Abstract
Focal adhesions are large protein complexes organized at the basal surface of cells, which physically connect the extracellular matrix to the cytoskeleton and have long been speculated to mediate cell migration. However, whether clustering of these molecular components into focal adhesions is actually required for these proteins to regulate cell motility is unclear. Here we use quantitative microscopy to characterize descriptors of focal adhesion and cell motility for mouse embryonic fibroblasts and human fibrosarcoma cells, across a wide range of matrix compliance and following genetic manipulations of focal adhesion proteins (vinculin, talin, zyxin, FAK, and paxilin). This analysis reveals a tight, biphasic gaussian relationship between mean size of focal adhesions (not their number, surface density, or shape) and cell speed. The predictive power of this relationship is comprehensively validated by disrupting nonfocal adhesion proteins (α-actinin, F-actin, and myosin II) and subcellular organelles (mitochondria, nuclear DNA, etc.) not known to affect either focal adhesions or cell migration. This study suggests that the mean size of focal adhesions robustly and precisely predicts cell speed independently of focal adhesion surface density and molecular composition.—Kim, D.-H., Wirtz, D. Focal adhesion size uniquely predicts cell migration.
Keywords: motility, mechanosensing, high-throughput phenotyping, systems biology
Cell migration is a highly integrated process resulting from multiple, complex interactions among a myriad of proteins and intracellular organelles (nucleus, microtubule organizing center, endoplasmic reticulum, etc.) in response to the rapidly changing extracellular environment (1–12) Focal adhesions are organized aggregates of specialized proteins (including cytoskeletal, signaling, regulatory, mechanosensing, structural, and scaffolding proteins) distributed at the basal surface of adherent cells (4, 7, 13). These proteins are involved in physical connections between the extracellular matrix and the actin cytoskeleton through transmembrane receptor integrins and mediate signaling between cell and environment (4, 7). Focal adhesions have long been speculated to play a critical role in many cell functions, in particular, cell migration (8). The presumption that focal adhesions play a role in cell migration stems in part from the fact that rapidly moving cells, such as Dictyostelium discoideum and neutrophils, display vanishingly small focal adhesions (14), while slowly moving cells such as fibroblasts show prominent focal adhesions. On the other hand, theoretical modeling and manipulation of ligand density on the substrate have also shown that the cell migration speed changes biphasically depending on the adhesion strength between cell and substratum (15, 16).
The apparent role of individual focal adhesion proteins in cell migration has been extensively studied through genetic manipulations and pharmacological interventions affecting the expresssion or activity of focal adhesion proteins (1–5, 7, 8, 13). However, whether a subset or all focal adhesion-specific proteins need to cluster into focal adhesion complexes in order to mediate cell migration is unknown, i.e., whether any change in the clustering of focal adhesion proteins induced by a change in expression/activation of a known or yet unidentified regulator of focal adhesions or biophysical/biochemical changes in the microenvironment necessarily can predict a change in cell migration is unknown. This is partly due to the fact that a functional relationship between focal adhesions (size, shape, number, turnover dynamics, etc.) and cell motility, if it exists, is largely missing.
That any change in cell speed could be predicted by a change in focal adhesions is a priori unlikely. We first determined functional relationships among descriptors of focal adhesion morphology and descriptors of cell motility, and then assessed the predictive power of relationships between these two families of descriptors through comprehensive blind tests affecting known and previously unknown regulators of cell speed and focal adhesions.
MATERIALS AND METHODS
Cell culture and drug treatments
Mouse embryonic fibroblasts (MEFs) and HT-1080 cells were cultured in DMEM [American Type Culture Collection (ATCC), Manassas, VA, USA] supplemented with 10% FBS (ATCC). Penicillin (100 U/ml) and 100 μg/ml streptomycin (Sigma, St. Louis, MO, USA) for MEFs and 0.1% gentimicin (Sigma) for HT-1080 were added, respectively. Cells infected with small hairpin RNA (shRNA) constructs were initially selected with medium containing 4 μg/ml puromycin (Sigma) for 3 d and then maintained in medium with 3 μg/ml puromycin added. Cells were maintained at 37°C with 5% CO2 in a humidified incubator and passaged every 3–4 d. F-actin depolymerizing drug latrunculin B (Sigma), mitochondrial complex I inhibitor rotenone (Sigma), and cell cycle inhibitor bleomycin (Sigma) were diluted to final concentrations of 0.1 μM (or 1 μM for high-dose treatment), 1 μM, and 1 mM, respectively. Cells were incubated with each drug in culture medium for 1 h before fixation.
Substrate preparation
Following the established method (1, 17), soft substrates denoted by stiff gel or soft gel were prepared by synthesizing polyacrylamide gel onto the 3-aminopropyl-trimethoxysilane and 10% glutaraldehyde-treated glass slides. Acrylamide and N,N-methylenebisacrylamide were mixed at a final concentration of 5 and 0.15% (for stiff gel or 0.015% for soft gel) in distilled H2O (w/v), respectively. Ammonium persulfate (10%) and N,N,N′,N′-tetramethylethylenediamine (Invitrogen, Carlsbad, CA, USA) were introduced as 5 and 0.5% (v/v) of total mixture. The hydrogels were then treated with the UV-activable cross-linker, N-sulfosuccinimidyl-6-(4′-azido-2′-nitrophenylamino) hexanoate (Pierce, Rockford, IL, USA) to bind collagen to the surface where 0.2 mg/ml type I collagen (BD Biosciences, San Jose, CA, USA) diluted in 0.2 N acetic acid was coated for 6 h at 4°C. Glass slides were also prepared by coating the same collagen. All substrates were soaked into medium and kept in the incubator for 30 min to adapt substrates to the cell culture environment before plating cells.
