the P. redivivus 64.4-Mb assembly had an N50 of 262.4 kb, with a maximum scaffold size of 2318 kb. The assembly has a GC content of 44.25%, and 7.01% of the genome was repeat-masked (Table 1). The Caenorhabditis genomes have a lower average GC content of 37%, which makes the genome of P. redivivus more similar to the necromenic nematode Pristionchus pacificus (43% GC) with respect to GC content.
Table 1. Features of the P. redivivus genome and transcriptome.
| Genome characteristics | P. redivivus | C. elegans | B. xylophilus | A. suum |
|---|---|---|---|---|
| Estimated genome size (Mb) | 64.4 | 100 | 74.6 | 272 |
| N50 (bp) | 26,2414 | a | 1,158,000 | 407,899 |
| GC content (%) | 44.25 | 35.4 | 40.4 | 37.9 |
| Repetitive sequences (%) | 7.1 | 22 | 4.4 | |
| Average intron length (bp) | 163 | 320 | 153 | 1,081 |
| Average exon length (bp) | 288 | 201.6 | 288.9 | 153 |
| Average no. of exons per gene | 4 | 6.5 | 4.5 | 6 |
The total estimated genome size of P. redivivus is 64.4 Mb based on our sequence data from the genome and transcriptome.
Fully sequenced genome, end-to-end.
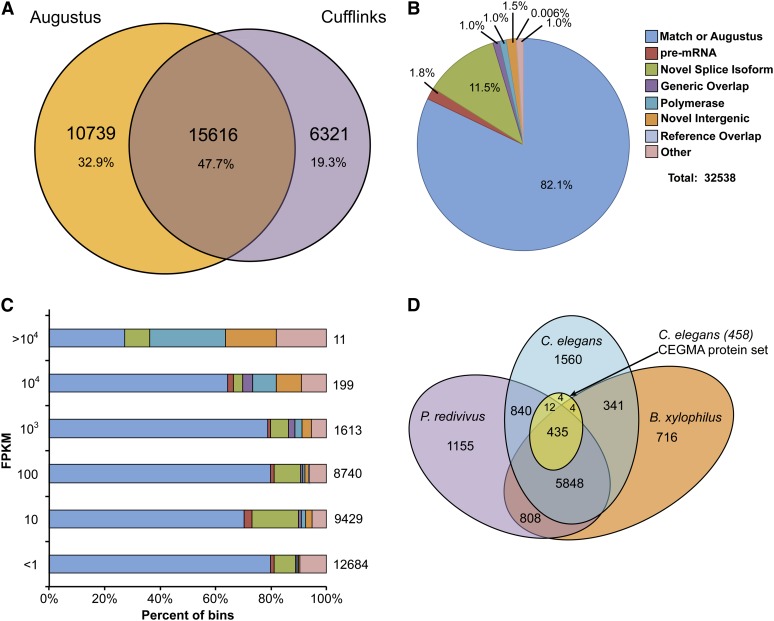
We collected RNA from ∼100,000 mixed-stage worms and sequenced 35 million, 200-bp cDNA fragments using paired-end 75-bp reads that were assembled into 18,298 distinct cDNAs with an N50 of 2.0 kb (Table S1) that were mapped onto the genome to assist the Augustus gene finder. Augustus identified 26,372 transcripts in 24,249 genes with 78,945 splice junctions. To extend the Augustus predictions of protein-coding genes, we used TopHat and Cufflinks (Trapnell et al. 2010) to map the RNA-seq data set onto the assembled genome. Cufflinks assembled 32,676 transcripts in 24,178 genes. Augustus predicted 32.9% of the consolidated gene models, whereas 19.3% of the models came from Cufflinks only (Figure 2A). Novel splice isoforms represented the bulk (11.5%) of the new transcripts identified by Cufflinks, while novel intergenic transcripts accounted for only 1.5% (Figure 2B). A survey of expression levels revealed that the novel splice isoform and non-Augustus gene models were highly expressed (Figure 2C). Thus, de novo protein-coding gene prediction (Augustus) and de novo transcript assembly on the genome (Cufflinks) are complementary methods that can be combined to obtain a more complete annotation of genes. We estimate that this draft of the P. redivivus genome is ∼98.2% complete, based on protein-clustering analysis of the P. redivivus proteome with the C. elegans Core Eukaryotic Genes Mapping Approach (CEGMA) protein set (Parra et al. 2007) (Figure 2D).
Figure 2.
Improved gene annotations using RNA sequencing. (A) Venn diagram capturing the differences between gene-finder-based annotations (Augustus) and RNA-seq-based transcript models (Cufflinks). All percentages are based on 32,676 consolidated transcript models that do not include the small categories in B. (B) Different match classes for Cufflinks + Augustus consolidated annotations to the original Augustus transcripts. Representative population size corresponds to all 30,601 models reported by cufflinks. (C) The distribution of transcripts detected in, or specific to, each fragments per kilobase of exon per million fragments mapped (FPKM) range and cumulative totals for all corresponding class annotations. (D) Venn Diagram capturing protein clusters between P. redivivus and the CEGMA protein set (Parra et al. 2007).
Analysis of small RNAs
A small RNA library was prepared by first isolating <200 nucleotide RNAs from whole mixed-stage animals by column chromatography. 5′ RNA and 3′ DNA adaptors were added using T4 RNA ligase I and T4 RNA ligase II (New England Biolabs), respectively. Both enzymes require 5′ phosphate present in the donor molecule and 3′ hydroxyl (OH) group in the acceptor for activity. The library was amplified and size selected for ∼90–120 bp fragments corresponding to inserts ∼20–50 bp in size. We sequenced 24 million reads from a small RNA library generated from mixed-stage animals (Table S2). We identified 248 miRNA precursors with at least 10 reads that support the presence of a mature miRNA derived from the hairpin precursor (Table 2 and File S2). For 218 hairpins, both 5′ and 3′ mature miRNAs were present with at least one read, and for 116 hairpins both were supported with at least 10 reads. In 157 miRNA genes, the dominantly expressed mature miRNA is located in the 3′ arm of the hairpin, a phenomenon that has also been observed in other nematodes (de Wit et al. 2009). In a few cases, there were two miRNAs expressed from the same miRNA loci, one from the plus strand and the other from the minus strand, suggesting the existence of antisense miRNA transcription (Ruby et al. 2007). We considered miRNA hairpins located within 500 bp from each other to be clustered; thus we found 14 miRNA clusters each containing two to seven miRNAs (Figure S2 and Figure S3C). In total, 42 miRNAs were located in these clusters and were likely derived from multicistronic precursors. Seventeen miRNAs came from multiple loci (Table 2).
Table 2. Summary of miRNAs discovered in P. redivivus.
| 248 | Confirmed miRNAs: criteria 1, computationally predicted hairpin; criteria 2, >10 reads |
| 116 | miRNAa sequences with >10 reads |
| 63 | Orthologs in related species |
| 16 | Located in exons |
| 2 | Located in gene UTR |
| 3 | Pairs of hairpins expressed from both strands |
| 14 | miRNA clusters |
| 17 | miRNAs have multiple origins in the genome |
“star” sequence, or less abundant mature miRNA molecule processed from the hairpin in miRNA nomenclature.
