Summary
Enveloped viruses have developed various adroit mechanisms to invade their host cells. This process requires one or more viral envelope glycoprotein to achieve cell attachment and membrane fusion. Members of the Flaviviridae such as flaviviruses possess only one envelope glycoprotein, E, whereas pestiviruses and hepacivirus encode two glycoproteins, E1 and E2. Although E2 is involved in cell attachment, it has been unclear which protein is responsible for membrane fusion. We report the crystal structures of the homodimeric glycoprotein E2 from the pestivirus bovine viral diarrhea virus 1 (BVDV1) at both neutral and low pH. Unexpectedly, BVDV1 E2 does not have a class II fusion protein fold, and at low pH the N-terminal domain is disordered, similarly to the intermediate postfusion state of E2 from sindbis virus, an alphavirus. Our results suggest that the pestivirus and possibly the hepacivirus fusion machinery are unlike any previously observed.
Graphical Abstract
Highlights
► Structure of the major antigenically dominant protein of BVDV ► The overall fold of BVDV E2 shows no similarity to the class II fusion proteins ► At low pH, BVDV E2 N-terminal domain is disordered ► Entry mechanism of BVDV is probably applicable to hepatitis C virus
Stuart and colleagues have determined the atomic structure of the ectodomain of bovine viral diarrhea virus E2 glycoprotein, the major, antigenically dominant protein on the virus surface. The structure was expected to resemble the fusion molecules found on the surface of viruses such as dengue virus, but it is unlike anything previously seen. E2 itself is not, in fact, the fusion protein but binds the cell receptor and directs fusion via a pH-dependent conformational switch.
Introduction
The Flaviviridae family comprises three genera: flavivirus, hepacivirus, and pestivirus. The most notable pestiviruses are bovine viral diarrhea virus (BVDV), classical swine fever virus (CSFV), and border disease virus (BDV), infecting cattle, pigs, and sheep, respectively. Pestiviruses have global economic impact, with clinical symptoms ranging from respiratory disorders to abortions (Lindenbach and Rice, 2001). Efforts are being made to detect outbreaks at early stages to prevent disasters such as that which occurred in the Netherlands in 1997 where CSFV was responsible for the death of more than 11 million pigs (Stegeman et al., 2000), with an estimated cost of US $2.3 billion (Meuwissen et al., 1999). Because of their close relationship to human pathogens of the hepacivirus genus, pestiviruses are used as a model for hepatitis C virus (HCV) (Buckwold et al., 2003), a major public health threat for which there is no vaccine currently available and therapeutic treatments have limited success (De Francesco and Migliaccio, 2005).
BVDV is a positive-sense single-stranded RNA enveloped virus. Its 12.5 kb genome encodes a single polyprotein, which is cleaved by viral and cellular proteases into structural and nonstructural proteins (Lindenbach and Rice, 2001). Of the three structural glycoproteins (Erns, E1, and E2) located on the outer surface of the virion, only the structure of Erns has been determined (Krey et al., 2012). E1 and E2 are type I transmembrane proteins with an N-terminal ectodomain and a C-terminal helix anchored in the viral membrane, whereas Erns lacks a typical transmembrane domain and is not required for cell entry. Unusually, pestiviral glycoproteins form two types of disulfide-linked dimers, E2-E2 and E2-E1, which are found on the virion surface (Durantel et al., 2001; Weiland et al., 1990). During virus assembly, E2 homodimers are formed early, whereas E1-E2 heterodimers are formed later after the release of E1 from the endoplasmic reticulum chaperone calnexin (Branza-Nichita et al., 2001). The heterodimer is known to be the functional fusion complex in mature virions (Wang et al., 2004). In the absence of firm data to identify which protein is directly responsible for fusion, it has been predicted that E2 fulfills this role and possesses the class II fusion fold (Garry and Dash, 2003; Kielian, 2006) that harbors a membrane distal fusion loop (rich in hydrophobic residues) and is composed of three globular domains. All known structures of flavivirus E glycoprotein and alphavirus E1 glycoproteins, share this fold. In their prefusion conformation, these class II fusion proteins form dimers (E-E for flaviviruses or E1-E2 for alphaviruses) that dissociate in the acidic pH environment of the endosome, exposing the fusion loops to the target membrane and facilitating their rearrangement into homotrimers (Kielian and Rey, 2006).
To clarify the role of pestivirus glycoprotein E2 in viral fusion and gain insights into the HCV fusion mechanism, we have determined the crystal structures of the BVDV1 glycoprotein E2 at neutral and low pH (at 2.6 Å and 3.3 Å resolution, respectively), providing structural insight into the pre- and postfusion state of the protein.
