Abstract
RNA replication of dengue virus (DENV) requires an RNA-RNA mediated circularization of the viral genome, which includes at least three sets of complementary RNA sequences on both ends of the genome. The 5′ and the 3′ untranslated regions form several additional RNA elements that are involved in regulation of translation and required for RNA replication. Communication between the genomic termini results in a structural reorganization of the RNA elements, forming a functional RNA panhandle structure. Here we report that the sequence composition downstream of the 5′ CS element in the capsid gene, designated as downstream CS (dCS) sequence -- but not the capsid protein -- also influences the ability of the viral genome to circularize and hence replicate by modulating the topology of the 5′ end. These results provide insights for the design of reporter sub-genomic and genomic mosquito-borne flavivirus constructs and contribute to the understanding of viral RNA replication.
Introduction
Dengue, the most common mosquito-borne viral disease in humans, is caused by the four serotypes of dengue virus (DENV 1-4), a member of the Flaviviridae family (Lindenbach et al., 2007). The small enveloped virus contains a plus-strand RNA genome that encodes the viral polyprotein and serves as a template for genome replication. RNA translation and replication are regulated by RNA elements located in the viral 5′ and 3′ untranslated regions (UTRs) and within the capsid-coding region immediately adjacent to the 5′ UTR (Alvarez et al., 2008; Alvarez et al., 2005a; Alvarez et al., 2006; Alvarez et al., 2005b; Chiu et al., 2005; Clyde et al., 2008; Clyde and Harris, 2006; Filomatori et al., 2011; Filomatori et al., 2006; Harris et al., 2006; Holden and Harris, 2004; Holden et al., 2006; Lodeiro et al., 2009; Polacek et al., 2009b; Villordo and Gamarnik, 2009; Wei et al., 2009).
Several RNA elements located within the 3′UTR are believed to regulate 5′-cap-dependent translation (Alvarez et al., 2005a; Holden and Harris, 2004; Holden et al., 2006; Manzano et al., 2011; Polacek et al., 2009b; Wei et al., 2009). Besides modulating translation, RNA elements at the viral genomic 3′ end are also essential for RNA replication as well as cytopathogenicity and pathogenicity (Funk et al., 2010; Pijlman et al., 2008; Silva et al., 2010). For initiation of minus-strand synthesis, the 3′SL, the HP-3′SL and three RNA sequences – the 3′ cyclization sequence (CS), the 3′ upstream of AUG region (UAR) element, and the 3′ downstream of AUG region (DAR) – are absolutely necessary (Alvarez et al., 2008; Alvarez et al., 2005b; Friebe and Harris, 2010; Friebe et al., 2011; Villordo and Gamarnik, 2009). Several studies have shown that these three sequences can interact with their complementary 5′ elements - the 5′ CS, the 5′ UAR, and the 5′ DAR elements – resulting in an RNA-RNA-mediated circularization of the genome and a reorganization of the 3′ RNA structure, a prerequisite for initiation of minus-strand RNA synthesis (Filomatori et al., 2011; Friebe and Harris, 2010; Villordo and Gamarnik, 2009).
The formation of the 5′-3′ panhandle structure brings the 5′ stem loop A (SL-A), which has been demonstrated to bind the viral RNA-dependent RNA polymerase (RdRp) NS5, in close proximity to the structurally-reorganized 3′ end, allowing initiation of minus-strand synthesis (Filomatori et al., 2011; Filomatori et al., 2006; Friebe and Harris, 2010). While SL-A and the 5′ UAR element are located within the 5′ UTR, the 5′ DAR and 5′ CS elements reside within the capsid-coding region. Thus far, one additional element within the viral coding region has been identified – the capsid-coding region hairpin (cHP), which is involved in RNA replication and also directs start codon selection (Clyde et al., 2008; Clyde and Harris, 2006). As the function of the cHP is position-dependent but sequence-independent, it is believed that its role in translation is to stall the scanning initiation complex over the first AUG, favoring its recognition. The exact involvement of cHP in RNA replication is less well understood, but recent work indicates a role in the formation of a functional 5′-3′ panhandle structure, a prerequisite for RNA replication. Although the cHP is not required to initiate basepairing between the viral termini, it must be part of the panhandle structure, forming a functional unit with the 5′ UAR, 5′ DAR and 5′ CS elements (Friebe and Harris, 2010). In contrast to SL-A, which is 5′-end-dependent, the position of the remaining RNA elements are not absolutely position-dependent, as long as the elements form a single unit in close proximity to the viral 5′ terminus (Friebe and Harris, 2010).
The viability of sub-genomic DENV reporter replicons, containing the 5′ UTR and the first 72 nucleotides (nt) of the capsid-coding region but lacking the remaining open reading frame (ORF) of the structural proteins, demonstrates that all RNA elements essential for RNA replication are located within this region, including the 5′ DAR, cHP and 5′ CS elements (Alvarez et al., 2005a; Clyde et al., 2008). The ability to produce reporter viral particles (RVPs) by packaging the replicon RNA into infectious viral particles further demonstrates that any essential packaging signals must be located in this region, outside of the missing coding region for capsid, prM/M and E (Ansarah-Sobrinho et al., 2008).
For hepatitis C virus (HCV), a closely related virus within the Flaviviridae family, additional regulatory RNA elements have been identified within the core-coding region, the protein homologous to DENV capsid protein. An RNA-RNA interaction between nucleotides in the 5′ UTR and the core-coding region was shown to affect viral translation (Kim et al., 2003). Furthermore, two RNA stem-loop structures within the core-coding region contribute to HCV genome translation and replication (Vassilaki et al., 2008). Besides non-essential RNA elements, an impact of the core protein on HCV RNA translation has been reported (Lourenco et al., 2008; Shimoike et al., 2006). Furthermore, the capsid protein of other RNA viruses has also been implicated in modulation of RNA replication (Tzeng et al., 2006). In the case of flaviviruses, one report suggested a role for the capsid protein in either unpackaging and/or early RNA synthesis (Schrauf et al., 2009). However, a study using DENV showed no difference in RNA replication in the presence or absence of wild-type (WT) capsid protein, but expression of a mutated capsid protein with an altered subcellular localization pattern had a negative effect on RNA accumulation (Samsa et al., 2009).
We therefore analyzed the impact of the capsid-coding region – focusing on the RNA sequence and role of the capsid protein – on viral translation and RNA replication, using DENV as a model and West Nile Virus (WNV) to confirm the results with another flavivirus. Our data show that the RNA sequence composition between nucleotide positions 170-200 of the viral RNA, rather than the capsid protein, contributes to RNA replication but has no effect on RNA translation. Furthermore, enhancement of viral RNA replication is not caused by a new RNA element located downstream of the 5′CS element, designated the dCS sequence, but rather by its sequence composition and its effect on the ability of the 5′ UAR, 5′ DAR and 5′ CS element to interact with their 3′ counterparts. This then modulates the affinity between the viral termini, affecting the ability of the viral genome to circularize, a prerequisite for RNA replication.
Results
The Downstream of 5′CS (dCS) sequence affects RNA replication
To evaluate whether non-essential regulatory RNA elements are embedded within the capsid-coding region, we included as many silent point mutations as possible in the region downstream of the DENV 5′ UTR and the first 72 nts of the capsid gene. This region, spanning nucleotide positions 171-438, was designated as the downstream CS (dCS) sequence, and the mutated sequence was designated as “SYN”. An alignment of “SYN” with the WT sequence is shown in Supplementary Figure 1. Care was taken to avoid introducing rarely-used codons. The “SYN” sequence was then introduced into a replicon with a similar design as 5′UTR-Cap-tr-SPACER (Friebe and Harris, 2010) but with the “SPACER” sequence replaced by the capsid-coding region. The new replicon backbone was designated biRep-5′UTR-CapFL (Fig. 1A), harboring either the WT or the SYN capsid-coding region (resulting in biRep-5′UTR-CapFL-WT or biRep-5′UTR-CapFL-SYN, respectively).
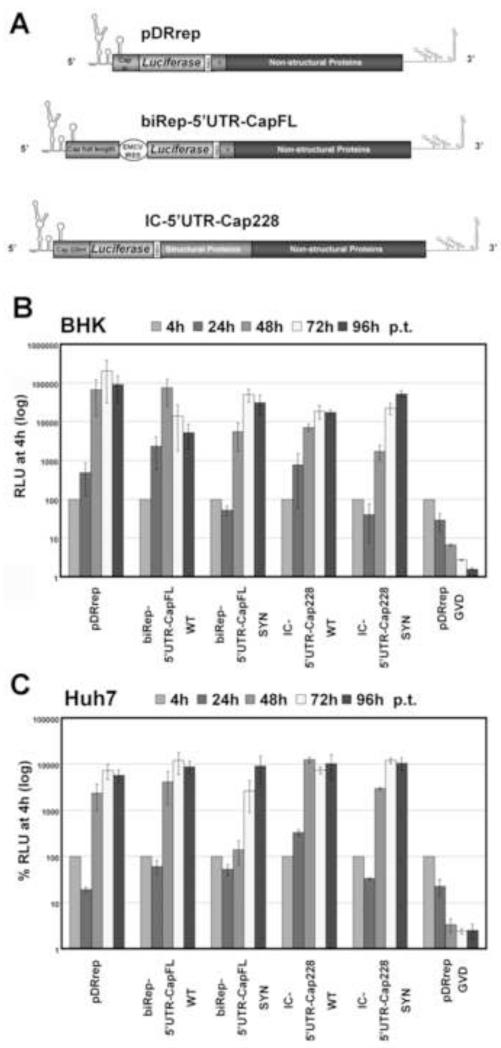
Fig. 1. Silent point mutations in the DENV capsid-coding sequence affect RNA replication.