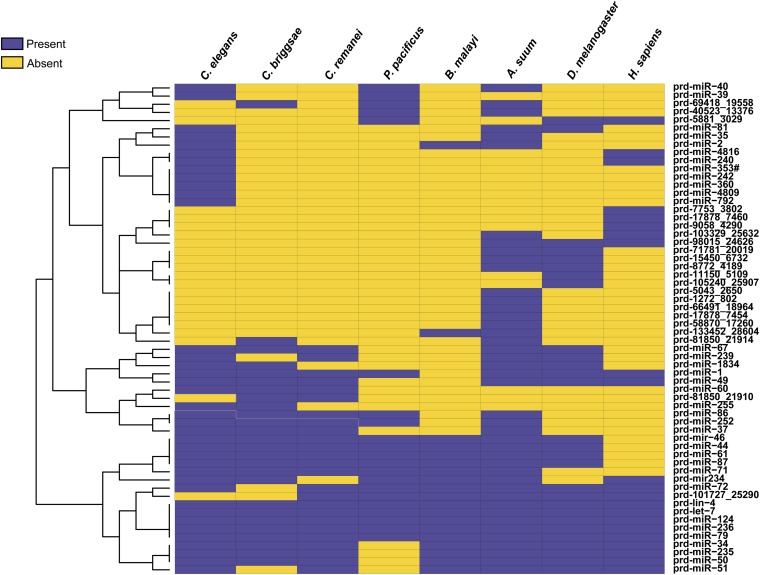
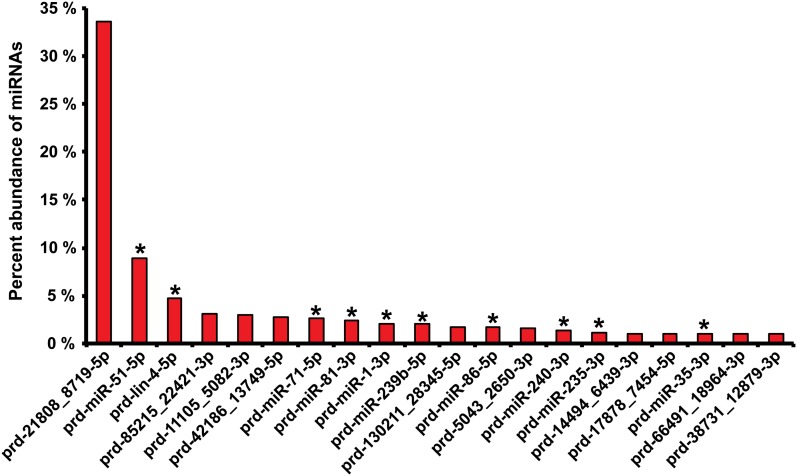
Using conservation of both mature miRNA and its hairpin sequence as criteria, we found orthologs for P. redivivus miRNAs in humans (46 of the 1527 miRBase miRNAs), Drosophila (31/240), C. elegans (46/223), Caenorhabditis briggsae (28/140), Caenorhabditis remanei (29/109), P. pacificus (20/124), B. malayi (20/32), and Ascaris suum (50/97) (Table S3). Among these were the well-studied and broadly conserved miRNAs let-7, miR-1, and miR-124 and the first miRNA identified, lin-4 (Lee et al. 1993). Altogether, 63 P. redivivus miRNAs have at least one ortholog among the species studied. Hierarchical clustering was used to visualize the distribution and conservation of these miRNAs, separating those highly conserved from miRNAs with only one or two orthologs (Figure 3). The most highly expressed miRNA was prd-21808-8719-5p (34%), for which we found no orthologs, whereas the second, prd-miR-51-5p, was conserved in six species (C. elegans, C. remanei, B. malayi, A. suum, D. melanogaster, and Homo sapiens). In addition, prd5043_2650-3p and prd17878_7454-5p were conserved only in A. suum. In all, 10 of the 20 most abundant miRNAs from P. redivivus had an ortholog in C. elegans (Figure 4). lin-4 (4.7% of all miRNA reads) and miR-1 (2.1%) were also among the 20 most abundantly expressed miRNAs in the data set (Figure 4).
Figure 3.
Clustered heat map of orthologous miRNAs from different species of nematodes, Drosophila and humans. The figure shows the spread of conservation of the P. redivivus miRNAs in the studied species. miRNAs with more orthologs are located in clusters at the bottom of the map, while those miRNAs with only a few orthologs are found at the top.
Figure 4.
Relative abundance of different miRNAs in the P. redivivus and their conservation in C. elegans. The proportions of the 20 most highly expressed P. redivivus miRNAs in our analysis. Ten of these miRNAs are conserved in C. elegans and are marked with asterisk at the top of the bar.
In addition to miRNAs, we also found evidence for the presence of endogenous small interfering RNAs (siRNAs) through identification of a cluster of nonhairpin-derived small RNAs. These consisted of reads that we tentatively identify as belonging to 21U, 22G, and 26G classes. The cluster in contig Pred1187 spanning nucleotides 15–486, consisted of 132 21U RNA reads (U first nucleotide, 21 nucleotides in length), 94 22G RNA reads (G first nucleotide, 22 nucleotides in length), and 78 26G RNA reads (G first nucleotide, 26 nucleotides in length) complementary to a 1-kb region of the predicted gene pred1_g624, a 221-amino-acid protein with two transmembrane domains and no obvious orthologs in other species. These RNAs were spaced at varying distances in both 5′ and 3′ directions (Figure S3A). We were not able to find large clusters of 21U-RNAs, similar to those described in C. elegans (15,722 unique 21U-RNA species expressed over a 200-kb region) (Ruby et al. 2007; Batista et al. 2008). C. elegans 21U-RNA species are expressed in the germline, are bound by Argonaut subfamily piwi-related protein PRG-1, and are thought to be the nematode equivalent of Piwi-interacting RNAs (piRNAs) found in Drosophila and humans. We were also unable to identify PRG-1 or PRG-2 orthologs in the transcriptome study (Table S4), suggesting that this class of small RNA may not be utilized in P. redivivus. By contrast, we did find evidence for the 22G-RNA class of small RNAs. We observed 18 clusters of 22G-RNA, which were defined by at least 20 22G-RNA reads (criteria for a cluster were multiple reads with <10 copies each, length 21–23 nt, and spaced by at most 200 nucleotides). Sixteen of these clusters were located on the opposite strand of the target gene. Within these clusters were only 22G-RNAs (G first nucleotide, 21–23 nucleotides in length). An example of a 22G cluster from P. redivivus is shown in Figure S2. The 22G-RNA class is further divided into two subclasses, one bound by CSR-1 and required for holocentric chromosome segregation (Claycomb et al. 2009), and the other subclass bound by worm-specific AGOs and playing an important role in transposon, pseudogene, cryptic locus, and protein-coding gene silencing (Gu et al. 2009). The P. redivivus genome has five CSR-1 orthologs (Table S4), suggesting that the CSR-1 pathway may be more elaborated in P. redivivus compared to C. elegans.
Orthology analysis
We analyzed protein orthology to explore the architecture of the P. redivivus proteome. We compared 24,249 P. redivivus proteins to the proteomes of seven other nematode species and an insect outgroup: P. redivivus, C. elegans, P. pacificus, Meloidogyne hapla, Bursaphelenchus xylophilus, Brugia malayi, A. suum, T. spiralis, and the parasitoid wasp Nasonia vitripennis (C. elegans Genome Sequencing Consortium 1998; Ghedin et al. 2007; Dieterich et al. 2008; Opperman et al. 2008; Werren et al. 2010; Jex et al. 2011; Kikuchi et al. 2011; Mitreva et al. 2011) (Table 3 and File S1). An important caveat of such orthology analyses is that the accuracy of the results relies on the quality and completeness of the proteomes used. We found a total of 9156 orthology clusters that included 17,415 P. redivivus proteins; 281 of these were found exclusively in nematodes. A total of 1,664 orthology clusters included at least one protein from each of the nine taxa that we analyzed (N:N), 521 of which were strictly conserved at a 1:1 ratio across all taxa (Table 3). These highly conserved proteins provide a candidate list of additional potential phylogenetic markers that could increase the signal-to-noise ratio in future phylum-wide phylogenetic analyses (Holterman et al. 2006; van Megan et al. 2009). P. redivivus had 6834 orphan proteins that did not cluster with any examined proteins, suggesting that they are uniquely derived in P. redivivus or that they are sufficiently divergent from their orthologs so as to not be recognizably related by sequence similarity alone. We find it remarkable that, despite using eight nematode proteomes, representing only 4 of the 12 clades (Figure 1), we still find that >20% of the protein-coding genes in each species are orphans, with little-to-no sequence homology with other proteins in the analysis (Table S5). This suggests that a tremendous diversity of proteins underlies the superficial similarity of nematode morphology and that many novel proteins may yet remain to be discovered with additional genome sequencing of these wonderfully adaptable worms.
Table 3. P. redivivus orthology statistics.
| Predicted proteins in P. redivivus | 24,249 |
| P. redivivus proteins in clusters | 17,415 |
| Clusters with P. redivivus proteins | 9,156 |
| Clusters without P. redivivus proteins | 8,794 |
| P. redivivus orphan proteins (unclustered) | 6,834 |
| N:N orthologous protein clusters | 1,664 |
| 1:1 orthologous protein clusters | 521 |
| Orthologous protein clusters including all nematode taxa but no insect ortholog | 281 |
The P. redivivus genome and transcriptome reveal 27,266 proteins. Of these, 443 proteins are conserved at a 1:1 ratio across seven other nematodes, listed in Table 6, and the insect outgroup N. vitripennis. Only 266 orthologous protein clusters in this data set were exclusively nematode proteins.