Results and Discussion
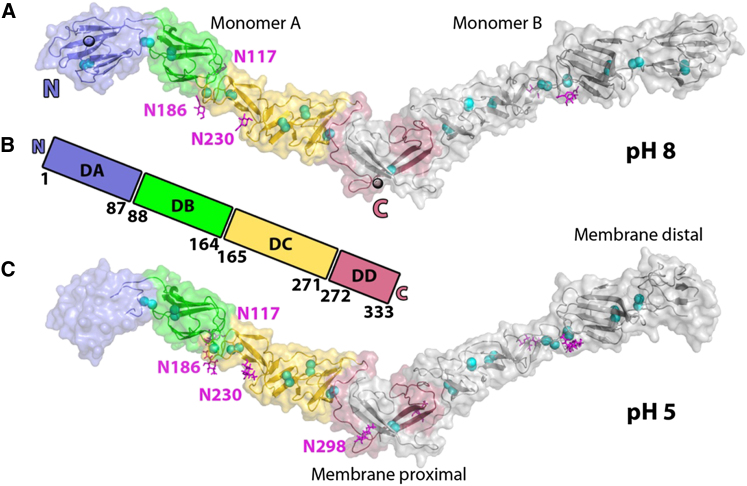
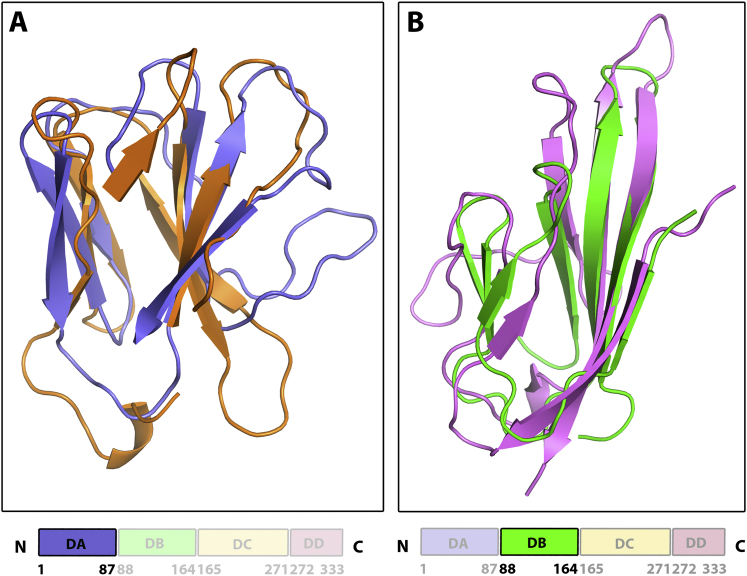
The ectodomain of BVDV1 E2 (residues 4–334, although residue 334 is not visible in the electron-density maps), bearing a mutation on the C-terminal glycosylation site (N298Q), was purified and crystallized as a covalently linked homodimer (Iourin et al., 2012). E2 is an elongated molecule consisting of four domains, DA, DB, DC, DD, arranged linearly from N to C terminus (Figure 1). All 17 cysteines are involved in disulfide bridges establishing one inter- and eight intramolecular bonds (Figure S1A). As expected from the deglycosylation process, electron density for a single N-acetylglucosamine-linked asparagine is clearly defined for residues N117, N186, N230, and N298 in the structure obtained at pH 5 where N298 has not been mutated (Figure 1C). Whereas for the pH 8 structure, as a result of the mutation N298Q, no glycan is seen at this position (Figure 1A). The overall fold, despite being composed primarily of β strands, shows no similarity to class II fusion proteins of flaviviruses or alphaviruses that do not have the domains organized linearly along the polypeptide chain.
Figure 1.
Overall Fold of BVDV1 E2
(A) Cartoon representation of the crystal structure of BVDV1 E2 obtained at pH8. A dimer of BVDV1 E2 is seen in the crystal. Domains of monomer A starting from the N terminus are colored in blue, green, yellow and red, respectively. The sugars are shown as magenta sticks and disulfide bridges are represented as cyan spheres.
(B) Linear representation of BVDV1 E2 showing the four domains DA, DB, DC and DD with their domain boundaries. Coloring as in a.
(C) Cartoon representation of the crystal structure of BVDV1 E2 obtained at pH5. Domains DA of monomers A and B are disordered (the transparent surface rendered shows the position of the domains when ordered at higher pH).
See also Figures S1, S2, S3, and S4.
Figure S1.
Amino Acid Conservation between Pestiviruses E2 Glycoprotein, Related to Figures 1, 2, and 3
(A) Sequence alignment of BVDV1 E2 with E2 proteins from other pestiviruses: BVDV2, BVDV3, Classical Swine Fever virus (CSFV), Border Disease virus (BDV), Pestivirus of giraffe (gir) and Bungowannah virus (bung). Conserved residues are drawn in red boxes, similar residues in red type. The secondary structure assignment for BVDV1 E2 is shown at the top and domains DA, DB, DC and DD are colored in blue, green, yellow and red, respectively. Cysteines involved in intra-chain disulfide bridges are marked by a green number, the single cysteine involved in the inter-chain disulfide bridge is marked by a green square, the four glycosylation sites of BVDV1 E2 are marked with magenta squares and the conserved histidine is marked with a cyan square.
(B) Surface representation of a monomer of BVDV E2 colored according to sequence conservation from white (non-conserved) to red (conserved).
