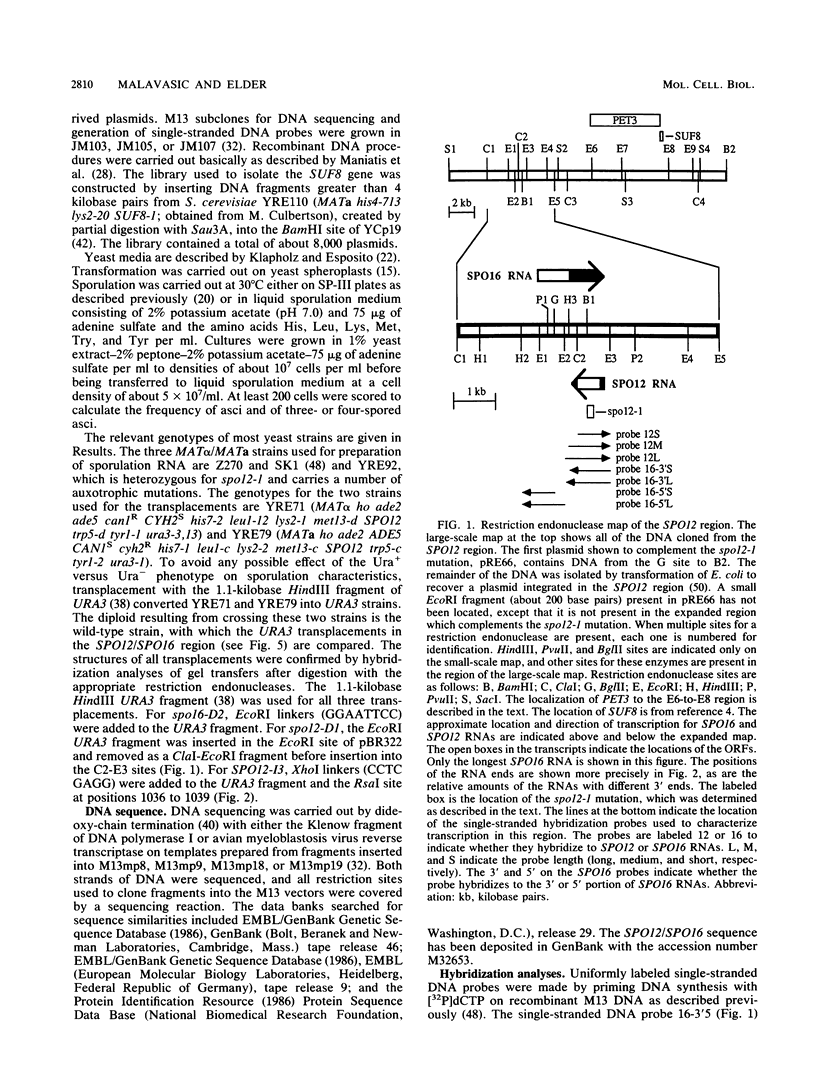
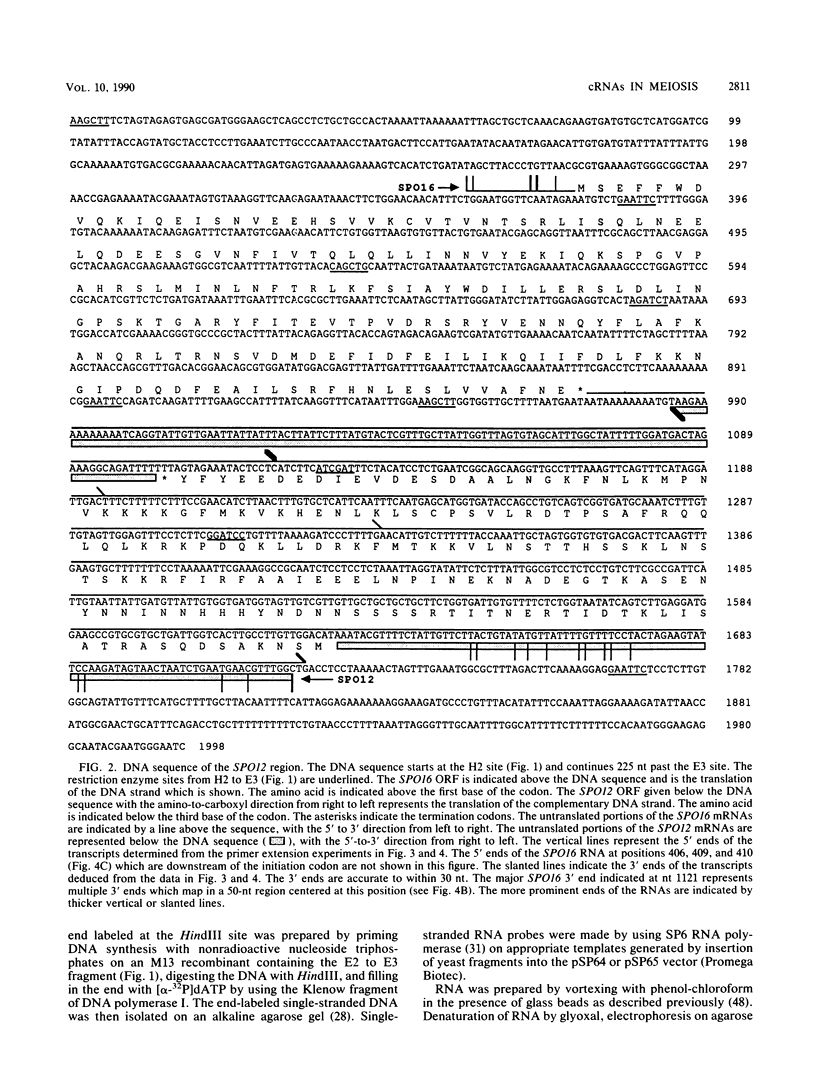
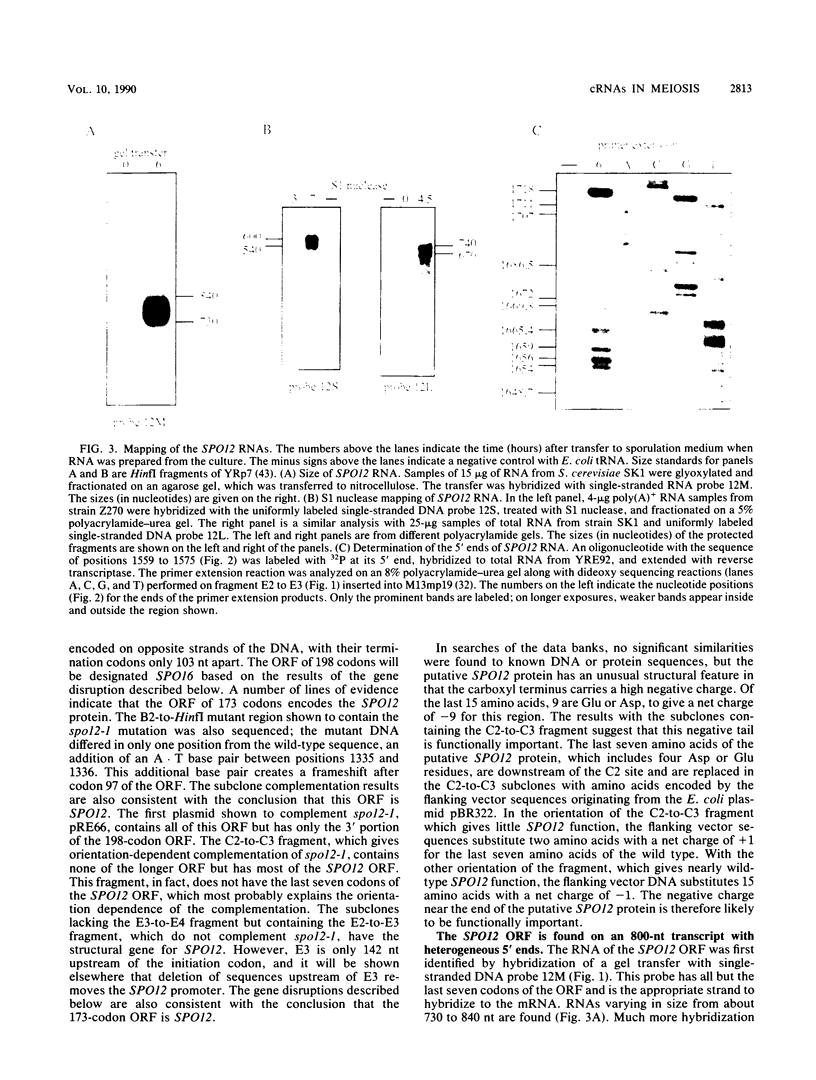
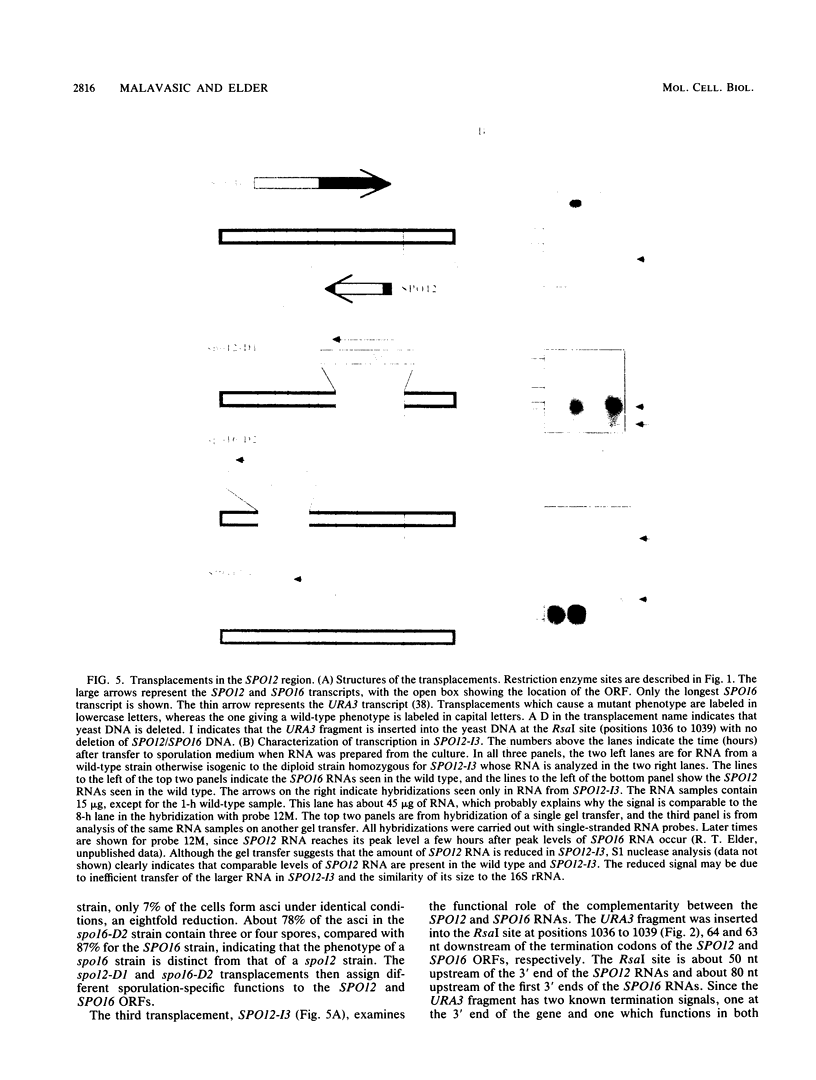
Abstract
The SPO12 gene, which is required for meiosis I chromosome division during sporulation of the yeast Saccharomyces cerevisiae, has been isolated. DNA sequencing has identified an open reading frame of 173 codons that encodes the putative SPO12 protein and has no significant sequence similarities to known genes. The last 15 amino acids of this putative protein have a high negative charge, which appears to be required for function. A second sporulation-specific gene, designated SPO16, was found adjacent to SPO12 and shown to be necessary for efficient spore formation. The two genes are encoded on opposite DNA strands with only 103 nucleotides between the termination codons. Up to 700 nucleotides of the SPO12 and SPO16 transcripts are complementary, and the 3' untranslated region of the longest SPO16 transcript is complementary to all or nearly all of the SPO12 mRNA. A strain homozygous for an insertion which removes the complementarity between the SPO12 and SPO16 mRNAs has an efficiency of sporulation, number of spores per ascus, and spore viability identical to those of a wild-type strain. The complementarity therefore has either no function or only a subtle function in meiosis and sporulation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman J. P., Bond C. T., Douglass J., Herbert E. Two mammalian genes transcribed from opposite strands of the same DNA locus. Science. 1987 Mar 20;235(4795):1514–1517. doi: 10.1126/science.3547652. [DOI] [PubMed] [Google Scholar]