Protein depletion
Following RNAi, sequences targeting mRNA were selected as described previously (17, 18). After treating cells with lentiviral-mediated RNAi, cells showing > 90% knockdown efficiency were selected. They include the following (number after the sequence denotes the targeting position in mRNA): mh-Talin1,2, GTGGATGAGAAGACCAAGGA (1372); mh-Paxillin, gTCAAGGAGCAGAACGACAA (1770); mh-FAK, GGGCATCATTCAGAAGATA (507); m-ACTN1,4, GGAGGACTTCCGAGACTATA (1164); and m-Zyxin, GCCTGTGTCTTCTGCTAATA (1004). A firefly luciferase shRNA was used as a control (5′-GCTTACGCTGAGTACTTCGA). Using the predicted sequences above, the shRNA expression cassettes were constructed by joint PCR, as described. shRNA-induced protein depletion was fully validated by Western blots before use (17, 18).
Immunofluorescence and morphometric analysis
Cells were incubated for 6 h after plating, fixed with 2% paraformaldehyde (Sigma) for 10 min, permeabilized with 0.1% Triton X-100 (Fisher Biotech, Hampton, NH, USA) for 10 min, and then blocked with PBS supplemented with FBS (10%, v/v) for 20 min. For immunostaining of focal adhesions, anti-vinculin antibody (Sigma) at 1:200, anti-focal adhesion kinase (FAK) antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1:50, anti-paxillin antibody (G. D. Longmore, Washington University in St. Louis, St. Louis, MO, USA) at 1:100, and anti-zyxin antibody (Sigma) at 1:100 were used. Actin filaments and nuclear DNA were stained using Alexa-Fluor phalloidin 488 and 300 nM 4′,6-diamidino-2-phenylindole (both from Invitrogen), respectively. For quantitative analysis of focal adhesions, fluorescent images were collected using a Cascade 1K CCD camera (Roper Scientific, Tucson, AZ, USA) mounted on a fluorescence microscope (TE2000E; Nikon, Melville, NY, USA) or a confocal laser microscope (A1; Nikon) equipped with an ×60 Plan Fluor lens (N.A. 1.4). Morphometric analysis (area, perimeter, length, breadth, and shape factor) of focal adhesions was conducted using image analysis software, Metamorph (Molecular Devices, Downingtown, PA, USA), and images collected by confocal microscope were processed using NIS-Elements (Nikon).
Analysis of cell motility
After incubating cells for 6 h, cells were moved to the live cell chamber, mounted on a fluorescence microscope (TE2000E; Nikon) and monitored using an ×10 lens for 8 h. Single cells were tracked using Metamorph Offline, and their x, y coordinates were recorded every 2 min. Cell speed was defined as root-mean-squared displacement calculated every 2 min of time interval divided by 2 min. Custom-made MatLab code was used to calculate mean squared displacement (MSD). Final distance was the displacement that a cell made for 8 h. To introduce persistence distance, persistence time, and number of turns, persistence vectors were calculated from cell tracking data (x, y coordinates, distance, and time) using an Excel macro (Microsoft, Redmond, WA, USA) as described previously (18).A persistent move was defined as the traveling length (≥10 μm) of a cell before it changed a moving direction significantly (>70°). Accordingly, persistent distance and persistent time were defined as the distance and duration that a cell traveled during a persistent move. The number of turns that defines the changes of persistent moves for 8 h of tracking interval was also counted. At least 50 cells were analyzed per condition.
Data processing and statistical analysis
To calculate and plot means ± sem of measured quantities, GraphPad Prism (GraphPad Software, San Diego, CA, USA) was used. Significances were assessed by 2-tailed unpaired t-tests. For multiple comparisons, 1-way ANOVA was applied, using Bonferroni's test to compare every pair of data or using Dunnette's test to compare all other conditions to control, respectively. Degree of correlation of compared data sets was assessed by Pearson product moment correlation, and data were regressed by gaussian fit or linear fit. See individual figure captions to see the selected method.
RESULTS
Size and shape of focal adhesions are closely related
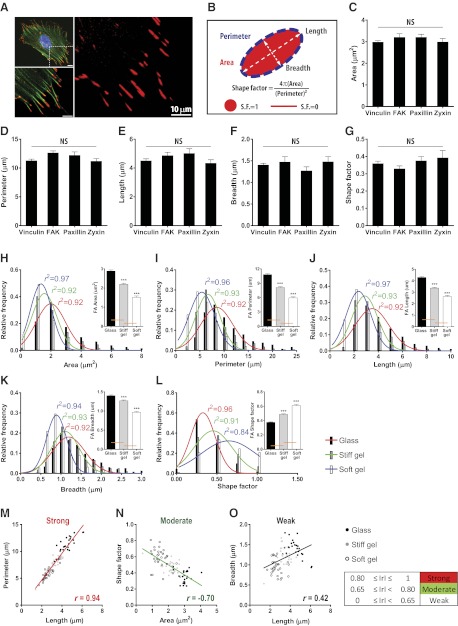
Focal adhesions localized at the basal surface of MEFs and terminating actin stress fibers typically displayed an ellipsoidal shape (Fig. 1A). Fibrillar adhesions, which evolve from mature focal adhesions, are located toward the center of the cell and contain very little focal adhesion components, such as paxillin and vinculin (19, 20). Therefore, we focused on vinculin-containing focal adhesions (21). Size and shape of individual focal adhesions were characterized by surface area, perimeter, length, breadth, and shape factor, defined as 4π(area)/(perimeter)2, approaching 1 for a rounded and 0 for an elongated focal adhesion (Fig. 1B). Notably, we observed no significant difference in measuring morphological parameters from staining with various antifocal adhesion antibodies, including vinculin, FAK, paxillin, and zyxin (Fig. 1C–G). To ensure sufficient distribution in the values of the measured morphological parameters, cells were placed on substrates of different mechanical compliance, where size and shape of focal adhesions readily changed (refs. 1, 4, 22, 23 and Fig. 1H–L). We found that all considered morphological parameters approximately followed gaussian statistics (see color-coded normal distribution curves and corresponding r2 values in Fig. 1H–L).