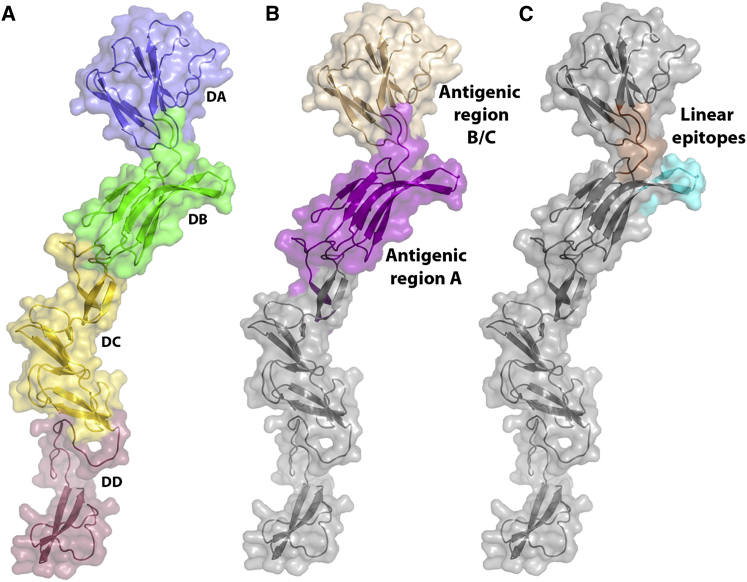
Domains DA and DB (residues 4–87 and 88–164, respectively) are the most distal from the viral membrane and are likely to be the most exposed on the virus surface, indeed the CSFV antigenic regions A and B/C map to exposed patches on the surface of these domains (Figure 2). Both domains possess Ig-like folds (Figure S2), consistent with a cell-receptor binding function, but no structure resembling a fusion loop or hydrophobic patch could be identified. CSFV E2 (65% sequence identity) binds to host cells, inhibiting infection by both CSFV and BVDV, implying a common cell receptor for pestiviruses attaching through E2 (Hulst and Moormann, 1997). Its role in cell binding is corroborated by the fact that E2 determines cell tropism of the virus (Liang et al., 2003). A CSFV host cell binding peptide (Li et al., 2011) maps to domain DB of BVDV1 E2 (Figure 2B), more precisely to a solvent exposed β-hairpin extrusion. CD46 has been shown to be a cellular receptor of BVDV (Maurer et al., 2004), and is a candidate for attachment to the β-hairpin.
Figure 2.
Mapping of Epitopes of Classical Swine Fever Virus E2 onto the Structure of BVDV E2
(A) Cartoon representation of a monomer of BVDV1 E2 colored by domain as in Figure 1.
(B) Positions of the antigenic regions A and B/C.
(C) Location of the linear epitopes LFDGTNP (orange) and TAVSPTTLR (cyan). LFDGTNP is at the border of the two antigenic regions, whereas TAVSPTTLR is located in the antigenic region A.
See also Figure S1.
Figure S2.
Domains DA and DB Have Ig-fold, Related to Figure 1
(A) Domain DA of BVDV1 E2 (neutral pH form) is colored in blue and is superimposed on a T cell receptor alpha chain (PDB: 2Z31, Z = 5.3, rmsd = 2.8Å) colored in orange.
(B) Domain DB is colored in green and is superimposed on the C-terminal domain of semaphorin 4D (PDB: 1OLZ, Z = 6.7, rmsd = 2.3Å) colored in pink.
