Abstract
Objective
The presence of Six1 mRNA gene portends a poor prognosis in ovarian cancer. We describe validation of a Six1 specific antibody and evaluate its association with tumorigenicity and prognosis in ovarian cancer.
Methods
A Six1 antibody (Six1cTerm) was raised to residues downstream of the Six1 homeodomain, representing its unique C-terminus as compared to other Six family members. Cells were transfected with Six1–Six6 and Western blot was performed to demonstrate Six1 specificity. Ovarian cancer cell lines were analyzed for Six1 mRNA and Six1cTerm and tumorigenicity was evaluated. Ovarian cancer tissue microarrays (OTMA) were analyzed for Six1cTerm by immunohistochemistry and scored by two blinded observers. The metastatic tumors of 15 stage IIIC high grade serous ovarian cancers were analyzed with Six1 mRNA and Six1cTerm and expression was compared to clinical factors and survival.
Results
The Six1cTerm antibody is specific for Six1. Cell line tumorigenicity in SCID mice correlates with Six1 levels both by mRNA(p=0.001, Mann–Whitney U test) and by protein (presence vs. absence, p=0.05 Fischer's Exact test). Six1 protein was present in up to 54% of OTMA specimens. Six1 protein expression in omental/peritoneal metastases correlated with worsened survival in a sample (n=15) of high grade serous stage IIIC ovarian cancers (p=0.001).
Conclusions
The Six1cTerm antibody is specific and able to detect Six1 in cell lines and tumor tissue. Six1 protein detection is common in ovarian cancer and is associated with tumorigenicity and poor prognosis in this group of patient samples. Six1cTerm antibody should be further validated as prognostic tool.
Keywords: Ovarian cancer, Homeoprotein, Homeobox gene, Six1, Prognosis
Introduction
Study of the expression of homeobox genes and their protein products has gained increasing importance in understanding the pathogenesis of human malignancies. Homeodomain-containing proteins act as transcription factors regulating the coordinated expression of a variety of genes involved in development and differentiation [1–10]. Interestingly, these genes are frequently inappropriately expressed in cancer [9–11]. Six homeobox1(Sine Oculis (so) homolog1), also called Six1, belongs to a subfamily of the Six class of homeodomain-containing transcription factors and is an important developmental regulator necessary for the proliferation of precursor cell populations during formation of the muscle, kidney, and inner ear, among other organs [12–14]. Six family genes are thought to have arisen from the multiplication of an ancestral Six gene that occurred prior to the evolution of the Bilateria (animals with bilateral symmetry as opposed to radial symmetry) [15]). The three Drosophila six genes so, optix and Dsix4 have distinct functions, and these genes are further duplicated in the vertebrate lineage, resulting in the orthologs Six1/Six2(so), Six3/Six6 (optix) and Six4/Six5 (Dsix4). The entire Six family of proteins share a highly conserved co-factor interacting Six domain (SD) and a highly conserved DNA binding Six-type homeodomain (HD), but otherwise have unique sequences and a non-conserved C-terminus [16]. Within Six family subclasses, the C-terminus may be important in that it can confer functional specificity [17].
Overexpression of Six1 mRNA is observed in human cancers including breast, ovarian, uterine cervical, rhabdomyosarcoma, and hepatocellular carcinoma [9,18–21]. When Six1 is expressed outside of normal development, it appears to impart developmental properties on adult cells causing increases in proliferation and metastasis and a decrease in basal and TRAIL-mediated apoptosis [10,12,18,19,22]. In ovarian cancer, Six1 mRNA is over-expressed in metastatic cancers as opposed to early stage cancers and postmenopausal normal ovaries and confers a poor prognosis independent of stage [18]. However, mRNA is not very stable and its use as a prognostic marker requires fresh tissue. Analysis of Six1 homeoprotein expression would be preferable since protein is stable and can be easily analyzed in archival tissue, but has been hampered by the lack of a Six1 specific antibody. Due to the significant sequence homology in Six family members, antibodies that are raised to antigens containing the SD and/or HD regions are likely to cross-identify other family members, particularly within the same Six family subclass [23]. We report the development of a Six1 specific antibody raised to the unique C-terminal region of Six1. We show that this antibody can be used for immunohistochemistry and demonstrate its prognostic utility in that detection of Six1 protein correlates with ovarian cancer cell line tumorigenicity and with poor prognosis in advanced stage high grade serous ovarian cancers.
Materials and methods
Antibody production
The C-term anti-Six1 antibody was generated and purified as previously described [24]. 2 mg of protein was sent to Proteintech (Chicago, IL) for antibody production in rabbits.
Cell culture, authentication and transfections
Cell lines were obtained from the gynecologic tumor bank at the University of Colorado Denver (UCD) (references listed in Table 1). The PECOC167 cell line was developed at the UCD from the omental metastasis of a patient with serous ovarian cancer. Cell line authentication and generation of Six family overexpressing cells and Six1 immunohistochemistry are detailed in the Supplemental materials and methods.