Figure 1.
Relationships among descriptors of focal adhesion morphology. A) Organization of actin filaments and focal adhesions in an MEF lying on a collagen-coated glass substrate. F-actin (green), nucleus (blue), and vinculin-marked focal adhesions (red) are imaged with a confocal microscope ×60. Details of actin filaments and associated focal adhesions in the region of interest are shown. Focal adhesions are elongated in the same direction of corresponding actin stress fibers. Scale bars = 10 μm. B) Schematic defining the morphological descriptors of focal adhesions used in this study. C–G) Area (C), perimeter (D), length (E), breadth (F), and shape factor (G) of focal adhesions stained with anti-vinculin, anti-FAK, anti-paxillin, and anti-zyxin antibodies. More than 20 cells were analyzed for each condition; values were averaged per cell. Error bars represent sem of averaged values; 1-way ANOVA using Bonferroni post test was applied to compare all possible pairs of conditions. No significant difference in morphological parameters from different focal adhesion staining was detected. H–L) Changes in area (H), perimeter (I), length (J), breadth (K), and shape factor (L) of focal adhesions in response to changes in substrate stiffness: rigid glass (black), a stiff gel (gray), and a soft gel (white). Distributions of these descriptors for different substrate compliances are plotted with color-coded gaussian fits: glass (red), stiff gel (green), and soft gel (blue), respectively, and the same color-coded coefficients of determination (r2) are shown in each plot. Higher average values show broader distributions. More than 900, 600, and 200 focal adhesions were analyzed for glass, stiff gel, and soft gel, respectively, from ≥30 cells/condition. Error bars indicate sem, and statistical differences were calculated using the unpaired t test. ***P < 0.001. M–O) Representative strong (M), moderate (N), and weak (O) correlations among descriptors of focal adhesion morphology. Length of focal adhesions is positively correlated with perimeter and breadth but to different extents (M, O). Size (i.e., area) of focal adhesions in cells is anticorrelated with shape factor (N). Data points are the averaged values from each cell. Degree of correlation is assessed by the Pearson product moment correlation coefficient (denoted by r). Straight line in each plot shows a linear regression of analyzed data set. Red, green, and black colors represent strong (0.80≤|r|≤1), moderate (0.65≤|r|<0.80), and weak (|r|<0.65) correlations; + and − denote positive and negative correlations, respectively. Correlation assessment and linear regression are applied to the merged data sets regardless of substrate stiffness. Correlation coefficients among all pairs of morphological parameters are summarized in Supplemental Fig. S1.
To assess relationships among descriptors of focal adhesion morphology, we calculated the Pearson product-moment correlation coefficients r between any 2 parameters, which were averaged per cell and merged independently of substrate stiffness, and determined the degree of correlation through calculated r values. Degrees of correlation among focal adhesion descriptors were denoted as strong, moderate, and weak for r values: 0.80 ≤ |r| ≤ 1, 0.65 ≤ |r| < 0.80, and |r| < 0.65, respectively (see summary in Supplemental Fig. S1). As expected, the perimeter of focal adhesions increased with the length of long axis (Fig. 1M), while the shape factor of focal adhesions decreased as their size (denoted by area) increased (Fig. 1N). Length and breadth of focal adhesions were weakly correlated (Fig. 1O). Confocal microscopy indicated that focal adhesions were elongated in the direction of their associated actin stress fibers (Fig. 1A). These results are consistent with the growth model of force-induced focal adhesion driven by actomyosin-mediated tension (24–27) Consequently, large focal adhesions tended to be elongated, while small focal adhesions tended to be round over a wide range of substrate compliance.
Cell speed weakly correlates with persistent motion of the cell
To characterize randomly migrating cells, we measured cell speed as well as persistence time and distance, number of turns, and final distance (defined as the total displacement that a cell made during the observation time; Fig. 2A, B). Cell speed was calculated from the root mean square displacement, which is a more rigorous measure of distance than a direct use of segment length, as it eliminates sources of noises stemming from erratically meandering cell motion (see more details on defining parameters in Materials and Methods). Mean values and distributions of cell motility descriptors readily changed in cells placed on substrates of different compliance (refs. 28, 29 and Fig. 2C–G). Similarly to focal adhesion descriptors (Fig. 1H–L), cell motility descriptors were well distributed. (Fig. 2C–G).
Figure 2.
Relationships among descriptors of cell motility. A) Defining cell motility. A trajectory (green), persistence vectors (yellow), and final distance (blue) in a MEF moving on a collagen-coated glass substrate. When a cell changes both its step size and direction significantly (>10 μm and >70°, respectively) a new persistence move is defined. Change of persistent move between neighboring persistence vectors is denoted by a turn. Persistence distances are not uniform during the tracking period. B) Schematic defining cell-motility parameters used in this study. Cell speed was defined as the root MSD calculated every 2 min divided by 2 min. C–G) Changes in speed (C), final distance (D), persistence distance (E), persistence time (F), and number of turns (G) of migrating cells in response to substrate stiffness, from a rigid glass (black), to a stiff gel (gray) and a soft gel (white). Relative frequency distributions of measured quantities for different substrate stiffness are plotted with color-coded gaussian fits: glass (red), stiff gel (green), and soft gel (blue), respectively, and the same color-coded coefficients of determination (r2) are shown in each plot. Error bars indicate sem, and statistical differences were calculated using the unpaired t test. NS, not significant (P>0.05). *P < 0.05, **P < 0.01, ***P < 0.001. H–J) Representative strong (H), moderate (I), and weak (J) correlations among descriptors of cell motility. While the number of turns and final distance that cells made correlate with persistence time and distance in strong and moderate manners, respectively (H, I), cell migration speed weakly correlated with final distance (J). Degree of correlation is assessed by the Pearson product moment correlation coefficient (denoted by r). Straight line in each plot shows linear regression of analyzed data set. Red, green, and black represent strong (0.80≤|r|≤ 1), moderate (0.65≤|r|<0.80), and weak (|r|<0.65) correlations; + and − denote positive and negative correlations, respectively. Correlation assessment and linear regression are applied to the merged data sets regardless of substrate stiffness. In panels C–J, ≥50 cells were monitored for 8 h per condition. Persistence vectors were calculated with a custom Excel macro (see Materials and Methods). Correlation coefficients among all pairs of motility parameters are summarized in Supplemental Fig. S1.