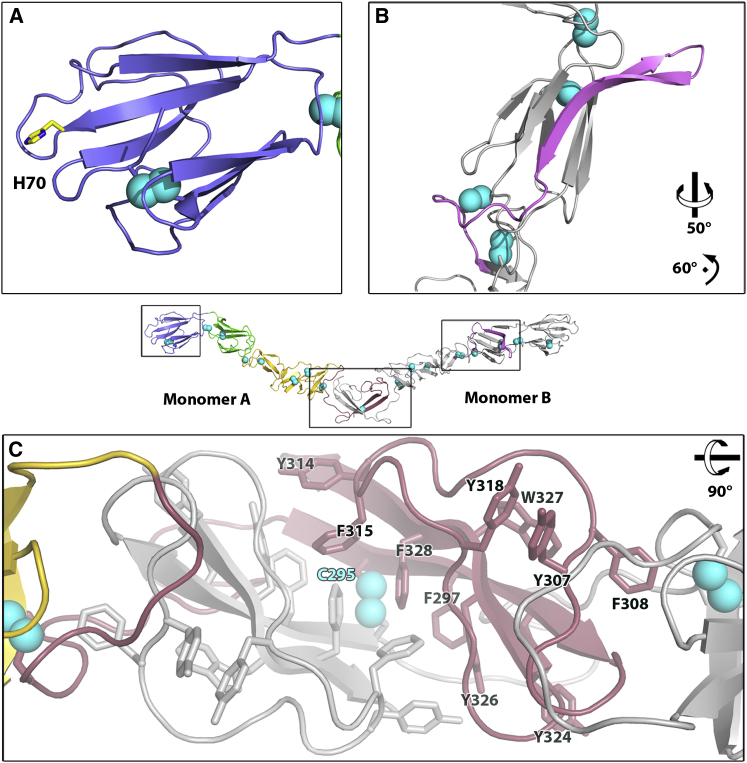
Domain DC (residues 165–271) could not be matched significantly to any known structure using the DALI server (Holm and Rosenström, 2010); it is a highly extended disulfide-rich structure, composed of loops and antiparallel β strands, containing two glycosylation sites (N186 and N230). Among pestivirus E2 glycoproteins, there are two conserved glycosylation sites (N117 and N186) (Figure S1A) that lie close together in the three-dimensional structure, and mutation of either of these leads to a marked reduction or loss of expression in baculovirus infected cells (Pande et al., 2005). Homodimerization occurs through domain DD (residues 272–333), which is the most conserved domain among pestiviruses (Figures S1A and S1B). Dimers are covalently stabilized by a disulfide bridge and, surprisingly, by a domain swap between monomers (Figure 3C), with DD, which is connected to the other domains via a long tether, being embedded in the adjacent subunit. Domain DD is composed of an extended loop that links domain DC to a β-hairpin. The hairpin loop folds back onto the β strands to form an hydrophobic pocket. Similarly to domain DC, no structural match could be found in the protein data bank. All disulfide bonds, apart from the C-terminal one, are involved in intramolecular bonds, so it is highly likely that the heterodimer E1-E2 is also formed via the cysteine involved in the homodimerization at the C terminus of E2 near the viral membrane. Moreover, the two swapped domains interact strongly with each other through hydrophobic patches rich in phenylalanines and tyrosines (Figure 2C). During biogenesis, both glycoproteins E1 and E2 associate with the chaperone protein calnexin (Branza-Nichita et al., 2001). The well-conserved hydrophobic patch of E2 is likely to be concealed temporarily through dimerization with chaperone proteins, and once released, used for homodimerization and heterodimerization, with both dimers being further stabilized by a disulfide bridge.
Figure 3.
Interactions of BVDV1 E2 and Their Locations
(A) Location of histidine (H70) of domain DA, which is conserved among pestiviruses and that may trigger an order-disorder transition of this domain at low pH.
(B) The potential host cell receptor binding site in domain DB colored in pink. Domain DB is rotated as shown.
(C) The interface of monomers A (shown colored) and B (shown in gray) contains a strand swap and hydrophobic residues such as phenylalanines, tyrosines and tryptophanes drawn as sticks. Domains DD of molecules A and B are linked by a disulfide bridge (cyan sphere) to form a covalent homodimer.
See also Figure S1.
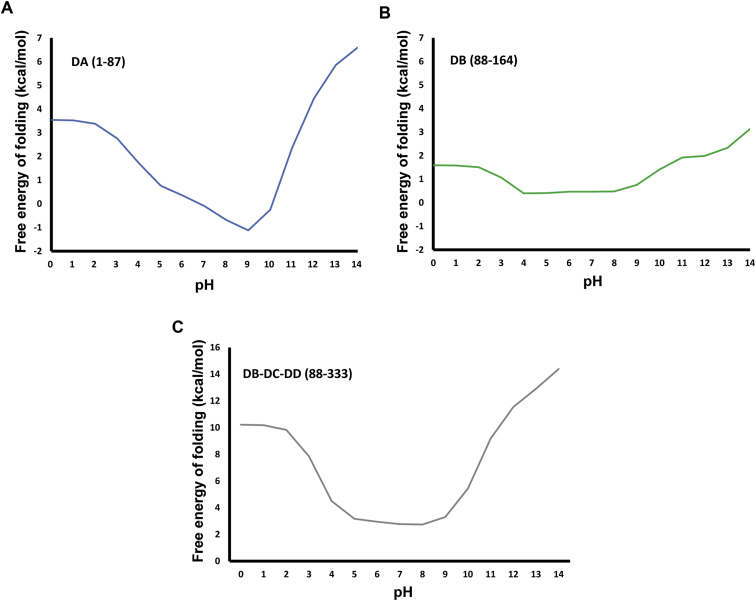
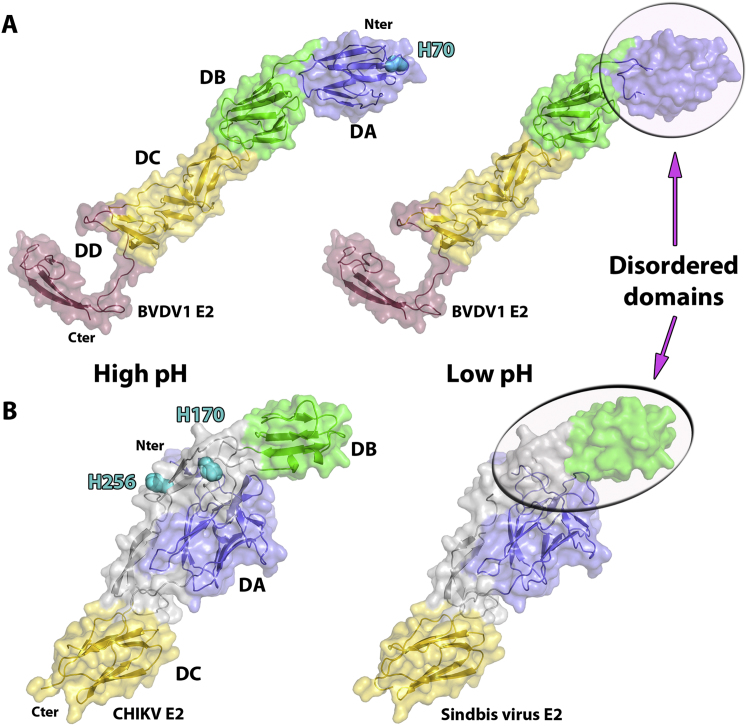
The low pH structure of BVDV1 E2 (residues 1–339) is similar to the one at neutral pH apart from a rigid body movement of one of the monomers of some 12° and, intriguingly, by disordering of domain DA (Figure 1C). In fusion proteins, histidine is believed to play a role in pH-induced conformational changes because its pKa is between the physiological and endosomal pH (Kampmann et al., 2006). A drop in local pH on entry into the endosome would consequently change the protonation state of histidines and their local interactions. Pestivirus E2 