Summary
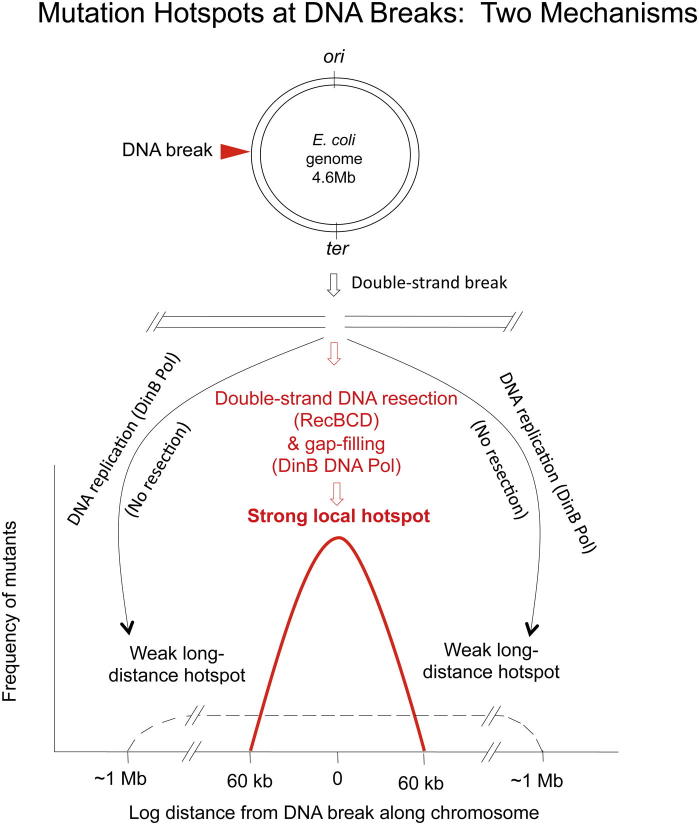
Mutation hotspots and showers occur across phylogeny and profoundly influence genome evolution, yet the mechanisms that produce hotspots remain obscure. We report that DNA double-strand breaks (DSBs) provoke mutation hotspots via stress-induced mutation in Escherichia coli. With tet reporters placed 2 kb to 2 Mb (half the genome) away from an I-SceI site, RpoS/DinB-dependent mutations occur maximally within the first 2 kb and decrease logarithmically to ∼60 kb. A weak mutation tail extends to 1 Mb. Hotspotting occurs independently of I-site/tet-reporter-pair position in the genome, upstream and downstream in the replication path. RecD, which allows RecBCD DSB-exonuclease activity, is required for strong local but not long-distance hotspotting, indicating that double-strand resection and gap-filling synthesis underlie local hotspotting, and newly illuminating DSB resection in vivo. Hotspotting near DSBs opens the possibility that specific genomic regions could be targeted for mutagenesis, and could also promote concerted evolution (coincident mutations) within genes/gene clusters, an important issue in the evolution of protein functions.
Graphical Abstract
Highlights
► Spontaneous mutation pathway in Escherichia coli causes hotpots at double-strand breaks ► Strong local (2–60 kb) hotspot mechanism double-strand resection and gap-fill ► Weak long-distance (1 Mb) mutagenesis by break-induced replication ► Break-induced replication and length of DNA-end resection in natural repair with sister chromosomes
Mutation hotspots promote cancer and genome evolution, yet how they occur remains obscure. Rosenberg and colleagues used targeted endonucleolytic cleavages in the Escherichia coli chromosome to show that double-strand breaks cause mutation hotspots. Strong local and weak distant hotspots are caused by two mutation mechanisms that accelerate evolution in stressed cells. Hotspotting at breaks raises the possibility that specific genomic regions can be targeted for mutagenesis and can promote concerted evolution within genes, an important issue in protein evolution.
Introduction
Evolutionary theory assumes that mutations fall randomly in genomic space (e.g., Mayr, 1985); however, mutation hotspots, clusters, and showers occur in organisms ranging from phage to human (Drake, 2007a, 2007b), and are very probably important forces in human tumor and organismal evolution. A recent study of Escherichia coli genomes revealed nonrandom distributions of mutations with hot and cold zones (Martincorena et al., 2012). Spontaneous mutations in mice fall in ∼30 kb showers of simultaneous multiple mutations (Drake, 2007b; Wang et al., 2007). Both chemically mutagenized yeast (Burch et al., 2011) and E. coli (Parkhomchuk et al., 2009) show local clusters of mutations, as do the genomes of human breast (Nik-Zainal et al., 2012) and colon (Roberts et al., 2012) cancer cells, and chemically treated yeast (Ma et al., 2012). These and other observations (Caporale, 2006; Drake, 2007a, 2007b) indicate that the processes of mutagenesis themselves, and not just the sites in which mutations are tolerated, can be localized in genomes and are not distributed randomly.
Mutational hotspotting can promote evolution, including evolution of tumors and pathogens, in important ways. First, hotspotting mechanisms may target regions in which variability might provide a growth advantage, as in somatic hypermutation of immunoglobulin genes (Di Noia and Neuberger, 2007), pathogen contingency genes (Moxon et al., 1994), and the cancer-driving Philadelphia chromosome (Albano et al., 2010). Second, restriction of mutagenesis to small zones, even if randomly chosen, could promote high-level multiple mutations (concerted evolution) within genes without causing deleterious mutations throughout the genome (Ninio, 1996; Ponder et al., 2005; Yang et al., 2008). The evolution of new protein functions usually requires multiple base substitutions (Romero and Arnold, 2009), and how this occurs is a significant issue in protein evolution.
Although mutational hotspotting is widespread, striking, and important, the molecular mechanisms that cause hotspots remain largely obscure. Various studies have hinted that mutation hotspots might be related to DNA double-strand breaks (DSBs), but their results were open to multiple interpretations. DSB-dependent mutation was first found in E. coli (Harris et al., 1994; Rosenberg et al., 1994) and then in yeast (Deem et al., 2011; Hicks et al., 2010; Strathern et al., 1995; Yang et al., 2008), both caused by DNA polymerase errors during DSB repair by homologous recombination (HR). In E. coli, DSB repair is nonmutagenic and uses the high-fidelity DNA polymerase (pol) III (Motamedi et al., 1999) in unstressed cells, but then is switched to a mutagenic mode using error-prone DNA polymerase DinB, which causes mutations, only under stress, under the control of the general stress response (Ponder et al., 2005; Shee et al., 2011a). Two kinds of mechanisms could underlie DSB-dependent mutagenesis: one that could produce hotspots and another that would not be expected to. If the DSB repair mechanism that recruits an error-prone polymerase is localized, then mutation hotspots might be expected. If the mutagenic repair mechanism is break-induced replication (BIR), which can prime processive replication from a DSB site to the telomere (observed in yeast [reviewed by Symington and Gautier, 2011]) or the replication terminus in E. coli (proposed by Kuzminov, 1995, and supported by data on recombination of phage λ DNA by Motamedi et al., 1999), then DSB-dependent mutagenesis might affect whole chromosome arms and not form hotspots. In the sole study to address this question to date, Deem et al. (2011) found robust mutagenesis in yeast as far away from a DSB site as they assayed (36 kb), in repair reactions that could proceed only by BIR and thus would not be expected to form hotspots. By contrast, DSBs were proposed as an explanation for the particular symmetrical patterns of mutations found in ∼100 kb mutation clusters in human cancer genomes and chemically mutagenized yeast (Nik-Zainal et al., 2012; Roberts et al., 2012), although as noted (Nik-Zainal et al., 2012), other repair/mutation mechanisms might be responsible.