As expected, when cells spent more time in each persistent move, they had fewer chances of switching into a next persistent move in the limited observation time, which resulted in fewer numbers of turns (r=−0.87; Fig. 2H). Interestingly, the final distance was moderately correlated with persistence distance (r=0.68; Fig. 2I), but weakly correlated with cell speed (r=0.40; Fig. 2J). These results indicate that fast-moving cells do not guarantee that they can move farther (Fig. 2J) but rather cells that can maintain their directional movement (thus higher persistence distance) move farther (Fig. 2I); cell speed is weakly correlated with persistence of migration (Supplemental Fig. S1A).
Functional relationship between focal adhesion morphology and cell migration
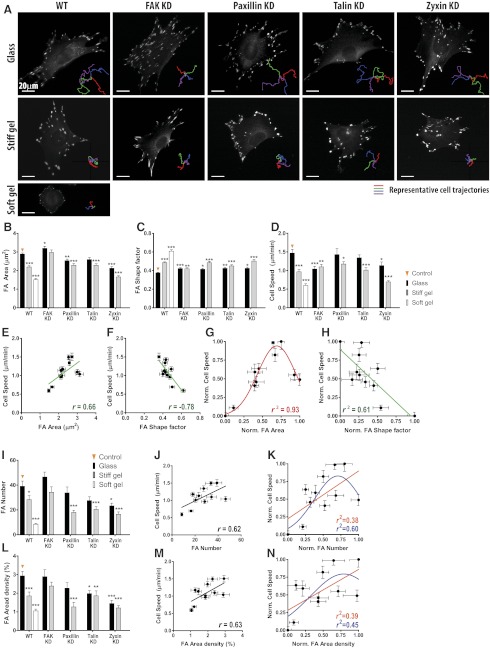
Now we can begin to test whether focal adhesion morphology can predict cell motility. To determine global correlations between focal adhesion morphology and cell motility, genetic manipulations of the cells were added to the extracellular physical variations discussed so far so as to expand the data sets. Major focal adhesion proteins (FAK, paxillin, talin, and zyxin) were depleted using shRNA technology as reported previously (17), and resulting cells were placed on substrates of controlled stiffness (Fig. 3A). Although individual effects of depletion of a focal adhesion protein or changes in substrate compliance on focal adhesion morphology or cell motility have been extensively studied (1, 2, 4, 30, 31), effects of combined disturbances (genetic and physical) are hard to interpret, since deviation of measured quantities from the control condition is not simply sum of the two individual effects, but the results of synergistic or compensatory effects that may be triggered by unrevealed or less related signal pathways. Therefore, instead of identifying the effects of individual disturbances, we compared the combined effect of 2 variations (i.e., substrate stiffness and genetic manipulations of focal adhesion proteins) to the control condition (i.e., wild type [WT] cells on glass slide) using 1-way ANOVA (Fig. 3B–D and Supplemental Fig. S2A, B).
Figure 3.
Functional relationship between focal adhesion morphology and cell speed: primary data set. A) Immunofluorescence images of vinculin-stained focal adhesions and representative cell trajectories reveal a wide range of focal adhesion morphology and cell speed in primary data set. B–D) Effect of changes in substrate compliance and depletion of focal adhesion proteins on the size (B) and shape (C) of focal adhesions and cell migration speed (D). E, F) Correlations of size and shape of focal adhesions with cell speed. Size (i.e., area) and shape factor of focal adhesions are positively (E) and negatively (F) correlated with cell speed, respectively. Straight line in each plot shows linear regression of analyzed data set. Degree of correlation between focal adhesion size and cell speed is not very strong (r=0.66), and cell speed changes nonmonotonously with focal adhesion size (E). Pearson coefficient r, r2, and P values and corresponding plots of all pairs of descriptors of focal adhesion morphology and cell motility are summarized in Supplemental Fig. S2 and Fig. 4. G, H) Gaussian (G) and linear (H) regressions of size and shape factor of focal adhesions vs. cell speed. Goodness of the fit of the gaussian relationship between focal adhesion size and cell speed (r2=0.93) was greater than the linear relationship between focal adhesion shape factor and cell speed (r2=0.61). I, L) Changes in number of focal adhesions per cell (I) and focal adhesion area density, defined as the fraction of cell area occupied by focal adhesions (L), in response to changes in substrate stiffness: rigid glass (black), a stiff gel (gray), and a soft gel (white) and depletion of focal adhesion proteins (FAK, paxillin, talin, and zyxin). J, M) Correlations of number of focal adhesions per cell (J) and area density (%) of focal adhesions (M) with cell speed. Both number and area density of focal adhesions per cell weakly correlate with cell speed (i.e., |r|<0.65). K, N) Assessment of regressed relationships of number of focal adhesions per cell (K) and area density of focal adhesions (N) with cell speed using either linear (orange) or gaussian (blue) models. goodness of fits for both linear and gaussian relationships was relatively weak compared to focal adhesion size vs. cell speed (G). In panels B–D, I, and L, focal adhesions were analyzed in >30 cells, and >50 cells were tracked per condition. Error bars represent sem of averaged values; 1-way ANOVA using Dunnett's post test was applied based on the values in the WT cells on stiff substrate (control) for multiple comparisons. Only comparisons with significant statistical difference (P<0.05) are shown. In panels E, F, J, and M, criteria for assessing the degree of correlation is the same as those in Fig. 1M–O and Fig. 2H–J. In panels G, H, K, and N, data are normalized as (x − xmin)/(xmax − xmin) so that all data ranges between 0 (min) and 1 (max). *P < 0.05, **P < 0.01, ***P < 0.001.