glycoproteins possess only one strictly conserved histidine (H70) located in domain DA (Figures 3A and S1A). H70 is surrounded by aliphatic chains from E70, R72, A73, and Y8 thus protonation of this conserved histidine is likely to destabilize domain DA whose stability is predicted to be sensitive to a drop of pH from 9 to 5 whereas the rest of the protein remains stable (Figure S3). Remarkably, the distal domain of sindbis virus E2, which caps the fusion peptide of E1, also becomes disordered in low pH environment (Figure S4), via a proposed histidine switch, to expose the fusion loop of E1 (Li et al., 2010). HCV E2 likewise has a conserved histidine (H445) toward its N terminus acting as a pH sensor (Boo et al., 2012), suggesting a similar mechanism.
Figure S3.
Stability of BVDV1 E2 Domains over pH, Related to Figure 1
(A) domain DA stability depends on pH, the results would suggest that domain become less stable when moving from physiological pH (7.4) to the endosome pH 5.
(B) For comparison, domain DB, a similarly sized Ig-like domain to domain DA, would not be destabilized by a change of pH from 7.4 to 5.
(C) Domains DB-DC-DD would not be destabilized by a change of pH from 7.4 to 5.
Figure S4.
Structural Comparison of Pestivirus and Alphavirus E2 Glycoproteins at High and Low pH, Related to Figure 1
(A) Cartoon representation of the crystal structures of BVDV E2 at high (left) and low pH (right) colored as defined in Figure 1.
(B) Cartoon representation of the crystal structures of CHIKV E2 obtained at high pH (left) and sindbis virus E2 obtained at low pH (right). The Ig domains A, B, C are colored in blue, green and yellow respectively. The linker domain is colored in gray.
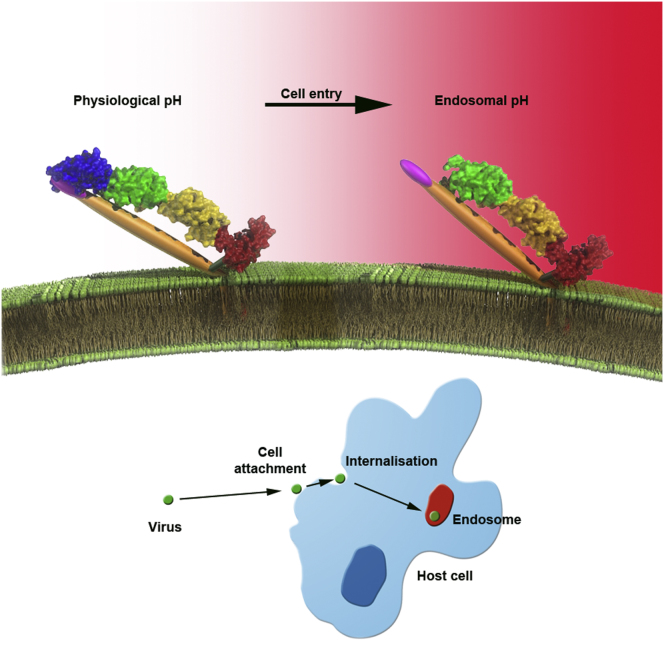
Because the membrane distal domains of BVDV1 E2 do not harbor a hydrophic peptide we propose that E2 is not the fusion protein. Furthermore BVDV1 E2 does not have a class II fusion protein fold and its shape instead resembles the alphavirus E2 attachment glycoprotein that also contains Ig-like domains and partially unfolds at low pH but is not fusogenic (Figure S4). We suggest instead that E1 is responsible for fusion. Consistent with this proposition, a fusion peptide-like motif located in the middle of the HCV E1 sequence (residues 264–290) has been proposed to play a role fusion (Drummer et al., 2007; Li et al., 2009). In pestivirus E1 proteins, a sequence similarly rich in hydrophobic amino acids can also be found between residues 57 and 85. However, E1 is about half the size of typical class II fusion proteins, so either it has a different fold, or, as suggested previously for HCV E1, a truncated class II fold (Garry and Dash, 2003). In any event, E1 must span the distance to the host cell membrane, perhaps 100 Å to 150 Å and is therefore likely to be a thin, extended protein, possibly using E2 as a scaffold (the domain swap organization we observe would allow E2 to partly substitute for the E1 domain proximal to the viral membrane). This model is in line with the observation that the heterodimer is essential for membrane fusion, in contrast to alphaviruses where E2 is not required once the fusion loop is exposed. Our model for the stages of pestivirus cell entry leading up to membrane fusion is shown in Figure 4.