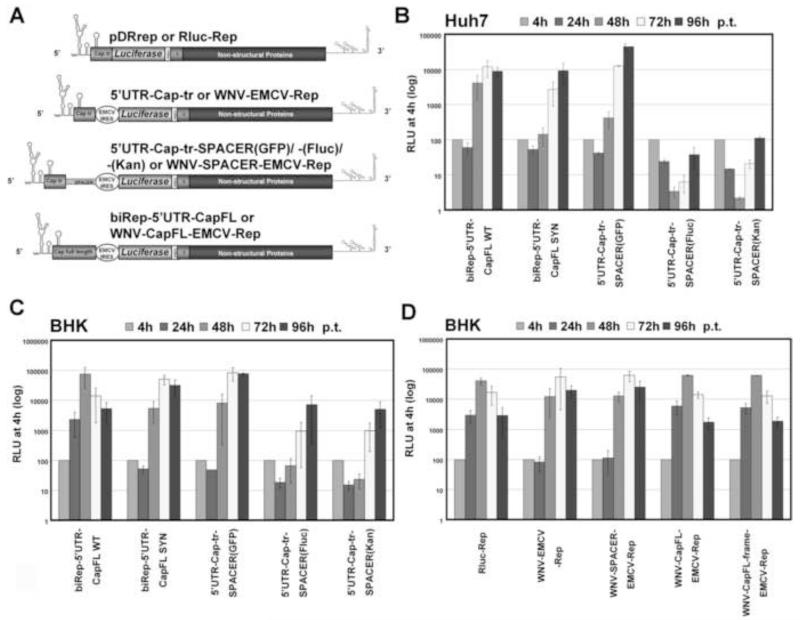
(A) Schematic presentation of the Renilla luciferase-expressing reporter constructs used in this study. pDRrep has been described previously (Friebe and Harris, 2010). Replicon biRep-5′UTR-CapFL carries either the WT or the “SYN” sequence of the full capsid-coding region (Cap full length), resulting in biRep-5′UTR-CapFL-WT or -SYN. The EMCV IRES mediates translation of the Renilla luciferase (Luciferase) reporter gene and the viral ORF spanning the C-terminal sequence of E and NS1 to NS5. Luciferase is cleaved from the viral proteins by an engineered FMDV2A (FMDV) cleavage site. For IC-5′UTR-Cap228, the first 228 nucleotides (Cap 228nt) of the capsid-coding region are present downstream of the 5′ UTR, fused to the Renilla luciferase (Luciferase). Luciferase is cleaved from the viral polyprotein (Structural Proteins, Non-structural Proteins) by an engineered FMDV2A (FMDV) cleavage site. The capsid-coding region either contains the WT (IC-5′UTR-Cap228-WT) or the “SYN” sequence (IC-5′UTR-Cap228-SYN). RNA structures in the 5′ and 3′ ends are indicated schematically. (B) Replication competence of indicated DENV reporter RNAs over a timecourse of 96h post-transfection (p.t.) in BHK cells. Timepoints are indicated on top, and RLU is expressed as percentage of the value measured 4h p.t., which was set to 100%. The GVD mutant (Friebe and Harris, 2010) served as a negative control. Results from at least three independent experiments are shown. Error bars reflect standard deviation. (C) Replication competence of indicated DENV reporter RNAs over a timecourse of 96h post-transfection (p.t.) in Huh7 cells. For details, refer to (B).
The replication phenotype of the new replicons was tested in BHK and Huh7 cells and compared to pDRrep by transfecting the same amount of RNA of each construct into either BHK or Huh7 cells. Luciferase activity was monitored over a timecourse of 4 hours (h), 24h, 48h, 72h and 96h. The 4h value, reflecting translation of the input RNA, served as an indicator of transfection efficiency and was set to 100%, and all later values were normalized to the 4h value to account for transfection efficiency. Luciferase activity (relative luciferase units, RLU) was comparable among all three replicon RNAs 4h post-transfection (p.t.) (Supplementary Figure 2A and data not shown), revealing no difference in EMCV IRES-mediated translation efficiency between biRep-5′UTR-CapFL-WT and -SYN. As a negative control, a replicon with an inactivating mutation (GDD to GVD) in the catalytic site of the NS5 RdRp was used (Friebe and Harris, 2010).
Comparison of the replication pattern in BHK cells showed that replicon biRep-5′UTR-CapFL-WT showed ~1.5 logs higher RNA accumulation levels than the mutant biRep-5′UTR-CapFL-SYN as early as 24h p.t. (Fig. 1B), and the same difference was observed in Huh7 cells at 48h p.t. (Fig. 1C). In BHK cells, differences at later time points (72h and 96h p.t.) disappear, as replication of the WT replicon is limited by the cytopathic effect of viral RNA replication on the host cell, which results in a loss of cells that support replication and hence reduced luciferase activity (data not shown).
To further validate the negative effect on RNA replication of the silent point mutations harbored by biRep-5′UTR-CapFL-SYN, we constructed a full-length reporter virus in which the complete viral open reading frame is under the authentic DENV 5′ cap-dependent translational control (Fig. 1B, IC-5′UTR-Cap228), with a similar design to mDV-R (Samsa et al., 2009). Note that the 5′ end of IC-5′UTR-Cap228 consists of the 5′ UTR and the first 228 nt of the DENV ORF, which are fused to the Renilla luciferase reporter that is cleaved from the C-terminal DENV polyprotein by an engineered FMDV2A cleavage site. The results using the more authentic full-length reporter viruses confirm the observations made with reporter replicons; namely, IC-5′UTR-Cap228-SYN shows delayed RNA accumulation kinetics as compared to IC-5′UTR-Cap228-WT in BHK as well as in Huh7 cells (Fig. 1B and C). For both RNAs, RLUs 4h p.t. were similar (Supplementary Figure 2B and data not shown), indicating that the mutations present in the capsid-coding region do not affect overall translation efficiency. Furthermore, both viral RNAs harbor the WT sequence of the capsid-coding region downstream of the Renilla luciferase gene, indicating that effects of mutations in IC-5′UTR-Cap228-SYN cannot be compensated by internal positioning of the WT capsid-coding region and arguing for position dependency. It is worth mentioning that both RNAs retained their ability to produce infectious viral particles (data not shown). To simplify the presentation of results, replication kinetics will hereafter be shown by comparison of the luciferase activity 48h p.t. in Huh7 and 24h p.t. in BHK cells, normalized to the value measured 4h p.t., although the full kinetic timecourse was performed in both cell lines for all experiments.
An additional predicted 5′-3′ RNA-RNA interaction and stem-loop structure in the dCS sequence do not moduate RNA replication
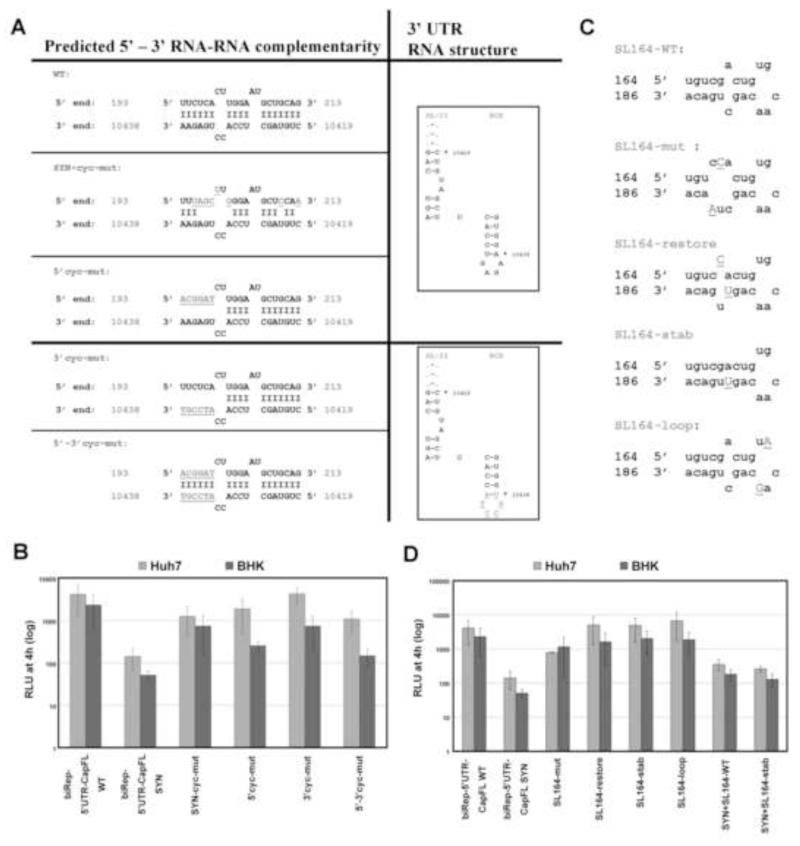
Using MPGAfold, a recent publication indicated a potential interaction between nucleotides 193-213 at the DENV-2 5′ end with a complementary sequence within the 3′ UTR (nucleotide positions 10419 to 10438 of the DENV-2 strain 16681 full-length sequence), although the interaction was only predicted as a suboptimal RNA folding structure (Manzano et al., 2011). Additional complementary sequences between the viral termini could assist the 5′-3′ UAR-, DAR- and CS-mediated circularization of the genome and therefore enhance RNA replication while not being absolutely required. The predicted interaction is depicted in Fig. 2A, revealing complementarity between 17 nucleotides in the viral 5′ and 3′ ends. The silent point mutations we introduced into this sequence reduce the number of base pairs from 17 to 10. To test whether RNA replication of biRep-5′UTR-CapFL-SYN was indeed reduced due to interference with the predicted 5′-3′ RNA-RNA interaction, we separated the mutations that interfere with complementarity to the 3′ end from the “SYN” context by inserting the “SYN” sequence between nucleotides 193-213 into the biRep-5′UTR-CapFL-WT replicon, resulting in construct biRep-5′UTR-CapFL-cyc-mut (Fig. 2A, SYN-cyc-mut). Replication of RNAs harboring only the SYN-cyc-mut mutations was slightly reduced as compared to the WT but did not reach the low-level replication of biRep-5′UTR-CapFL-SYN (Fig. 2B, SYN-cyc-mut), indicating that the potential 5′-3′ complementarity depicted in Fig. 2A does not account for the impaired replication phenotype observed with the “SYN” construct.