Signaling and regulatory pathways in P. redivivus
Organisms often display remarkable plasticity during their life cycle and are capable of adapting to different conditions by sensing their environment and physiological status. Behavioral and metabolic changes are the most common forms of plasticity in response to environmental changes (Fielenbach and Antebi 2008). Both these processes are rapid responses to the environment and help the organism maintain homeostasis. Since the nematode P. redivivus is free-living and appears to inhabit nutrient-rich environments, we examined whether changes in components of signaling or developmental pathways reflect its adaptation to such a lifestyle. We also screened the assembled genome for the conservation of important biological pathways including the dauer, cell-death, and RNA interference (RNAi) pathways.
Dauer formation pathway
One of the most extensively studied molecular pathways in C. elegans is the dauer formation pathway (Fielenbach and Antebi 2008). The dauer diapause represents a long-lived life stage, which is a developmental response to stressful environmental conditions such as low availability of food and high population density. Detailed molecular and genetic analyses in C. elegans have revealed how the worm senses its environment and reacts to changing environmental conditions by activating conserved signaling pathways to initiate entry into the dauer stage (Fielenbach and Antebi 2008; Schaedel et al. 2012).
Several researchers have suggested that P. redivivus does not form dauers (Hechler 1970; Stock and Nadler 2006). This observation is surprising as the genome of P. redivivus encodes nearly all the major components of the dauer pathway, with 21 of the 25 proteins that we examined being conserved (Table 4). Howerver, Panagrellus isolates do form dauers in nature (Félix and Duveau 2012), and it is possible that this species or this laboratory strain lost the ability to form dauers under standard laboraotory conditions. Several of the pathway components apparently absent in P. redivivus appear to be specific to the caenorhabditids.
Table 4. Conservation of the dauer pathway.
| C. elegans protein | Cele | Ppac | Pred | Bxyl | Mhap | Bmal | Asuu | Tspi | Nvit |
|---|---|---|---|---|---|---|---|---|---|
| Pheromone | |||||||||
| DAF-22 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | x | 1 |
| DAF-6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | x | x |
| Guanylyl cyclases | |||||||||
| DAF-11 | 1 | 1 | 1 | 1 | x | 1 | 1 | x | x |
| DAF-21 | 1 | 1 | 1 | x | 1 | 1 | 1 | 1 | 2 |
| DAF-10 | 1 | x | 1 | 1 | x | 1 | 1 | 1 | 1 |
| TGFβ-like pathway | |||||||||
| DAF-1 | 1 | 1 | 1 | 1 | x | 1 | 1 | 2 | 2 |
| DAF-4 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| DAF-7 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| DAF-8 | 3 | 1 | 3 | 3 | 1 | 3 | 3 | 2 | 3 |
| DAF-14 | 1 | x | x | x | x | x | x | x | x |
| DAF-3 | 2 | 5 | 1 | 2 | 1 | 1 | 2 | 2 | 1 |
| DAF-5 | 1 | x | x | x | x | x | x | x | x |
| Insulin-like pathway | |||||||||
| DAF-2 | 1 | 1 | 2 | 2 | x | 4 | 2 | 2 | 1 |
| DAF-16 | 1 | 1a | 1 | 2 | 1 | 2 | 1 | 1 | 2 |
| DAF-23/AGE-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | x | 1 |
| Steroid hormone | |||||||||
| DAF-9 | 1 | 1 | 2 | 1 | x | 1 | 1 | x | x |
| DAF-12 | 1 | 1 | 1 | 1 | x | 1 | 2 | x | x |
| Other effectors | |||||||||
| DAF-15 | 1 | 1 | 1 | 1 | x | 1 | 1 | 1 | 1 |
| DAF-19 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| DAF-18 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DAF-28 | 8 | x | x | x | x | x | x | x | x |
| DAF-36 | 1 | 1 | x | x | x | 4 | 1 | x | 3 |
| TAX-2 | 1 | 1 | 1 | 1 | 1 | x | 1 | x | x |
| TAX-4 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 |
| EGL-4 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
Shown are the number of proteins from each species analyzed that cluster as orthologs with the known C. elegans protein. “x” indicates that no proteins from the proteome used clustered with the known C. elegans protein. Descriptive labels for certain pathway components known in C. elegans are given in the first column. Results are based on an orthology analysis using the available proteomes and OrthoMCL (see Supporting Information).
Note that, while daf-16 was not present in the version of the proteome that we used, it is known to be present in the P. pacificus genome (Ogawa et al. 2011).
Cell death pathway
Cell death is a critical process during development because animals need to eliminate unwanted cells in a regulated and timely manner (Horvitz 2003). In C. elegans, programmed cell death is believed to be molecularly initiated by the activation of a core cell-death pathway, consisting of egl-1, ced-9, ced-4, and ced-3 (Metzstein et al. 1998). Cell death is highly abundant during P. redivivus germline and somatic development (Sternberg and Horvitz 1981, 1982). In addition, P. redivivus development undergoes specific cell deaths that are evolutionarily derived (e.g., the female gonadal posterior distal tip cell). Given these observations, we examined whether core components of the cell-death pathway are present in P. redivivus. Most known effectors are conserved; however, CED-9 appears to be absent (Table S6). We assume that this is a result of the draft nature of the genome or possibly the result of divergent sequence rather than the actual absence of this gene. CED-9 is central in the regulation and prevention of cell death in many species and is highly conserved from C. elegans to humans (Metzstein et al. 1998).
RNAi pathway
The RNAi pathway has become a valuable experimental tool to perturb individual or groups of genes to uncover their specific function(s), although its application and reliability across different nematodes has been inconsistent (Urwin et al. 2002; Viney and Thompson 2008; Dalzell et al. 2010, 2011). An obvious potential explanation for this is a conspicuous lack of certain RNAi effectors in some nematode species. However, we note that other factors, such as culturing conditions, rather than the disparity of RNAi effectors across species may better explain RNAi competencies among nematodes (Dalzell et al. 2011). We found that many RNAi effector genes are conserved in P. redivivus (Table S4 and Table S7). We found 56 RNAi effector proteins that cluster with known effectors in C. elegans, including at least 16 Argonaute-like proteins, in the P. redivivus genome (Table S4 and Table S7), suggesting that P. redivivus has more of the known RNAi pathway conserved with C. elegans than any other noncaenorhabditid nematode that has been sequenced; however, this is due in part to P. redivivus expansions in certain orthologous clusters such as CSR-1- and EKL-1-like proteins (Dalzell et al. 2011). Despite the high number of orthologous effectors in C. elegans and P. redivivus, 21 of the 73 RNAi effectors that we examined appear to be specific to the C. elegans lineage, having no apparent orthologs in any of the taxa that we analyzed (Table S4 and Table S7). We are hopeful that P. redivivus will be susceptible to experimental RNAi, given the apparent conservation of so many effectors that need to be tested.
A novel small RNA pathway required for the production and/or function of germline small RNA(s) in C. elegans includes four regulator genes (csr-1, drh-3, ego-1, and ekl-1) (Rocheleau et al. 2008). We found that three of these genes have expanded to small families in the P. redivivus lineage, with five paralogs of CSR-1, three paralogs of DRH-3, and six paralogs of EKL-1. All three genes are required for RNAi in the C. elegans germline and share a chromosome segregation-defective and embryonic-lethal phenotype (Grishok et al. 2001, 2005; Kim et al. 2005; Robert et al. 2005; Duchaine et al. 2006). An unusual group of retrotransposons, named PAT elements, was previously identified in P. redivivus. These retrotransposons have contributed to a higher spontaneous mutation rate in P. redivivus compared to C. elegans (Link et al. 1987; de Chastonay et al. 1992). Although we could not precisely determine the number of PAT element copies in the P. redivivus genome, an HMMER Pfam analysis indicates the presence of at least 65 copies of reverse transcriptase and 23 copies of integrase, suggesting at least 23–65 retroelements (Table S8) (Finn et al. 2011). The apparent expansion of csr-1, drh-3, and ekl-1 and the abundance of retrotransposons in the genome suggest pronounced regulation of transposons in the germline of P. redivivus.