Pearson correlation analysis revealed that cell speed and persistence of migration were correlated with a subset of focal adhesion parameters (Figs. 3E, F and 4 and Supplemental Fig. S2). Specifically, focal adhesion size moderately correlated with cell speed (Fig. 3E) but weakly correlated with descriptors of migratory persistence (Supplemental Fig. S2C), while the shape factor of focal adhesions moderately correlated with cell speed (Fig. 3F) as well as final distance traveled and persistence distance (Supplemental Fig. S2D). Notably, focal adhesions were larger and more elongated in fast-moving cells (Fig. 3E, F). However, we also noted that the magnitude of correlation coefficient between focal adhesion size and cell speed was not high, which suggested that focal adhesion size and cell speed were not well described by a linear relationship, as assumed by Pearson correlation analysis.
Figure 4.
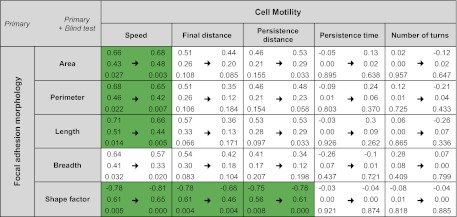
Summary of correlations between focal adhesion morphology and cell motility. Pearson correlation coefficient r, r2, and P values are calculated (top, middle, and bottom, respectively) between focal adhesion morphology (area, perimeter, length, breadth, and shape factor) and cell motility (speed, final distance, persistence distance, persistence time, and number of turns) in primary data set (left) and blind test results included data set (right). Data in green represent moderate (0.65≤ |r| <0.80) correlations; + and − denote positive and negative correlations, respectively. Focal adhesion size (denoted by area) correlates with cell speed, but the magnitude is not very high (r=0.66 or 0.68). Blind test results do not substantially disturb the degree of correlations determined by the primary data set.
Indeed, while focal adhesion shape and cell speed were monotonically related (Fig. 3F), we observed a prominent biphasic relationship between focal adhesion size and cell speed (Fig. 3E). Thus, gaussian (i.e., nonlinear) and linear models were applied to the normalized data set ranged between 0 to 1 to assess the nonmonotonic response of cell speed to the changes in focal adhesion size (Fig. 3G, H). We found that a gaussian fit of focal adhesion size vs. cell speed was excellent (r2=0.93; Fig. 3G), revealing a tight, nonmonotonic relationship between focal adhesion size and cell speed. This result indicated that, over a wide range of conditions (∼70% of the total range in measured focal adhesion size), cell speed increased when focal adhesions increase in size, until an optimum was reached. Moreover, a gaussian relationship between focal adhesion size and cell speed (r2=0.93; Fig. 3G) was much tighter than the linear relationship between focal adhesion shape factor and cell speed (r2=0.61; Fig. 3H), which suggests that focal adhesion size, rather than shape, was highly predictive of cell migration speed.
We asked whether not only the mean size of individual focal adhesions but also cumulative effect of all focal adhesions present in a cell predicted cell speed. To test this hypothesis, we measured the total number of focal adhesions per cell and focal adhesion area density, defined as the fraction of the cell area occupied by focal adhesions (Fig. 3I, L). These parameters did not show considerable correlations with cell speed (i.e., at least, |r|≥0.65) and they were poorly approximated by linear and nonlinear fits (Fig. 3J, K, M, N). These results reinforce the notion that focal adhesion size is a parameter that uniquely predicts cell speed, independently of the surface coverage of focal adhesions in a cell.
Predictive power of focal adhesion size for cell migration speed
So far, we have assessed the relationship between focal adhesion morphology and cell motility through genetic manipulation of focal adhesion components and controlled changes in substrate compliance, which are known to affect focal adhesions (which we shall call the primary data set). Through 5 blind validating tests, we next tested disturbances progressively more remote (physically and functionally) from focal adhesions to assess whether aggregation of focal adhesion proteins was the functional unit required to modulate cell migration and to determine whether focal adhesion size indeed predicted cell speed.
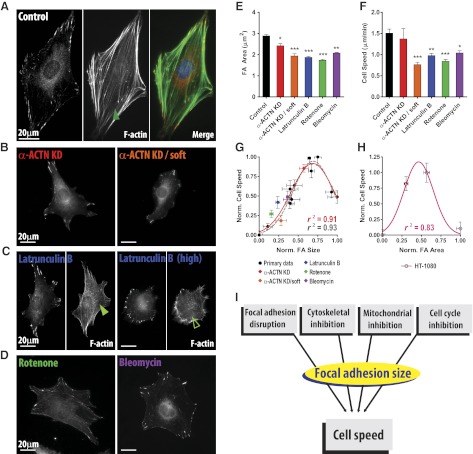
First, α-actinin-1,4, a major F-actin cross-linking/bundling protein (32–36), which is associated with, but is not a component of, focal adhesions, was shRNA depleted, and resulting cells were tested on substrates of low and high compliance. Compared to control WT cells (Fig. 5A), the depletion of α-actinin in cells on stiff substrates slightly reduced the size of focal adhesions, which led to little change in cell speed (red bars, Fig. 5E, F), as correctly predicted by the primary data set (red diamond, Fig. 5G). When these cells were placed on soft substrates, however, the size of focal adhesions changed significantly (Fig. 5B), which in turn changed cell speed (Supplemental Movie S1). These coordinated changes in focal adhesion size and cell speed were again correctly predicted by the primary data set (Fig. 5E–G, orange).