Figure 4.
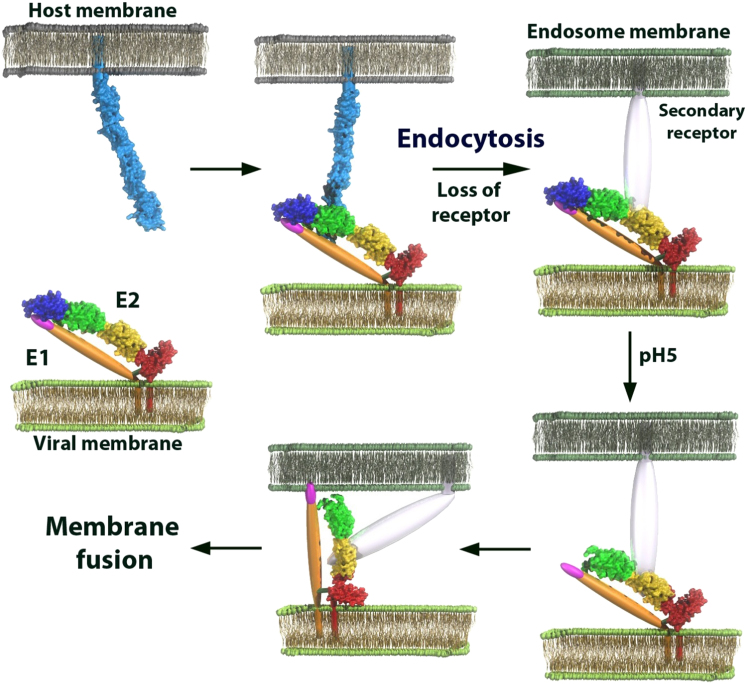
Proposed Model of the Fusion Mechanism of Pestiviruses
A covalently linked E1-E2 heterodimer is present on the viral membrane. E2 binds to the host receptor (possibly CD46, shown in light blue) through domain DB prior to endocytosis. As CD46 is not internalized, a putative secondary receptor (transparent) presumably replaces CD46. Throughout this process the putative fusion loop of E1 (magenta), is concealed by domain DA of E2. At low pH, domain DA of E2 becomes disordered and exposes the fusion loop that will then be able to insert into the endosome membrane and trigger membrane fusion. It is as yet unknown whether E1 forms trimers as seen for other fusion proteins.
The pestivirus genus was moved from the Togaviridae (containing the alphaviruses) to the Flaviviridae because of similarity in genome organization (Collett et al., 1988). The presence of heterodimers E1-E2 and our observation that BVDV1 E2 is reminiscent of alphavirus E2, suggest that the cell entry machineries of the pestivirus and probably hepacivirus genera are in fact closer to the Togaviridae than to flaviviruses.
Although BVDV has been used as a surrogate for HCV, there is no direct evidence that the glycoproteins are similar between these viruses. However, there is also no detectable sequence similarity between that the fusion proteins of the flaviviruses and alphaviruses although they share the same fold. Indeed a series of observations support the contention that HCV envelope glycoproteins are more similar to pestiviruses than other viruses: (1) BVDV and HCV both require an extra step for membrane fusion beyond a simple pH drop, suggesting a similar entry mechanism (Krey et al., 2005), (2) their genome organization is very similar: HCV has two glycoproteins E1 and E2 like alphaviruses and pestiviruses; however, the size of E1 is about half of E2, as seen in pestiviruses, (3) BVDV and HCV E2 are both the immunodominant proteins that generate neutralizing antibodies, and (4) they both bind the cell receptors (Hulst and Moormann, 1997; Pileri et al., 1998). Together, these findings suggest that HCV will likely follow the unexpected pathway for cell attachment that we propose for BVDV1 and, more specifically that the BVDV1 E2 structures presented in this article should provide a useful model for HCV.