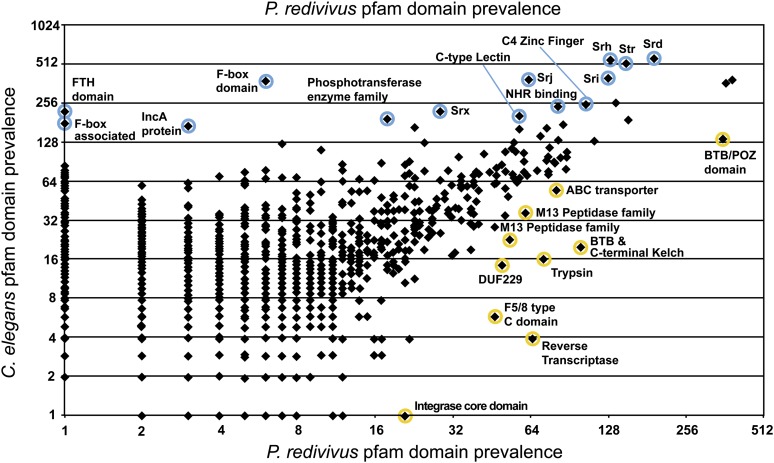
Protein family domain abundance
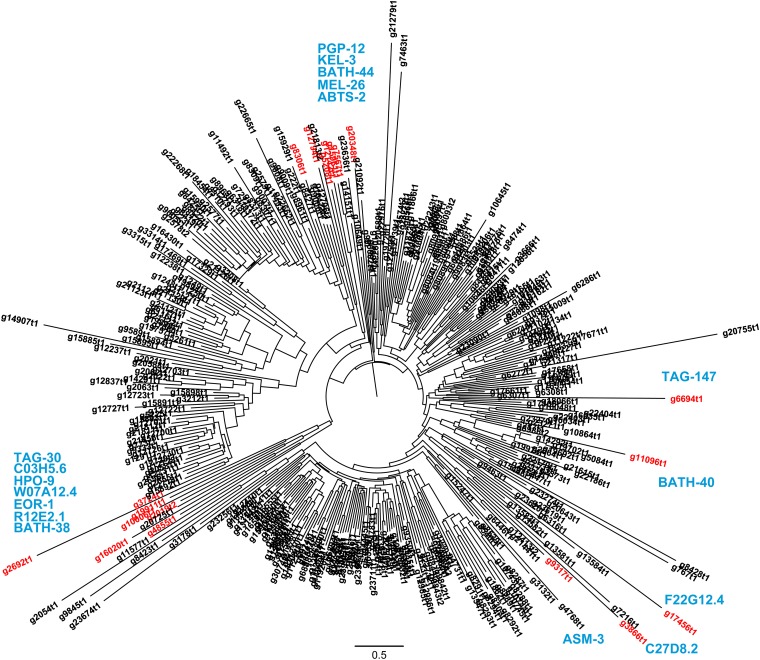
An analysis of domain abundance of various protein families provides an unbiased approach to exploring the vast sea of genomic data and reveals striking differences between the free-living nematodes C. elegans and P. redivivus (Figure 5; Figure S5). The C. elegans genome is greatly enriched in F-box, F-box-associated, FTH, and C-type lectin domains, among others, when compared to the P. redivivus genome. By contrast, the P. redivivus genome is enriched in BTB/POZ, BTB and C-terminal Kelch, trypsin, reverse transcriptase, and integrase core domains, among others (Figure 5). Both F-box and BTB domains are structural motifs that mediate protein–protein interactions, and proteins containing these domains are associated with signal transduction, cell-cycle regulation, and other cellular functions (Craig and Tyers 1999; Kipreos and Pagano 2000; Pintard et al. 2003; Stogios et al. 2005). The few members of these protein families that have been well-studied function as adaptors that determine the substrate specificity of E3 ubiquitin ligases, targeting substrates for proteolysis (Bai et al. 1996; Craig and Tyers 1999; Gagne et al. 2002; Furukawa et al. 2003; Pintard et al. 2003). Both F-box and BTB proteins are thought to play an important role in nematode immunity, with certain substrate-binding motifs having undergone heavy positive selection to target bacterial and viral peptides in the ever-escalating host–pathogen arms race (Dawkins and Krebs 1979; Thomas 2006). A detailed examination of the family of BTB domain-containing proteins in the P. redivivus genome revealed that there are large lineage-specific clades that appear to be rapidly evolving, suggestive of their involvement in immune responses (Tables 5 and 6; Figure 6). In addition, the presence of smaller, conserved orthology clusters of BTB/POZ and BTB/C-terminal Kelch proteins suggests that these likely target endogenous proteins, possibly for degradation in an E3 ligase proteolysis pathway (Figure 6; Figure S4) (Petroski and Deshaies 2005). This pattern of expansion and conservation of P. redivivus BTB domain-containing proteins, with many seemingly fast-evolving lineage-specific clusters, is consistent with observations from the C. elegans genome that F-box and BTB domain-containing proteins likely function in immunity and proteolysis (Thomas 2006).
Figure 5.
A scatterplot showing the abundance of Pfam protein family domains in the P. redivivus and C. elegans genomes. The 14 most enriched Pfam domains in C. elegans relative to P. redivivus are highlighted in blue while those seemingly enriched in P. redivivus relative to C. elegans are highlighted in yellow. The genome of C. elegans is enriched in serpentine family domain GPCRs and F-box domains. In contrast, P. redivivus is highly enriched in BTB domains.
Table 5. Selected domain prevalence among nematodes.
| Species | F-box/region/associated | SOCS/BC Box | BTB/POZ/C-terminal Kelch | Cullin |
|---|---|---|---|---|
| M. hapla | 10 | 2 | 54 | 3 |
| B. xylophilus | 15 | 1 | 51 | 9 |
| P. redivivus | 7 | 4 | 368 | 16 |
| C. elegans | 299 | 5 | 107 | 7 |
| P. pacificus | 17 | 0 | 78 | 7 |
| A. suum | 10 | 5 | 39 | 8 |
| B. malayi | 15 | 5 | 47 | 8 |
| T. spiralis | 11 | 1 | 15 | 8 |
Free-living nematodes have a dramatic expansion of these domains throughout their genomes.
Table 6. Selected domain prevalence among animals.
| Species | F-box/region/associated | SOCS/BC BOX | BTB/POZ/C-terminal Kelch | Cullin |
|---|---|---|---|---|
| H. sapiens | 68 | 59 | 194 | 9 |
| Mus musculus | 82 | 45 | 202 | 9 |
| Tribolium castaneum | 15 | 11 | 66 | 7 |
| N. vitripennis | 27 | 6 | 148 | 7 |
| D. melanogaster | 26 | 21 | 219 | 7 |
| Arabidopsis thaliana | 307 | 0 | 51 | 10 |
| Saccharomyces cerevisiae | 4 | 0 | 1 | 4 |
Pfam domain abundance of F-box, SOCS, BTB, and cullin domain-containing proteins across eukaryotes. Most eukaryotes have more BTB-domain-containing proteins than F-box-domain-containing proteins.
Figure 6.
Protein neighbor-joining tree of the BTB-domain-containing proteins in P. redivivus. This neighbor-joining gene tree shows all of the 367 BTB-domain-containing proteins in the P. redivivus proteome. Only 21 of these are conserved in at least two other nematode species, forming a total of 17 orthology clusters. These conserved proteins are highlighted in red, with the C. elegans ortholog names written in blue, where known. There are 22 genes conserved in at least one other nematode species, forming a total of 18 orthology clusters (see File S1). The 354 lineage-specific BTB proteins are suggested to be rapdily evolving and could function in binding non-endogenous proteins such as bacterial pattern recognition proteins.
The extent of variation in the number of F-box and BTB domains between P redivivus and C. elegans is striking. We pursued this observation further by evaluating the prevalence of F-box and BTB proteins across many nematodes and found that the free-living nematodes are outliers, having far more of either of these protein domains than any other nematodes, including the necromenic nematode P. pacificus as well as B. xylophilus, a plant-parasitic member of clade 10 along with P. redivivus (Figure 1 and Table 5). We also note that the trend across nematodes seems to favor BTBs over F-box proteins, with the exception of C. elegans, which has far more F-box proteins. Expanding this analysis across eukaryotes reveals that metazoans generally have more BTB proteins than F-box proteins, with plants and C. elegans being the exceptions (Table 5 and Table 6).