Table 1.
Six1 mRNA expression and SCID mouse tumorigenicity in a panel of ovarian cancer cell lines with a range of mRNA expression.
| Cell line | Description/reference | Six1 mRNA (% of SKOV3 reference) | cTerm protein | Tumors in SCID mice |
|---|---|---|---|---|
| OV429 | Serous ovarian cancer [36] | 0.005±0.004 | Negative | No |
| OV420 | Serous ovarian cancer [36] | 0.02±0.02 | Negative | No |
| OVCAR5 | Ovarian cancer ascites [37] | 0.07±0.04 | Negative | No |
| OV432 | Serous ovarian cancer [36] | 0.14±0.13 | Negative | No |
| 2008 | Ovarian cancer [38] | 0.20±0.09 | Negative | No |
| DOV-13 | Ovarian cancer [39] | 0.25±.16 | Positive | No |
| OV1847 | Ovarian cancer | 0.37±.17 | Negative | Yes |
| CaOV3 | Ovarian cancer [40] | 0.42±.14 | Positive | Yes |
| OVCAR2 | Ovarian cancer ascites [37] | 0.60±.13 | Positive | Yes |
| PECOC167 | Serous Ovarian Cancer | 0.63±.06 | Negative | Yes |
| HeyC2 | Serous ovarian cancer [41] | 0.81±.06 | Positive | Yes |
| Hey | Serous ovarian cancer [40] | 1.0±0.16 | Positive | Yes |
| SKOV3 | Grade 2 ovarian cancer [40] | 1.0 (reference) | Positive | Yes |
| A2780 | Ovarian cancer [42] | 10.0±3.0 | Positive | Yes |
Western blot analysis
48 h post-transfection, nuclear extracts were collected from the transfected cells according to Jamieson et al. [25]. Bradford assays were performed to determine the protein concentration, and 15 μg of protein from each extract was run on a 10% SDS-polyacrylamide gel. Western blotting was performed as described [24] using the following primary antibodies: 1:1000 C-term anti-Six1 (antibody described in this manuscript), 1:250 anti-Six1 (Sigma), 1:200 anti-Six4 (Abnova), 1:500 anti-Six5 (Bioworld), and 1:200 anti-Six6 (Affinity BioReagents). Secondary antibodies were 1:10,000 anti-rabbit or anti-mouse IgG horseradish peroxidase (Sigma). Chemiluminescence with SuperSignal West Pico (Thermo Scientific) was used to detect signal.
Immunohistochemistry and ovarian tissue microarrays
Human tissues were obtained by an IRB approved protocol (COMIRB 05-1081) and processed as described in the Supplemental materials and methods with Six1cTerm (1:500). Ovarian tissue microarrays (OTMA) were obtained from the BioChain Institute (OTMA1, Catalog #T8235725-5, lot #A912112 and OTMA2, Catalog# Z7020086, www.biochain.com, Hayward, CA, USA). Slides were stained using Autostainer Universal Staining System (DakoCytomation) along with the Vectastain elite ABC kit (Vector Laboratories, Burlingame, CA). Stain intensity was scored on a scale of 0–3 (0—negative, 1—slight, 2—moderate, 3—intense) and a composite score was calculated by multiplying percent stained nuclei by the intensity score (range 0–300). Two blinded observers (K.B. and M.P.) scored the slides independently and inter-observer reliability (κ statistic) was calculated. Image acquisition data is described in the Supplemental materials and methods.
Quantitative real-time RT-PCR
Total RNA was extracted from patient specimens (COMIRB 05-1081) stored in RNAlater and analyzed using Six1-specific primers and Taqman probes as described in Supplemental materials and methods. Results are measured as fg Six1/ng 18S rRNA sample divided by fg/Six1/ng 18S rRNA of log phase growing SKOV3 cells and reported as percentage of (SKOV3) reference from the same experiment (Relative to SKOV3 Reference, RSR).
Tumorigenicity analysis
10 million log phase growing cells were injected into each flank of triplicate CB-17 SCID mice (Taconic) using an IACUC approved protocol. Mice were evaluated daily and three dimensional tumor measurements were obtained three times weekly using calipers. Each experiment was repeated at least once. Mice were euthanized if they became moribund or lost greater than 15% of starting weight.
Patient tumor data collection
Between January 2003 and June 2008, fresh omental or peritoneal metastases of 15 patients with stage IIIC high grade serous [26] ovarian cancer were obtained under an IRB approved protocol and highquality RNA was extracted as above. All patients underwent an exploratory laparotomy with the intent of performing total abdominal hysterectomy (when the uterus was present), bilateral salpingooophorectomy, omentectomy, and aggressive tumor cytoreduction. All patients were treated with post-operative platinum-based chemotherapy. All pathology was reviewed by the University of Colorado Anschutz Medical Campus gynecologic pathologists and reviewed again by a single pathologist at the institution's weekly Gynecologic Oncology Tumor Board. Archival tumors from the same specimen as the extracted RNA above were obtained and evaluated by H&E and immunohistochemistry on 5–8 μm sections. Clinical variables abstracted included age, stage of disease, tumor histology, pre-operative CA125 levels, extent of residual disease and tumor response to platinum chemotherapy as measured by a recurrence free interval of greater than 6 months. Variables were compared with the t-test, Mann–Whitney U test and χ2 and Fischer's Exact test. Survival was analyzed by the Kaplan–Meier Method with comparisons via the Log Rank test.