- Atcheson C. L., DiDomenico B., Frackman S., Esposito R. E., Elder R. T. Isolation, DNA sequence, and regulation of a meiosis-specific eukaryotic recombination gene. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8035–8039. doi: 10.1073/pnas.84.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Cummins C. M., Culbertson M. R., Knapp G. Frameshift suppressor mutations outside the anticodon in yeast proline tRNAs containing an intervening sequence. Mol Cell Biol. 1985 Jul;5(7):1760–1771. doi: 10.1128/mcb.5.7.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C. M., Gaber R. F., Culbertson M. R., Mann R., Fink G. R. Frameshift suppression in Saccharomyces cerevisiae. III. Isolation and genetic properties of group III suppressors. Genetics. 1980 Aug;95(4):855–879. doi: 10.1093/genetics/95.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C. Anionic regions in nuclear proteins. J Cell Biol. 1987 Oct;105(4):1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber A. T., Segall J. The SPS4 gene of Saccharomyces cerevisiae encodes a major sporulation-specific mRNA. Mol Cell Biol. 1986 Dec;6(12):4478–4485. doi: 10.1128/mcb.6.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M. G., Petes T. D. Most of the yeast genomic sequences are not essential for cell growth and division. Cell. 1986 Sep 26;46(7):983–992. doi: 10.1016/0092-8674(86)90697-5. [DOI] [PubMed] [Google Scholar]
- Gottlin-Ninfa E., Kaback D. B. Isolation and functional analysis of sporulation-induced transcribed sequences from Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2185–2197. doi: 10.1128/mcb.6.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Eghtedarzadeh M. K. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics. 1987 Dec;117(4):711–725. doi: 10.1093/genetics/117.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Keene M. A., Fechtel K., Fristrom J. W. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 1986 Jan 17;44(1):33–42. doi: 10.1016/0092-8674(86)90482-4. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Delihas N. Small RNAs in the prokaryotes: a growing list of diverse roles. Cell. 1988 Apr 8;53(1):5–7. doi: 10.1016/0092-8674(88)90480-1. [DOI] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Kaback D. B., Feldberg L. R. Saccharomyces cerevisiae exhibits a sporulation-specific temporal pattern of transcript accumulation. Mol Cell Biol. 1985 Apr;5(4):751–761. doi: 10.1128/mcb.5.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D., Kirschner M. W. An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell. 1989 Nov 17;59(4):687–696. doi: 10.1016/0092-8674(89)90015-9. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. A new mapping method employing a meiotic rec-mutant of yeast. Genetics. 1982 Mar;100(3):387–412. doi: 10.1093/genetics/100.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics. 1980 Nov;96(3):567–588. doi: 10.1093/genetics/96.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics. 1980 Nov;96(3):589–611. doi: 10.1093/genetics/96.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Waddell C. S., Esposito R. E. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985 Jun;110(2):187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes S., Ma H., Overbye K., Fox M. S., Botstein D. Fine structure recombinational analysis of cloned genes using yeast transformation. Genetics. 1987 Jan;115(1):73–81. doi: 10.1093/genetics/115.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law D. T., Segall J. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol Cell Biol. 1988 Feb;8(2):912–922. doi: 10.1128/mcb.8.2.