Abstract
Sorghum (Sorghum bicolor) is one of the world's most important cereal crops. S. propinquum is a perennial wild relative of S. bicolor with well-developed rhizomes. Functional genomics analysis of S. propinquum, especially with respect to molecular mechanisms related to rhizome growth and development, can contribute to the development of more sustainable grain, forage, and bioenergy cropping systems. In this study, we used a whole rice genome oligonucleotide microarray to obtain tissue-specific gene expression profiles of S. propinquum with special emphasis on rhizome development. A total of 548 tissue-enriched genes were detected, including 31 and 114 unique genes that were expressed predominantly in the rhizome tips (RT) and internodes (RI), respectively. Further GO analysis indicated that the functions of these tissue-enriched genes corresponded to their characteristic biological processes. A few distinct cis-elements, including ABA-responsive RY repeat CATGCA, sugar-repressive TTATCC, and GA-responsive TAACAA, were found to be prevalent in RT-enriched genes, implying an important role in rhizome growth and development. Comprehensive comparative analysis of these rhizome-enriched genes and rhizome-specific genes previously identified in Oryza longistaminata and S. propinquum indicated that phytohormones, including ABA, GA, and SA, are key regulators of gene expression during rhizome development. Co-localization of rhizome-enriched genes with rhizome-related QTLs in rice and sorghum generated functional candidates for future cloning of genes associated with rhizome growth and development.
Introduction
Sorghum [Sorghum bicolor (L.) Moench], which is widely grown throughout arid and semi-arid tropical regions, is the world's fifth most important cereal crop [1]. Because it is more resistant to drought, extreme temperature, and nutrient deficiency than maize, soybeans, wheat, and other crops, phenomena such as C4 photosynthesis, drought tolerance, signaling compound response, and aphid and high salinity resistance have been extensively investigated in sorghum [2], [3], [4], [5], [6], [7]. In addition, the use of sorghum in biofuel production promises to further increase the economic impact of this species.
Sorghum bicolor is the most economically important of the approximately 30 species in the genus. Sorghum bicolor is cultivated for grain and forage, while a wild relative native from Asia [8], S. propinquum (Kunth) Hitchc., is cultivated only for forage. Sorghum propinquum is a perennial with small seeds, high levels of tillering, narrow leaves, and well-developed rhizomes [9]. Rhizomes, which are underground stems, are associated with both perenniality and biomass partitioning; in Sorghum, their growth and development is controlled by multiple genes, as revealed by genetic analysis using a S. bicolor × S. propinquum mapping population [10].
Further elucidation of the genetic control of rhizome growth and development may contribute to the development of more sustainable grain, forage, and bioenergy cropping systems. A few efforts have been made to characterize the regulatory mechanisms of rhizome development. For example, using cDNA macroarray analysis, a number of genes rhizome-enriched in S. propinquum were implicated in secondary and hormone metabolism, abiotic stimuli, and development [11]. In another study, comparative analysis of coding regions and regulatory sequences for 54 rhizome-enriched genes in S. propinquum and S. bicolor indicated that several important cis-elements were more abundant in S. propinquum promoters than in those of non-rhizomatous S. bicolor or Oryza sativa [12]. Using O. longistaminata and Phyllostachys praecox, many efforts have been made to elucidate the genes and molecular mechanisms underlying the rhizomatous trait [13], [14], [15], [16], [17], [18], [19], [20]. Hu et al. [16] reported that the rhizome phenotype in O. longistaminata is controlled by two dominant-complementary genes, Rhz2 and Rhz3, and comparative mapping studies indicated that each gene closely corresponds to a major quantitative trait locus (QTL) controlling rhizomatousness in S. propinquum. In addition, a set of rhizome-specific genes were identified by genome-wide differential expression analysis in O. longistaminata, suggesting a complex gene regulatory network underlying rhizome development and growth [18]. These results collectively provide a foundation for cloning genes governing rhizome-related traits.
Because of its high-throughput capability, the microarray platform has been widely used for transcriptome analysis. Previous studies have demonstrated that heterologous microarrays can be used to efficiently profile gene expressions when species-specific microarrays are not available [21], [22], [23], [24], [25], [26], [27]. For such cross-species gene expression studies, there is evidence suggesting that a long oligonucleotide-based (cDNA or 60-mer) microarray platform may be more suitable than a short oligonucleotide-based (25-mer) one [28]. Although the sorghum genome has been published [29], few transcriptome data are available for this species.
It has been estimated that rice diverged from the common ancestor of sorghum and maize approximately 50 million years ago [30], [31]. Sorghum-rice alignments based on the completely-sequenced S. bicolor and O. sativa genomes demonstrate high levels of DNA conservation between the two species. In addition, the number and sizes of sorghum gene families are similar to those of Arabidopsis and rice. It has been observed that 39.9% of rice-sorghum aligned sequences are conserved at the 70%/100 bp level, and 77.5% of the length of sorghum exon sequences overlap with those of rice [29], [32], [33]. Because sorghum and rice are members of the same plant family and, based on sequence similarity, are closely related to each other, we chose to hybridize sorghum RNA to a rice microarray. Oryza sativa (unlike O. longistaminata) is not rhizomatous, but the use of a rice microarray for an S. propinquum rhizome study is still worthwhile. Although some S. propinquum specific genes would be missed, any detected ones would probably represent multiple, evolutionarily conserved genes shared by S. propinquum and O. sativa [28]. Given the close phylogenetic relationship between the two species, the well-annotated rice genome and its known genome history [34], [35] can be exploited when profiling the tissue-specific genome expression of S. propinquum. In this manner, we can discover and characterize genes and putative pathways specifically responsible for rhizome initiation and elongation in sorghum.