Here we show that DSBs tightly focus stress-inducible mutations to small zones or hotspots in the E. coli chromosome. We show two kinds of hotspots: strong local hotspots that occur up to ∼60 kb away from a DSB, and weak long-distance hotspots that extend to ∼1 Mb away. Moreover, we show that the strong local and weak long-distance hotspots occur by distinct mechanisms. The data indicate that one way by which mutation hotspots can occur is via mechanisms that couple mutagenesis to DSB repair, and illuminate the molecular basis of one of those mechanisms.
Results
Mutations Focused in Hotspots near DSBs
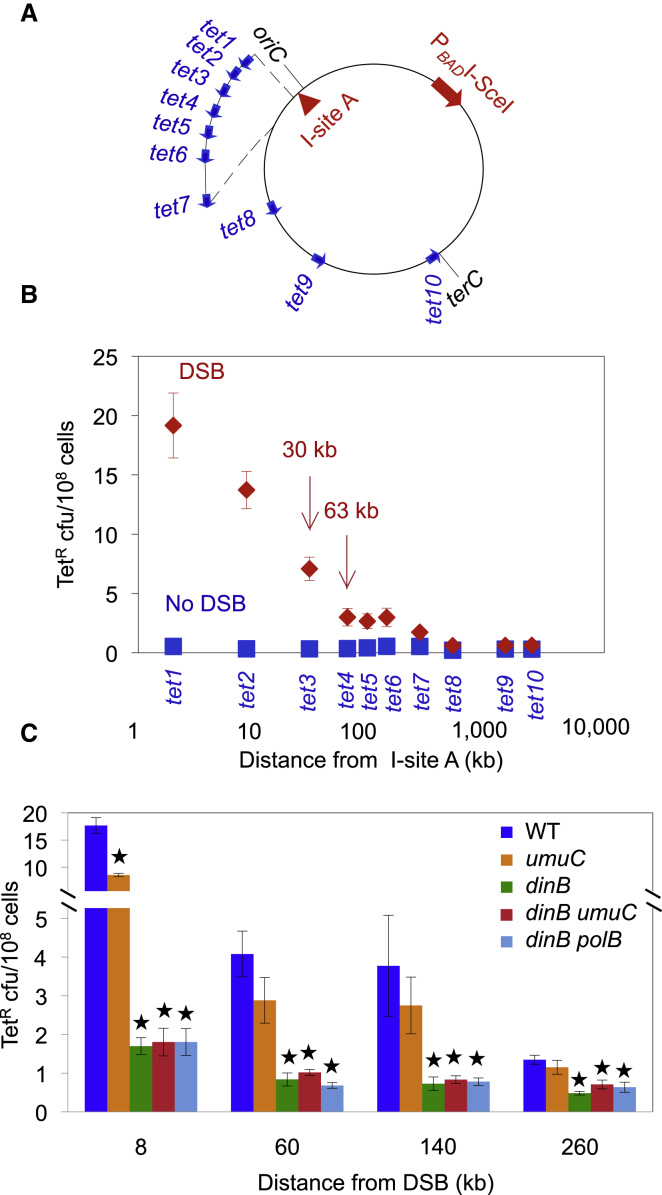
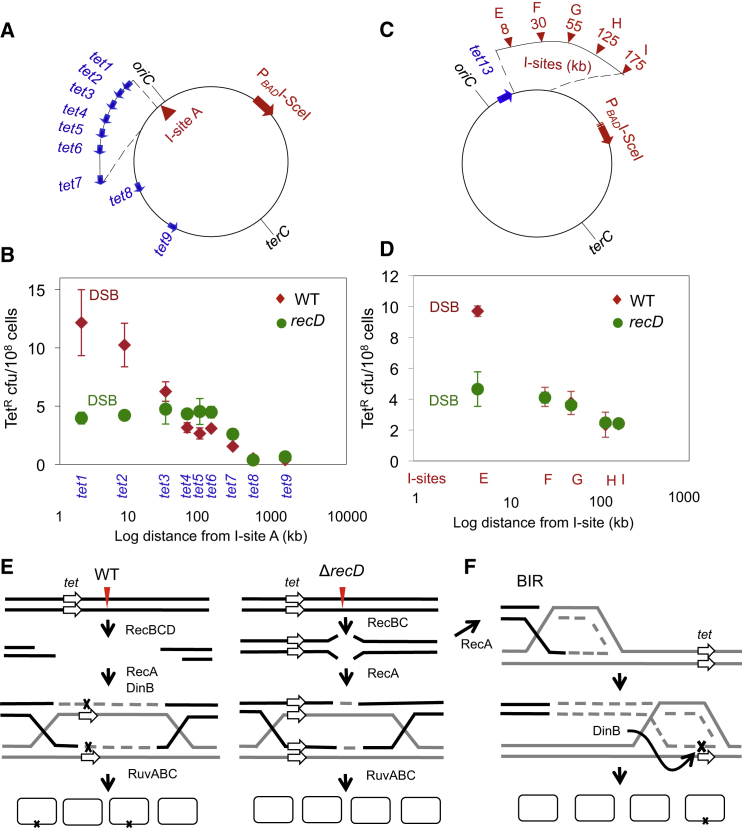
We constructed a movable tet +1 bp mutation-reporter gene cassette that reverts to wild-type (WT) function, and confers tetracycline resistance, by a −1 bp deletion mutation (Shee et al., 2011a). We used the movable tet reporter to construct strains with this cassette inserted at ten different sites between 2 kb and 2.4 MB away from an I-SceI double-strand endonuclease (restriction) site (I-site A, tet1–tet10 cassettes; Figure 1A). Each tet cassette resides in two different strains, one with and one without the I-SceI-endonuclease-encoding gene cloned under a regulatable promoter in the E. coli chromosome. Thus, each strain pair reports on tetracycline-resistant (TetR) mutant frequencies in a cell with and without an I-SceI-induced DSB in the same DNA molecule as the tet reporter. These breaks are repaired by HR with either an uncleaved sister chromosome (present in ∼40% of starving E. coli; Akerlund et al., 1995) or an uncleaved spontaneous tandem DNA duplication (present in ∼10−3 of cells; reviewed in Rosenberg et al., 2012). We measured reversion in starvation-stressed cells as previously described (Shee et al., 2011a), under conditions in which the formation of nearby mutations (at tet2; Figure 1A) requires the RpoS and SOS stress responses, DinB error-prone DNA polymerase, and HR/DSB-repair enzymes RecBC, RecA, and RuvABC, and the mutations arise in acts of DSB repair (Ponder et al., 2005; Shee et al., 2011a). Although the tet reporter used here captures frameshift or indel (insertion/deletion) mutations, base substitutions are also promoted (Shee et al., 2011a) and outnumber (Petrosino et al., 2009) indels in DSB-dependent stress-induced mutagenesis. Thus, the data obtained pertain to both base-substitution and indel mutagenesis.
Figure 1.