Results
Because of the high degree of conservation between Six family members, we verified the specificity of the Six1cTerm antibody for Six1 using Western blot analysis and after transfecting each Six family member into MCF7 cells. Fig. 1 (cropped) shows that the Six1cTerm antibody detects mouse six1 (msix1) while not cross-reacting with any other mouse six family members, and detects human Six1 (hSix1) while not cross-reacting with the most conserved (as compared to Six1) hSix2 protein. Human Six3–Six6 do not contain the C-terminus used to generate this antibody, have sequence homology with mouse six3–six6, and were not analyzed.Supplemental Fig. 1 shows the full blots from which Fig. 1 was generated. Additionally in Supplemental Fig. 1C, a commercially available Six1 antibody, the Sigma anti-Six1 antibody, is shown to detect not only human Six1 (marker 37), but also human Six2, and mouse six1 as well as mouse six2 and mouse six3 (2nd blot from left). A repeat experiment redemonstrated Sigma anti-Six1 binding to both human and mouse six2 (3rd blot from left). In the fourth, fifth and sixth blots, presence of each of the other mouse six family members (six4, six5, six6) in the MCF7 extracts is detected with the specific Six family antibody.
Fig. 1.
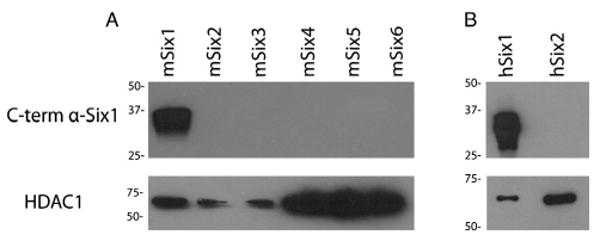
Western blot analysis on nuclear extracts from MCF7 cells transfected with mouse (A) and human (B) Six family members. For the analysis, an anti-Six1 antibody developed against the C-terminus of the protein was used. Images have been cropped for inclusion in this figure. Full-length blots, as well as verification of the presence of all Six family members on the membranes (using specific Six family antibodies and antibodies that cross-react with several Six family members), are included in Supplemental Fig. 1. Bottom panel demonstrates probing of the same membrane with an HDAC antibody as a loading control.
To determine whether the Six1 specific antibody could detect Six1 using immunocytochemical methods, we utilized our previously generated Caov3/Six1 overexpressing clones and our Caov3/CAT control clones [18], which express high and low levels of Six1 respectively. Fig. 2 demonstrates that Six1cTerm antibody detects increased levels of exogenous Six1 protein (nuclear staining), in corroboration with the level of mRNA detected. Transgenic six1 knockout and six1 expressing embryonic mice were used as immunohistochemical positive control (Supplemental Fig. 2).
Fig. 2.
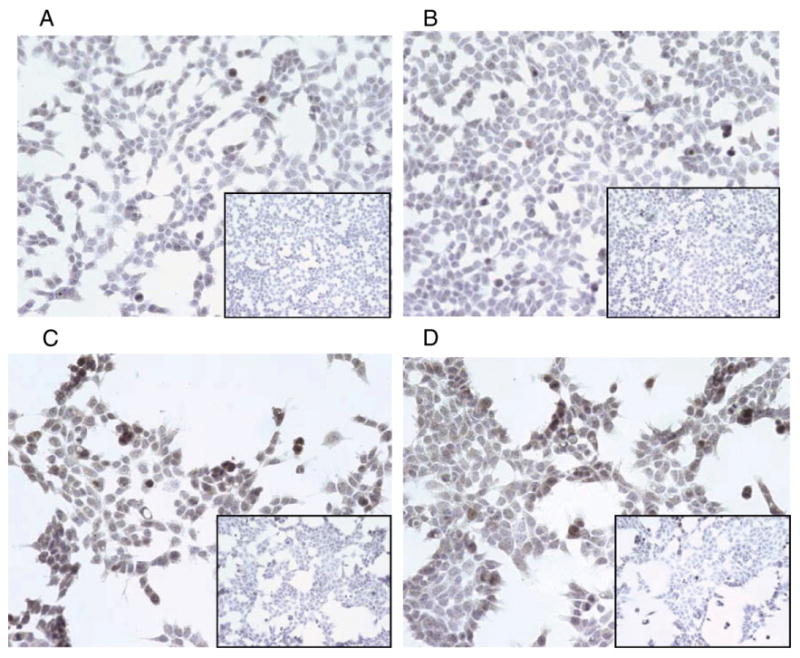
Six1 immunohistochemistry with the cTerm antibody at 1:500 of the CaOV3-CAT clones (panels A and B) and CaOV3-Six1 clones (panels C and D). Magnification is 200×. Exposure was 68 ms for all pictures and none was altered. Insets for each panel are negative controls.