Materials and Methods
Plant material and total RNA extraction
For this study, plants of a S. propinquum vegetative clone (unnamed accession) with vigorous rhizomes were cultured in the greenhouse. We collected samples at the vegetative growth stage as described by Hu et al. [18]. Five different tissues—rhizome tips (RT), rhizome internodes (RI), shoot tips (ST), shoot internodes (SI), and young leaves (YL)—were sampled for total RNA extraction. Three independent biological replicates for each sample from individual sorghum plants were collected and snap-frozen in liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen), and then purified and concentrated using an RNeasy MinElute cleanup kit (Qiagen). RNA quality and concentrations were determined using a Bioanalyzer (Agilent).
Microarray hybridization
Because no microarray platform is available for any Sorghum species, an Agilent rice gene expression microarray (product number: G2519F, 44K) was used in this study. The array contained 45,220 independent probes (60-mer) corresponding to 21,495 O. sativa mRNA sequences available in GenBank [34], [35], [36]. RNA amplification, labeling, and hybridization, and microarray imaging were carried out by a specialized biotech company, CapitalBio Corporation (Beijing, China). Equal amounts of RNA samples from five tissues (RT, RI, ST, SI or YL) were pooled and used as a common control. Each experimental sample and control (mix of the five experimental samples) labeled with Cy5-dCTP and Cy3-dCTP separately were produced by Eberwine's linear RNA amplification method and subsequent enzymatic reactions [37], [38]. Array hybridization was performed overnight at 42°C with 8 rpm rotation in a CapitalBio BioMixer II Hybridization Station, followed by two washes. Images were obtained with a confocal LuxScan scanner and then analyzed using LuxScan 3.0 software.
Data analysis
For individual channel data extraction, faint spots with intensities below 400 units after background subtraction in both channels (Cy3 and Cy5) were removed, and a space- and intensity-dependent normalization based on the LOWESS algorithm [39] was then carried out. Significance analysis of microarrays (SAM, version 3.02, [40]) was performed to determine significantly differentially expressed genes. To identify tissue-enriched genes, the microarray data were first subjected to preliminary screening using the multiclass method in SAM with a false discovery rate (FDR) <5% used as a cutoff. The remaining data were then further screened to include only those genes with expression values for a given tissue showing more than 1.5-fold change compared with other tissues (p <0.05) in Wilcoxon rank-sum tests. The whole set of original microarray data has been deposited in NCBI's Gene Expression Omnibus and can be freely accessed through GEO Series number GSE40380.
Functional classification and prediction of cis-acting regulatory elements for the tissue-specific genes
Hierarchical clustering was conducted by Cluster 3.0 and the ratio between experimental sample and control for the fifteen arrays were log transformed. Functional enrichment/overrepresentation analysis was carried out using the agriGO database (http://bioinfo.cau.edu.cn/agriGO/, [41]). GO Slim, representing a reduced version of the GO ontologies containing a subset of the terms in the whole GO were used. For overrepresentation determination, the FDR-adjusted significance level cutoff was set at 0.05. Mapping of rhizome-enriched genes to the 338 currently defined metabolic pathways in the RiceCyc database (http://www.gramene.org/pathway/) was accomplished using the Pathway Tools software package (version 15.5; [42]). Cis-elements of tissue-enriched genes were identified from both strands of upstream 1-kb promoter sequences retrieved from the rice homologous genes with the aid of the PLACE cis-element database (http://www.dna.affrc.go.jp/PLACE/) and a Perl program (‘regulatory’) provided by CapitalBio (CapitalBio Corporation, Beijing). To determine overrepresentation of putative cis-regulatory elements between two groups of genes, two-sample tests of proportion were conducted and the significance threshold was 0.05.
Quantitative RT-PCR and RNA in situ hybridization
Quantitative real-time PCR was performed using an ABI Prism 7900 Sequence Detection System (Applied Biosystems). Diluted cDNA was amplified using gene specific primers and SYBR Green Master Mix (Applied Biosystems); expression levels of tissue-enriched transcripts were normalized with endogenous Actin transcripts. Each set of experiment was performed three times, and the delta-delta Ct (ddCt) relative quantification strategy was used to evaluate quantitative variation. Primers used are listed in Table S1.