Fig. 2. Predicted RNA-RNA interactions do not contribute significantly to RNA replication.
(A) Left column: schematic overview of predicted interaction between nucleotides in the capsid-coding region (positions 193 to 213) and the 3′ UTR (positions 10419 to 10438) for DENV2 16681 (WT), as published recently (Manzano et al., 2011). Complementary nucleotides between the 5′ end (top) and 3′ end (bottom) are indicated by a dash. Names are indicated above each sequence. Mutants with nucleotide changes introduced into the 3′ UTR are separated by a line (lower panel) from mutants with changes only at the viral 5′ end (upper panel). All mutations are underlined. Right column: predicted RNA structures in the 3′ UTR (SL-II and RCS) as published previously (Proutski et al., 1997). Borders of the 3′ sequence predicted to be complementary to the 5′ motif are indicated with an asterisk, and nucleotide positions are indicated. Introduced mutations are underlined (lower panel). (B) Replication competence of indicated DENV reporter RNAs in Huh7 and BHK cells. As indicated in the text, only the luciferase activity 48h p.t. in Huh7 and 24h p.t. in BHK cells, normalized to the value measured 4h p.t., are shown, although the full kinetic timecourse was performed in both cell lines. Results from at least three independent experiments are shown. Error bars reflect standard deviation. (C) Schematic overview of predicted stem-loop structure between nucleotide position 164 and 186, designated as SL164-WT. Mutated nucleotides are capitalized and underlined. The names of the mutants are indicated above each struture. (D) Replication competence of indicated DENV reporter RNAs in Huh7 and BHK cells. For details, refer to (B).
As the mutations in biRep-5′UTR-CapFL-cyc-mut nonetheless affected RNA replication, we next wanted to analyze whether the additional complementarity shown in Fig. 2A between the 5′-3′ termini nevertheless enhanced RNA replication. However, introducing compensatory mutations into the 3′ sequence that would restore complementarity to the biRep-5′UTR-CapFL-cyc-mut mutation was hampered by the fact that the 3′ sequence is predicted to form two functional RNA structures within the 3′ UTR in the absence of 5′ RNA: SL-II and RCS (as indicated in Fig. 2A, right panel). In the case of WNVKun, SL-II was reported to be a key element for the production of the subgenomic flavivirus RNA (sfRNA), which is required for flavivirus cytopathogenicity and pathogenicity (Funk et al., 2010; Pijlman et al., 2008). As the exact requirements and effects of DENV sfRNA have not been defined yet and deletion of the variable region (VR) affects DENV RNA replication at least in BHK cells (Alvarez et al., 2005a), it is important to keep the RNA structures SL-II and RCS intact. We therefore substituted six nucleotides in the 5′ motif between nucleotide positions 193 and 198 so as to allow restoration of complementary to the 3′ sequence by introducing mutations into the loop and in one position on both sides of the RCS stem, thereby maintaining the RCS RNA structure as predicted using Mfold (Zuker, 2003) (Fig. 2A, 5′-cyc-mut, 3′-cyc-mut and 5′-3′cyc-mut). Note that mutations introduced into 5′-cyc-mut are not silent and result in a FS to TD change. When tested for RNA replication kinetics, all mutants showed a reduced RNA replication pattern as compared to the WT, which was more pronounced in BHK cells than in Huh7 cells (Fig. 2B). The fact that restoring complementarity did not restore but rather worsened RNA replication implies that 5′-3′ complementarity alone is not sufficient to support high level-RNA replication. Furthermore, none of the mutants displayed a delayed RNA amplification phenotype as severe as that observed with biRep-5′UTR-CapFL-SYN. Together, these data indicate the presence of at least one more regulatory RNA sequence affected by the mutations introduced into the dCS sequence that is not mediated by the analyzed complementarity we investigated.
Another recent publication identified a replication enhancer element (REE) within the capsid-coding region of tick-borne encephalitis virus, forming a stable stem-loop structure designated as SL6 (Tuplin et al., 2011). Mfold-based structure prediction revealed an RNA element between nucleotide positions 164 and 186 that seems to be conserved between different DENV serotypes, which we designated as SL164. The structure predicted for the DENV-2 16681 sequence is shown in Fig. 2C and contains the loop sequence “UGCAA”, which is very similar to the conserved “UGCCAA” motif of tick-borne encephalitis virus that was implicated in the REE function of SL6 (Tuplin et al., 2011). However, this sequence is not conserved within other DENV serotypes. To analyze whether the predicted RNA structure in DENV-2 also acts as an REE, we constructed a panel of mutants targeting the region between nucleotide positions 164 and 186. We inserted silent point mutations to disrupt or stabilize the predicted stem structure, and we targeted the loop sequence (Fig. 2C, SL164-mut, SL164-restore, SL164-stab, and SL164-loop). When tested for its ability to undergo RNA replication, the SL164-mut displayed somewhat reduced RNA replication but not as severe as the SYN mutant (Fig. 2D, compare biRep-5′UTR-CapFL-WT, SL164-mut, and SYN). Restoring or stabilizing the predicted SL164 RNA structure or mutations introduced into the loop region did not affect RNA replication (Fig. 2D, SL164-restore, SL164-stab and SL164-loop). However, replacing the sequence between 164 and 186 with the WT sequence or the stabilized WT sequence in the backbone of biRep-5′UTR-CapFL-SYN did not restore RNA replication levels (Fig. 2D, SYN+SL164-WT and SYN+SL164-stab). Taken together, these results do not indicate an REE function for the predicted RNA element between nucleotide positions 164-186 in DENV-2, which is consistent with the fact that the loop sequence is not conserved among different DENV serotypes.