DSBs Promote Strong Local and Weak Long-Distance Mutation Hotspots
(A) Blue arrows: sites and direction of the tet +1 bp frameshift-mutation-reporter gene placed at various locations in the E. coli chromosome in different strains, one with and one without a chromosomal inducible I-SceI gene (PBADISceI). oriC and terC, chromosomal reference points. Approximate distances between the I-SceI cutsite (I-site A) and tet cassettes: tet1, 2 kb; tet2, 8.5 kb; tet3, 29.5 kb; tet4, 62.5 kb; tet5, 92.5 kb; tet6, 136 kb; tet7, 261 kb; tet8, 500 kb; tet9, 1.4 Mb; and tet10, 2.4 Mb. See Tables S1, S2, and S3 for the exact chromosomal locations of each tet reporter and I-site, the strains that carry them, and the PCR primers used to construct them, for this and all figures.
(B) Mutant frequency is highest near the I-site and decreases logarithmically to ∼60 kb from the DSB. A weak but significant hotspot extends from ∼60 kb to 1 Mb (see text). DSB (♦) and No-DSB (I-SceI cutsite-only control, ▪) strains for each tet allele are indicated. Points show the mean ± SEM for three or more independent experiments.
(C) DinB is required for strong DSB local and weak long-distance hotspotting. Each genotype has a null mutation in the gene(s) indicated. dinB encodes DinB/DNA Pol IV; umuC, an essential subunit of DNA Pol V; polB, DNA Pol II. ∗Significantly different from the WT strain at the same distance (p ≥ 0.05). Error bars represent 1 SEM for n = 3 experiments.
See also Figure S1.
Here we observed that the DSB-dependent TetR mutant frequency was highest at the tet1 cassette, ∼2 kb from I-site A (Figure 1B), at which the DSBs induced 65 ± 14-fold more mutations than were observed in the cutsite-only (no-DSB) control. Mutant frequencies decreased logarithmically to ∼60 kb from the break and then gradually tapered off for up to a megabase from the break (Figure 1B). Thus, mutations localize tightly in a hotspot near the DSB site, mostly in the first 2 kb, and then fall off logarithmically to ∼60 kb. Additionally, from ∼60 kb to 1 Mb, DSBs promote mutagenesis weakly but significantly above the level observed in the no-DSB control strains (60 kb, 8.8 ± 2.5; 90 kb, 4.6 ± 1.1; 130 kb, 5.6 ± 1.6; 260 kb, 3.7 ± 0.58; 500 kb, 2.8 ± 0.42; 1.4 Mb, 3.0 ± 0.51-fold; p = 0.0027, 0.0109, 0.0112, 0.00007, 0.0003, and 0.0007, respectively). The weak long-distance DSB-dependent mutagenesis requires DinB (Figure 1C), indicating DSB-dependent DNA polymerase errors, similarly to mutagenesis close to a DSB (Ponder et al., 2005; Shee et al., 2011a; Figure 1C).
DSBs Focus Mutations Independently of the Specific Genomic Position
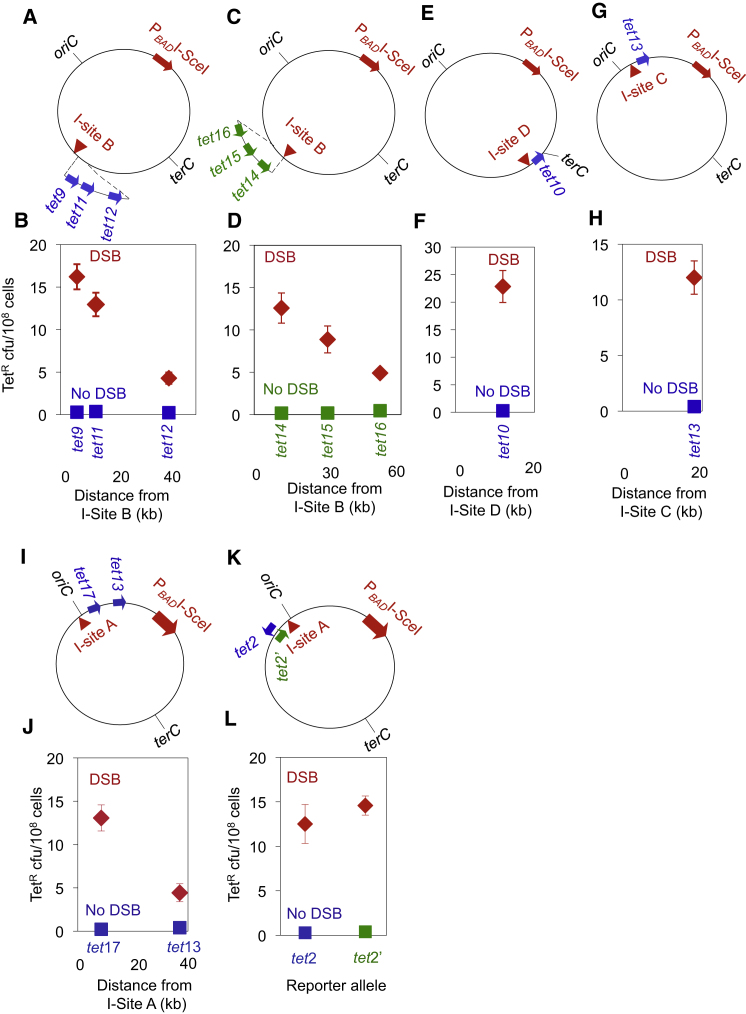
Neither the specific location of the tet reporter genes nor I-sites in the chromosome (e.g., near the replication origin; Figure 1) causes DSB-proximal hotspotting of mutagenesis. Rather, mutational hotspotting appears to be a general effect of the proximity of tet to a DSB, as follows. With I-site B placed about halfway between oriC and ter, the tet9, tet11, and tet12 genes are activated for mutation proportionally to their proximity to the DSB (6, 13, and 40 kb, respectively; Figures 2A and 2B). At tet9, 6 kb from I-site B, DSBs at I-site B increased mutation 90 ± 19-fold relative to the cutsite-only no-DSB control (Figure 2B). By comparison, with DSBs at I-site A, tet9 was almost inactive (Figures 1A and 1B). Therefore, proximity to the DSB, rather than the absolute genomic position, dictates mutability.
Figure 2.
Mutation Hotspots at DSBs Reflect Distance from the DSB Independently of Chromosomal Location
(A, C, E, G, I, and K) Diagrams of the constructs used. Symbols as in Figure 1A.
(B) oriC-distal I-site B promotes a strong local mutation hotspot downstream in the replicore at tet9, tet11, and tet12, 6 kb, 13 kb, and 40 kb away.
(D) I-site B promotes a strong local hotspot upstream in the replicore at tet14, tet15, and tet16, 12 kb, 35 kb, and 60 kb upstream.
(F) Mutation at tet10 near the replication terminus (terC) is activated 108 ± 29-fold by I-site D cleavage 12 kb away.
(H) I-site C cleavage activates mutation at tet13, 18 kb downstream in the right replicore.
(J) Hotspots cross the replication origin. I-site A stimulates mutations at nearby tet17 and tet13, across oriC in the opposite replicore.
(L) Equal stimulation of mutation by I-site A cleavage at tet2 (blue arrow) and tet2′ (green arrow), 8.5 kb away in opposite transcriptional orientations.
Points show the mean ± SEM for three or more independent experiments. For the relationship between mutant frequency and the log of the distance between tet reporters and I-sites, see Figure S1.