To assess the ability of the Six1cTerm antibody to detect endogenous Six1, ovarian cancer cells lines available from the gynecologic tumor bank at the University of Colorado and with varied levels of Six1 mRNA (Table 1) were selected for Six1cTerm staining. Additionally, given previous data showing that expression of Six1 in nontumorigenic MCF12A breast cancer cells can induce tumor formation [27], cell lines were assayed for tumorigenicity in SCID mice (Table 1). Median cell line Six1 expression was 0.39 RSR (Relative to SKOV3 Reference) with a range of 0–10.0 RSR. All cell lines with Six1≥0.37 RSR were tumorigenic while all cell lines with Six1 expression below 0.37 RSR were not tumorigenic. Six1 mRNA expression was significantly correlated with tumorigenicity (p=0.001 Mann–Whitney U test). Six1cTerm detection (any nuclear staining, composite score>0 with agreement by both evaluators) also correlated with tumorigenicity (p=0.05 Fischer's Exact test). The A2780 cell line displayed intense staining with composite score of 300 (3+, 100%) and Six1 mRNA RSR of 10.0±3.0. The next highest Six1 mRNA RSR level of 1.0 (SKOV3) expressed 2+, 5% of cells nuclear staining (composite score=10) as compared to negative control and the remainder of the Six1cTerm positive cell lines had similar staining. Representative photographs of Six1 staining for the OV429 (Six1cTerm negative), DOV13 (Six1cTerm positive), CaOV3 (Six1cTerm positive), OVCAR2 (Six1cTerm positive), SKOV3 (Six1cTerm positive) and A2780 (cTerm positive) cell lines are shown in Fig. 3.
Fig. 3.
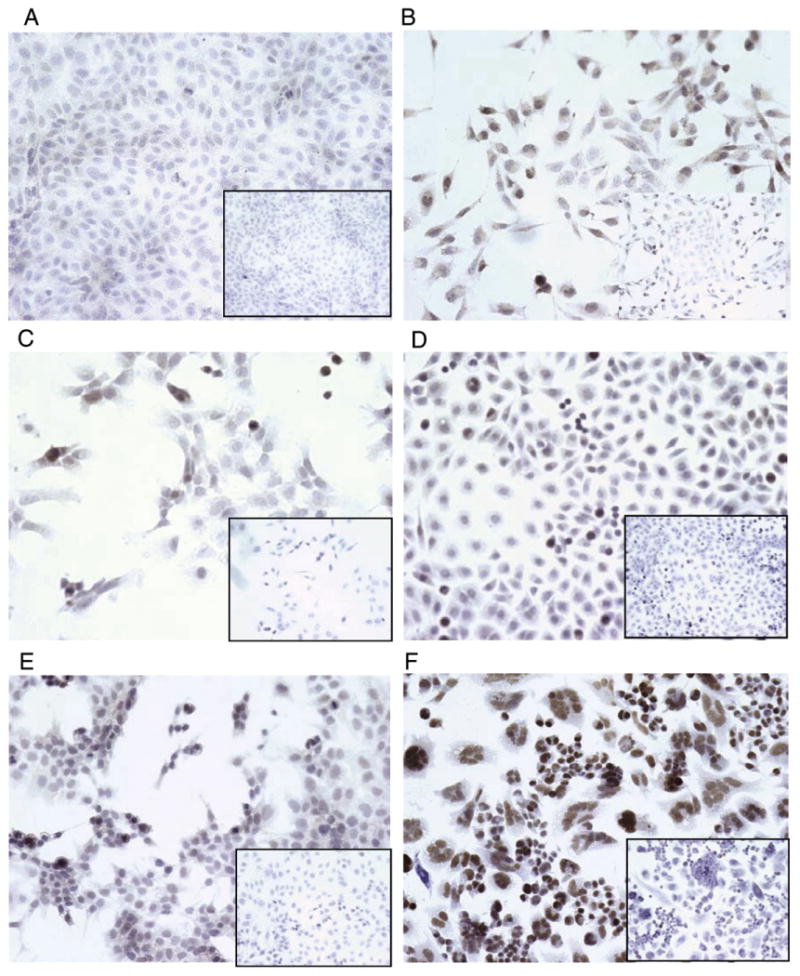
Six1 immunohistochemistry with the cTerm antibody at 1:500 OV429 (Six1 mRNA=0 RSR, panel A), DOV13 (Six1 mRNA 0.25 RSR, panel B), CaOV3 (Six1 mRNA=0.42 RSR, panel C), OVCAR2 (Six1 mRNA 0.6 RSR, panel D), SKOV3 (Six1 mRNA= 1.0 RSR, panel E) and A2780 cell lines (Six1 mRNA=10.0 RSR, panel F). Insets for each panel are negative controls.