In situ hybridization was carried out using the method described by Jackson [43]. Apical portions (1 cm long) of rhizomes were excised and fixed without RNase contamination. Two different templates were constructed by separately cloning Sb01g047010 and Sb06g028820 DNA coding sequences into pBluescript plasmids (Invitrogen). Gene primers used were Sb01g047010F (5′-CCCAGTGTTTTACGTGTATTGG-3′), Sb01g047010R (5′-CTTCATCTTTTTAACCTTGCTT-3′), Sb06g028820F (5′-TGGCAT CGTTGAGCACTGGGTG-3′), and Sb06g028820R (5′-GCCTGGGCAGGTTCATGTCTGG-3′). Following linearization of plasmids, antisense and sense RNA probes were transcribed by T3 and T7 RNA polymerase, respectively, in the presence of dig-UTP (Roche). Each experiment was performed three times using independent samples.
Results
Hybridization of S. propinquum cDNA to the rice microarray
As sufficient genomic data are available for both sorghum (S. bicolor) and rice (O. sativa), we remapped the microarray probes to the sorghum genome and found S. bicolor homologs for all of the rice genes on the microarray. The microarray data could therefore be used to provide information about gene expression in S. propinquum. After removing control probes, the remaining 43,803 independent probes corresponded to 21,495 O. sativa mRNA sequences in Genbank, with their homologs in sorghum then identified using the database at the Sorghum Genome Project (Department of Energy Joint Genome Institute, www.phytozome.net). Finally, 12,842 rice genes were found to be homologous to genes in S. bicolor; 10,147 (79.0%) of these possessed exactly one orthologous gene and 2,695 were in a one-to-many relation (data not shown). Of these, 3,406 independent probes on the rice array were mapped to the S. bicolor genome with 100% sequence identity, resulting in 1,703 O. sativa genes corresponding to 2,199 S. bicolor annotated genes (416 probes mapped to 2 or 3 S. bicolor genes, data not shown).
In this study, tissue from rhizome tips (RT), rhizome internodes (RI), shoot tips (ST), shoot internodes (SI), and young leaves (YL) of S. propinquum was chosen to discover specific sets of genes responsible for unique tissue development and growth. Of the genes on the rice microarray, 4,234 (10.1%) were found to be expressed in at least one tissue in this experiment (3,405 in RT, 3,803 in RI, 3,870 in ST, 3,509 in SI, and 3,452 in YL). Among these expressed genes, 2,760 were homologous to genes in S. bicolor, with 2,182 (79.1%) possessing exactly one orthologous gene and 578 in a one-to-many relation (data not shown). GO analysis demonstrated that genes detected in each tissue were all well-represented in every GO category, except for cellular nitrogen compound metabolic processes. In addition, 2,322 (5.1%) were present in all tissues, and 548 were specifically expressed in only one of the five tissues (Table S2, S3).
Identification of tissue-specific gene expression in S. propinquum
A total of 548 tissue-enriched genes were identified using SAM, including 31, 26, 114, 159, and 218 unique genes specifically enriched in RT, ST, RI, SI, and YL, respectively (Table 1; Table S3, S4, S5, S6, S7). Based on the Sorghum Genome Project, these rice genes on the array corresponded to 23, 14, 76, 106, and 133 annotated sorghum genes. With the exception of RT and ST tissues, which had nearly identical transcriptomic patterns, hierarchical clustering of gene expression profiles revealed that expression was tissue-driven (Figure 1 and Table S2). These results indicate that the identity of specific tissues is derived from their respective transcriptomes.
Table 1. The list of genes enriched specifically in rhizome tips relative to other tissues.
| Oryza GI | FCa | q-value | Best Sorghum BLAST hit | Function Annotation |
| LOC_Os10g38020 | 1.66 | 0 | Sb01g031130 | expressed protein |
| LOC_Os03g07480 | 1.84 | 0 | Sb01g045720 | sucrose transporter |
| LOC_Os03g05480 | 2.54 | 0 | Sb01g047010 | Zinc finger, C2H2 type family protein |
| LOC_Os05g08600 | 1.79 | 0 | Sb02g012970 | pre-mRNA splicing factor PRP38 family protein |
| LOC_Os09g19910 | 1.55 | 0 | Sb02g022990 | expressed protein |
| LOC_Os08g33640 | 1.76 | 0 | Sb02g024480 | expressed protein |
| LOC_Os09g25810 | 1.75 | 0 | Sb02g025200 | Integral membrane protein DUF6 containing protein |
| LOC_Os07g33660 | 1.54 | 0 | Sb02g034910 | expressed protein |
| LOC_Os02g46750 | 1.52 | 0 | Sb04g031140 | expressed protein |
| LOC_Os02g52780 | 1.52 | 0 | Sb04g034190 | bZIP transcription factor family protein |
| LOC_Os11g39000 | 1.59 | 0 | Sb05g023765 | Helix-loop-helix DNA-binding domain containing protein |
| LOC_Os11g10800 | 2.13 | 0 | Sb05g024940 | dirigent-like protein, expressed |
| LOC_Os11g41670 | 1.59 | 0 | Sb06g014780 | expressed protein |
| LOC_Os01g54080 | 1.56 | 0 | Sb06g019450 | kinesin motor domain containing protein |
| LOC_Os04g52830 | 4.88 | 0 | Sb06g028810 | kelch repeat-containing F-box family protein |
| LOC_Os08g42950 | 1.94 | 0 | Sb07g025430 | haloacid dehalogenase-like hydrolase family protein |
| LOC_Os12g42700 | 1.82 | 0 | Sb08g022070 | expressed protein |
| LOC_Os05g02520 | 1.65 | 0 | Sb09g001680 | legumin, putative |