Composition of the dCS sequence 25-55 nt downstream of the 5′ CS element influences RNA replication
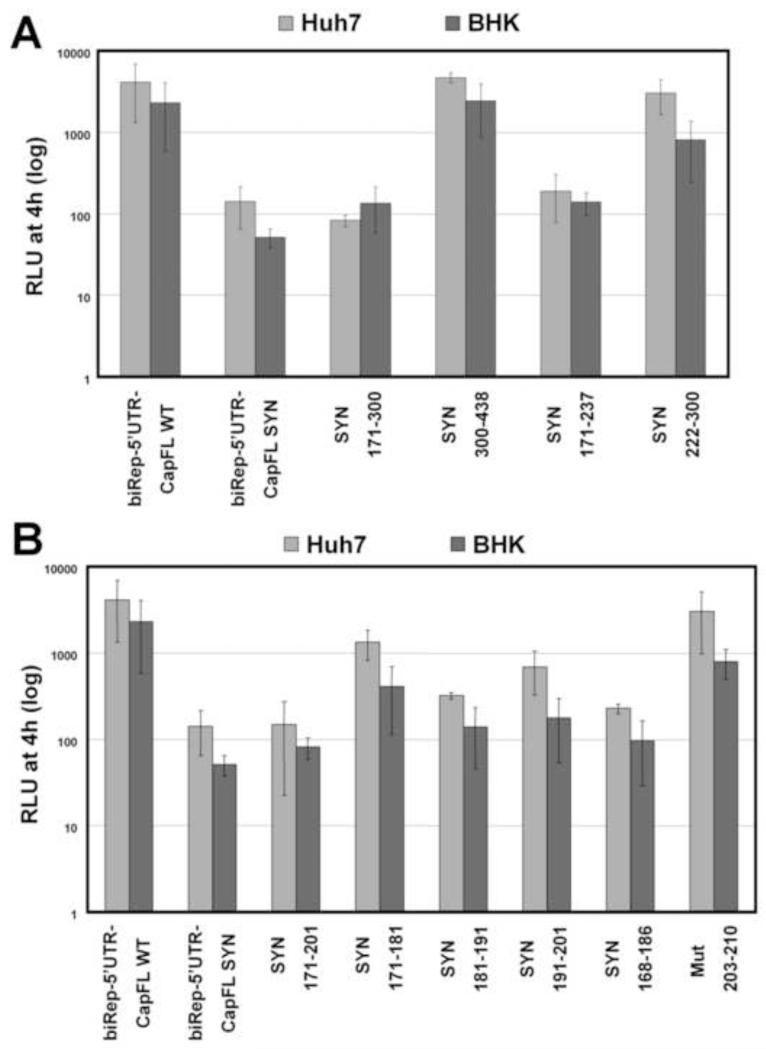
To further map the sequences responsible for the effect observed on RNA replication, we used the biRep-5′UTR-CapFL replicon backbone to construct a set of mutants with insertion of smaller regions harboring the silent point mutations. The replication patterns depicted in Fig. 3A demonstrate that the sequence impacting RNA replication is located between nucleotide positions 171 to 222, as mutations downstream of 222 did not affect RNA replication (Fig. 3A, compare SYN171-237 and SYN222-300 to either biRep-5′UTR-CapFL-WT or -SYN). Next, we continued to minimize the sequence analyzed by additional higher resolution mapping of the region of interest. We replaced the WT coding region with sequences harboring silent point mutations between nucleotide positions 171-201, 171-181, 181-191, 191-201, 168-186 and also mutated positions 203-210 (mut203-210: 5′GAATGCTG3′ to 5′CACACGAC3′). Note that mutations between nucleotide positions 203-210 are not silent and cause mutation within in the amino acid sequence (GML to AHD). Summarizing the RNA replication results derived from this set of mutants indicates that the main impact on RNA accumulation is caused by mutations introduced between nucleotide positions 170 and 200 (Fig. 3B).
Fig. 3. Silent point mutations between nucleotide positions 170-200 have the greatest effect on RNA replication.
(A) and (B) Replication competence of indicated DENV reporter RNAs in Huh7 (A) and BHK (B) cells. For more details, please refer to the legend of Figure 2B.
The dCS sequence does not affect RNA translation
Although Renilla luciferase activities 4h p.t. were similar between IC-5′UTR-Cap228-WT and -SYN (data not shown) and both RNAs express the full-length capsid protein, indicating no dramatic impact of the mutations on expression levels or availibility of the capsid protein, a more rigorous analysis is needed to fully rule out any effect on RNA translation. For instance, altered capsid expression levels or start codon usage, which could result in more dominant expression of an N-terminal truncated version of the capsid protein, could impact RNA levels over time. To analyze whether additional RNA elements located within the dCS sequences regulate RNA translation, we constructed a set of reporter RNAs fusing a 3XFLAG epitope either to the 5′ UTR and the first 270 nt or to the 5′ UTR and the full-length caspid coding-region, resulting in constructs biRep-5′UTR-Cap270-FLAG or biRep-5′UTR-CapFL-FLAG (Fig. 4A). As this backbone allows the expression of the viral NS proteins, it is important to note that the full-length capsid protein contains the dibasic cleavage site that is recognized by the viral NS2B-NS3 protease (Amberg and Rice, 1999) and could lead to cleavage of the 3XFLAG epitope over time. However, this internal cleavage site is not present in the biRep-5′UTR-Cap270-FLAG backbone. The WT capsid sequence of both backbones was then replaced with the coding sequence haboring silent point mutations.
Fig. 4. No effect of the capsid protein on RNA replication is observed.
(A) Schematic overview of the constructs used to analyze the impact of silent point mutations in the capsid-coding region on 5′-mediated RNA translation. A 3XFLAG sequence (FLAG) was fused either to the first 270 nucleotides (Cap 270) or to the full-length (Cap full length) capsid-coding region, using either the biRep-5′UTR-CapFL-WT or -SYN backbone. (B) Cell lysates of Huh7 cells transfected with the RNAs indicated on top were prepared 4h p.t. and analyzed by Western blot using anti-FLAG M2, anti-DENV-NS1 and anti-actin antibodies. Results displayed are representative of at least 3 independent experiments. (C) Replication competence of indicated DENV reporter RNAs in Huh7 and BHK cells. For more details, please refer to the legend of Figure 2B.
In vitro-transcribed RNAs of the constructs described above were transfected into Huh7 or BHK cells. Cell lysates harvested 4h p.t. were separated by SDS-PAGE electrophoresis, and the amount of protein expressed from the first or internal start codon was monitored as a shift in molecular weight on an anti-FLAG immunoblot. DENV NS1 levels were used to control for transfection efficiency, and actin served as loading control (Fig. 4B, NS1 and Actin panels). Initiation from the first start codon was favored in all cases, while leaky scanning occurred to a lesser degree in Huh7 (Fig. 4B) and BHK cells (data not shown). The same pattern was previously observed for a similar WT reporter RNA in Hep3B cells, whereas mutations interfering with the formation of the cHP element led to a higher initiation rate at the second start codon (Clyde and Harris, 2006). The overall amount of protein expression as well as the ratio between protein products initiated at the first or the second start codon showed no differences between the different RNAs tested, indicating that the dCS sequence does not affect RNA translation.
To further rule out any effect of differences in translation of the capsid protein on RNA replication, we constructed a panel of deletion mutants in which the first 201, 249, 300, 348, or 399 nt of the viral genome are present at the 5′ end, fused to the internal EMCV IRES. The biRep-5′UTR-CapFL-WT and -SYN, both harboring the first 430 nt of the DENV genome at the 5′ end, served as controls. Results depicted in Fig. 4C clearly demonstrate that the presence of the first 249 nt at the viral RNA 5′end promotes high-level RNA replication of the corresponding replicon RNA (Fig. 4C, compare biRep-5′UTR-CapFL-WT with 249nt) and even an RNA carrying only the first 201 nt showed Renilla luciferase activity kinetics very simlar to biRep-5′UTR-CapFL-WT, although neither of the two RNAs express more than the N-terminal part of the capsid protein. Taken together, our results show that the dCS region does not impact 5′cap-dependent RNA translation, nor does the presence or absence of the capsid protein affect RNA replication.
The dCS sequence affects 5′-3′ UAR-DAR-CS sequence-mediated genome circularization
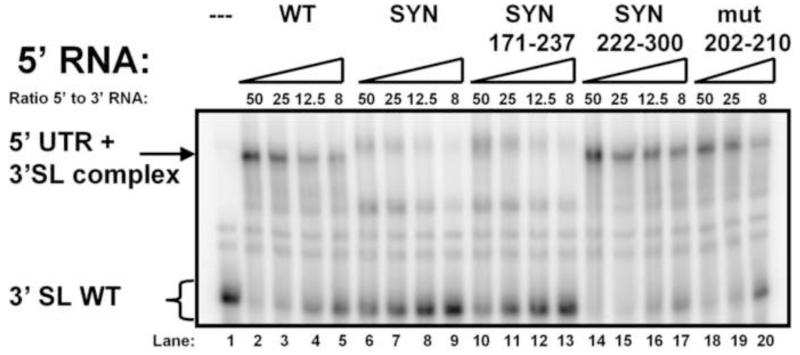
The observation that fusion of the EMCV IRES in close proximity to the DENV 5′ end resulted in a decrease in RNA replication levels due to altered topology of the 5′ UTR (Friebe and Harris, 2010) led us to the hypothesis that changes in the dCS sequence might have a similar effect on the DENV 5′ RNA elements. Therefore, we next investigated effects of the dCS region on UAR-DAR-CS sequence-mediated genome circularization, using a previously reported method to study DENV 5′-3′ RNA-RNA interaction (Alvarez et al., 2005b; Friebe and Harris, 2010). 5′ RNAs spanning the first 299 nt of the DENV genome were in vitro transcribed, as was a radiolabeled 3′ WT RNA consisting of the final 106 nt of the DENV genome. To investigate the affinity of the different 5′ RNAs in binding to the 3′ end, serial RNA titrations were utilized. As previously reported, the WT 5′ RNA was able to shift the 3′ RNA (Fig 5, lanes 2-5) (Friebe and Harris, 2010), whereas the affinity of the 5′ RNA “SYN” and “SYN171-237” for the 3′ RNA was considerably reduced and resulted in an altered mobility pattern (Fig. 5, lanes 6-9 and 10-13). In comparison, 5′ RNA “SYN222-300” and “mut203-210” showed a pattern and affinity similar to the 5′ WT RNA (Fig. 5, lanes 14-17 and 18-20). The results depicted in Fig. 5 reveal a correlation between RNA replication and 5′-3′ RNA-RNA affinity of mutations between nucleotide positions 170 and 200.
Fig. 5. Silent point mutations in the capsid-coding region between nucleotide position 170-200 affect genome circularization.