Due to the dramatic disparity of F-box domains between the free-living nematodes P. redivivus and C. elegans, we investigated the conservation of F-box domain-containing proteins across nematodes (Table 5 and Table S9). We found few F-box domain-containing proteins broadly conserved in nematodes and insects (Table S9) (Jin et al. 2004). We have yet to find the highly conserved SEL-10 (CDC4), known for its role in Skp, Cullin, F-box containing complex (or SCF complex)-mediated proteolysis in our genome or transciptome.
We suggest that the apparent evolution of F-box and BTB proteins in P. redivivus could be a response to viral susceptibility. Both BTB and F-box domain-containing proteins are traditionally known for their roles as the substrate-specifying subunits of multisubunit cullin-RING ubiquitin ligases (CRLs) (Feldman et al. 1997; Michel and Xiong 1998; Pintard et al. 2003; Petroski and Deshaies 2005; Sarikas et al. 2011). These ligases are modular and are responsible for targeting a wide variety of substrates for proteolysis by ubiquitylating them. These complexes are assembled on a cullin scaffold, which tethers a RING protein to a substrate-specifying subunit, usually through an adaptor protein as in the case of the canonical SCFCdc4 CRL. There are a variety of different CRLs, each associating with a specific cullin protein and possessing different specificity, depending on the adaptor and/or substrate-specifying subunit used (Petroski and Deshaies 2005). For example, C. elegans is known to have seven CRLs with ubiquitin-ligase activity, each built on a distinguishing cullin scaffold (cul-1 through cul-6 and apc-2) (Sarikas et al. 2011). CUL-1 CRLs use F-box proteins for substrate specificity, while those with a CUL-3 cullin scaffold use BTB proteins for substrate specificity. CRL machinery is widely exploited by viruses as a method of immune evasion, with most known examples targeting aspects of the CUL-1 CRL (Barry and Fruh 2006).
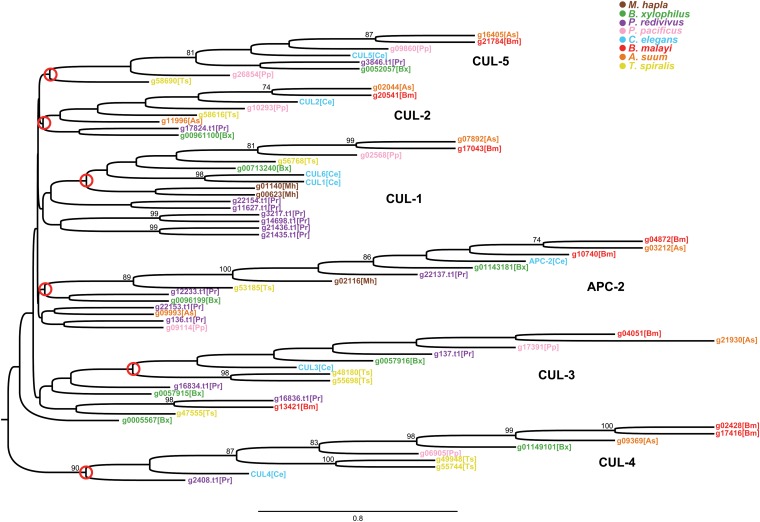
In support of this hypothesis, we observed an unprecedented expansion of cullin proteins in the P. redivivus genome (Table 5 and Figure 7). We explored this expansion by constructing a gene tree of all cullin homology domain-containing proteins across sequenced nematode genomes (Figure 7). Because of the dramatic expansion of BTB proteins, we expected an accompanying expansion of the CUL-3 family in P. redivivus, but paradoxically found an expansion of CUL-1-like proteins, which are known to use F-box proteins for substrate specificity in other metazoans (Petroski and Deshaies 2005; Sarikas et al. 2011). We also identified a number of novel cullin proteins, with little similarity to any described families, including several that appear to have arisen due to recent tandem duplications (Figure 7). The apparent absence of any CUL-2 ortholog and the abundance of novel cullin proteins suggest a surprising amount of regulatory and proteolytic plasticity in P. redivivus, which may be shaped by the stressful demands of the free-living lifestyle, immunological or otherwise. It is not uncommon for components of the ubiquitin system to be adapted to expand the immune system (e.g., Han et al. 2011; Yewdell 2005). Although beyond the scope of this current work, our data suggest that exploring the role of these cullins and the function of the CRL complexes that they form would increase our understanding of the adaptative changes that P. redivivus has undergone to cope with the stresses of its ecological niche.
Figure 7.
Distance-based protein tree of cullin homology domain-containing proteins among nematodes. All five major nematode cullin families are monophyletic with their putative origins circled in red. P. redivivus has six CUL-1-like proteins, two APC-2-like proteins, and four other cullin homology domain-containing proteins with little sequence similarity to known families.
G-protein-coupled receptors
Genomes of different organisms encode families of chemoreceptors, the size and diversity of which can reflect the niche inhabited by the organism (Thomas and Robertson 2008). These proteins mediate the first step in the transduction of chemical and other types of stimuli such as taste and pheromone signals. We screened the G-protein-coupled receptor (GPCR) repertoire in the genome of P. redivivus to better understand how the evolution of this large family of receptors might reflect its life history and how it compares to C. elegans. Not unexpectedly, both C. elegans and P. redivivus have an abundance of serpentine family domains (Srh, Str, Srd, etc.) belonging to the GPCR superfamily (Table 7). We found that, although the P. redivivus genome possesses a variety of GPCR proteins, it is far less abundant than what we found in C. elegans (Table 7). We observed that P. redivivus had 1075 serpentine domains compared to the 3259 domains of C. elegans. It has been suggested that C. elegans requires an abundance of chemoreceptors to navigate and interpret the nutrient-rich environments in which it lives (Robertson and Thomas 2006).
Table 7. pfam GPCR abundance and diversity.
| Serpentine receptor class | Pfam domain | Cele | Pred | Bxyl | Ppac | Mhap | Asuu | Bmal | Tspi |
|---|---|---|---|---|---|---|---|---|---|
| 7TM GPCR chemoreceptor Srh | PF10318.2 | 593 | 196 | 221 | 154 | 2 | 9 | 0 | 0 |
| 7TM GPCR chemoreceptor Srd | PF10317.2 | 446 | 131 | 206 | 146 | 28 | 16 | 0 | 0 |
| 7TM GPCR chemoreceptor Str | PF10326.2 | 441 | 152 | 238 | 176 | 20 | 13 | 0 | 0 |
| 7TM GPCR chemoreceptor Sri | PF10327.2 | 367 | 129 | 90 | 61 | 0 | 1 | 0 | 0 |
| 7TM GPCR chemoreceptor Srj | PF10319.2 | 338 | 63 | 143 | 131 | 2 | 6 | 0 | 0 |
| 7TM GPCR chemoreceptor Srx | PF10328.2 | 138 | 28 | 23 | 50 | 9 | 20 | 9 | 2 |
| 7TM GPCR chemoreceptor Srw | PF10324.2 | 170 | 22 | 27 | 15 | 5 | 23 | 7 | 4 |
| 7TM GPCR chemoreceptor Srsx | PF10320.2 | 82 | 70 | 62 | 41 | 60 | 35 | 2 | 18 |
| Srg family chemoreceptor | PF02118.14 | 101 | 82 | 13 | 39 | 8 | 1 | 0 | 0 |
| 7TM GPCR chemoreceptor Srbc | PF10316.2 | 88 | 9 | 4 | 4 | 0 | 4 | 0 | 0 |
| 7TM GPCR receptor class ab chemoreceptor | PF10292.2 | 80 | 42 | 51 | 25 | 13 | 5 | 5 | 1 |
| Sre G-protein-coupled chemoreceptor | PF03125.11 | 68 | 51 | 30 | 38 | 9 | 5 | 2 | 0 |
| 7TM GPCR chemoreceptor Srv | PF10323.2 | 60 | 44 | 10 | 36 | 1 | 2 | 0 | 4 |
| 7TM GPCR chemoreceptor Srz | PF10325.2 | 72 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 7TM GPCR chemoreceptor Srt | PF10321.2 | 63 | 40 | 33 | 35 | 21 | 3 | 0 | 1 |
| 7TM GPCR chemoreceptor Sra | PF02117.9 | 59 | 4 | 6 | 5 | 2 | 1 | 3 | 0 |
| 7TM GPCR chemoreceptor Sru | PF10322.2 | 47 | 11 | 0 | 1 | 1 | 1 | 0 | 0 |
| 7TM GPCR chemoreceptor Srb | PF02175.9 | 30 | 0 | 0 | 4 | 0 | 1 | 0 | 0 |
| Serpentine receptor-like protein, class xa | PF03383.8 | 16 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total serpentine GPCR Domains | 3259 | 1075 | 1157 | 961 | 191 | 146 | 28 | 31 | |
| No. of proteins with multiple GPCR domains | 768 | 276 | 296 | 229 | 34 | 24 | 3 | 3 | |
| No. of proteins with only one GPCR domain | 746 | 290 | 198 | 307 | 115 | 78 | 20 | 25 | |
| Total no. of GPCR domain-containing proteins | 1514 | 566 | 494 | 536 | 149 | 102 | 23 | 28 |
Repertoire of seven transmembrane receptor domain families across various nematode species identified by hmmscan and pfam is shown (Finn et al. 2011). C. elegans exhibits a dramatic expansion of various subfamilies of these proteins. In contrast, the parasitic species do not display a large number of domains for this class of genes.