Figure 5.
Functional relationship between focal adhesions and cell speed: validation. A–D) Representative immunofluorescence images of focal adhesions in control cells (A), α-actinin-depleted cells on glass substrate and polyacrylamide gel (denoted by soft; B), and cells subjected to actin depolymerizing latrunculin B (C), or mitochondrial deactivator rotenone and cell cycle inhibitor bleomycin (D). Thick actin stress fibers in control cells (A) were gradually disorganized depending on the concentration of latrunculin B (C). Only small round punctuate focal adhesions were detected when cells were treated with high dose of latrunculin B. E, F) Changes in focal adhesion size (E) and cell speed (F) resulting from the blind tests. One-way ANOVA using Dunnett's post test was applied based on the values in the WT cells on stiff substrate (control) for multiple comparisons. Only comparisons with significant statistical difference (P<0.05) are shown. *P < 0.05, **P < 0.01, ***P < 0.001. G) Comparison of relationship between focal adhesion size and cell speed before and after merging blind test results with the primary data set (gray dotted line vs. red solid line), including depletion of α-actinin, treatment of latrunculin B, rotenone, and bleomycin. All the calculated Pearson coefficient r, r2, and P values and corresponding plots of all pairs of normalized values of focal adhesion morphology and cell motility including blind test results are summarized in Supplemental Fig. S3 and Fig. 4. Blind test results do not disturb the relationships calculated from primary data set. H) Confirmation of relationship between focal adhesion size and cell speed in human cells. A tight biphasic relationship was reconstructed using WT and FAK-, talin-, and zyxin-depleted human fibrosarcoma HT-1080 cells (r2=0.83). Focal adhesion size and cell speed of these cells are summarized in Supplemental Fig. S3C. In panels E–H, focal adhesions were analyzed >30 cells and >50 cells were tracked per condition. Error bars represent sem of averaged values. In panels G–H, data are normalized as (x − xmin)/(xmax − xmin), so that all data are ranged between 0 (min) and 1 (max). I) Schematic of cell speed prediction by focal adhesion size. Regardless of the type of cell disturbance, focal adhesion size directly predicts cell speed.
Next, cells were treated with latrunculin B, which inhibits the assembly of actin filaments (17, 37–39). Actin stress fibers were gradually disassembled depending on the concentration of latrunculin B (Fig. 5C). In cells treated with a high dose of latrunculin B (1 μM), actin filaments were dismantled, and a majority of focal adhesions disappeared or diminished to small round punctuate focal adhesions (Fig. 5C), which restricted cell migration. By reducing the concentration of latrunculin B to 0.1 μM to avoid such an extreme response, we observed significant decrease in the size of focal adhesions, which quantitatively decreased migration speed to the extent predicted by the primary data set (Fig. 5E–G, blue).
Finally, in order to further test the predictive power of the relationship between focal adhesion size and cell speed and to verify whether clustering of focal adhesion components into focal adhesions actually controlled cell motility, we next deactivated cell components that had not previously been reported as regulators of focal adhesions or cell migration. Here we used rotenone, which inhibits mitochondrial complexes by interfering with electron transport chain in the mitochondria (40), and bleomycin, which disrupts cell cycle by forming free radicals that break DNA strands and inhibiting thymidine incorporation into DNA (41, 42). Cells treated with these drugs simultaneously reduced focal adhesion size and cell speed in accordance to the primary data set (Fig. 5D, green and purple; E–G).
To test the robustness of the above relationship between focal adhesions and cell migration, we merged the primary data set (Supplemental Fig. S2A, B) based on the depletion of focal adhesion components and changes in substrate compliance with the blind test results (Supplemental Fig. S3A, B) generated by the depletion of α-actinin on stiff and soft substrates, the disruption of F-actin, the deactivation of mitochondria, and the inhibition of cell cycle through release of free radicals (data summarized in Supplemental Fig. S3 and Fig. 4). The functional relationships obtained from the primary data set were remarkably well maintained, even after addition of the validating tests (Supplemental Figs. S2 vs. S3 and Fig. 4), and the characteristic biphasic relationship between focal adhesion size and cell speed was preserved (r2=0.93 and r2=0.91, before and after merging blind test results, Fig. 5G).
Finally, we tested human fibrosarcoma cells to confirm the relationship between focal adhesion size and cell migration speed across species and types of cells. Using previously characterized control HT-1080 cells and FAK-, talin-, and zyxin-depleted HT-1080 cells (18, 43), we quantified focal adhesion size and cell speed as discussed so far (Supplemental Fig. 3C). A tight biphasic relationship between focal adhesion size and cell speed was maintained in human cancer cells (r2=0.83, Fig. 5H).
Together, these results reveal that the mean size of focal adhesions is highly predictive of cell migration speed; i.e., any observed change in cell speed following a modulation of a known or unknown regulator of cell migration can be quantitatively predicted by a change in the mean size of focal adhesions in these cells (see schematic, Fig. 5I).
DISCUSSION
The existence of a robustly predictive, functional relationship between cell migration and focal adhesions has not been previously established. Prior to this work, disparate observations across cell types (e.g., fast-moving Dictyostelium discoideum and neutrophils vs. slowly moving fibroblasts) suggested an inverse relation between focal adhesion size and cell migration speed (14, 44, 45). In contrast, our results reveal that cell speed largely changes biphasically with focal adhesion size. More precisely, MEF speed steadily increases with focal adhesion size, until a threshold value of ∼0.7 (corresponding to ∼2.6 μm2) in normalized focal adhesion size (Figs. 3G and 5G) beyond which cell speed declines. The late stage is mainly due to the FAK-depleted cells whose focal adhesions outgrew and thus cell motility was retarded. This result is in concordance with a previous study showing that inhibition of FAK activity decreased focal adhesion turnover and significantly reduced cell speed (44, 46).