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. Recombinationless meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Oct;1(10):891–901. doi: 10.1128/mcb.1.10.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin C. S., Warner J. R., Edmonds M., Nakazato H., Vaughan M. H. Polyadenylic acid sequences in yeast messenger ribonucleic acid. J Biol Chem. 1973 Feb 25;248(4):1466–1471. [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Transcription initiation of the Saccharomyces cerevisiae iso-1-cytochrome c gene. Multiple, independent T-A-T-A sequences. J Mol Biol. 1986 Feb 5;187(3):363–378. doi: 10.1016/0022-2836(86)90439-0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miyajima N., Horiuchi R., Shibuya Y., Fukushige S., Matsubara K., Toyoshima K., Yamamoto T. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell. 1989 Apr 7;57(1):31–39. doi: 10.1016/0092-8674(89)90169-4. [DOI] [PubMed] [Google Scholar]
- Morrison A., Miller E. J., Prakash L. Domain structure and functional analysis of the carboxyl-terminal polyacidic sequence of the RAD6 protein of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Mar;8(3):1179–1185. doi: 10.1128/mcb.8.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Percival-Smith A., Segall J. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2443–2451. doi: 10.1128/mcb.6.7.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988 Dec 23;55(6):929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D. Structure and function of the yeast URA3 gene. Differentially regulated expression of hybrid beta-galactosidase from overlapping coding sequences in yeast. J Mol Biol. 1983 Nov 15;170(4):883–904. doi: 10.1016/s0022-2836(83)80193-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A., Gietz R. D., Hodgetts R. B. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature. 