Mutations are generated both upstream and downstream of the DSB at I-site B in the chromosome’s unidirectional replication paths (replicores; Figures 2A–2D). When tet genes are placed at three positions on either side of I-site B (in 12 different isogenic strains, one for each cutsite with and one for each cutsite without the I-SceI endonuclease), the mutant frequencies reflect the distance from the break regardless of the upstream or downstream position (Figures 2A–2D).
Further, tet10, in the ter region of the genome, is inactive when I-site A is placed 2.4 MB away, but is subject to robust DSB-promoted mutation (108 ± 29-fold increase relative to no-DSB control) when I-site D is engineered 12 kb away (Figures 2E and 2F). Finally, the stimulatory effect of DSBs on mutation seen in the left replicore (Figures 1A, 1B, and 2A–2F) also occurs in the right replicore; tet13, 18 kb from I-site C in the right replicore, shows 44 ± 12-fold enhancement of mutant frequency by I-SceI cleavage (Figures 2G and 2H), compared with the 60 ± 14- and 22 ± 3.7-fold enhancement at tet2 and tet3, respectively, located 8.5 and 29.5 kb from I-site A in the left replicore (Figures 1A and 1B).
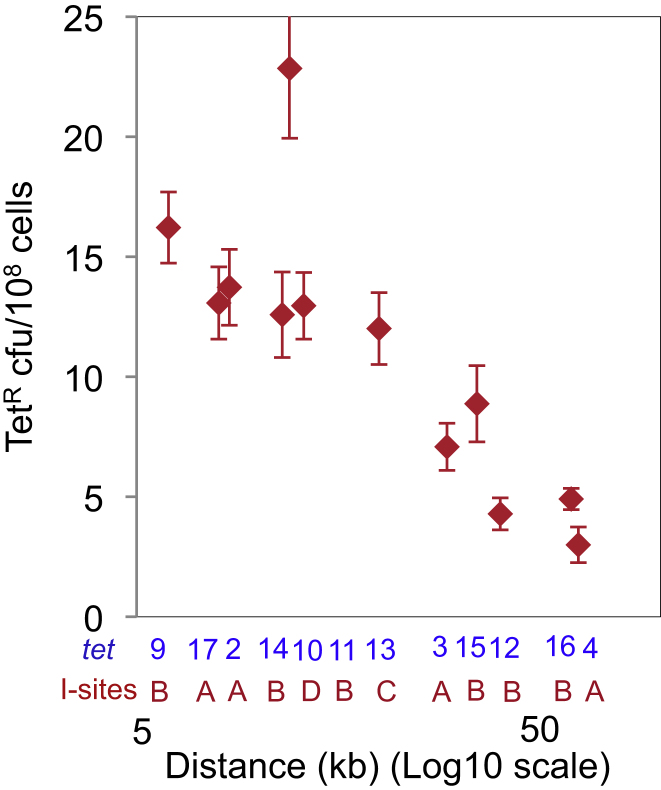
Within the ∼60 kb strong hotspots, DSB-dependent mutant frequencies are related roughly to the log of the distance between the tet reporter and each I-site (Figure S1), with the exception of tet10, 12 kb from I-site D, which is located in the dif (replication-terminus-proximal) region (Figures 2E, 2F, and Figure S1). The mutant frequency at this site was 2-fold higher than that of tet11, located 13 kb from I-site B, which is not in the dif region (Figures 2A and 2B). The higher RecBCD-mediated HR observed near dif (Louarn et al., 1991) might contribute to the higher DSB-repair-coupled mutation observed here.
Figure S1.
Mutant Frequencies in Hotspots Decline Logarithmically with Distance from DSBs at Multiple Genomic Sites, Related to Figures 1, 2, and 3
Data from tet reporter genes and I-sites at multiple chromosomal positions from experiments in Figures 1–3. The single outlying tet reporter/I-site pair is the terminus-proximal tet10 and I-site D (Figures 2E and 2F) as discussed in the text.
The Direction of Replication or Transcription Does Not Affect DSB-Coupled Mutation
The E. coli chromosome is arranged in two unidirectional replication paths, or replicores (a left arm and a right arm), extending from oriC to the replication terminus. We find that the local mutational hotspotting at a DSB can extend from one replicore to the other, across oriC. The mutant frequency reflects the distance from the DSB, regardless of whether mutagenesis is assayed on the same or opposite side of oriC (compare the mutant frequencies of tet11 and tet12, 13 and 40 kb from their I-site [Figures 2A and 2B] with those of tet17 and tet13, 9.5 and 37 kb from their I-site [Figures 2I and 2J]). oriC does not appear to block local DSB-dependent mutation tracts. Similarly, we find no orientation dependence or strand bias of mutagenesis relative to the direction of transcription of the reporter gene (Figure 2L). The tet2 and tet2′ alleles, in opposite orientations at the same site 8.5 kb from I-site A, are affected similarly by I-site A cleavage (46 ± 10- and 40 ± 4-fold, respectively) relative to the no-DSB controls.
Strong Local Hotspotting at DSBs Requires RecBCD-Mediated Degradation from DSB Ends
Double-strand digestion of DSB ends prior to repair in E. coli is carried out by the RecBCD enzyme in a manner that depends on the RecD subunit (Dillingham and Kowalczykowski, 2008). Mutants that lack RecD are repair proficient, even hyperrecombinagenic (Biek and Cohen, 1986; Chaudhury and Smith, 1985), but the repair occurs immediately at the DSB end (Thaler et al., 1989), not at a distance from the end dictated by double-strand DNA (dsDNA) degradation (per the model of Rosenberg and Hastings, 1991). We find that the strong local hotspotting between 2 and 8 to under 30 kb from the DSB requires RecD (Figure 3). The two tet reporters nearest I-site A, tet1 and tet2, display much reduced mutant frequencies in the recD null mutant, with mutagenesis reduced to the level observed for more distant sites (Figures 3A and 3B). No significant difference in frequency was observed at tet cassettes farther away (Figures 3A and 3B). Similarly, in the right replicore at tet13, mutagenesis that was provoked by cutting at the 8-kb proximal I-site E was reduced in the recD background to levels similar to those seen at tet13 when the more-distant I-sites F (30 kb), G (55 kb), H (125 kb), and I (175 kb) were used (Figures 3C and 3D). Because I-site A activates mutagenesis at tet1 and tet2 from upstream of those reporters in the replicore, whereas I-site E activates tet13 mutagenesis from downstream of the reporter, these data imply that dsDNA resection by RecBCD on both sides of the DSB causes the upstream and downstream mutation hotspots near DSBs.
Figure 3.
The RecD Subunit of RecBCD DSB-Resection Exonuclease Is Required for Strong Local Hotspotting at DSBs
(A) Approximate distances between I-site A and tet cassettes. Symbols as in Figure 1A.
(B) Loss of the strong local mutation hotspot downstream of the DSB in recD exonuclease-defective but repair-proficient mutant cells.
(C) Positions of I-sites E-I relative to tet13 in the right chromosome arm.
(D) Loss of the I-site E-promoted strong local hotspot at tet13, 8 kb from I-site E, in recD resection-exonuclease-deficient but repair-proficient cells.