| LOC_Os05g10840 | 1.53 | 0.01 | Sb09g006130 | calmodulin-binding family protein |
| LOC_Os05g12474 | 1.55 | 0.03 | Sb09g006935 | expressed protein |
| LOC_Os05g39840 | 1.58 | 0 | Sb09g023350 | expressed protein |
| LOC_Os03g28960 | 1.75 | 0 | Sb10g006995 | DNA-directed RNA polymerase III 130 kDa polypeptide |
| LOC_Os02g10350 | 1.62 | 0 | Sb10g023955 | Mlo family protein, expressed |
| LOC_Os01g16250 | 1.55 | 0 | unknown | expressed protein |
| LOC_Os09g07640 | 1.59 | 0 | unknown | retrotransposon protein |
| LOC_Os01g01520 | 2.08 | 0 | unknown | Transferase family protein |
| LOC_Os07g26100 | 1.86 | 0 | unknown | expressed protein |
| LOC_Os02g33840 | 1.59 | 0 | unknown | F-box domain containing protein |
| LOC_Os05g41870 | 1.77 | 0 | unknown | glycine-rich cell wall protein |
| LOC_Os12g08760 | 1.55 | 0.07 | unknown | Carboxyvinyl-carboxyphosphonate phosphorylmutase |
| LOC_OS03g05620 | 1.75 | 0 | unknown | inorganic phosphate transporter |
FC:Fold Change, represents the ratio of Avg_RT vs. MAX (Avg_ST, Avg_RI, Avg_SI, and Avg_YL), and q-value (%) ≤5%, while Avg_x represents the average ratio of the three biological replicates while RT for Rhizome tips/control, ST for Shoot tips/control, RI for Rhizome internodes/control, SI for Stem internodes/control and YL for Young leaves/control.
Figure 1. Dendrogram of 548 tissue-specifically expressed genes in five tissues of Sorghum propinquum.
1. Rhizome tips, 2. Shoot tips, 3. Rhizome internodes, 4. Stem internodes, 5. Young leaves. The suffixes a, b, and c indicate the three biological replicates. In the color panels, each horizontal line represents a single gene and the color of the line indicates the expression level (in a log scale) of the gene relative to the median in a specific sample: high expression in red, low expression in green. The raw data represented here are detailed in Table S3.
Of the 218 genes highly enriched in the leaves, most were related to stimulus response, biological regulation, localization, metabolic processes, and cellular processes (Table S4). These included 17 genes encoding transcription factor proteins, such as WLIM1, auxin response factor, ethylene responsive element binding factor 1, and TCP family transcription factor, whose homologous genes were responsible for leaf differentiation in Arabidopsis [44]. In addition, a few genes, such as JMJ706, FMO1, HRB1, FT, and GAMMA_CA2, were found to be enriched in YL. Of these genes, JMJ706, which encodes heterochromatin-associated H3K9 demethylase, is involved in regulation of flower development in rice [45]. Functions for three genes have been identified in Arabidopsis: FMO1, encoding flavin-containing monooxygenase family protein, is critical for the development of systemic acquired resistance (SAR)[46], and HRB1 and GAMMA_CA2 have a modulatory role in the flowering pathway mediated by phyB and in photorespiration [47], [48].
A total of 26 enriched genes in ST were identified in our study (Table S5). These genes were found to be functionally associated with catalytic and binding activity. Among them, the LHC-related gene LIL3:1 (Sb04g002190) plays an essential role in chlorophyll and tocopherol biosynthesis [49]. In addition, KEG (Sb09g019370), a regulator of abscisic acid signaling, and CaS (Sb04g029100), encoding a calcium-sensing receptor protein, are essential for plant growth and development [50], [51].
We detected 159 genes enriched in SI (Table S6); these genes included those involved in phytohormone signaling and metabolism, such as SAUR10, IAA9, the gibberellin receptor GID1L2, an auxin efflux carrier component (Sb03g029320), and two genes coding for AUX/IAA family proteins. Two circadian clock-related genes were also enriched in SI: an LHY-related gene (Sb06g026500) and Sb09g003090, a homolog of the Arabidopsis gene PIF3 that functions in early phytochrome signaling at the dark-to-light transition [52]. In addition, based on near homologs reported in Arabidopsis and rice, a few genes related to plant growth and development, such as WRKY70, PRMT11, APL2, GAPCP-2, PDAT, SAMC1, PTP1, and OsSub31, were also determined to be highly enriched in SI, indicating their important role in shoot growth and development.
There were 113 genes identified as RI-enriched (Table S7). These included five genes functionally related to transport, including ATCHX17, MST6, SPK1, TIM17-2, and ClCa; seven genes related to cell wall biogenesis and the cell cycle, including EXT3, WAK1, IRX10, ECI1, BTF3, VIM1, and PHS1; two genes involved in oxidative damage and disease response (GolS2 and SGT1); eight functioning in plant growth and development, including CYP707As, NAM, GH3.9, OsLOG, NAC1, SEU, PTR2, and SSII; and three genes related to photosynthesis, including DAL1, CYP97A3, and PHOT2. CYP707As encodes a cytochrome P450 family protein and is essential for proper control of seed dormancy and germination [53]. The rice LONELY GUY (LOG) gene is required to maintain meristem activity; its loss of function causes premature termination of the shoot meristem [54]. Previous reports have demonstrated that molecular mechanisms underlying cold temperature regulation of flowering time in Arabidopsis are controlled by NAC1 [55].