In previous studies (15, 16, 47), cell migration speed biphasically changed depending on the adhesion strength between cell and underlying substrates where ligand density was referred to the adhesion strength and biphasic behavior of cell speed was observed when plotted to the logarithmic scale of the adhesion strength. This may suggest that adhesion strength increases nonlinearly with focal adhesion size. Initially focal adhesion molecules nucleate and grow in size, where adhesion strength rapidly increases to the available maximum value, but adhesion strength might not be further increased even though apparent size of focal adhesions keeps growing, and this focal adhesion enters the decaying phase, where the size of focal adhesions would progressively decrease due to the dissipation of focal adhesion molecules. This notion is supported by the finding that adhesion strength increases nonlinearly with adhesive area controlled by micropatterned adhesive island size (48).
Changes in focal adhesion size seem to serve as a necessary funnel through which intracellular or extracellular disturbances (including changes in substrate compliance, depletion of focal adhesion proteins, disruption of cytoskeleton, deactivation of nonfocal adhesion proteins, e.g., mitochondria, inhibition of cell cycle, etc.) have to go through to regulate cell speed (Fig. 5I). Future work will determine whether the characteristic correlation between focal adhesion protein clustering and cell migration speed demonstrated in this study applies to cells in settings of different dimensionality, such as cancers cells fully embedded in a three-dimensional extracellular matrix (12, 43).
Acknowledgments
The authors thank Dr. Sean X. Sun (Johns Hopkins University) for helpful discussions about the interpretation of correlations discussed in this paper. The authors thank G. D. Longmore and Y. Feng (Washington University in St. Louis, School of Medicine, St. Louis, MO, USA) for using their knockdown cells.
This work was supported by U.S. National Institutes of Health grants U54CA143868 and R01CA174388.
The authors declare no conflicts of interest.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- FAK
- focal adhesion kinase
- MEF
- mouse embryonic fibroblast
- MSD
- mean squared displacement
- shRNA
- small hairpin RNA
- WT
- wild-type
REFERENCES
- 1. Pelham R. J., Jr., Wang Y. (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. U. S. A. 94, 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Discher D. E., Janmey P., Wang Y. L. (2005) Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 [DOI] [PubMed] [Google Scholar]
- 3. Small J. V., Kaverina I., Krylyshkina O., Rottner K. (1999) Cytoskeleton cross-talk during cell motility. FEBS Lett. 452, 96–99 [DOI] [PubMed] [Google Scholar]
- 4. Prager-Khoutorsky M., Lichtenstein A., Krishnan R., Rajendran K., Mayo A., Kam Z., Geiger B., Bershadsky A. D. (2011) Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 13, 1457–1465 [DOI] [PubMed] [Google Scholar]
- 5. Webb D. J., Parsons J. T., Horwitz A. F. (2002) Adhesion assembly, disassembly and turnover in migrating cells: over and over and over again. Nat. Cell Biol. 4, E97–100 [DOI] [PubMed] [Google Scholar]
- 6. Lock J. G., Wehrle-Haller B., Stromblad S. (2008) Cell-matrix adhesion complexes: master control machinery of cell migration. Sem. Cancer Biol. 18, 65–76 [DOI] [PubMed] [Google Scholar]
- 7. Geiger B., Spatz J. P., Bershadsky A. D. (2009) Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 [DOI] [PubMed] [Google Scholar]
- 8. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Cell migration: integrating signals from front to back. Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 9. Lauffenburger D. A., Horwitz A. F. (1996) Cell migration: a physically integrated molecular process. Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 10. Lee J. S. H., Chang M. I., Tseng Y., Wirtz D. (2005) Cdc42 mediates nucleus movement and MTOC polarization in Swiss 3T3 fibroblasts under mechanical shear stress. Mol. Biol. Cell 16, 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hale C. M., Chen W. C., Khatau S. B., Daniels B. R., Lee J. S. H., Wirtz D. (2011) SMRT analysis of MTOC and nuclear positioning reveals the role of EB1 and LIC1 in single-cell polarization. J. Cell Sci. 124, 4267–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khatau S. B., Bloom R. J., Bajpai S., Razafsky D., Zang S., Giri A., Wu P. H., Marchand J., Celedon A., Hale C. M., Sun S. X., Hodzic D., Wirtz D. (2012) The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci. Rep. 2, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patla I., Volberg T., Elad N., Hirschfeld-Warneken V., Grashoff C., Fassler R., Spatz J. P., Geiger B., Medalia O. (2010) Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat. Cell Biol. 12, 909–915 [DOI] [PubMed] [Google Scholar]
- 14. Nagasaki A., Kanada M., Uyeda T. Q. P. (2009) Cell adhesion molecules regulate contractile ring-independent cytokinesis in Dictyostelium discoideum. Cell Res. 19, 236–246 [DOI] [PubMed] [Google Scholar]
- 15. DiMilla P. A., Barbee K., Lauffenburger D. A. (1991) Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys. J. 60, 15–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maheshwari G., Brown G., Lauffenburger D. A., Wells A., Griffith L. G. (2000) Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 113(Pt. 10), 1677–1686 [DOI] [PubMed] [Google Scholar]
- 17. Katz B. Z., Zamir E., Bershadsky A., Kam Z., Yamada K. M., Geiger B. (2000) Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Mol. Biol. Cell 11, 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scales T. M., Parsons M. (2011) Spatial and temporal regulation of integrin signalling during cell migration. Curr. Opin. Cell Biol. 23, 562–568 [DOI] [PubMed] [Google Scholar]