(E) Model for RecBCD-nuclease-promoted DNA resection, repair synthesis, and strong local mutation hotspotting in WT but not recD exonuclease-defective mutant cells. Lines: strands of DNA. Dashed lines: newly synthesized DNA. Red arrowhead: DNA DSB. Left: Our data imply that dsDNA resection by RecBCD double-strand exonuclease occurs equally well on either side of a DSB and decreases with distance from the break. Degradation may stop at a site where a productive HR event occurs that repairs the break. Our data suggest that this length is ≤2–60 kb. Mutagenesis results from error-prone DSB repair synthesis caused by DinB, which occurs when the RpoS stress response is activated (Ponder et al., 2005; Shee et al., 2011a). Confinement of repair synthesis (dashed gray lines) to the resected area can explain strong local hotspotting of mutations (black X) near DSBs in WT cells. We suggest that in recD null cells (right), there is a window of double-strand degradation and repair smaller than the 2 kb distance at which mutations were assayed here, consistent with previous results regarding the extreme proximity of HR to a DSB end in recD cells (Thaler et al., 1989).
(F) Model for weak long-distance hotspotting by BIR. We suggest that occasional extension of repair synthesis by BIR beyond the resection points underlies the weak long-distance mutation hotspotting in both WT and recD backgrounds, up to 260 kb to 1 Mb from a DSB. Because this mutagenesis is RecD independent, it (and BIR more generally) may require little or no resection. Previously, RecA/BC-mediated BIR was shown to have conservative segregation of new DNA strands, as shown here, and to require high-fidelity Pol III but not RuvABC (Motamedi et al., 1999). We do not know whether the BIR that generates mutations using DinB similarly requires Pol III.
Points show the means ± SEM for three or more independent experiments. See also Figure S1.
As illustrated in the model shown in Figure 3E, left, the data indicate that the strong local hotspots result from RecBCD-mediated DNA resection and gap-filling repair synthesis using error-prone DinB polymerase. As illustrated in the model shown in Figure 3E, right, the data imply that in resection-defective recD mutants, this degradation is less than the 2 kb that would be required to erode and then resynthesize the closest tet gene to the I-site, which therefore acquires few mutations. In support of our interpretation that the recD mutant repairs efficiently but with smaller tracts of resection/resynthesis, we note that survival of the DSB is only slightly reduced by I-SceI cutting in recD (30 ± 3% survival relative to the uncut control, I-site E, experiments; Figures 3C and 3D) compared with the WT (40 ± 3% survival relative to the uncut control, same site). Thus, as reported previously (Biek and Cohen, 1986; Chaudhury and Smith, 1985; Thaler et al., 1989), recD mutants are DSB-repair proficient.
Further, we conclude that the long-distance weak mutational hotspotting (Figure 1B) occurs independently of RecD-dependent exonucleolytic resection, because it is unaffected in recD mutants (Figures 3B, tet3-tet7 and Figure 3D, I-sites F–I). We suggest that this low-level mutagenesis, from 60 kb to ∼1 Mb, results from less frequent repair events that produce a processive replication fork that exceeds the window of degraded DNA and synthesizes long distances (BIR). BIR was observed in RecBC-dependent DSB repair in phage λ (Motamedi et al., 1999) and was hypothesized to occur in the E. coli chromosome (Cox et al., 2000; Kuzminov, 1995; Motamedi et al., 1999), but had not been documented there. The DinB dependence of the long-distance mutation (Figure 1C) implies that DinB, a low-processivity DNA Pol, participates, even if distributively, in long tracts of repair replication (illustrated in Figure 3F).
Mutagenesis up to 8 kb from the DSB differs significantly between the WT and recD strains with I-SceI cleavage. Beyond that distance, there is no significant difference (p = 0.0032 at 2 kb and 0.00006 at 8 kb, and 0.28, 0.1, 0.074, 0.075, 0.051, 0.362, and 0.16 for the remaining distances, respectively; Figure 3B), implying that in both WT and recD, low-level long-distance mutagenesis, which we propose results from processive BIR, is functional. In Figure 3D, p = 0.000075 at 5 kb, and 0.93, 0.4, 0.84, and 0.85 for the remaining distances, respectively.
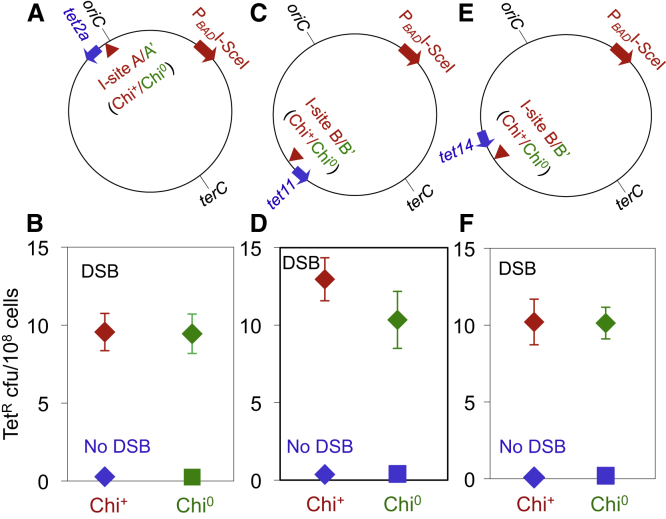
RecBCD-dependent double-strand exonuclease activity is reduced at Chi sites (Dillingham and Kowalczykowski, 2008). Whereas RecBCD-dependent DNA degradation is critical for the formation of DSB-proximal mutation hotspots (Figure 3), we find that addition of extra Chi sites at I-sites makes no difference to the distribution of mutations (Figure 4). This may be because the E. coli genome is already effectively saturated with Chi sites (discussed below).
Figure 4.
Chi Sites Engineered into I-Sites Have No Effect on DSB-Proximal Hotspotting
(A, C, and E) Approximate locations of tet mutation reporter genes and I-sites with and without three Chi sites in the terminus-proximal side of each I-site. tet2a is the same as tet2 except that its linked selectable cat gene was not removed, and therefore it is 9.5 kb from I-sites A (Chi+) and A′ (Chi°). The tet11 cassette is 13 kb from I-site B/B′. tet14 is also 13 kb from I-site B/B′, but upstream.
(B, D, and F) TetR mutant frequencies at (B) tet2a, (D) tet11, and (F) tet14 with and without additional Chi sites in active orientation at each I-site. All no-DSB strains are the cutsite-only control.
Points show the mean ± SEM for three or more independent experiments. For the distribution of active Chi sites at four of the I-sites used, see Figure S2.
Discussion
We found that DSBs produce two kinds of hotspots during stress-induced mutation: (1) strong local hotspots that form via RecD-dependent resection from DSBs and gap-filling synthesis, and (2) weak long-distance hot zones that extend to ∼1 Mb from a DSB and form independently of resection, presumably by BIR. Models for each of these mechanisms are illustrated in Figures 3E and 3F. Whereas cells that underwent stress-induced mutation appeared to be mutated genome-wide (Galhardo et al., 2007; Torkelson et al., 1997), our results suggest that in any given cell, the mutation(s) may be localized near spontaneous DSBs. Importantly, DSB-dependent stress-induced mutagenesis, requiring DSB repair proteins, stress-response activation, and DinB, underlies most spontaneous chromosomal base substitutions and frameshift mutations in starved cells, with no I-SceI, presumably at spontaneous DSBs (Shee et al., 2011a). Thus, the fact that hotspots occur at DSBs, as reported here, is likely to bear importantly on genome evolution.