A relatively small set of 31 genes were identified as enriched in RT (Table 1). These genes include RELATIVE OF EARLY FLOWERING 6 (REF6, Sb01g047010), AREB1,SUT1 (Sb01g045720), and SUC3 (Sb09g006130). REF6 functions as an FLC (FLOWERING LOCUS C) repressor in the regulation of Arabidopsis flowering [56], [57]. AREB1, a homologous gene of Sb04g034190, is a key positive regulator of ABA signaling in vegetative tissues of Arabidopsis under drought stress [58]. In addition, the two sucrose transporters SUT1 and SUC3 are involved in filling grain, germination, early seedling growth, and phloem loading of sucrose retrieved from the apoplast along the transport pathway [59], [60]. Furthermore, genes encoding kinesin protein, inorganic phosphate transporter, seven transmembrane MLO13, dirigent-like protein, glycine-rich cell wall protein, retrotransposon protein, and SRL1 (splicing factor) conferring biotic and abiotic stress tolerance, were also enriched.
Identification of distinct cis-regulatory elements in tissue-enriched genes
Using the PLACE cis-element database and a Perl program, the cis-elements of tissue-enriched genes were identified on both strands of upstream 1-kb promoter sequences of rice homologous genes. A cis-element comparative analysis was performed on 29, 17, 86, and 117 genes enriched in RT, ST, RI, and SI, respectively, and several distinct elements were found between RT and ST, RI, and SI (Table 2).
Table 2. Identification of distinct cis-regulatory elements in the tissue-enriched genes.
| Cis-element | RT (%) | ST (%) | p valueb | Function |
| No. of tested genes | 29 | 17 | ||
| C[ACGT]GTT[AG] | 65.5 | 35.3 | 0.023 | water stress |
| TGAC[CT] | 62.1 | 88.2 | 0.029 | wounding |
| AACGG | 55.2 | 23.5 | 0.018 | M phase |
| AACCAA | 44.8 | 17.6 | 0.031 | phytochrome |
| CATGCA | 41.4 | 17.6 | 0.049 | ABA |
| TTWTWTTWTTa | 41.4 | 11.8 | 0.018 | scaffold |
| ACGTG | 37.9 | 64.7 | 0.04 | ABRE |
| TTATCC | 37.9 | 11.8 | 0.029 | axillary |
| CATGCA[CT] | 34.5 | 5.9 | 0.014 | storage protein |
| TAACA[AG]A | 31 | 5.9 | 0.023 | amylase |
| TAACAA[AG] | 27.6 | 5.9 | 0.037 | GA |
| RI (%) | SI (%) | |||
| No. of tested genes | 86 | 117 | ||
| [CT]TCA[ACGT]T[CT][CT] | 67.4 | 53 | 0.019 | initiater |
| TTATCC | 26.7 | 16.2 | 0.034 | axillary |
| AGCAGC | 20.9 | 36.8 | 0.008 | anaerobic |
| TGGGC[CT] | 48.8 | 32.5 | 0.009 | cytochrome |
W stands for [AT], ie A or T. bP value represents the significance between RT and ST, or RI and SI.
Nine cis-elements were significantly more abundant in RT-enriched genes compared with ST, whereas two were more predominant in ST. The RT abundant cis-elements included the ABA-responsive RY repeat CATGCA, the sugar-repressive element TTATCC associated with axillary bud outgrowth, and the GA-responsive element TAACAA [AG] [61], [62], [63]. Interestingly, in a previous study [18] the RY repeat CATGCA was also found to be significantly more abundant in RT up-regulated genes than in the other four tissues of S. propinquum. In addition, three cis-elements were significantly more abundant in RI-enriched genes compared with SI: sugar-repressive element TTATCC, initiator element [CT] TCA [ACGT] T [CT] [CT], and Site II element TGGGC [CT] related to cytochrome [64].
Rhizome-enriched genes in QTL regions related to rhizome traits
To identify rhizome-enriched genes corresponding to rhizome-related QTLs previously reported in S. propinquum [10], all identified RT- and RI-enriched genes were mapped onto the sorghum chromosomes. In a similar manner, the rice homologs of these rhizome-enriched genes in S. propinquum were mapped onto rhizome-related QTL regions of rice chromosomes identified by Hu et al. [16].