The number of serpentine GPCR domains in P. redivivus is similar to that of its clade mate, the migratory endoparasitic B. xylophilus. We did find that, of the nematodes that we analyzed, the animal parasites (A. suum, B. malayi, and T. spiralis) have far fewer GPCR domains compared to their free-living counterparts. Our data suggest that, in the specialized environments that these worms inhabit inside their hosts, they do not need a large repertoire of receptors, whereas the free-living nematodes, and nematodes that spend more foraging time in complex soil environments (e.g., P. pacificus and B. xylophilus), require a larger set so that they can better navigate and interpret their environment (Table 7).
ABC transporters
ATP-binding cassette (ABC) transporters provide a means for a wide variety of substrates to be actively transported across membranes, hydrolyzing ATP in the process (Davidson et al. 2008; Sundaram et al. 2008). C. elegans has 61 ABC transporters, representing ∼0.3% of its protein-coding gene repertoire (Sheps et al. 2004). We found 94 putative ABC transporters in the P. redivivus genome, representing ∼0.4% of the total number of genes that we report in this draft genome (Table S10 and File S1). We found P. redivivus orthologs for 52 of the 61 C. elegans ABC transporters, indicating a high level of conservation (Table S10). In addition to having lineage-specific ABC transporters, we see expansions of hmt-1-like and pgp-like ABC transporters (Table S10). Unsurprisingly, both of these families of ABC transporters are involved in tolerance of heavy metals and other toxins. hmt-1 functions in heavy metal tolerance, mitigating the toxic effects of arsenic, cadmium, and copper on C. elegans, while pgp-5 is involved in resistance to heavy metals and bacterial toxins (Kurz et al. 2007; Schwartz et al. 2010). Expansions in these particular families of ABC transporters may explain the high level of copper tolerance reported in P. redivivus, which has been shown to have higher tolerance to copper than C. elegans or P. pacificus (Boyd and Williams 2003).
Eukaryotic release factor
The expanded protein domain families in P. redivivus compared to C. elegans include the eukaryotic release factor domains 1, 2, and 3. These three domains are found in the eukaryotic release factor 1 protein (eRF1), which is highly conserved from yeast to humans and plays a key role in translational termination (Frolova et al. 1994). eRF1 recognizes termination codons contained in messenger RNA (mRNA) and competes with suppressor transfer RNA(s) for the ribosomal A site (Drugeon et al. 1997). Most animals have 2–3 eRF1 orthologs while P. redivivus seems to have a striking expansion of 15 (Tables 5 and 6). While one of these is a putative ortholog of ETF-1 in C. elegans, the rest are quite diverse and appear to be scattered throughout the genome (Figure S6). How would a nematode or any other animal make use of an expansion of eRF1-like proteins and what might that reveal about the life history or natural ecology of P. redivivus?
Suppression of translational termination is a common strategy of animal and plant viruses and is necessary for the replication of some viruses (ten Dam et al. 1990). The expansion of eRF1 proteins in P. redivivus could represent an enhanced arsenal against viral assault, providing evidence of an historical or ongoing arms race between P. redivivus and viral antagonists. eRF1 levels are important for translational termination such that overexpressing eRF1 reduces readthrough (Drugeon et al. 1997; Le Goff et al. 1997) and depleting eRF1 increases readthrough (Stansfield et al. 1996). It is known that targeted depletion of eRF1 is a strategy employed by some viruses, such as the murine leukemia virus, whose reverse transcriptase interacts with eRF1 to increase translational readthrough, leading to efficient replication of the virus (Orlova et al. 2003). Perhaps additional copies of eRF1 ensure peptide chain termination, preventing or decreasing translational readthrough and/or ribosomal frameshifting and thus conferring resistance or immunity to some viruses. Little is known about the breadth and diversity of viruses that infect nematodes, especially noncaenorhabditid nematodes. There are no reports regarding viral infection of P. redivivus; however, P. redivivus is known to have a relatively high load of unusual retrotransposons, designated as PAT retroid elements and thought to be distantly related to the Gypsy family of retrotransposons (Link et al. 1987; de Chastonay et al. 1992). While most retroid elements produce GAG and Pol genes by translational readthrough or ribosomal framshifting from the same mRNA transcript, PAT elements are thought to generate separate transcripts for GAG and Pol genes, respectively (Link et al. 1987; de Chastonay et al. 1992). This implies that they have evolved to regulate GAG and Pol ratios at the transcriptional level, bypassing the need for translational readthrough or ribosomal frameshifting. We speculate that this could represent a vivid example of an evolutionary arms race, with P. redivivus evolving an expanded repertoire of eRF1 genes to ensure translational termination while PAT elements, the only known active retroelements in P. redivivus, have shifted to generate their genes in discrete transcripts, thus overcoming the host genome’s defenses, although this remains to be explored.
Concluding remarks
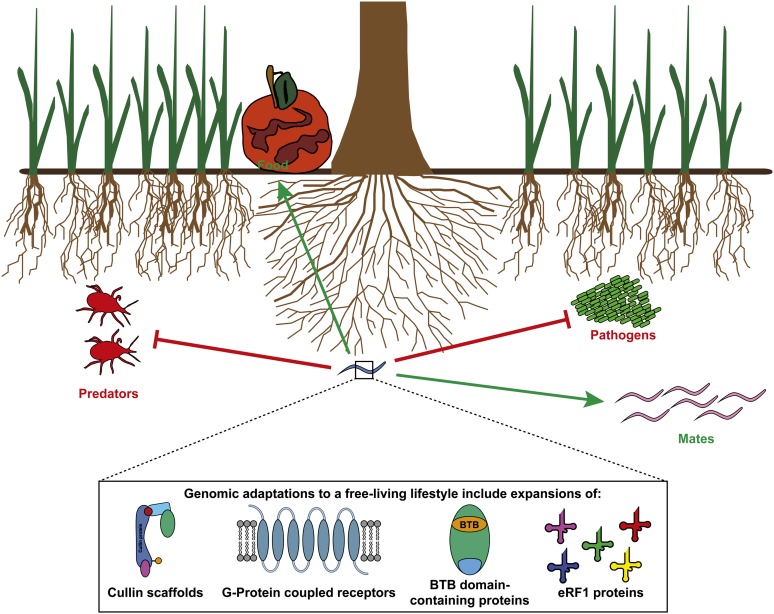
The annotated draft genome and transcriptome of P. redivivus provides a powerful resource in evolutionary and ecological comparative genomics. As it is the first free-living genome outside of the Caenorhabditis family to be sequenced, the genome highlights features that are common with the C. elegans genome. This may reflect common constraints and adaptations resulting from the free-living lifestyle. Free-living worms live in a complex and dynamic environment and must be able to generate appropriate responses to different stimuli and protect themselves against exogenous threats such as predators and pathogens, while still managing to find food and mates (Figure 8). Our analyses suggest some common genomic and transcriptomic features between the P. redivivus and C. elegans genomes. These include a large complement of GPCRs for interpreting and navigating nutrient-rich environments and an expansion of immune-related proteins to combat the abundant pathogens found in such environments. We also observe unexpected novelties, such as an unprecedented expansion of cullin scaffold proteins in P. redivivus and an unprecedented expansion of eRF1 orthologs. Some of the genomic features that we have described such as expansions in certain ABC transporters and eRF1 proteins may explain previous observations regarding toxin tolerance and the unusual PAT retroelements present in the P. redivivus genome (Link et al. 1987; de Chastonay et al. 1992; Boyd and Williams 2003) . These findings potentially encourage the development of new avenues in nematode research (Figure 8).