Our results provide a plausible mechanistic explanation for mutational hotspotting and possibly showers in genomes: hot regions could occur at DNA break sites. Hotspots are areas with higher mutation rates/frequencies (as observed here), and showers or clusters are hotspots with multiple mutations (not assayed here, but shown previously to occur in E. coli DSB-dependent stress-induced mutation [Bull et al., 2000]; discussed below). In E. coli, single-base differences are nonrandomly distributed across sequenced genomes with higher frequencies in poorly expressed genes, and with hot and cold regions spanning entire operons (Martincorena et al., 2012), about the size of the strong hotspots mapped here (2 to <60 kb). We hypothesize that poorly expressed genes may be DSB prone and thus mutagenic. For example, poorly expressed genes are often oriented oppositely to replication paths (Brewer, 1988; Nomura and Morgan, 1977; Price et al., 2005), which could produce head-on collisions of transcription with replication. Such collisions generate DSBs in bacteria (Tehranchi et al., 2010) and eukaryotes (Bermejo et al., 2012). Other mechanisms are possible. Mutation showers in mice are ∼30 kb (Wang et al., 2007), similar to our DSB-provoked local hotspots (e.g., Figure 1B). Mice and humans possess homologs and analogs of bacterial HR-DSB-repair proteins (Krejci et al., 2012), and three homologs and an ortholog of DinB (Nohmi, 2006); therefore, it is plausible that mouse and human mutation showers (Nik-Zainal et al., 2012; Roberts et al., 2012) could be caused by the resection/gap-filling mechanism demonstrated here.
Mutational hotspotting at DSBs could contribute to the rapid evolution of pathogens with hosts, and to cancer development, potentially by targeting specific genomic regions and, we hypothesize, by promoting mutation clusters that facilitate concerted evolution. Although rates of spontaneous DNA breakage are being quantified (Pennington and Rosenberg, 2007), the break positions remain obscure. Hotspotting could facilitate concerted evolution (Ninio, 1996; Ponder et al., 2005; Yang et al., 2008), an important problem in protein evolution (Romero and Arnold, 2009). In previous studies of DSB-dependent mutation, we observed mutation clustering (i.e., more linked double mutants than would be expected for independent events) using a plasmid-based assay (Bull et al., 2000) with very high mutant frequencies, probably because the higher copy number allowed more efficient repair (reviewed in Rosenberg et al., 2012; Shee et al., 2011b). With the chromosomal assay used here, the mutant frequencies were too low (∼10−9) to measure coincident double mutants, which occur ≥3 logs less frequently (Bull et al., 2000). However, the chromosomal and plasmid-based assays behave similarly in nearly all ways measured (reviewed in Rosenberg et al., 2012; Shee et al., 2011b), suggesting that concerted evolution is likely to be promoted at DSBs in the E. coli chromosome, and in other organisms that utilize similar mutation mechanisms.
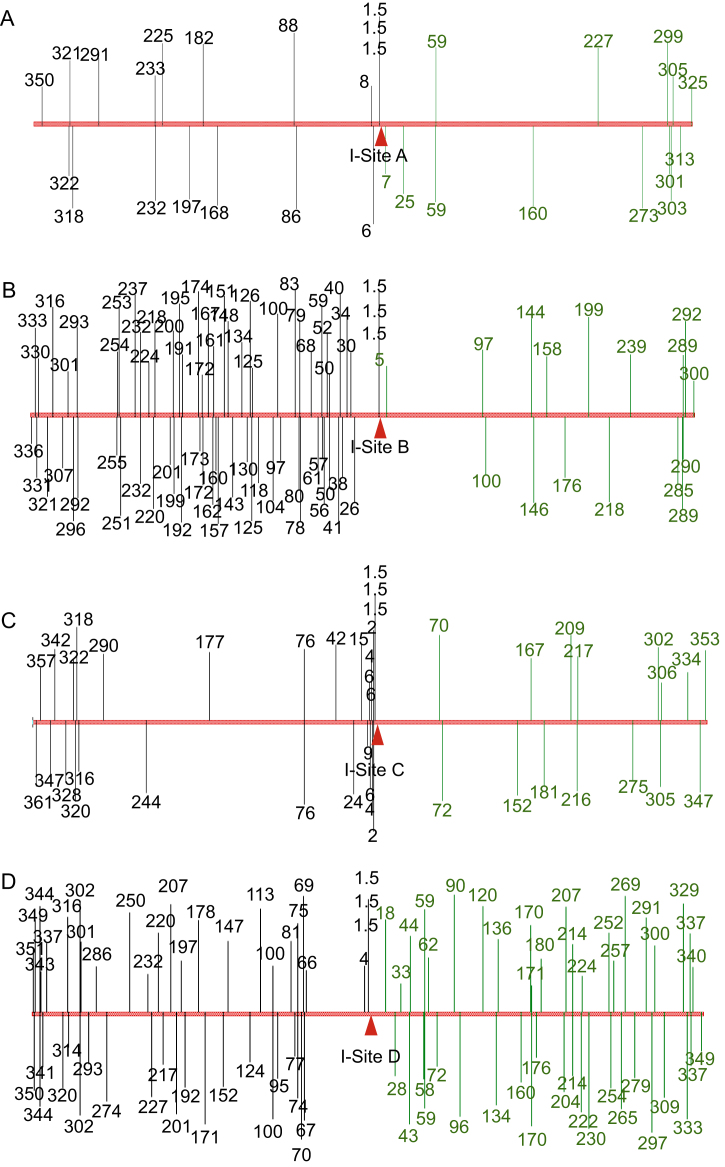
One surprise in this study is the small size and symmetry of strong hotspots upstream and downstream in the replicores (Figures 2A–2D). The small size is surprising because DSB repair synthesis tracts were predicted to run from a DSB to the replication terminus, potentially megabases away (e.g., Cox et al., 2000; Kuzminov, 1995). The symmetry is surprising because Chi sites, which inhibit RecBCD resection exonuclease activity, are predicted to cause asymmetrical resection in the chromosome (Kuzminov, 1995). RecBCD recognizes Chi from only one side of the Chi sequence, causing cessation of resection (Dillingham and Kowalczykowski, 2008), and there are more active Chis upstream than downstream in the replicores (Kuzminov, 1995). This predicted long degradation tracts downstream and short ones upstream (Kuzminov, 1995), which is not what our data indicate (Figures 2A–2D). Perhaps, although Chis are distributed asymmetrically, the genome is nevertheless effectively saturated with Chis in both orientations (the distribution of active Chi sites at four of the I-sites we used is shown in Figure S2). In support of this possibility, we found that additional Chi sites added at the I-site had no additional effect on the distribution of mutations (Figure 4), despite the demonstrated dependence of that distribution on RecBCD-mediated DNA degradation (Figure 3). The high frequency of Chi sites in both orientations in the genome (e.g., Figure S2) may be sufficient to create small symmetrical degradation and resynthesis tracts that cause the strong local hotspots at DSBs.
Figure S2.