RNA mobility shift analysis showing the effect of mutations within the capsid-coding region on 5′-3′ RNA-RNA interaction. The 5′ RNA consists of the first 299 nts of the DENV genome, containing the mutations indicated along the top of the gel. The uniformly-labeled 3′SL RNA includes the final 106 nts of the DENV RNA genome. The ratio between the 5′ and 3′ RNAs is indicated on top. The mobility of the 3′SL alone or in complex with the 5′ RNA is indicated on the left. The gel displayed is representative of at least 3 independent experiments.
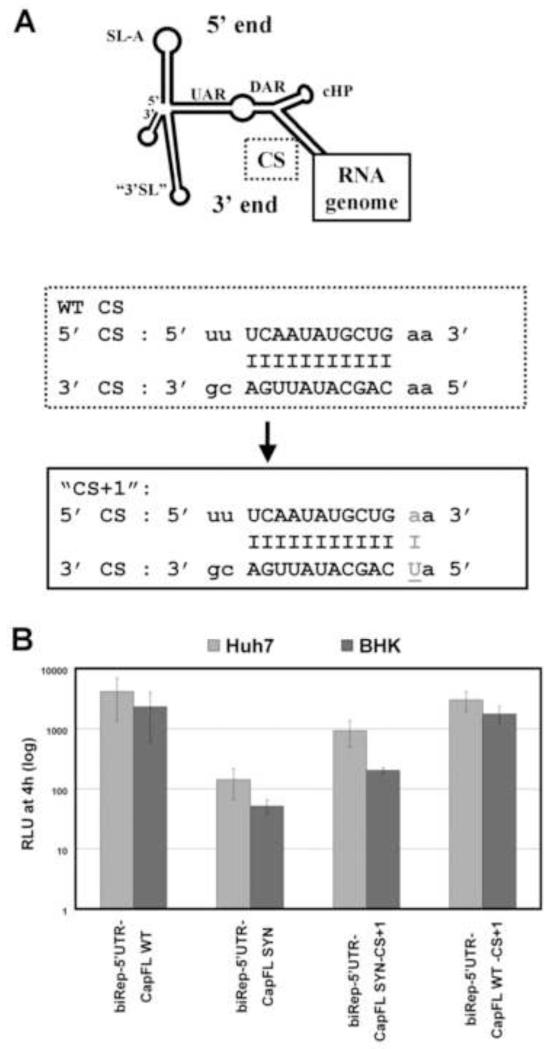
To further prove that mutations within the dCS sequence affect the affinity of the 5′-3′ RNA-RNA interaction, thereby impacting genome cirularization, we pursued a reverse genetics approach. A mutation was introduced into the 3′ end extending the region of complementarity between the 5′ and 3′ CS element by one nucleotide (Fig. 6A), which is predicted to increase the stability of the RNA-RNA interaction between the viral termini according to Mfold (data not shown). This was the only mutation recovered in a full-length DENV harboring the silent point mutations in the caspid-coding region after several weeks of passaging in BHK cells (data not shown). The results derived using biRep-5′UTR-CapFL-SYN-CS+1 RNA are consistent with the hypothesis that silent point mutations in the dCS sequence affect the affinity between the viral termini. RNA replication levels were enhanced as compared to the parental biRep-5′UTR-CapFL-SYN replicon (Fig. 6B). The replication kinetics of the biRep-5′UTR-CapFL-WT RNA were not enhanced by increasing the length of the CS elements complementarity, indicating that the WT sequences already enable high affinity interaction between the viral genomic ends (Fig. 6B, biRep-5′UTR-CapFL-WT-CS+1). However, RNA replication levels of biRep-5′UTR-CapFL-SYN-CS+1 are not completely restored to WT levels, and extension of the 5′-3′ CS compementarity did not alter the results of the RNA-RNA binding affinity assay described above, using a 3′ radiolabeled RNA including the mutation to extend CS complementarity (data not shown).
Fig. 6. Restoring 5′-3′ RNA-RNA affinity enhances RNA replication of RNAs carrying silent point mutations in the capsid-coding region.
(A) Upper Panel: schematic overview of the DENV 5′-3′ panhandle structure, with names of known RNA elements indicated. The 5′-3′ CS interaction is indicated with a dotted box and magnified below (WT CS). Complementarity between nucleotides is indicated by a dash. The mutation introduced is shown in the box with solid line below (“CS+1”), and the nucleotide change is underlined and capitalized. (B) Replication competence of indicated DENV reporter RNAs in Huh7 and BHK cells. For details, refer to the legend of Figure 2B.
Mfold prediction of the 5′-3′ panhandle structure, using either the WT or the “SYN”-derived sequence of the first 300 nt of the viral 5′ end in combination with the last 100 nt of the 3′ end resulted in proper formation of the known RNA elements: SL-A, 5′-3′ UAR, DAR and CS interactions, cHP, and reorganized 3′SL (data not shown). The 5′ “SYN” sequence is predicted to form a less stable 5′-3′ panhandle structure and even extension of the 5′-3′ CS complementarity does not restore the free energy levels of the RNA-RNA structure predicted for 5′-3′ WT sequence (data not shown).
The sequence downstream of the CS interferes with RNA replication by affecting the topology of the 5′ end
To further address whether the capsid-coding region contains a specific sequence contributing positively to genome circularization and RNA replication or whether the sequence composition downstream of the CS sterically constrains the 5′ RNA elements, we tested different reporter constructs based on the parental 5′UTR-Cap-tr-SPACER (Friebe and Harris, 2010) with a GFP coding region-derived “spacer” between the DENV 5′ RNA sequence and the internal EMCV IRES, which will be referred to as 5′UTR-Cap-tr-SPACER(GFP). We replaced the GFP-derived spacer with a sequence derived from the Firefly luciferase gene, yielding 5′UTR-Cap-tr-SPACER(Fluc), or with a sequence from the Kanamycin resistance gene, resulting in 5′UTR-Cap-tr-SPACER(Kan) (Fig. 7A). We then evaluated the RNA replication pattern of the different RNAs in Huh7 and BHK cells (Fig. 7B and C). Replicon RNAs with heterologus sequences in the region downstream of the CS, replacing the dCS region, showed very different RNA replication phenotypes. For the construct in which the sequence downstream of the CS was derived from the GFP coding region, replication kinetics slightly better than biRep-5′UTR-CapFL-SYN were observed, whereas sequences derived from the Firefly luciferase coding region or the Kanamycin resistance gene replacing the natural capsid-coding sequences downstream of the CS rendered the RNA almost replication-incompetent in Huh7 cells (Fig. 7B) and reduced RNA replication levels dramatically in BHK cells (Fig. 7C).
Fig. 7. The sequence composition downstream of the 5′ CS element affects DENV and WNV RNA replication.
(A) Schematic overview of replicons used. The design of DENV replicons pDRrep, 5′UTR-Cap-tr and 5′UTR-Cap-tr-SPACER(GFP) and WNV replicons Rluc-Rep, WNV-EMCV-Rep and WNV-SPACER(GFP)-EMCV-Rep have been described previously (Friebe and Harris, 2010). Please refer to the text for differences between the sequences of -SPACER(GFP), -(Fluc) and -(Kan) constructs. WNV-CapFL-EMCV-Rep is the analogous WNV replicon to biRep-5′UTR-CapFL (please refer to the legend of Figure 1 for more details). (B) and (C) Replication competence of indicated DENV reporter RNAs in Huh7 (B) and BHK (C) cells. For more details, please refer to the legend of Figure 1C. (D) Replication competence of indicated WNV reporter RNAs in BHK cells. For more details, please refer to the legend of Figure 1C.
Based on the dramatic effect caused by inserting a sequence derived from the Kanamycin resistance gene, we used Mfold (Zuker, 2003) to monitor effects of the dCS(Kan) sequence on the formation of DENV 5′ RNA elements. Contrary to other sequences downstream of the CS that were tested, the most stable Mfold prediction using the viral 5′ sequence with the dCS(Kan) sequence showed a major reorganization of the viral 5′ RNA structures, indicating potential effects of the composition of the dCS not only on the accessibility of the 5′ UAR, 5′ DAR and 5′ CS sequence but also on the formation of SL-A (Supplementary Figure 3). Interestingly, Mfold predicts a WT-like 5′-3′ panhandle structure for the dCS(Kan) sequence, which falls energetically between the WT and the “SYN” panhandle prediction (data not shown).
The dCS sequence is also important for West Nile Virus RNA replication
To investigate whether the dCS sequence is specific to DENV or can be found in mosquito-borne flaviviruses, we used WNV as another model system. We took advantage of a previously published reporter replicon WNV-SPACER-EMCV-Rep (Friebe et al., 2011) to create replicons similar to biRep-5′UTR-CapFL by replacing the “SPACER” of WNV-SPACER-EMCV-Rep with the WNV capsid-coding region (Fig. 7A, WNV-CapFL-EMCV-Rep). To control for the impact of the capsid protein on RNA replication, we constructed a frameshift variant, with an insertion of a cytidine at nucleotide position 198, resulting in the expression of a frameshift protein (WNV-CapFL-frame-EMCV-Rep). We tested the ability of all constructs to replicate in BHK cells. In case of WNV, the two replicons showing the highest RNA replication phenotypes were Rluc-Rep (Friebe et al., 2011) and WNV-CapFL-EMCV-Rep (Fig. 7D), whereas WNV-SPACER-EMCV-Rep showed an intermediate replication phenotype, indicating that the sequence downstream of the CS region also has an impact on viral RNA replication in case of WNV. WNV-CapFL-frame-EMCV-Rep replicated to comparable levels as WNV-CapFL-EMCV-Rep, demonstarting that the presence or absence of capsid protein does not affect WNV RNA replication either (Fig. 7D).