Figure 8.
Summary graphic showing the free-living lifestyle of P. redivivus and several genomic adaptations that facilitate it. As a free-living nematode, P. redivivus must seek out mates and find food resources while avoiding pathogens and predators in the complex environments in which it lives. The sequenced genome shows expansions of cullin proteins, GPCRs, BTB-domain-containing proteins, and immune effectors. These features of the P. redivivus genome appear to be adaptations to the free-living niche it occupies.
Comparative nematode genomics has come a long way since the release of the first whole nematode genome sequence (C. elegans Genome Sequencing Consortium 1998). Many additional nematode genomes have been sequenced, and the continuing drop in cost will ensure that even more will be sequenced (Stein et al. 2003; Ghedin et al. 2007; Abad et al. 2008; Dieterich et al. 2008; Mortazavi et al. 2010; Jex et al. 2011; Kikuchi et al. 2011; Mitreva et al. 2011; Sommer and Streit 2011). In addition, sequencing the genomes of nematode pests is providing researchers an avenue for identifying pharmacological targets that could be useful in the development of novel drugs against these parasitic nematodes (e.g., Jex et al. 2011). Comparison of genes involved in parasitism across various nematode clades expands our knowledge of the role played by processes such as horizontal gene transfer in the evolution of parasitism by nematodes (Mayer et al. 2011). Although there are 13 nematode genome sequences available, with many more in preparation, sequencing efforts have focused primarily on the crown clades of Chromadoria, heavily covering clade 9, and many of these projects have focused (appropriately) on parasites; however, we believe that our understanding of development, gene regulation, and niche partitioning among nematodes, as well as parasitism, will be greatly enhanced by studying the free-living ancestors from which parasites evolved (Dillman et al. 2012). This comparative analysis highlights some of the potential selective pressures on free-living nematodes and the adaptations that allow them to thrive in the natural world.
Acknowledgments
We thank Hillel Schwartz, Margaret C. Ho, Alexei Aravin, Claire C. de la Cova, Iva Greenwald, Kay Hofmann, and Raymond Deshaies for discussions. We thank Alicia Rogers and Zane Goodwin for their patience and help with some of the computations. This work was supported by a National Institutes of Health U.S. Public Health Service Training Grant (T32GM07616) to A.R.D. and by the Howard Hughes Medical Institute (with which P.W.S. is an Investigator). Sequencing was carried out at the Millard and Muriel Jacobs Genome Facility at the California Institute of Technology.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Abad P., Gouzy J., Aury J. M., Castagnone-Sereno P., Danchin E. G., et al. , 2008. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26: 909–915. [DOI] [PubMed] [Google Scholar]
- Ager D., Samoiloff M. R., Petkau A., 1984. Effect of dose-rate on the frequency of X-linked lethal mutation in the nematode Panagrellus redivivus. Mutat. Res. 140: 107–110. [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H. R., 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416. [DOI] [PubMed] [Google Scholar]
- Ayala F. J., Rzhetsky A., 1998. Origin of the metazoan phyla: molecular clocks confirm paleontological estimates. Proc. Natl. Acad. Sci. USA 95: 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., et al. , 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274. [DOI] [PubMed] [Google Scholar]
- Barry M., Fruh K., 2006. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE 2006: pe21. [DOI] [PubMed] [Google Scholar]
- Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N., et al. , 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. L., De Ley P., Garey J. R., Liu L. X., Scheldeman P., et al. , 1998. A molecular evolutionary framework for the phylum Nematoda. Nature 392: 71–75. [DOI] [PubMed] [Google Scholar]
- Boyd W. A., Williams P. L., 2003. Comparison of the sensitivity of three nematode species to copper and their utility in aquatic and soil toxicity tests. Environ. Toxicol. Chem. 22: 2768–2774. [DOI] [PubMed] [Google Scholar]
- C. elegans Genome Sequencing Consortium , 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Choe A., Chuman T., von Reuss S. H., Dossey A. T., Yim J. J., et al. , 2012. Sex-specific mating pheromones in the nematode Panagrellus redivivus. Proc. Natl. Acad. Sci. USA 109: 20949–20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig K. L., Tyers M., 1999. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog. Biophys. Mol. Biol. 72: 299–328. [DOI] [PubMed] [Google Scholar]
- da Cruz D. G., Araujo F. B., Molento M. B., Damatta R. A., de Paula Santos C., 2011. Kinetics of capture and infection of infective larvae of trichostrongylides and free-living nematodes Panagrellus sp. by Duddingtonia flagrans. Parasitol. Res. 109: 1085–1091. [DOI] [PubMed] [Google Scholar]
- Dalzell J. J., McMaster S., Fleming C. C., Maule A. G., 2010. Short interfering RNA-mediated gene silencing in Globodera pallida and Meloidogyne incognita infective stage juveniles. Int. J. Parasitol. 40: 91–100. [DOI] [PubMed] [Google Scholar]
- Dalzell J. J., McVeigh P., Warnock N. D., Mitreva M., Bird D. M., et al. , 2011. RNAi effector diversity in nematodes. PLoS Negl. Trop. Dis. 5: e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. L., Dassa E., Orelle C., Chen J., 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72: 317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R., Krebs J. R., 1979. Arms races between and within species. Proc. R. Soc. Lond. B Biol. Sci. 205: 489–511. [DOI] [PubMed]
- Debus R., Niemann R., 1994. Nematode test to estimate the hazard potential of solved contaminations. Chemosphere 29: 611–621. [DOI] [PubMed] [Google Scholar]
- de Chastonay Y., Felder H., Link C., Aeby P., Tobler H., et al. , 1992. Unusual features of the retroid element PAT from the nematode Panagrellus redivivus. Nucleic Acids Res. 20: 1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Linsen S. E., Cuppen E., Berezikov E., 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 19: 2064–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., et al. , 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman A. R., Mortazavi A., Sternberg P. W., 2012. Incorporating genomics into the toolkit of nematology. J. Nematol. 44(2): 191–205. [PMC free article] [PubMed] [Google Scholar]
- Drugeon G., Jean-Jean O., Frolova L., Le Goff X., Philippe M., et al. , 1997. Eukaryotic release factor 1 (eRF1) abolishes readthrough and competes with suppressor tRNAs at all three termination codons in messenger RNA. Nucleic Acids Res. 25: 2254–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr, et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Feldman R. M., Correll C. C., Kaplan K. B., Deshaies R. J., 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230. [DOI] [PubMed] [Google Scholar]
- Félix M. A., Duveau F., 2012. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012 10: 59 doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris H., 2009. The beer mat nematode, Panagrellus redivivus: a study of the connectedness of scientific discovery. J. Nematode Morphol. Syst. 12: 19–25. [Google Scholar]
- Fielenbach N., Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22: 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R., 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39: W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Frolova L., Le Goff X., Rasmussen H. H., Cheperegin S., Drugeon G., et al. , 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703. [DOI] [PubMed] [Google Scholar]
- Furukawa M., He Y. J., Borchers C., Xiong Y., 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Gagne J. M., Downes B. P., Shiu S. H., Durski A. M., Vierstra R. D., 2002. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., et al. , 2007. Draft genome of the filarial nematode parasite Brugia malayi. Science 317: 1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., et al. , 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Grishok A., Sinskey J. L., Sharp P. A., 2005. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 19: 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Jr, Vasale J., Batista P. J., et al. , 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Lou D. I., Sawyer S. L., 2011. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 7: e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechler H. C., 1970. Reproduction, chromosome number, and postembryonic development of Panagrellus redivivus (Nematoda: Cephalobidae). J. Nematol. 2: 355–361. [PMC free article] [PubMed] [Google Scholar]
- Hempel M., Chau Y. K., Dutka B. J., McInnis R., Kwan K. K., et al. , 1995. Toxicity of organomercury compounds: bioassay results as a basis for risk assessment. Analyst (Lond.) 120: 721–724. [DOI] [PubMed] [Google Scholar]
- Holterman M., van der Wurff A., van den Elsen S., van Megen H., Bongers T., et al. , 2006. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Mol. Biol. Evol. 23: 1792–1800. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., 2003. Nobel lecture. Worms, life and death. Biosci. Rep. 23: 239–303. [DOI] [PubMed] [Google Scholar]