Experimental Procedures
Cloning, Purification, and Crystallization
Cloning, purification, and crystallization have been described (Iourin et al., 2012). In brief, DNA coding the ectodomain of BVDV1 E2 (residues 1–339) or a truncated version BVDV1 E2tr (residues 4–334) were cloned with a C-terminal His6 tag. Site-directed mutagenesis was performed on BVDV1 E2tr to remove N-linked glycosylation by replacing asparagine 298 with a glutamine (BVDV1 E2tr N298Q). Both constructs were transiently expressed in HEK293T cells in the presence of the N-glycosylation inhibitor kifunensine (Toronto Research Chemicals, North York, ON) (Aricescu et al., 2006). Purification used affinity and size exclusion chromatography, following deglycosylation with Endo F1 and His-tag removed using Bovine Pancreas Carboxypeptidase A-Agarose (Sigma) from BVDV1 E2tr N298Q. Both BVDV1 E2 and BVDV1 E2tr constructs formed covalently-linked homodimers. To facilitate protein concentration, 3-(1-Pyridino)-1-propane sulfonate (NDSB 201) was added to a final concentration of 300 mM. BVDV1 E2 was crystallized in 16% PEG 6000, 79 mM citric buffer pH 5.0, 20 mM MgSO4. Selenomethionine-labeled BVDV1 E2tr N298Q crystallized in 52.5% methyl-2,4-pentanediol (MPD) and 0.1 M Tris pH 8. Crystals were flash frozen using 25% (v/v) glycerol/reservoir solution as cryo-protectant, or for selenomethionine-labeled E2tr N298Q frozen without additional cryo-protection.
Data Collection and Structure Determination
Diffraction data were recorded at the Diamond Light Source I02, I03, and I24 beamlines and processed with HKL2000 (Otwinowski and Minor, 1997). The structure was determined with the program PHENIX AUTOSOL (Adams et al., 2002) by multiwavelength anomalous dispersion (MAD) phasing using selenomethionine-labeled crystals grown at neutral pH. The low pH conformation was solved by molecular replacement with MOLREP (Vagin and Teplyakov, 1997) using the neutral pH form as a search model. Manual building was performed with the program COOT (Emsley and Cowtan, 2004) and restrained refinement (with TLS) with AUTOBUSTER (Bricogne et al., 2008). Final models geometry were checked with MolProbity (Davis et al., 2007). For data collection and refinement statistics see Table S1.
Structure Analysis
The superimpositions of the neutral and low pH forms were performed using SHP (Stuart et al., 1979). Figure S1 was generated with CLUSTALW (Larkin et al., 2007) and ESPript (Gouet et al., 1999). The PROPKA server (Bas et al., 2008) was used to determine the free energy of folding of domains of BVDV1 E2 presented in Figure S4.
Acknowledgments
We are grateful to Dr. Geoff Sutton, Dr. Thomas Bowden, and Prof. Yvonne Jones for discussions and Weixian Lu for technical assistance. We thank the staff of beamlines I02, I03, and I24 at the Diamond Light Source synchrotron for technical support. This work was supported by the Medical Research Council (MRC) and the Wellcome Trust provided administrative support (grant 075491/Z/04). K.E. collected X-ray diffraction data, solved, and refined the structures. O.I. carried out cloning, purification and crystallization. K.H. performed crystal mounting and collected X-ray diffraction data. K.E., J.M.G., and D.I.S. analyzed the structures and wrote the manuscript. D.I.S. supervised the project.
Published: December 27, 2012
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.12.001.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
Coordinates and structure factors of BVDV1 E2 at neutral and low pH have been deposited in the Protein Data Bank under ID codes 2yq2 and 2yq3, respectively.