1986 Jul 17;322(6076):279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash S., Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988 Nov;2(11):1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Wise J. A., Guthrie C. A U4-like small nuclear RNA is dispensable in yeast. Cell. 1983 Dec;35(3 Pt 2):753–762. doi: 10.1016/0092-8674(83)90108-3. [DOI] [PubMed] [Google Scholar]
- Tsuboi M. The isolation and genetic analysis of sporulation-deficient mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1983;191(1):17–21. doi: 10.1007/BF00330883. [DOI] [PubMed] [Google Scholar]
- Wagstaff J. E., Klapholz S., Esposito R. E. Meiosis in haploid yeast. Proc Natl Acad Sci U S A. 1982 May;79(9):2986–2990. doi: 10.1073/pnas.79.9.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. T., Frackman S., Kowalisyn J., Esposito R. E., Elder R. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol Cell Biol. 1987 Apr;7(4):1425–1435. doi: 10.1128/mcb.7.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- Winston F., Chumley F., Fink G. R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- Yamashita I., Fukui S. Transcriptional control of the sporulation-specific glucoamylase gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3069–3073. doi: 10.1128/mcb.5.11.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarger J. G., Armilei G., Gorman M. C. Transcription terminator-like element within a Saccharomyces cerevisiae promoter region. Mol Cell Biol. 1986 Apr;6(4):1095–1101. doi: 10.1128/mcb.6.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Carbon J., Bloom K. Tightly centromere-linked gene (SPO15) essential for meiosis in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):158–167. doi: 10.1128/mcb.6.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]