Discussion
Based on our results, we conclude that the DENV dCS sequence, especially nucleotide positions 25-55 downstream of the 5′ CS sequence (nucleotide positions 170 to 200 in the viral RNA), affects the formation and functionality of the 5′ RNA elements by modulating the topology of the 5′ end. Similar results were observed with WNV. The presence of the full capsid-coding region downstream of the viral 5′ UTR benefits RNA replication by increasing the affinity between the viral 5′ and 3′ ends to optimize circularization of the viral genome, a prerequisite for RNA replication.
Effects of the flavivirus capsid gene on RNA replication have been observed previously in the literature (Basu and Brinton, 2011; Clyde et al., 2008; Filomatori et al., 2011; Rouha et al., 2011; Samsa et al., 2009; Tuplin et al., 2011; Zhang et al., 2010; Zhou et al., 2007). However, no clear differences between DENV RNA replication levels in the presence or absence of WT capsid protein have been reported, although the presence of a mutated capsid protein was shown to be detrimental (Samsa et al., 2009). A recent study showed that SL-A was the only specific DENV RNA element necessary for binding the NS5 polymerase, but non-specific contacts with nucleotides downstream of SL-A were also found to be required for NS5-RNA binding (Filomatori et al., 2011). Although the dCS sequence composition could therefore contribute to NS5 binding and thereby affect RNA replication, the SL-A is predicted to form at the 5′ end of the genome in the presence of the WT as well as the “SYN” dCS sequence. Interestingly, NS5-RNA binding requirements differ between WNV and DENV; for instance, there is evidence in WNV for genetic interaction between the methyltransferase (MTase), RdRp and the 5′ SL of the genomic RNA (Zhang et al., 2008), whereas the MTase domain of the DENV NS5 protein does not contribute to SL-A binding (Filomatori et al., 2011; Filomatori et al., 2006; Lodeiro et al., 2009). Nonetheless, RNA sequences downstream of the 5′ SL were also shown to impact WNV NS5 binding to the viral RNA (Dong et al., 2008), which could potentially explain the dCS effect we observed for WNV.
Interestingly, the presence or absence of the structural protein coding region has been shown to impact the function of the flavivirus 2′-O methyltransferase. Ray and co-workers reported that Ala substitutions of the 2′-O methyltransferase active site (K61A, K182A, and E218A) rendered a WNV replicon almost replication-incompetent (Ray et al., 2006), whereas in a full-length RNA, the same mutations were replication-competent, albeit with reduced efficiency as compared to the WT (Zhou et al., 2007). These results indicate that either the RNA sequences themselves or the structural proteins benefit viral replication, compensating for the mutation in the 2′-O methyltransferase active site. It is tempting to speculate that in this case, the presence of the WT dCS sequence positively affects NS5 binding to the viral RNA, thereby partially compensating for the 2′-O methyltransferase mutations and enabling greater replication.
In line with the assumption that the presence of the capsid ORF modulates replication efficiency are reports showing that mutations affecting the complementarity between 5′-3′ CS sequences have different phenotypes when present in replicons (Alvarez et al., 2005a; Corver et al., 2003; Khromykh et al., 2001; Kofler et al., 2006; Lo et al., 2003) or full-length RNAs (Basu and Brinton, 2011; Zhang et al., 2010). WNV can tolerate up to three adjacent mismatches disrupting long-distance 5′-3′ cyclization sequence basepairs (Basu and Brinton, 2011) or even tolerate a deletion of the complete 3′ CS sequence (Zhang et al., 2010) if the capsid-coding region is present, whereas the 3′CS deletion rendered a replicon lacking the majority of the capsid-coding region RNA replication-incompetent. A similar impact of the capsid-coding region on the effect of mismatches introduced into the 5′-3′ CS basepairs was also observed for DENV (P. Friebe and E. Harris, unpublished data).
Additionally, a recent report revealed a replication enhancer element (REE) within the capsid ORF in tick-borne encephalitis virus, forming a stable stem-loop structure designated as SL6 (Tuplin et al., 2011). In another study, nucleotides involved in the formation of the REE SL6 were found to form a different stem loop structure, designated as SL4, which was also associated with RNA replication (Rouha et al., 2011). However, using RNA structure prediction algorithms in combination with a reverse genetics approach, we were unable to identify an REE homologous to that of tick-borne encephalitis virus for DENV. Furthermore, replacing the capsid-coding region with different heterologous non-viral sequences showed very distinct effects on RNA replication, demonstrating that the sequence downstream of the 5′ end causes an effect on RNA replication by a mechanism unrelated to a specific RNA structure.
Recently, Pu and co-workers (Pu et al., 2011) identified a region containing a cis-acting sequence within the central portion of the DENV capsid-coding region that modulated RNA replication. Effects on RNA replication were evaluated after inserting silent point mutations between nucleotide positions 160 and 243 of the viral genome. The greatest effect was caused by silent point mutations between nucleotide positions 160-205, whereas changes between 198-243 resulted in only a minor reduction in RNA replication (Pu et al., 2011). These results are consistent with our findings. However, the underlying mechanism was not further investigated.
Our results demonstrate that mutations in the capsid-coding region can modulate the topology of the viral 5′ end, affecting its functionality. No effect of the dCS sequence composition on RNA translation was observed, nor did the presence or absence of the capsid protein alter DENV or WNV RNA replication. Nonetheless, a clear correlation of changes in dCS sequence composition on genome circularization was found, whereby the dCS sequence affected the formation of the 5′-3′ panhandle structure by altering the functionality of the 5′ UAR, DAR and CS sequences. The following data support this conclusion: a) our RNA-RNA interaction experiments showed that dCS composition affects interaction between the DENV 5′ end and the last 106 nucleotides of the 3′ end, which only contain complementary sequences that interact with the 5′ UAR, DAR and CS elements (Hahn et al., 1987; Khromykh et al., 2001; Polacek et al., 2009a; Shi et al., 1996); b) serial cell culture passages of a dengue virus containing silent point mutations in the capsid gene resulted in only one adaptive change at the 3′ end, increasing the length of complementarity between the CS elements by one nucleotide, thus restoring RNA replication to some extent; and c) adaptive mutations in a WNV RNA lacking the 3′ CS sequence only affected the complementarity between 5′-3′ UAR and DAR elements, and no evidence for additional base-pairing between the dCS and the 3′ UTR was found (Zhang et al., 2010). The fact that replicons with heterologous sequence composition downstream of the CS element display very different replication phenotypes, ranging from WT to severely impaired RNA replication, further strengthens the point that the capsid-coding region does not contain specific cis-acting RNA sequences but rather modulates RNA replication by allowing the formation of a topologically functional structure of the 5′ RNA elements. Further work is needed to determine whether mutations in the dCS sequence affect the topology of the 5′ RNA elements by a direct RNA-RNA interaction or by modifying interactions with host or viral proteins.
Our findings contribute to the understanding of viral RNA replication and provide new insights for the design of sub-genomic and genomic mosquito-borne flavivirus reporter constructs. Recent studies reported the use of genome-length reporter viruses expressing Renilla luciferase, which showed a slower replication kinetic as compared to the WT virus (Samsa et al., 2009; Zou et al., 2011). In both studies, the Renilla luciferase gene was fused to first 104 or 114 nucleotides of the capsid gene, designated as cis-acting element (CAE) as it harbors the DAR and CS sequences and the cHP element. Although the reporter viruses harboring the CAE contain the dCS sequence most crucial for affecting viral RNA replication, replacement of Renilla luciferase by the GFP reporter gene rendered the virus unstable over time in Vero cells (Zou et al., 2011). Interestingly, comparison between reporter replicons expressing either the Renilla luciferase or the Firefly luciferase gene revealed a delayed RNA replication phenotype for the latter (P. Friebe and E. Harris, unpublished data). Together, these studies further demonstrate the importance of the sequence composition downstream the CS element for RNA replication.
We have demonstrated that the sequence composition downstream of the DENV CS element (specifically, nucleotide positions 170-200) can affect RNA replication by modulating the topology of the 5′ end, affecting genome circularization and thereby RNA replication. Our results further indicate that in viral reporter RNAs in which the majority of the dCS sequence is replaced by a heterologous non-viral coding region, the position of the sequence affecting the viral 5′ RNA elements is not limited to nucleotide positions 170-200, as sequences downstream of nucleotide 200 can also affect RNA replication by modulating the topology of the 5′ end. We showed similar results with WNV, and it is likely that these observations are applicable to mosquito-borne flaviviruses in general.
Material and Methods
Cell culture
Baby hamster kidney cells 21 clone 15 (BHK) were grown in minimal essential medium-alpha (MEM-α, Gibco, Carlsbad, CA) with 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% fetal bovine serum (FBS; HyClone, Logan, UT) at 37°C in 5% CO2. Huh7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 10 mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2.