Supplemental Information
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Bas D.C., Rogers D.M., Jensen J.H. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- Boo I., teWierik K., Douam F., Lavillette D., Poumbourios P., Drummer H.E. Distinct roles in folding, CD81 receptor binding and viral entry for conserved histidine residues of hepatitis C virus glycoprotein E1 and E2. Biochem. J. 2012;443:85–94. doi: 10.1042/BJ20110868. [DOI] [PubMed] [Google Scholar]
- Branza-Nichita N., Durantel D., Carrouée-Durantel S., Dwek R.A., Zitzmann N. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J. Virol. 2001;75:3527–3536. doi: 10.1128/JVI.75.8.3527-3536.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Smart O.S., Vonrhein C., Womack T. Global Phasing Ltd; Cambridge: 2008. BUSTER-TNT 2.5.1 and autoBUSTER 1.3.1. [Google Scholar]
- Buckwold V.E., Beer B.E., Donis R.O. Bovine viral diarrhea virus as a surrogate model of hepatitis C virus for the evaluation of antiviral agents. Antiviral Res. 2003;60:1–15. doi: 10.1016/s0166-3542(03)00174-8. [DOI] [PubMed] [Google Scholar]
- Collett M.S., Anderson D.K., Retzel E. Comparisons of the pestivirus bovine viral diarrhoea virus with members of the flaviviridae. J. Gen. Virol. 1988;69:2637–2643. doi: 10.1099/0022-1317-69-10-2637. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., 3rd, Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Francesco R., Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- Drummer H.E., Boo I., Poumbourios P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 2007;88:1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- Durantel D., Branza-Nichita N., Carrouée-Durantel S., Butters T.D., Dwek R.A., Zitzmann N. Study of the mechanism of antiviral action of iminosugar derivatives against bovine viral diarrhea virus. J. Virol. 2001;75:8987–8998. doi: 10.1128/JVI.75.19.8987-8998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Garry R.F., Dash S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology. 2003;307:255–265. doi: 10.1016/s0042-6822(02)00065-x. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Métoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulst M.M., Moormann R.J. Inhibition of pestivirus infection in cell culture by envelope proteins E(rns) and E2 of classical swine fever virus: E(rns) and E2 interact with different receptors. J. Gen. Virol. 1997;78:2779–2787. doi: 10.1099/0022-1317-78-11-2779. [DOI] [PubMed] [Google Scholar]
- Iourin O., Harlos K., El Omari K., Lu W., Kadlec J., Iqbal M., Meier C., Palmer A., Ian J., Brownlie J., Grimes J.M. Expression, purification and crystallization of the ectodomain of the envelope glycoprotein E2 from Bovine viral diarrhoea virus. Acta Crystallogr. F. 2012 doi: 10.1107/S1744309112049184. Published online December 15, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann T., Mueller D.S., Mark A.E., Young P.R., Kobe B. The Role of histidine residues in low-pH-mediated viral membrane fusion. Structure. 2006;14:1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Kielian M. Class II virus membrane fusion proteins. Virology. 2006;344:38–47. doi: 10.1016/j.virol.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey T., Thiel H.J., Rümenapf T. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 2005;79:4191–4200. doi: 10.1128/JVI.79.7.4191-4200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey T., Bontems F., Vonrhein C., Vaney M.C., Bricogne G., Rümenapf T., Rey F.A. Crystal structure of the pestivirus envelope glycoprotein E(rns) and mechanistic analysis of its ribonuclease activity. Structure. 2012;20:862–873. doi: 10.1016/j.str.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li H.F., Huang C.H., Ai L.S., Chuang C.K., Chen S.S. Mutagenesis of the fusion peptide-like domain of hepatitis C virus E1 glycoprotein: involvement in cell fusion and virus entry. J. Biomed. Sci. 2009;16:89. doi: 10.1186/1423-0127-16-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Jose J., Xiang Y., Kuhn R.J., Rossmann M.G. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang L., Zhao D., Zhang G., Luo J., Deng R., Yang Y. Identification of host cell binding peptide from an overlapping peptide library for inhibition of classical swine fever virus infection. Virus Genes. 2011;43:33–40. doi: 10.1007/s11262-011-0595-7. [DOI] [PubMed] [Google Scholar]
- Liang D., Sainz I.F., Ansari I.H., Gil L.H., Vassilev V., Donis R.O. The envelope glycoprotein E2 is a determinant of cell culture tropism in ruminant pestiviruses. J. Gen. Virol. 2003;84:1269–1274. doi: 10.1099/vir.0.18557-0. [DOI] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. Volume 1. Lippincott Williams & Wilkins; Philadelphia, PA: 2001. (The Pestiviruses). [Google Scholar]
- Maurer K., Krey T., Moennig V., Thiel H.J., Rümenapf T. CD46 is a cellular receptor for bovine viral diarrhea virus. J. Virol. 2004;78:1792–1799. doi: 10.1128/JVI.78.4.1792-1799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen M.P., Horst S.H., Huirne R.B., Dijkhuizen A.A. A model to estimate the financial consequences of classical swine fever outbreaks: principles and outcomes. Prev. Vet. Med. 1999;42:249–270. doi: 10.1016/s0167-5877(99)00079-3. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pande A., Carr B.V., Wong S.Y., Dalton K., Jones I.M., McCauley J.W., Charleston B. The glycosylation pattern of baculovirus expressed envelope protein E2 affects its ability to prevent infection with bovine viral diarrhoea virus. Virus Res. 2005;114:54–62. doi: 10.1016/j.virusres.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A.J., Houghton M., Rosa D., Grandi G., Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Stegeman A., Elbers A., de Smit H., Moser H., Smak J., Pluimers F. The 1997-1998 epidemic of classical swine fever in the Netherlands. Vet. Microbiol. 2000;73:183–196. doi: 10.1016/s0378-1135(00)00144-9. [DOI] [PubMed] [Google Scholar]
- Stuart D.I., Levine M., Muirhead H., Stammers D.K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J. Mol. Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Wang Z., Nie Y., Wang P., Ding M., Deng H. Characterization of classical swine fever virus entry by using pseudotyped viruses: E1 and E2 are sufficient to mediate viral entry. Virology. 2004;330:332–341. doi: 10.1016/j.virol.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Weiland E., Stark R., Haas B., Rümenapf T., Meyers G., Thiel H.J. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J. Virol. 1990;64:3563–3569. doi: 10.1128/jvi.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.