OMTA1 and OMTA2 were stained using the Six1cTerm antibody, and scored by two independent blinded reviewers (KB and MDP). Both reviewers had to agree to the presence of staining before it was called positive and interosberver correlation κ was 0.63 (substantial). OMTA1 contains 63 ovarian specimens, 51 from the primary ovarian tumors of patients with stages II–IV epithelial ovarian cancer as well as 4 benign thecomas. Mean age of the patients with epithelial ovarian cancer whose specimens were represented on OTMA1 was 53 years (range 33–81 years) and 12/51 (23.5%) were scored as positive whereas none of the benign thecomas were scored as positive. Representative sections of tumors are included in Supplemental Fig. 3 and the entire OTMA1 as well as scoring by the two blinded observers is available as Supplemental Fig. 4. As contrasted with OTMA1, OTMA2 contains 75 ovarian specimens in duplicate. These include 65 primary epithelial ovarian cancer specimens, stages I–III, as well as 4 benign ovarian tumors. Intra-observer correlation between the duplicates was 0.71 (Spearman) for KB and 0.63 for MDP, whereas inter-observer correlation κ was 0.63. Of 65 epithelial cancers, 35 (54%) were Six1 positive. These included 17/26 (65%) serous cancers, 7/11 (64%) mucinous cancers, 10/25 (40%) endometrioid cancers and 1/3 (33%) classified as adenocarcinoma unspecified. All benign tumors were Six1 negative. Representative sections of tumors are included in Supplemental Fig. 5 and the entire OTMA2 as well as scoring by the two blinded observers is available as Supplemental Fig. 6.
To explore whether Six1 protein levels could be useful as a prognostic indicator, archived tumor of 15 patients with stage IIIC high grade serous ovarian cancer were obtained under an IRB approved protocol, analyzed with Six1cTerm, and compared to qRT-PCR results available from the same patients. Patients were included if they had stage IIIC high grade serous ovarian cancer and had been part of an earlier study to analyze the relationship between Six1 mRNA and prognosis in ovarian cancer, and had consented to have their archived tumor blocks retrieved for the comparison between mRNA and protein. These were not consecutive patients treated at this institution nor a randomized sample of patients, but there was no other selection criteria applied and no patients were excluded. Patients, demographic and tumor data and survival data are presented in Table 2. The mean age of patients enrolled was 60 (range, 48–76). All patients had high grade serous stage IIIC ovarian cancer and elevated (our institution cut-off value) CA125>20mIU/ml (mean 831 mIU/ml, range 25–3907 mIU/ml). Mean tumor Six1 mRNA from omental or peritoneal metastases was 0.69 RSR (range 0–4.2 RSR) and 3/15 specimens had detectable Six1 protein by Six1cTerm (score>0 with agreement by both slide reviewers). Interobserver correlation between the two blinded slide reviewers (K.B., M.D.P.) was substantial (κ=0.76). Age, preoperative CA125, extent of residual disease and months disease free interval (DFI) were not different between patients whose tumors expressed Six1 protein versus those whose tumors did not express Six1 protein. Fig. 4 shows Kaplan–Meier survival curves by Six1 protein and mRNA level. At 34 months of median follow-up, median survival for the patients whose tumors did not have Six1cTerm staining had not been reached as compared to 22 months (95% C.I. 14–31 months) for those whose tumors were positive for Six1cTerm (p=0.001 log rank test). Mean survivals were 59 months (95% C.I. 50–69 months) for those without staining as opposed to 26 months (95% C.I. 13–40 months) for those positive for Six1. mRNA data from the same patient data set revealed a similar but non-significant trend (Fig. 4B).
Table 2.
Patient demographics, tumor data and follow-up data. Only patients with stage IIIC high grade serous tumors were included to provide a uniform data set. Specimens were from omental or peritoneal metastases. RSR = Relative to SKOV3 Reference, DFI = Disease Free Interval, AWD = Alive with disease, DOD = Dead of Disease, NED = no evidence of disease. For patients that are NED, DFI after primary therapy=months follow-up. *Patient with persistent disease, DFI=0 since the patient was never disease free.