Construction of DNA plasmids
Unless otherwise stated, standard recombinant DNA technologies were used for all cloning procedures. The basic DENV constructs pDRrep, pDEN-5′UTR-Cap-tr and pDEN-5′UTR-Cap-tr and WNV constructs Rluc-Rep, WNV-EMCV-Rep and WNV-SPACER-EMCV-Rep have been described previously (Clyde et al., 2008; Friebe and Harris, 2010; Friebe et al., 2011). To construct biRep-5′UTR-CapFL-WT, the 5′UTR and capsid-coding sequence was amplified by PCR and inserted into pDEN-5′UTR-Cap-tr, using MluI and XbaI. The sequence downstream of the capsid stop codon is identical to the sequence in pDEN-5′UTR-Cap-tr, keeping the multiple stop codons for the various DENV 5′ start codons and a multiple cloning site that contains unique restriction sites for XbaI, PmeI, AgeI, SnaBI and AscI (Friebe and Harris, 2010). For biRep-5′UTR-CapFL-SYN, the capsid-coding region between nucleotide positions 171-438 was replaced by a sequence harboring silent point mutations (a gift from Theodore C. Pierson, NIH). All chimeric capsid-coding region sequences in the biRep-5′UTR-CapFL replicon backbone were generated by PCR and inserted into biRep-5′UTR-CapFL-WT using MluI/XbaI. The 3XFLAG sequence was derived from the C-FLAG construct (Clyde and Harris, 2006) and fused to the indicated capsid-coding region via PCR, then inserted into biRep-5′UTR-CapFL-WT using MluI/XbaI. 5′UTR-Cap-tr-SPACER(Kan) was constructed by inserting a PCR product of ~500 nucleotides derived from the kanamycin resistance gene (using primer pairs S_SnaBI-Kan (5′ CCGGTTACGTAGGAGGAACAAGATGGATTGCACGC 3′) and A_AscI-Kan (5′ GAGATGGCGCGCCCTATTCTATTCTATGCCTTGAGCCTGGCGAAC AGTTC 3′)) into pDEN-5′UTR-Cap-tr using SnaBI/AscI. 5′UTR-Cap-tr-SPACER(Fluc) was constructed by inserting a ~600 nucleotide-long PCR fragment with a sequence derived from the Firefly luciferase coding region (using primer pairs S_F-luc_SPACER (5′ CGGTTACGTACTATCCGCTGGAAGATGGAACCGC 3′) and A_F-luc_SPACER (5′ GAGATGGCGCGCCGTTTATCTATGAGGCAGAGCGACACC 3′)) into pDEN-5′UTR-Cap-tr using SnaBI/AscI. Constructs IC-5′UTR-Cap228-WT and IC-5′UTR-Cap228-SYN were derived by replacing the nucleotides upstream of the Renilla luciferase gene of pD2IC-Renilla (P. Friebe and E. Harris, unpublished) by either the first 228 nucleotides of biRep-5′UTR-CapFL-WT or biRep-5′UTR-CapFL-SYN using PCR techniques. WNV-CapFL-EMCV-Rep or WNV-CapFL-frame-EMCV-Rep were constructed by inserting the WNV capsid-coding region or the WNV capsid-coding region with an additional cytidine at nucleotide position 198 into WNV-EMCV-Rep using BamHI/AscI. Primer sequences are available upon request.
In vitro transcription
DENV replicon DNA templates were generated by ClaI digestion of the corresponding plasmid (for pDRrep, XbaI was used), followed by phenol-chloroform extraction and ethanol precipitation. RNAs were generated by in vitro transcription using the RiboMax Large Scale RNA Production System (T7; Promega) with the following modifications to the manufacturer’s protocol: 3mM each GTP, CTP and UTP; 1mM ATP; and 6mM m7G(5′)ppp(5′)A cap analog (New England Biolabs, Beverly, MA). Samples were incubated for 4 hours (h) at 37°C with the addition of 2mM ATP after 30 minutes (min). Transcription was terminated by the addition of 2-3U of RNase-free DNase (Promega) per μg of plasmid DNA and incubation for 30 min at 37°C, followed by acidic phenol-chloroform extraction and isopropanol precipitation. WNV replicon RNAs were generated similarly as described above, with the exception that DNA templates were digested with XbaI, and in vitro transcription was performed using 3mM each ATP, CTP and UTP; 1mM GTP; and 6mM m7G(5′)ppp(5′)G cap analog (New England Biolabs, Beverly, MA) with the addition of 2mM GTP after 30 min. RNAs corresponding to the 5′ or 3′ viral ends were generated as described above, with the exception that 3mM of each rNTP without m7G(5′)ppp(5′)A cap analog was used. Different DNA templates were derived by PCR based on the corresponding plasmid, using primer pair S_T7-50_MluI (5′CATGCGCACCCGTGGCCAGG3′) and A-Cap-Seq-WT (5′CTCTTCAATATCCCTGCTGTTGGTGGG3′) for templates harboring a WT sequence between nucleotide positions 280-300 or A-Cap-Seq-SYN (5′ CGTTTGAGGATGCCGGCGGTGGGGGG3′) for templates containing the SYN sequence. For 3′SL RNA, templates were generated via PCR using primer pairs S_EcoRI-T7-3′CS-3′SL (5′AAGCTTGATATCGAATTCTAATACGACTCACTATAGACCCCCCCGAAACAAAAAACAGC3′) and P_A-3′end (5′AGAACCTGTTGATTCAACAGCACC3′). All RNAs were quantified spectrophotometrically, and the integrity was verified by electrophoresis on agarose gels.
RNA transfection and transient luciferase replication assays
Transfection of RNAs into BHK or Huh7 cells was performed as described previously (Friebe and Harris, 2010). Briefly, 5-10μg of RNA transcript generated in vitro from DNA templates was mixed with 400μl of a suspension of 107 BHK or Huh7 cells per ml in a cuvette with a gap width of 0.4cm (Bio-Rad). After one pulse at 975μF and 270V with a Gene Pulser System II (Bio-Rad), cells were immediately transferred into 14ml of the corresponding growth medium. Two ml of the cell suspension were seeded per well of a 6-well plate and harvested at various timepoints. Cells were harvested using 500μl 1X Renilla Luciferase Assay Lysis Buffer per well, and luciferase activity was measured using the Renilla Luciferase Assay System (Promega) and a GloMax™ 96 Microplate Luminometer (Promega) according to the manufacturer’s instructions with slight modifications; namely, 50μl cell lysate and 50μl 1X Renilla Luciferase Assay Substrate were used per sample. For immunoblot analysis, cells were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions with slight modifications as described previously (Clyde et al., 2008).
RNA binding assays
Uniformly 32P-labeled RNA probes were in vitro transcribed as described above with minor modifications, using 1mM rCTP and 50μCi α-32P CTP. RNAs were heat-denatured for 5 min at 95°C and slowly cooled to 70°C, followed by incubation on ice. RNAs (0.1nM radioactively-labeled 3′ RNA and indicated amounts of 5′ RNAs) were subsequently incubated in binding reaction buffer (5mM HEPES (pH7.5), 100mM KCl, 5mM MgCl2, 3.8% glycerol and 2.5μg freshly added tRNA) at 37°C for 30 min to allow RNA-RNA complexes to form. RNA-RNA complexes were analyzed by electrophoresis through native 4.5% polyacrylamide gels supplemented with 5% glycerol. Gels were pre-run for 30 min at 4°C at 125V prior to sample loading using 0.5X TBE running buffer. Electrophoresis was carried out for 4-5h at 4°C with constant voltage. Gels were dried at 80°C for 2h and visualized by exposure on a PhosphorImager plate.
Western blot
Cell lysates were prepared using lysis buffer (1% Triton X-100, 20 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol) containing complete EDTA-free protease inhibitor (Roche Applied Science) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 14% acrylamide gel, then transferred to 0.2 μm Protran nitrocellulose (Whatmann). The immunoblots were incubated with anti-FLAG M2 monoclonal antibody (Sigma), followed by incubation with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (Jackson Immunoresearch, West Grove, PA) as described previously (Peña and Harris, 2011). Detection of NS1 was performed using anti-NS1 7E11 monoclonal antibody (Peña and Harris, 2011) followed by incubation with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (Jackson Immunoresearch, West Grove, PA). Actin was detected as described previously (Pena and Harris, 2011). The immunoblots were imaged on a Chemi-Doc EQ system (Bio-Rad).
Supplementary Material
Suppl. Fig. 1. Alignment between the first 438 nucleotides of WT (top) and “SYN” (bottom) sequences. Positions are indicated in 80-nucleotide intervals. Nucleotide differences are underlined and highlighted in bold.
Suppl. Fig. 2. No effect of the SYN mutation on viral RNA translation. (A) No difference in RNA translation efficiency between the indicated DENV reporter RNAs in BHK cells. 5μg of in vitro transcribed replicon RNAs were transfected into BHK cells and cells were harvested 4h p.t. to measure Luciferase activities. Relative Luciferase counts (RLU) are shown. (B) No differences in RNA translation between the IC-5′UTR-Cap228-WT and IC-5′UTR-Cap228-SYN RNA in BHK cells. For more details see (A).