A total of 74 rhizome-enriched genes were physically mapped onto 11 rhizome-related QTL regions of sorghum. Among these, 9 genes were mapped onto rice rhizome-related QTLs (Table S8). Thirty-one rhizome-enriched genes were mapped onto the LAR QTL interval influencing the number of rhizomes producing above-ground shoots; these genes encode cell wall biogenesis and cell cycle related proteins such as BTF3, BAHD, SPIKE1, sucrose transporter (SUT1), and a protein required for starch breakdown (BAM4) [59], [65]. There were 59 rhizome-enriched genes mapped onto 8 QTL intervals (LSR) affecting subterranean rhizomatousness. Among these, there were three transporter protein encoding genes, including the sucrose transporter SUC3, the K+ transporter CHX17, and the silicon influx transporter Lsi6 identified in rice [66]. Additional genes involved in plant growth and development, including CYP707A1, OPR3, and SSII, as well as PHS1 related to the cell cycle, were also identified to be involved in LSR.
In addition, the rice homologues of 26 rhizome-enriched genes in S. propinquum were uniquely mapped onto 7 rice rhizome-related QTLs including the Rhz3 interval. These genes include NAM-B1 (Os07g37920), encoding a no apical meristem (NAM) protein, which was isolated as a QTL gene accelerating senescence and increasing nutrient remobilization from leaves to developing grains in wheat [67]. Another identified gene was PTR4 (Os07g41330), encoding a peptide transporter, the homolog of which in Arabidopsis is the protein importer AtTIM17-2 [68], [69]. All these genes should provide putative functional candidates for identified rhizome-related QTLs.
Validation of tissue-enriched gene expression by quantitative RT-PCR and in situ hybridization
To confirm the microarray data, 12 tissue-enriched genes were selected for quantitative RT-PCR analysis (Figure 2). This set included 5 RT-, 3 ST-, 1 RI-, 1 SI- and 2 YL-enriched genes. Overall, gene expression profiles of the five tissues detected by the microarray experiments were very similar to those obtained from the qRT-PCR analyses. Correlation coefficients (r) between 0.74 and 0.94 were calculated, thereby indicating the reliability and robustness of the microarray data.
Figure 2. Real time PCR profiles of 12 selected tissue-enriched genes.
RT, ST, RI, SI, and YL represent the rhizome tip, shoot tip, rhizome internodes, shoot internodes, and young leaves, respectively. Expression levels were calculated based on the expression level of YL genes set to 1. Expression profiles obtained by real time PCR for one gene, Sb01g036550, were not consistent with data obtained from microarray analysis. Correlation coefficients (r) for the remaining 11 genes were 0.74, 0.74, 0.74, 0.74, 0.77, 0.74, 0.82, 0.76, 0.76, 0.94, and 0.76, from left to right, respectively. Bars donate standard deviation.
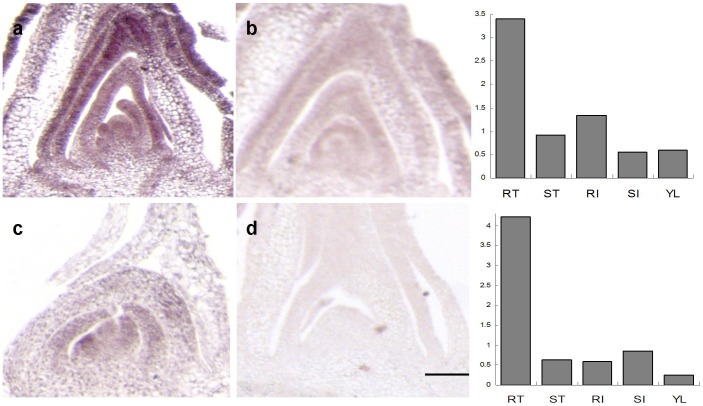
We additionally validated our microarray expression results by in situ hybridization analysis of two S. propinquum RT-enriched genes (Figure 3). The transcript of Sb01g047010, which codes for a C2H2-type family protein, was highly expressed in the RT apical meristem. Another gene (Sb06g028820), encoding a kelch repeat-containing F-box family protein, exhibited slightly lower expression in the RT apical meristem. The in situ expression patterns of these two genes confirm the S. propinquum RT-enriched profiles obtained from the microarray analysis.
Figure 3. Validation of microarray data by in situ hybridization.
In situ localization of transcripts corresponding to the genes (a) Sb01g047010 and (c) Sb06g028820 in S. propinquum rhizome tips are illustrated; (b) and (d) represent the sense probe for control. Corresponding microarray-based expression profiles of these two genes are also shown as bar graphs for comparison.
Comparative analysis of rhizome-enriched genes in O. longistaminata and S. propinquum
We comparatively analyzed the rhizome-enriched genes identified in this study along with genes characterized as rhizome-specifically or differentially expressed genes (DEGs) in previous studies. Genes used for comparison were those found in O. longistaminata using transcriptome sequencing (Li et al., unpublished) and Affymetrix microarray analysis [18], and in S. propinquum based on a cDNA macroarray analysis [11].