| Patient | Age | CA125 mIU/ml | Six1 mRNA level (RSR) | Six1 protein (intensity, percent positive cells = score) | Residual disease after debulking | Months DFI after primary therapy | Months follow-up | Patient status at last follow-up |
|---|---|---|---|---|---|---|---|---|
| 1 | 48 | 408 | 0.05 | 0 | 0 | 17 | 39 | AWD |
| 2 | 48 | Unknown | 0.08 | 0 | 0.5 cm | 23 | 62 | AWD |
| 3 | 76 | 389 | 0.29 | 0 | 0.3 cm | 45 | 52 | DOD |
| 4 | 51 | 692 | 0.56 | 0 | 0 | 28 | 28 | NED |
| 5 | 61 | 834 | 0.47 | 0 | 0.5 cm | 5.4 | 43 | AWD |
| 6 | 66 | 315 | 1.09 | 2+, 50%=100 | 0.3 cm | 19 | 40 | DOD |
| 7 | 51 | 25 | 2.30 | 0 | 0.2 cm | 67 | 67 | NED |
| 8 | 65 | 900 | 0.08 | 0 | 1.5 cm | 45 | 45 | NED |
| 9 | 67 | 3907 | 0.41 | 0 | 1.5 cm | 43 | 61 | AWD |
| 10 | 60 | 770 | 4.20 | 1+, 50%=50 | 1.0 cm | 0* | 17 | DOD |
| 11 | 75 | 300 | 0.60 | 0 | 0 | 26 | 26 | NED |
| 12 | 51 | 68 | 0.04 | 2+, 50%=100 | 0.1 cm | 3.4 | 24 | DOD |
| 13 | 51 | 867 | 0.15 | 0 | 1.0 cm | 5.5 | 32 | DOD |
| 14 | 61 | 170 | 0 | 0 | 6.0 cm | 17 | 17 | NED |
| 15 | 71 | 2000 | 0 | 0 | 1.0 cm | 18 | 18 | NED |
Fig. 4.
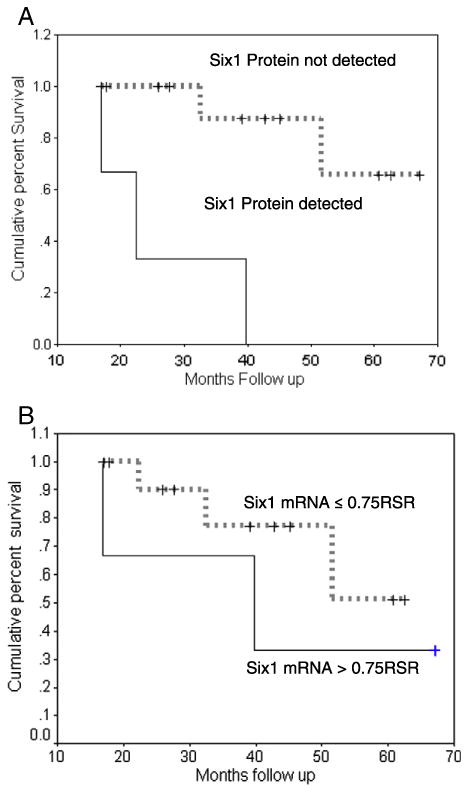
A. Six1 protein expression and the survival of patients with high grade serous Stage IIIC Ovarian Cancer. The survival difference is statistically significant (Log Rank p=0.003). B. Six1 mRNA expression from the same patients predicts a survival trend, but is less powerful than protein analysis.
Discussion
We have developed a Six1 specific C-terminal antibody that can detect Six1 reliably and specifically in archived tissue samples and whose expression can yield prognostic information. Our findings are significant in that prior publications using anti-Six1 antibodies for immunohistochemistry have described the possibility of cross-reactivity to other Six family members when antibodies used were raised to fragments containing the Six SD or HD regions [23]. Expression of other Six family members can be mistaken for Six1 expression, confounding results. For example, Six6 is overexpressed in acute T-Cell leukemias [28] while Six3 is downregulated in gastric cancer, yet overexpressed in extraskeletal myxoid chondrosarcoma [29,30]. Additionally, Six5 is overexpressed in low malignant potential ovarian tumors [31]. As a subset of invasive low grade ovarian tumors share genetic aberrations with their low malignant potential precursors, a non-specific antibody may erroneously identify Six1 expression when one of the other Six family members, such as Six5, is elevated. Furthermore, as seen in the mammary glands of six1-/six1- knockout mice, inducing loss of six1 is associated with increase in other family members such as six2 and six4 [32]. A non-specific Six1 protein marker would not be able to discern the successful knockdown of Six1 in this scenario, picking up Six2 or Six4 staining in error.
Six1 expression is correlated with worse prognosis in cancers as diverse as B cell lymphoma and oligodendroglioma [33]. However, mRNA over-expression is commonly less than 3 fold [23]. In a larger sample of patients with ovarian cancer, Six1 mRNA was over-expressed in metastatic cancers compared to early stage cancers and postmenopausal normal ovaries and conferred a poor prognosis independent of stage [18]. However, the use of mRNA as a prognostic marker is limited by the need for fresh tissue for analysis, the inability to accurately separate tumor and normal tissue, and by the need for an extremely sensitive PCR test that can accurately detect small changes in mRNA expression. Thus, a Six1 specific antibody that can be utilized for immunohistochemical analysis of paraffin embedded samples allows the use of readily available archival tissue and facilitates visualization of tumor versus normal tissue staining. While in a larger sample of tumors from ovarian cancer patients, we have previously shown that Six1 mRNA is prognostically important, here we have shown prognostic importance of Six1cTerm. While our study was not designed for a direct prognostic comparison of Six1 mRNA versus protein, it does suggest that protein analysis by Six1cTerm is possible and prognostic.