Maps of Chi Sites on Either Side of I-Sites A–D in Active Orientation with Respect to Each I-Site, Related to Figure 4
Chi sites are recognized by RecBCD enzyme as it degrades DNA from a DSB end, and are recognized only if RecBCD encounters them from the 3′GG side of the 5′GCTGGTGG3′ Chi sequence (Dillingham and Kowalczykowski, 2008). Positions are shown of Chi sites in active orientation with respect to the DSB that is created upon cleavage of the I-site (that is, the 3′ side of the 5′GCTGGTGG3′ Chi sequence toward the I site) for I-sites A–D. Numbers represent the distance in kilobases from the I-site. In each map, left is origin-proximal and right is terminus-proximal. Positions of Chi sites that would be active with the I-site DSB downstream (those oriented 5′GCTGGTGG3′ from origin to terminus) are shown in black, and those that would be active with the I-site DSB upstream (oriented 3′GGTGGTCG5′ origin to terminus) are shown in green.
There are multiple mechanisms of spontaneous mutation (Drake, 1993). However, the DSB-dependent, stress-induced mutation mechanism studied here is a major contributor to spontaneous mutagenesis, at least in E. coli, in which it produces both base substitution and frameshift/indel mutations (Shee et al., 2011a), with base substitutions outnumbering the indels (Petrosino et al., 2009). Thus, DSB-dependent, stress-induced mutation is likely to contribute to evolution. DSB-dependent, stress-induced mutation is now shown to occur both nonrandomly in time, preferentially coupled to stress by its dependence on stress-response activation (Ponder et al., 2005; Shee et al., 2011a), and nonrandomly in genomic space, causing hotspots close to DSB sites (Figures 1, 2, 3, and 4). The coupling to stress responses increases mutations and potentially the ability to evolve, specifically when cells are maladapted to their environment, i.e., are stressed. Hotspotting could also speed evolution, as discussed above. Regardless of how they evolved, both of these layers of regulation of mutagenesis change part of our picture of evolution from a chaotic one to one in which the ability to evolve has evolved, is evolving, and is a real-time (not solely historical) biological property. The identification and eventual manipulation of the molecular determinants of the ability to evolve may be crucial to efforts to combat the evolution-based problems of cancer and infectious diseases (e.g., Rosenberg et al., 2012), and is certainly necessary for a mechanistic understanding of evolution.
Experimental Procedures
Strains, Media, and Growth
The E. coli strains used in this work are shown in Table S1. Bacteria were grown in LBH (Torkelson et al., 1997) or M9 minimal medium (Miller, 1992) supplemented with 10 μg/ml thiamine (vitamin B1) and 0.1% glucose as the carbon source. Other additives were used at the following concentrations (μg/ml): ampicillin, 100; chloramphenicol, 25; kanamycin, 50; tetracycline, 10; and sodium citrate 20 mM.
Starvation/Stress-Induced DSB-Dependent Mutation Assays
Assays were performed as previously described (Shee et al., 2011a). Single colonies from M9 glucose vitamin B1 (B1) plates that had been incubated for ∼22 h at 37°C were inoculated into 5 ml of M9 glucose B1 broth and grown for 12 h with shaking. These liquid cultures were diluted 1:100 into the same medium and grown for 8–10 h, diluted 1:100 and grown for 12 h to saturation, and then incubated further for 72 h. Three independent cultures per genotype were used for each experiment. Mutant frequencies were determined as colony-forming units (cfu) on LBH glucose tetracycline (TetR mutant cfu) and LBH glucose plates (total cfu), and the means ± SEMs for three or more independent experiments are displayed. The p values were determined by two-tailed Student’s t test.
Movable tet Reporter Gene
We used the movable tetA +1 bp mutation-reporter allele linked with a selectable cat cassette developed by Shee et al. (2011a). The precise location of each insertion is given in Table S2. We constructed the tet alleles and I-SceI-cutsite-carrying strains using the primer sets listed in Table S3.
Chromosomal I-SceI Cleavage System and Cutsites
We used the chromosomal I-SceI endonuclease expression system (Gumbiner-Russo et al., 2001), which was previously used to introduce DSBs into F’128 (Ponder et al., 2005), and in the chromosome (Shee et al., 2011a, 2001b). The 18bp I-SceI cutsite sequence was engineered into various loci by inclusion in primers for amplifying a Kan cassette (Table S1) and recombined into the genomes. The chromosomal I-SceI gene is expressed from the PBAD promoter and thus is induced strongly by arabinose and weakly in the absence of glucose (Ponder et al., 2005), the condition used here and in a previous work (Shee et al., 2011a). In all experiments measuring mutagenesis and/or efficiency of DSB formation by the I-SceI system, terminal cultures were shown to retain the functional I-SceI gene and cleavage site by quantitative measurement of arabinose sensitivity, comparing cfu titers on arabinose and glucose plates. The typical frequencies of arabinose-resistant mutants, which have acquired a mutation in the I-SceI cutsite or gene (Ponder et al., 2005), were between 10−4 and 10−5, as observed previously (Ponder et al., 2005; Shee et al., 2011a), demonstrating that most cells in our experiments were DSB competent. Arabinose-resistant mutants consist mostly of cutsite mutants, presumably from low-level, Ku-independent, nonhomologous end-joining (Ponder et al., 2005). The locations of the cutsites are given in Table S2.
Acknowledgments
This paper is dedicated to Frank Stahl, whose strategies we aimed to emulate. We thank D. Bates, H. Dierick, R. Frisch, R. Galhardo, J.E. Haber, J. Halliday, P.J. Hastings, C. Herman, G. Ira, A. Kuzminov, E. Rogers, J.D. Wang, and three anonymous reviewers for helpful comments on the manuscript. This work was supported by National Institutes of Health grant R01-GM53158.
Published online: October 4, 2012
Footnotes
Supplemental Information includes two figures, three tables, and a list of strains used in each figure and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.08.033.