- Huang X., Liu J., Ding J., He Q., Xiong R., et al. , 2009. The investigation of nematocidal activity in Stenotrophomonas maltophilia G2 and characterization of a novel virulence serine protease. Can. J. Microbiol. 55: 934–942. [DOI] [PubMed] [Google Scholar]
- Jex A. R., Liu S., Li B., Young N. D., Hall R. S., et al. , 2011. Ascaris suum draft genome. Nature 479: 529–533. [DOI] [PubMed] [Google Scholar]
- Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., et al. , 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18: 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T., Cotton J. A., Dalzell J. J., Hasegawa K., Kanzaki N., et al. , 2011. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7: e1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K., Gabel H. W., Kamath R. S., Tewari M., Pasquinelli A., et al. , 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. [DOI] [PubMed] [Google Scholar]
- Kiontke K., Sudhaus W., 2006. Ecology of Caenorhabditis species (January 9, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.37.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Kipreos E. T., Pagano M., 2000. The F-box protein family. Genome Biol. 1: REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz C. L., Shapira M., Chen K., Baillie D. L., Tan M. W., 2007. Caenorhabditis elegans pgp-5 is involved in resistance to bacterial infection and heavy metal and its regulation requires TIR-1 and a p38 map kinase cascade. Biochem. Biophys. Res. Commun. 363: 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws T. R., Smith S. A., Smith M. P., Harding S. V., Atkins T. P., et al. , 2005. The nematode Panagrellus redivivus is susceptible to killing by human pathogens at 37 degrees C. FEMS Microbiol. Lett. 250: 77–83. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V., 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Li G. H., Shen Y. M., Zhang K. Q., 2005. Nematicidal activity and chemical component of Poria cocos. J. Microbiol. 43: 17–20. [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. 1000 Genome Project Data Processing Subgroup. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C. D., Graf-Whitsel J., Wood W. B., 1987. Isolation and characterization of a nematode transposable element from Panagrellus redivivus. Proc. Natl. Acad. Sci. USA 84: 5325–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhov V. V., Hope W. D., 1994. Nematodes: Structure, Development, Classification, and Phylogeny. Smithsonian Institution Press, Washington, DC. [Google Scholar]
- Mayer W. E., Schuster L. N., Bartelmes G., Dieterich C., Sommer R. J., 2011. Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol. Biol. 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzstein M. M., Stanfield G. M., Horvitz H. R., 1998. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 14: 410–416. [DOI] [PubMed] [Google Scholar]
- Michel J. J., Xiong Y., 1998. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 9: 435–449. [PubMed] [Google Scholar]
- Mitreva M., Jasmer D. P., Zarlenga D. S., Wang Z., Abubucker S., et al. , 2011. The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 43: 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Schwarz E. M., Williams B., Schaeffer L., Antoshechkin I., et al. , 2010. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res. 20: 1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X. M., Wang Y. L., Chu Y. S., Xue H. X., Li N., et al. , 2010. Nematodetoxic aurovertin-type metabolites from a root-knot nematode parasitic fungus Pochonia chlamydosporia. J. Agric. Food Chem. 58: 828–834. [DOI] [PubMed] [Google Scholar]
- Ogawa A., Bento G., Bartelmes G., Dieterich C., Sommer R. J., 2011. Pristionchus pacificus daf-16 is essential for dauer formation but dispensable for mouth form dimorphism. Development 138: 1281–1284. [DOI] [PubMed] [Google Scholar]
- Opperman C. H., Bird D. M., Williamson V. M., Rokhsar D. S., Burke M., et al. , 2008. Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. USA 105: 14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M., Yueh A., Leung J., Goff S. P., 2003. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell 115: 319–331. [DOI] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I., 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J., 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6: 9–20. [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J. H., Willems A., Johnson J. L., Srayko M., et al. , 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316. [DOI] [PubMed] [Google Scholar]
- Robert V. J., Sijen T., van Wolfswinkel J., Plasterk R. H., 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19: 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Thomas J. H., 2006. The putative chemoreceptor families of C. elegans (January 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.66.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Rocheleau C. E., Cullison K., Huang K., Bernstein Y., Spilker A. C., et al. , 2008. The Caenorhabditis elegans ekl (enhancer of ksr-1 lethality) genes include putative components of a germline small RNA pathway. Genetics 178: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Trelles F., Tarrio R., Ayala F. J., 2002. A methodological bias toward overestimation of molecular evolutionary time scales. Proc. Natl. Acad. Sci. USA 99: 8112–8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P., 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikas A., Hartmann T., Pan Z. Q., 2011. The cullin protein family. Genome Biol. 12: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedel O. N., Gerisch B., Antebi A., Sternberg P. W., 2012. Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol. 10: e1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M. H., Zerbino D. R., Vingron M., Birney E., 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. S., Benci J. L., Selote D. S., Sharma A. K., Chen A. G., et al. , 2010. Detoxification of multiple heavy metals by a half-molecule ABC transporter, HMT-1, and coelomocytes of Caenorhabditis elegans. PLoS ONE 5: e9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheps J. A., Ralph S., Zhao Z., Baillie D. L., Ling V., 2004. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 5: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R. J., Streit A., 2011. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annu. Rev. Genet. 45: 1–20. [DOI] [PubMed] [Google Scholar]
- Srinivasan J., Durak O., Sternberg P. W., 2008. Evolution of a polymodal sensory response network. BMC Biol. 6: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., Haussler D., 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. [DOI] [PubMed] [Google Scholar]
- Stansfield I., Eurwilaichitr L., Akhmaloka, Tuite M. F., 1996. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 20: 1135–1143. [DOI] [PubMed] [Google Scholar]
- Stein L. D., Bao Z., Blasiar D., Blumenthal T., Brent M. R., et al. , 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1981. Gonadal cell lineages of the nematode Panagrellus redivivus and implications for evolution by the modification of cell lineage. Dev. Biol. 88: 147–166. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1982. Postembryonic nongonadal cell lineages of the nematode Panagrellus redivivus: description and comparison with those of Caenorhabditis elegans. Dev. Biol. 93: 181–205. [DOI] [PubMed] [Google Scholar]
- Stock P. S., Nadler S. A., 2006. Morphological and molecular characterisation of Panagrellus spp. (Cephalobina: Panagrolaimidae): taxonomic status and phylogenetic relationships. Nematology 8: 921–938. [Google Scholar]
- Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Prive G. G., 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Sundaram P., Han W., Cohen N., Echalier B., Albin J., et al. , 2008. Caenorhabditis elegans ABCRNAi transporters interact genetically with rde-2 and mut-7. Genetics 178: 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W. A., Bosch L., 1990. RNA pseudoknots: translational frameshifting and readthrough of viral RNAs. Virus Genes 4: 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., 2006. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 16: 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., Robertson H. M., 2008. The Caenorhabditis chemoreceptor gene families. BMC Biol. 6: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin P. E., Lilley C. J., Atkinson H. J., 2002. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Molecular plant-microbe interactions. Mol. Plant Microbe Interact. 15: 747–752. [DOI] [PubMed] [Google Scholar]
- van Megan H., van den Elsen S., Holterman M., Karssen G., Mooyman P., et al. , 2009. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 11: 927–950. [Google Scholar]
- Viney M. E., Thompson F. J., 2008. Two hypotheses to explain why RNA interference does not work in animal parasitic nematodes. Int. J. Parasitol. 38: 43–47. [DOI] [PubMed] [Google Scholar]
- Werren J. H., Richards S., Desjardins C. A., Niehuis O., Gadau J., et al. , 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell J. W., 2005. Immunoproteasomes: regulating the regulator. Proc. Natl. Acad. Sci. USA 102: 9089–9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Shaham S., Ledoux S., Ellis H. M., Horvitz H. R., 1993. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75: 641–652. [DOI] [PubMed] [Google Scholar]