- 19. Dumbauld D. W., Michael K. E., Hanks S. K., Garcia A. J. (2010) Focal adhesion kinase-dependent regulation of adhesive forces involves vinculin recruitment to focal adhesions. Biol. Cell 102, 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goffin J. M., Pittet P., Csucs G., Lussi J. W., Meister J. J., Hinz B. (2006) Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibers. J. Cell Biol. 172, 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo W. H., Frey M. T., Burnham N. A., Wang Y. L. (2006) Substrate rigidity regulates the formation and maintenance of tissues. Biophys. J. 90, 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan C. E., Odde D. J. (2008) Traction dynamics of Filopodia on compliant substrates. Science 322, 1687–1691 [DOI] [PubMed] [Google Scholar]
- 23. Galbraith C. G., Yamada K. M., Sheetz M. P. (2002) The relationship between force and focal complex development. J. Cell Biol. 159, 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Besser A., Safran S. A. (2006) Force-induced adsorption and anisotropic growth of focal adhesions. Biophys. J. 90, 3469–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parsons J. T., Horwitz A. R., Schwartz M. A. (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khatiwala C. B., Peyton S. R., Putnam A. J. (2006) Intrinsic mechanical properties of the extracellular matrix affect the behavior of pre-osteoblastic MC3T3-E1 cells. Am. J. Physiol. Cell Physiol. 290, C1640–C1650 [DOI] [PubMed] [Google Scholar]
- 27. Ulrich T. A., Pardo E. M. D., Kumar S. (2009) The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 69, 4167–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim D. H., Khatau S. B., Feng Y., Walcott S., Sun S. X., Longmore G. D., Wirtz D. (2012) Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci. Rep. 2, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang H. B., Dembo M., Hanks S. K., Wang Y. L. (2001) Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc. Natl. Acad. Sci. U. S. A. 98, 11295–11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu W. M., Baribault H., Adamson E. D. (1998) Vinculin knockout results in heart and brain defects during embryonic development. Development 125, 327–337 [DOI] [PubMed] [Google Scholar]
- 31. Honda K., Yamada T., Endo R., Ino Y., Gotoh M., Tsuda H., Yamada Y., Chiba H., Hirohashi S. (1998) Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J. Cell Biol. 140, 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi C. K., Vicente-Manzanares M., Zareno J., Whitmore L. A., Mogilner A., Horwitz A. R. (2008) Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu J. Y., Tseng Y., Wirtz D. (2000) Strain hardening of actin filament networks: regulation by the dynamic cross-linking protein alpha-actinin. J. Biol. Chem. 275, 35886–35892 [DOI] [PubMed] [Google Scholar]
- 34. Tseng Y., Fedorov E., McCaffery J. M., Almo S. C., Wirtz D. (2001) Micromechanics and ultrastructure of actin filament networks crosslinked by human fascin: a comparison with alpha-actinin. J. Mol. Biol. 310, 351–366 [DOI] [PubMed] [Google Scholar]
- 35. Esue O., Tseng Y., Wirtz D. (2009) Alpha-actinin and filamin cooperatively enhance the stiffness of actin filament networks. PloS ONE 4, e4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wakatsuki T., Schwab B., Thompson N. C., Elson E. L. (2001) Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 114, 1025–1036 [DOI] [PubMed] [Google Scholar]
- 37. Khatau S. B., Hale C. M., Stewart-Hutchinson P. J., Patel M. S., Stewart C. L., Searson P. C., Hodzic D., Wirtz D. (2009) A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. U. S. A. 106, 19017–19022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hale C. M., Sun S. X., Wirtz D. (2009) Resolving the role of actoymyosin contractility in cell microrheology. PloS ONE 4, e7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J. A., Robinson J. P. (2003) Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278, 8516–8525 [DOI] [PubMed] [Google Scholar]
- 40. Byrnes R. W., Petering D. H. (1994) DNA double-strand breakage by bleomycin in Ehrlich ascites tumor cells as measured by nondenaturing filter elution. Rad. Res. 137, 162–170 [PubMed] [Google Scholar]
- 41. Cloos J., Reid C. B., van der Sterre M. L., Tobi H., Leemans C. R., Snow G. B., Braakhuis B. J. (1999) A comparison of bleomycin-induced damage in lymphocytes and primary oral fibroblasts and keratinocytes in 30 subjects. Mutagenesis 14, 87–93 [DOI] [PubMed] [Google Scholar]
- 42. Fraley S. I., Feng Y., Krishnamurthy R., Kim D. H., Celedon A., Longmore G. D., Wirtz D. (2010) A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fraley S. I., Feng Y. F., Giri A., Longmore G. D., Wirtz D. (2012) Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat. Commun. 3, 719–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ilic D., Furuta Y., Kanazawa S., Takeda N., Sobue K., Nakatsuji N., Nomura S., Fujimoto J., Okada M., Yamamoto T. (1995) Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377, 539–544 [DOI] [PubMed] [Google Scholar]
- 45. Thompson O., Moore C. J., Hussain S. A., Kleino I., Peckham M., Hohenester E., Ayscough K. R., Saksela K., Winder S. J. (2010) Modulation of cell spreading and cell-substrate adhesion dynamics by dystroglycan. J. Cell Sci. 123, 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iwanicki M. P., Vomastek T., Tilghman R. W., Martin K. H., Banerjee J., Wedegaertner P. B., Parsons J. T. (2008) FAK, PDZ-RhoGEF and ROCKII cooperate to regulate adhesion movement and trailing-edge retraction in fibroblasts. J. Cell Sci. 121, 895–905 [DOI] [PubMed] [Google Scholar]
- 47. Palecek S. P., Loftus J. C., Ginsberg M. H., Lauffenburger D. A., Horwitz A. F. (1997) Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 385, 537–540 [DOI] [PubMed] [Google Scholar]
- 48. Gallant N. D., Michael K. E., Garcia A. J. (2005) Cell adhesion strengthening: contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol. Biol. Cell 16, 4329–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]