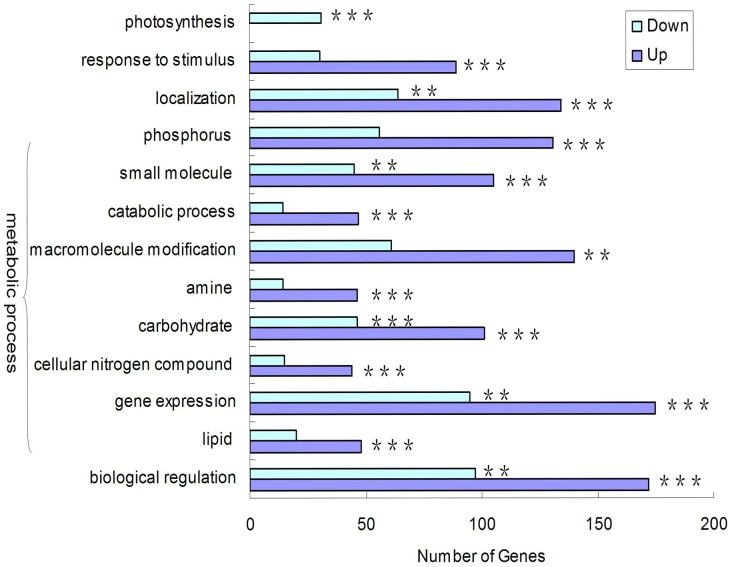
All these DEGs, comprising 1,856 up-regulated and 1,172 down-regulated genes in rhizomes, were first combined for GO analysis (Figure 4). Results indicated that genes related to photosynthesis were significantly abundant only in the down-regulated set, while genes related to stimulus response and metabolic processes, including cellular nitrogen compound metabolic processes and lipid metabolic processes, were significantly represented only in the up-regulated group. GO Slim terms such as localization, metabolic processes (including small molecule metabolic processes and carbohydrate metabolic processes, gene expression), and biological regulation, were significant in both up- and down-regulated genes. In addition to the GO analysis, all rhizome up-regulated genes were mapped to currently defined metabolic pathways in the RiceCyc database (http://www.gramene.org/pathway/). Many different metabolic pathways were broadly represented at every step by rhizome up-regulated genes. In contrast, secondary metabolic pathways and cofactors, prosthetic groups, and electron carrier biosynthesis showed the least representation by RT up-regulated genes (Figure S1). This suggests that major metabolic pathways are involved in RT formation and development.
Figure 4. GO slim categories in up- and down-regulated RT genes combined from this experiment and three other reported studies.
Bars show number of genes with significantly higher relative transcript abundance. All GO slim categories significantly over- or underrepresented are calculated based on a hypergeometric distribution. Significant over- or under-represented categories are indicated by * for p ≤ 0.05, ** for p ≤ 0.01, and *** for p ≤ 0.001.
Hormone-related genes possibly involved in cross-talk during rhizome formation and development were identified using HORMONOMETER analyses (http://genome.weizmann.ac.il/hormonometer/, [70]). Because HORMONOMETER is based upon a list of hormone indexes available for Arabidopsis, we aligned all DEGs to the Arabidopsis genome using Phytozome v7.0 before starting the analysis. A heat map was produced by analyzing the correlation between rhizome up- and down-regulated genes in all studied gene sets and a curated set of ATH1 arrays for different hormone treatments (Figure 5). Significant correlation was found between up-regulated genes and transcriptional response to abscisic acid (ABA), auxin, gibberellic acid (GA), and salicylic acid (SA).
Figure 5. Heat map showing the relationship between rhizome related differentially expressed genes (DEGs) and hormone target genes.
The heat map was produced by analyzing genes comprising rhizome related DEGs for methyl jasmonate (MJ), ethylene (C2H4), abscisic acid (ABA), auxin (AUX), gibberellic acid (GA), zeatin, brassinosteroids (BR), and salicylic acid (SA). Subfigures a–g represent the seven gene sets analyzed: DEGs of RT vs. ST in O. longistaminata from (a) transcriptome sequencing and (c) microarray analysis, and (e) results of the present study in S. propinquum; DEGs of underground tissues (RT and RI) vs. above-ground tissues (ST, SI, and YL) in O. longistaminata from (b) transcriptome sequencing and (d) microarray analysis, and (f) results of the present study in S. propinquum; and (g) candidate rhizome-enriched genes in S. halepense (pSH) and S. propinquum. In the HORMONOMETER analysis, orange (1) = complete correlation, white (0) = no correlation, and blue (-1) = anti-correlation.
Using the comparative genomics tool Phytozome v7.0 (http://www.phytozome.net/), all the rhizome-enriched genes identified in our heterologous microarray study were compared with rhizome-enriched genes or DEGs detected with the other three platforms. There were a number of genes showing the same expression pattern in at least two different platforms (Table 3, S9); 43 of these exhibited rhizome-enriched expression patters (Table 3) and 61 were repressed (Table S9). As shown in Table 3, two genes encoding NAM proteins (Os01g64310 and Os07g37920) were found to be highly rhizome-enriched in the three gene sets. Genes related to transcription regulation and signal transduction, including WRKY TF, eIF6, protein kinase, and HSP, were identified as rhizome-enriched genes in at least two experiments, indicating their important role in rhizome growth and development.