Suppl. Fig. 3. RNA structure prediction using Mfold. (A) The most stable Mfold prediction using the first 300 nucleotides of the DENV 5′ sequence. The 5′ and 3′ ends of the RNA are designated. RNA structures are indicated (SLA, SLB, cHP). The predicted free energy is given at the bottom. (B) The most stable Mfold prediction using the first 300 nucleotides of the 5′UTR-Cap-tr-SPACER(SYN) 5′ sequence. The 5′ and 3′ ends of the RNA as well as SLA and SLB are designated, and the predicted free energy is shown at the bottom. (C) The most stable Mfold prediction using the first 300 nucleotides of the 5′UTR-Cap-tr-SPACER(Kan) 5′ sequence. Effects on RNA structure formation of SLA, SLB and cHP are highlighted by annotating the predicted RNA structures based on their position and nucleotide composition with “SLA”, “SLB” and “cHP”. For more details see (A).
Highlights.
-
➢
Mosquito-borne flavivirus capsid-coding sequence composition influences the ability of the viral genome to circularize and replicate by modulating the topology of the 5′ end.
-
➢
Important new insights are revealed for the design of reporter sub-genomic and genomic mosquito-borne flavivirus constructs
Acknowlegments
We thank Theodore C. Pierson for the “SYN” sequence. This work was supported by NIH grant R01AI52324 (to EH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez DE, Filomatori CV, Gamarnik AV. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375:223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005a;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Filomatori CV, Fucito S, Mondotte JA, Gamarnik AV. Structural and functional analysis of dengue virus RNA. Novartis.Found.Symp. 2006;277:120–132. [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J.Virol. 2005b;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg SM, Rice CM. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J.Virol. 1999;73:8083–8094. doi: 10.1128/jvi.73.10.8083-8094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansarah-Sobrinho C, Nelson S, Jost CA, Whitehead SS, Pierson TC. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology. 2008;381:67–74. doi: 10.1016/j.virol.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M, Brinton MA. West Nile virus (WNV) genome RNAs with up to three adjacent mutations that disrupt long distance 5′-3′ cyclization sequence basepairs are viable. Virology. 2011;412:220–232. doi: 10.1016/j.virol.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WW, Kinney RM, Dreher TW. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J.Virol. 2005;79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Barrera J, Harris E. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology. 2008;379:314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J.Virol. 2006;80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J.Virol. 2003;77:2265–2270. doi: 10.1128/JVI.77.3.2265-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi PY. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology. 2008;381:123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J.Biol.Chem. 2011;286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Harris E. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J. Virol. 2010;84:6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Shi PY, Harris E. The 5′ and 3′ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J. Virol. 2011;85:1900–1905. doi: 10.1128/JVI.02037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori ME, Edmonds J, Dong H, Shi PY, Khromykh AA. RNA structures required for production of subgenomic flavivirus RNA. J.Virol. 2010;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J.Mol.Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Harris E, Holden KL, Edgil D, Polacek C, Clyde K. Molecular biology of flaviviruses. Novartis Found.Symp. 2006;277:23–39. [PubMed] [Google Scholar]
- Holden KL, Harris E. Enhancement of dengue virus translation: role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology. 2004;329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Holden KL, Stein DA, Pierson TC, Ahmed AA, Clyde K, Iversen PL, Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem-loop structure. Virology. 2006;344:439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J.Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler RM, Hoenninger VM, Thurner C, Mandl CW. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J.Virol. 2006;80:4099–4113. doi: 10.1128/JVI.80.8.4099-4113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel HJ, Rice CM, Knipe DM, Howley PM. Flaviviridae: The viruses and their replication, Fields Virology. Lippincott-Raven Publishers; Philadelphia: 2007. pp. 1101–1152. [Google Scholar]
- Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J.Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodeiro MF, Filomatori CV, Gamarnik AV. Structural and functional studies of the promoter element for dengue virus RNA replication. J.Virol. 2009;83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco S, Costa F, Debarges B, Andrieu T, Cahour A. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by cis-acting RNA elements and transacting viral factors. FEBS J. 2008;275:4179–4197. doi: 10.1111/j.1742-4658.2008.06566.x. [DOI] [PubMed] [Google Scholar]
- Manzano M, Reichert ED, Polo S, Falgout B, Kasprzak W, Shapiro BA, Padmanabhan R. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J.Biol.Chem. 2011;286:22521–22534. doi: 10.1074/jbc.M111.234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña J, Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J.Biol.Chem. 2011;286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der AL, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Polacek C, Foley JE, Harris E. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J.Virol. 2009a;83:1161–1166. doi: 10.1128/JVI.01362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek C, Friebe P, Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J.Gen.Virol. 2009b;90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- Proutski V, Gould EA, Holmes EC. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 1997;25:1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu SY, Wu RH, Yang CC, Jao TM, Tsai MH, Wang JC, Lin HM, Chao YS, Yueh A. Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. J.Virol. 2011;85:2927–2941. doi: 10.1128/JVI.01986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J.Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouha H, Hoenninger VM, Thurner C, Mandl CW. Mutational analysis of three predicted 5′-proximal stem-loop structures in the genome of tick-borne encephalitis virus indicates different roles in RNA replication and translation. Virology. 2011;417:79–86. doi: 10.1016/j.virol.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauf S, Mandl CW, Bell-Sakyi L, Skern T. Extension of flavivirus protein C differentially affects early RNA synthesis and growth in mammalian and arthropod host cells. J.Virol. 2009;83:11201–11210. doi: 10.1128/JVI.01025-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Brinton MA, Veal JM, Zhong YY, Wilson WD. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- Shimoike T, Koyama C, Murakami K, Suzuki R, Matsuura Y, Miyamura T, Suzuki T. Down-regulation of the internal ribosome entry site (IRES)-mediated translation of the hepatitis C virus: critical role of binding of the stem-loop IIId domain of IRES and the viral core protein. Virology. 2006;345:434–445. doi: 10.1016/j.virol.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J.Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin A, Evans DJ, Buckley A, Jones IM, Gould EA, Gritsun TS. Replication enhancer elements within the open reading frame of tick-borne encephalitis virus and their evolution within the Flavivirus genus. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr237. doi:10.1093/nar/gkr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng WP, Matthews JD, Frey TK. Analysis of rubella virus capsid protein-mediated enhancement of replicon replication and mutant rescue. J.Virol. 2006;80:3966–3974. doi: 10.1128/JVI.80.8.3966-3974.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J.Virol. 2008;82:11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Qin C, Jiang T, Li X, Zhao H, Liu Z, Deng Y, Liu R, Chen S, Yu M, Qin E. Translational regulation by the 3′ untranslated region of the dengue type 2 virus genome. Am.J.Trop.Med.Hyg. 2009;81:817–824. doi: 10.4269/ajtmh.2009.08-0595. [DOI] [PubMed] [Google Scholar]
- Zhang B, Dong H, Ye H, Tilgner M, Shi PY. Genetic analysis of West Nile virus containing a complete 3′CSI RNA deletion. Virology. 2010;408:138–145. doi: 10.1016/j.virol.2010.09.033. [DOI] [PubMed] [Google Scholar]
- Zhang B, Dong H, Zhou Y, Shi PY. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5′ stem-loop of genomic RNA. J.Virol. 2008;82:7047–7058. doi: 10.1128/JVI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J.Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G, Xu HY, Qing M, Wang QY, Shi PY. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral Res. 2011;91:11–19. doi: 10.1016/j.antiviral.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1. Alignment between the first 438 nucleotides of WT (top) and “SYN” (bottom) sequences. Positions are indicated in 80-nucleotide intervals. Nucleotide differences are underlined and highlighted in bold.
Suppl. Fig. 2. No effect of the SYN mutation on viral RNA translation. (A) No difference in RNA translation efficiency between the indicated DENV reporter RNAs in BHK cells. 5μg of in vitro transcribed replicon RNAs were transfected into BHK cells and cells were harvested 4h p.t. to measure Luciferase activities. Relative Luciferase counts (RLU) are shown. (B) No differences in RNA translation between the IC-5′UTR-Cap228-WT and IC-5′UTR-Cap228-SYN RNA in BHK cells. For more details see (A).
Suppl. Fig. 3. RNA structure prediction using Mfold. (A) The most stable Mfold prediction using the first 300 nucleotides of the DENV 5′ sequence. The 5′ and 3′ ends of the RNA are designated. RNA structures are indicated (SLA, SLB, cHP). The predicted free energy is given at the bottom. (B) The most stable Mfold prediction using the first 300 nucleotides of the 5′UTR-Cap-tr-SPACER(SYN) 5′ sequence. The 5′ and 3′ ends of the RNA as well as SLA and SLB are designated, and the predicted free energy is shown at the bottom. (C) The most stable Mfold prediction using the first 300 nucleotides of the 5′UTR-Cap-tr-SPACER(Kan) 5′ sequence. Effects on RNA structure formation of SLA, SLB and cHP are highlighted by annotating the predicted RNA structures based on their position and nucleotide composition with “SLA”, “SLB” and “cHP”. For more details see (A).