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
Supplemental Information
References
- Akerlund T., Nordström K., Bernander R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J. Bacteriol. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano F., Anelli L., Zagaria A., Coccaro N., Casieri P., Rossi A.R., Vicari L., Liso V., Rocchi M., Specchia G. Non random distribution of genomic features in breakpoint regions involved in chronic myeloid leukemia cases with variant t(9;22) or additional chromosomal rearrangements. Mol. Cancer. 2010;9:120. doi: 10.1186/1476-4598-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Lai M.S., Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol. Cell. 2012;45:710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Biek D.P., Cohen S.N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J. Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B.J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Bull H.J., McKenzie G.J., Hastings P.J., Rosenberg S.M. Evidence that stationary-phase hypermutation in the Escherichia coli chromosome is promoted by recombination. Genetics. 2000;154:1427–1437. doi: 10.1093/genetics/154.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch L.H., Yang Y., Sterling J.F., Roberts S.A., Chao F.G., Xu H., Zhang L., Walsh J., Resnick M.A., Mieczkowski P.A., Gordenin D.A. Damage-induced localized hypermutability. Cell Cycle. 2011;10:1073–1085. doi: 10.4161/cc.10.7.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporale L.H. Oxford University Press; New York: 2006. The Implicit Genome. [Google Scholar]
- Chaudhury A.M., Smith G.R. Role of Escherichia coli RecBC enzyme in SOS induction. Mol. Gen. Genet. 1985;201:525–528. doi: 10.1007/BF00331350. [DOI] [PubMed] [Google Scholar]
- Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Deem A., Keszthelyi A., Blackgrove T., Vayl A., Coffey B., Mathur R., Chabes A., Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dillingham M.S., Kowalczykowski S.C. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J.W. General antimutators are improbable. J. Mol. Biol. 1993;229:8–13. doi: 10.1006/jmbi.1993.1002. [DOI] [PubMed] [Google Scholar]
- Drake J.W. Mutations in clusters and showers. Proc. Natl. Acad. Sci. USA. 2007;104:8203–8204. doi: 10.1073/pnas.0703089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J.W. Too many mutants with multiple mutations. Crit. Rev. Biochem. Mol. Biol. 2007;42:247–258. doi: 10.1080/10409230701495631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo R.S., Hastings P.J., Rosenberg S.M. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner-Russo L.M., Lombardo M.-J., Ponder R.G., Rosenberg S.M. The TGV transgenic vectors for single-copy gene expression from the Escherichia coli chromosome. Gene. 2001;273:97–104. doi: 10.1016/s0378-1119(01)00565-0. [DOI] [PubMed] [Google Scholar]
- Harris R.S., Longerich S., Rosenberg S.M. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Hicks W.M., Kim M., Haber J.E. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Altmannova V., Spirek M., Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- Louarn J.M., Louarn J., François V., Patte J. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J. Bacteriol. 1991;173:5097–5104. doi: 10.1128/jb.173.16.5097-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Rogacheva M.V., Nishant K.T., Zanders S., Bustamante C.D., Alani E. Mutation hot spots in yeast caused by long-range clustering of homopolymeric sequences. Cell Rep. 2012;1:36–42. doi: 10.1016/j.celrep.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I., Seshasayee A.S., Luscombe N.M. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485:95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, MA: 1985. The Growth of Biological Thought: Diversity, Evolution, and Inheritance. [Google Scholar]
- Miller J.H. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. A Short Course in Bacterial Genetics. [Google Scholar]
- Motamedi M.R., Szigety S.K., Rosenberg S.M. Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon E.R., Rainey P.B., Nowak M.A., Lenski R.E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Gene conversion as a focusing mechanism for correlated mutations: a hypothesis. Mol. Gen. Genet. 1996;251:503–508. doi: 10.1007/BF02173638. [DOI] [PubMed] [Google Scholar]
- Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu. Rev. Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- Nomura M., Morgan E.A. Genetics of bacterial ribosomes. Annu. Rev. Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D., Amstislavskiy V., Soldatov A., Ogryzko V. Use of high throughput sequencing to observe genome dynamics at a single cell level. Proc. Natl. Acad. Sci. USA. 2009;106:20830–20835. doi: 10.1073/pnas.0906681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington J.M., Rosenberg S.M. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino J.F., Galhardo R.S., Morales L.D., Rosenberg S.M. Stress-induced beta-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J. Bacteriol. 2009;191:5881–5889. doi: 10.1128/JB.00732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder R.G., Fonville N.C., Rosenberg S.M. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Price M.N., Alm E.J., Arkin A.P. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res. 2005;33:3224–3234. doi: 10.1093/nar/gki638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S.A., Sterling J., Thompson C., Harris S., Mav D., Shah R., Klimczak L.J., Kryukov G.V., Malc E., Mieczkowski P.A. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P.A., Arnold F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S.M., Hastings P.J. The split-end model for homologous recombination at double-strand breaks and at Chi. Biochimie. 1991;73:385–397. doi: 10.1016/0300-9084(91)90105-a. [DOI] [PubMed] [Google Scholar]
- Rosenberg S.M., Longerich S., Gee P., Harris R.S. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- Rosenberg S.M., Shee C., Frisch R.L., Hastings P.J. Stress-induced mutation via DNA breaks in Escherichia coli: a molecular mechanism with implications for evolution and medicine. Bioessays. 2012;34:885–892. doi: 10.1002/bies.201200050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Gibson J.L., Darrow M.C., Gonzalez C., Rosenberg S.M. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2011;108:13659–13664. doi: 10.1073/pnas.1104681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Ponder R., Gibson J.L., Rosenberg S.M. What limits the efficiency of double-strand break-dependent stress-induced mutation in Escherichia coli? J. Mol. Microbiol. Biotechnol. 2011;21:8–19. doi: 10.1159/000335354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J.N., Shafer B.K., McGill C.B. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tehranchi A.K., Blankschien M.D., Zhang Y., Halliday J.A., Srivatsan A., Peng J., Herman C., Wang J.D. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler D.S., Sampson E., Siddiqi I., Rosenberg S.M., Thomason L.C., Stahl F.W., Stahl M.M. Recombination of bacteriophage lambda in recD mutants of Escherichia coli. Genome. 1989;31:53–67. doi: 10.1139/g89-013. [DOI] [PubMed] [Google Scholar]
- Torkelson J., Harris R.S., Lombardo M.J., Nagendran J., Thulin C., Rosenberg S.M. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Gonzalez K.D., Scaringe W.A., Tsai K., Liu N., Gu D., Li W., Hill K.A., Sommer S.S. Evidence for mutation showers. Proc. Natl. Acad. Sci. USA. 2007;104:8403–8408. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sterling J., Storici F., Resnick M.A., Gordenin D.A. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K.A., Tomita, M., Wanner, B.L., and Mori, H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008. [DOI] [PMC free article] [PubMed]
- Bull, H.J., Lombardo, M.J., and Rosenberg, S.M. (2001). Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc. Natl. Acad. Sci. USA 98, 8334–8341. [DOI] [PMC free article] [PubMed]
- Cairns, J., and Foster, P.L. (1991). Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128, 695–701. [DOI] [PMC free article] [PubMed]
- Cherepanov, P.P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. [DOI] [PubMed]
- Chumley, F.G., Menzel, R., and Roth, J.R. (1979). Hfr formation directed by Tn10. Genetics 91, 639–655. [DOI] [PMC free article] [PubMed]
- Datsenko, K.A., and Wanner, B.L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645. [DOI] [PMC free article] [PubMed]
- Dillingham, M.S., and Kowalczykowski, S.C. (2008). RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671. [DOI] [PMC free article] [PubMed]
- Gumbiner-Russo, L.M., Lombardo, M.-J., Ponder, R.G., and Rosenberg, S.M. (2001). The TGV transgenic vectors for single-copy gene expression from the Escherichia coli chromosome. Gene 273, 97–104. [DOI] [PubMed]
- Kovach, M.E., Elzer, P.H., Hill, D.S., Robertson, G.T., Farris, M.A., Roop, R.M., 2nd, and Peterson, K.M. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. [DOI] [PubMed]
- McKenzie, G.J. (2002). The SOS response and low fidelity DNA polymerase IV in adaptive mutation in Escherichia coli. PhD thesis, Baylor College of Medicine, Houston, TX.
- McKenzie, G.J., Harris, R.S., Lee, P.L., and Rosenberg, S.M. (2000). The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97, 6646–6651. [DOI] [PMC free article] [PubMed]
- McKenzie, G.J., Magner, D.B., Lee, P.L., and Rosenberg, S.M. (2003). The dinB operon and spontaneous mutation in Escherichia coli. J. Bacteriol. 185, 3972–3977. [DOI] [PMC free article] [PubMed]
- Miller, J.H. (1992). A Short Course in Bacterial Genetics (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Ponder, R.G., Fonville, N.C., and Rosenberg, S.M. (2005). A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19, 791–804. [DOI] [PubMed]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning—A Laboratory Manual (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shee, C., Gibson, J.L., Darrow, M.C., Gonzalez, C., and Rosenberg, S.M. (2011). Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 108, 13659–13664. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.