We have characterized the Six1 cTerm antibody and have demonstrated Six1 specificity. The antibody was generated against the c-terminus of Six1 in an attempt to generate a Six1 specific antibody by excluding the highly similar SD and HD regions shared by the other Six family members. This was critical for generating an antibody that would not cross-react with Six1's subfamily member Six2, which shares 93% and 98% amino acid identity with Six1 in the SD and HD respectively. Six1 cTerm antibody was able to detect human/mouse Six1 by Western Blot analysis while not cross reacting with human/mouse Six2. We demonstrated the utility of Six1cTerm antibody by the Six1 staining of ovarian cancer cell lines with varied levels of Six1 mRNA and in ovarian tissue microarrays. Displaying an expected Six1 phenotype, both Six1 mRNA and Six1cTerm staining were associated with tumorigenicity in ovarian cancer cell lines and poor prognosis in a preliminary sample of ovarian cancer patients.
In further development, the Six1cTerm antibody could be used to give pre-operative or pre-chemotherapy prognostic information via a small volume biopsy or assessment of ascites commonly seen with ovarian cancer. Additionally, given the small cell number requirements of immunohistochemistry as compared to mRNA analysis, circulating tumor cells, which are known to be present in the blood of ovarian cancer patients [34], could be queried with Six1cTerm antibody to yield prognostic information. It is important to note that we have not directly compared the prognostic utility of various Six1 detecting antibodies, whether Six1 specific or Six1 non-specific, and only present prognostic information in ovarian cancers as assayed with Six1cTerm.
We have previously shown poor survival with Six1 expression in a series of advanced ovarian cancer patients with varied histologic subtypes [18]. Given information about significant variances in ovarian cancer molecular subtypes [35], we have selected the most common and deadly variety, high grade serous ovarian cancer, in the common substage, IIIC, for this analysis. We show that Six1 protein expression is associated with decreased survival, even in as few as a 15 patient cohort. However, we recognize that the small sample size (only 3 of 15 stage IIIC cancers were Six1 positive) while statistically significant, allows for possible errors. The Six1 cTerm polyclonal antibody could therefore be a useful tool as a prognostic indicator in high grade serous ovarian cancers, however our conclusions should be validated in a larger sample of ovarian cancers.
Supplementary Material
Footnotes
Financial support: The work described in this manuscript was supported by a Department of Defense Ovarian Cancer Idea Award # OC-06143 (K.B.), by a Pediatric Hematology/Oncology Postdoctoral Fellowship 2T32082086-11A1 (A.N.P.) and by an NIH R01 CA124545 (A.T., K.B. and H.L.F. triple P.I. grant).
Supplementary materials related to this article can be found online at doi:10.1016/j.ygyno.2012.02.007.
Conflict of interest statement: All of the authors have signed a conflict of interest declaration form. None of the authors has indicated a conflict of interest.
References
- 1.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 2.Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, et al. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–43. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai J, Montell D. Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development. 2002;129:5377–88. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- 4.Fabrizio JJ, Boyle M, DiNardo S. A somatic role for eyes absent (eya) and sine oculis (so) in Drosophila spermatocyte development. Dev Biol. 2003;258:117–28. doi: 10.1016/s0012-1606(03)00127-1. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–3. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- 6.Xu PX, Cheng J, Epstein JA, Maas RL. Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc Natl Acad Sci U S A. 1997;94:11974–9. doi: 10.1073/pnas.94.22.11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–7. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 8.Kurusu M, Nagao T, Walldorf U, Flister S, Gehring WJ, Furukubo-Tokunaga K. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc Natl Acad Sci U S A. 2000;97:2140–4. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford HL, Kabingu EN, Bump EA, Mutter GL, Pardee AB. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc Natl Acad Sci U S A. 1998;95:12608–13. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101:6478–83. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–85. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–54. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 13.Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozaki H, Nakamura K, Funahashi J, Ikeda K, Yamada G, Tokano H, et al. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–62. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- 15.Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009;66:565–83. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes—structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–26. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 17.Weasner BP, Kumar JP. The non-conserved C-terminal segments of Sine Oculis Homeobox (SIX) proteins confer functional specificity. Genesis. 2009;47:514–23. doi: 10.1002/dvg.20517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behbakht K, Qamar L, Aldridge CS, Coletta RD, Davidson SA, Thorburn A, et al. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67:3036–42. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 19.Yu Y, Khan J, Khanna C, Helman L, Meltzer PS, Merlino G. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med. 2004;10:175–81. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 20.Ng KT, Man K, Sun CK, Lee TK, Poon RT, Lo CM, et al. Clinicopathological significance of homeoprotein Six1 in hepatocellular carcinoma. Br J Cancer. 2006;95:1050–5. doi: 10.1038/sj.bjc.6603399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan F, Miao X, Quraishi I, Kennedy V, Creek KE, Pirisi L. Gene expression changes during HPV-mediated carcinogenesis: a comparison between an in vitro cell model and cervical cancer. Int J Cancer. 2008;123:32–40. doi: 10.1002/ijc.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reichenberger KJ, Coletta RD, Schulte AP, Varella-Garcia M, Ford HL. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res. 2005;65:2668–75. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 23.McCoy EL, Iwanaga R, Jedlicka P, Abbey NS, Chodosh LA, Heichman KA, et al. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J Clin Invest. 2009;119:2663–77. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford HL, Landesman-Bollag E, Dacwag CS, Stukenberg PT, Pardee AB, Seldin DC. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J Biol Chem. 2000;275:22245–54. doi: 10.1074/jbc.M002446200. [DOI] [PubMed] [Google Scholar]
- 25.Jamieson C, McCaffrey PG, Rao A, Sen R. Physiologic activation of T cells via the T cell receptor induces NF-kappa B. J Immunol. 1991;147:416–20. [PubMed] [Google Scholar]
- 26.Bodurka DC, Deavers MT, Tian C, Sun CC, Malpica A, Coleman RL, et al. Reclassification of serous ovarian carcinoma by a 2-tier system: A Gynecologic Oncology Group Study. Cancer. 2011 Nov 9; doi: 10.1002/cncr.26618. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coletta RD, Christensen KL, Micalizzi DS, Jedlicka P, Varella-Garcia M, Ford HL. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68:2204–13. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- 28.Soulier J, Clappier E, Cayuela JM, Regnault A, Garcia-Peydro M, Dombret H, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106:274–86. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 29.Rajkumar T, Vijayalakshmi N, Gopal G, Sabitha K, Shirley S, Raja UM, et al. Identification and validation of genes involved in gastric tumorigenesis. Cancer Cell Int. 2010;10:45. doi: 10.1186/1475-2867-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laflamme C, Filion C, Bridge JA, Ladanyi M, Goldring MB, Labelle Y. The homeotic protein Six3 is a coactivator of the nuclear receptor NOR-1 and a corepressor of the fusion protein EWS/NOR-1 in human extraskeletal myxoid chondrosarcomas. Cancer Res. 2003;63:449–54. [PubMed] [Google Scholar]
- 31.Winchester C, Robertson S, MacLeod T, Johnson K, Thomas M. Expression of a homeobox gene (SIX5) in borderline ovarian tumours. J Clin Pathol. 2000;53:212–7. doi: 10.1136/jcp.53.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coletta RD, McCoy EL, Burns V, Kawakami K, McManaman JL, Wysolmerski JJ, et al. Characterization of the Six1 homeobox gene in normal mammary gland morphogenesis. BMC Dev Biol. 2010;10:4. doi: 10.1186/1471-213X-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Baron AE, Harrell JC, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–90. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He W, Kularatne SA, Kalli KR, Prendergast FG, Amato RJ, Klee GG, et al. Quantitation of circulating tumor cells in blood samples from ovarian and prostate cancer patients using tumor-specific fluorescent ligands. Int J Cancer. 2008;123:1968–73. doi: 10.1002/ijc.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Raposo C, Mendiola M, Barriuso J, Hardisson D, Redondo A. Molecular characterization of ovarian cancer by gene-expression profiling. Gynecol Oncol. 2010;118:88–92. doi: 10.1016/j.ygyno.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Bast RC, Jr, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68:1331–7. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton TC, Young RC, Ozols RF. Experimental model systems of ovarian cancer: applications to the design and evaluation of new treatment approaches. Semin Oncol. 1984;11:285–98. [PubMed] [Google Scholar]
- 38.Orth K, Hung J, Gazdar A, Bowcock A, Mathis JM, Sambrook J. Genetic instability in human ovarian cancer cell lines. Proc Natl Acad Sci U S A. 1994;91:9495–9. doi: 10.1073/pnas.91.20.9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauh-Adelmann C, Lau KM, Sabeti N, Long JP, Mok SC, Ho SM. Altered expression of BRCA1, BRCA2, and a newly identified BRCA2 exon 12 deletion variant in malignant human ovarian, prostate, and breast cancer cell lines. Mol Carcinog. 2000;28:236–46. doi: 10.1002/1098-2744(200008)28:4<236::aid-mc6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 40.Buick RN, Pullano R, Trent JM. Comparative properties of five human ovarian adenocarcinoma cell lines. Cancer Res. 1985;45:3668–76. [PubMed] [Google Scholar]
- 41.Ahluwalia A, Hurteau JA, Bigsby RM, Nephew KP. DNA methylation in ovarian cancer. II. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol Oncol. 2001;82:299–304. doi: 10.1006/gyno.2001.6284. [DOI] [PubMed] [Google Scholar]
- 42.Perez RP, Hamilton TC, Ozols RF. Resistance to alkylating agents and cisplatin: insights from ovarian carcinoma model systems. Pharmacol Ther. 1990;48:19–27. doi: 10.1016/0163-7258(90)90015-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