Table 3. The list of up regulated genes in the comparison of the rhizome related DEGs.
| Oryza ID | Ea | F | A | B | C | G | Annotation |
| LOC_Os10g38850 | 4 | Up | Adrenodoxin reductase family protein | ||||
| LOC_Os05g06660 | 2.8 | 1.3 | Peptidase S10, serine carboxypeptidase family protein | ||||
| LOC_Os03g27590 | 2.21 | 2.45 | Peptidase S10, serine carboxypeptidase family protein | ||||
| LOC_Os02g44770 | 2.13 | 1.87 | MscS Mechanosensitive ion channel family protein | ||||
| LOC_Os06g25250 | 2.02 | 3.74 | Ribonuclease III domain containing protein | ||||
| LOC_Os02g56250 | 1.86 | 1.53 | Conserved hypothetical protein | ||||
| LOC_Os09g39960 | 1.83 | 1.74 | Dynamin-like protein 4 (ADL4) | ||||
| LOC_Os12g08760 | 1.77 | 1.63 | Isocitrate lyase and phosphorylmutase family protein | ||||
| LOC_Os09g28230 | 1.72 | 3.45 | 1.68 | Esterase/lipase/thioesterase domain containing protein | |||
| LOC_Os03g21710 | 1.7 | 2.61 | 1 | WRKY DNA binding protein | |||
| LOC_Os09g20000 | 1.59 | 1.63 | 1.86 | 1.24 | Heavy metal transport/detoxification protein | ||
| LOC_Os01g17330 | 1.59 | 4.45 | Eukaryotic translation initiation factor 6 (eIF-6) | ||||
| LOC_Os07g48100 | 1.58 | 1.16 | 2.3 | Up | Serine/threonine protein kinase | ||
| LOC_Os02g27760 | 1.57 | Up | 40S ribosomal protein S15a | ||||
| LOC_Os02g43430 | 1.53 | 1.42 | Protein serine/threonine kinase | ||||
| LOC_Os01g73470 | 1.53 | 1.17 | Conserved hypothetical protein | ||||
| LOC_Os07g37920 | 1.53 | 2.75 | 9.9 | No apical meristem (NAM) protein | |||
| LOC_Os04g32920 | 2.56 | 2.67 | Potassium transporter 5 (AtPOT5) | ||||
| LOC_Os06g07630 | 2.3 | Up | 26S protease regulatory subunit 6A | ||||
| LOC_Os02g05330 | 2.19 | Up | Eukaryotic initiation factor 4A (eIF4A) | ||||
| LOC_Os08g03020 | 2.18 | 3.53 | Resistance protein candidate (Fragment) | ||||
| LOC_Os04g39410 | 2.12 | 1.71 | TPR-like domain containing protein | ||||
| LOC_Os11g47830 | 2.09 | 1.28 | RNA-binding region RNP-1 | ||||
| LOC_Os05g01280 | 1.95 | 3.32 | AT.I.24-5 protein (Fragment) | ||||
| LOC_Os11g05190 | 1.9 | 1.79 | Phytosulfokines 2 precursor | ||||
| LOC_Os04g01740 | 1.89 | 5.3 | Heat shock protein 81-1 (HSP81-1) | ||||
| LOC_Os02g12420 | 1.87 | 1.15 | Protein kinase domain containing protein | ||||
| LOC_Os08g37520 | 1.86 | 1.51 | TPR-like domain containing protein | ||||
| LOC_Os12g31850 | 1.85 | 1.7 | Allantoin permease | ||||
| LOC_Os05g02060 | 1.8 | 1.34 | Amino acid selective channel protein | ||||
| LOC_Os01g59920 | 1.78 | 1.18 | Cysteine synthase, chloroplast precursor | ||||
| LOC_Os03g05260 | 1.78 | Up | Ankyrin repeat containing protein | ||||
| LOC_Os02g07680 | 1.69 | 1.24 | 1.41 | Cytochrome P450 family protein | |||
| LOC_Os08g30820 | 1.67 | 1.04 | Conserved hypothetical protein | ||||
| LOC_Os05g45350 | 1.65 | 3.86 | Heat shock protein DnaJ family protein | ||||
| LOC_Os05g43970 | 1.65 | 2.61 | 28 kDa heat- and acid-stable phosphoprotein | ||||
| LOC_Os03g10210 | 1.65 | 1.17 | 4.04 | Homeodomain leucine zipper protein CPHB-7 | |||
| LOC_Os01g64310 | 1.63 | 2.3 | 4.35 | No apical meristem (NAM) protein | |||
| LOC_Os08g36310 | 1.58 | 2.11 | 2.37 | E-class P450, group I family protein | |||
| LOC_Os06g46030 | 1.58 | 2.2 | Ribulose bisphosphate carboxylase | ||||
| LOC_Os04g35540 | 1.51 | 2.74 | 2.52 | Amino acid/polyamine transporter I family protein | |||
| LOC_Os07g14150 | 1.51 | 1.17 | Nitrogen fixation protein |
A∼G represent the expression level of the 7 gene sets including DEGs of rhizome tip verse shoot tip in O. longistaminata from transcriptome sequencing data (A), microarray analysis data (C) and results of the present study in Sorghum propinquum (E), DEGs of underground tissues (rhizome tip and rhizome internode) verse above ground tissues (shoot tip, shoot internode and young leaf) in O. longistaminata from transcriptome analysis (B), microarray analysis (D) and results of the present study in Sorghum propinquum (F), and candidate rhizome-enriched genes in S. Halepense (pSH) and S. propinquum (G).
We further analyzed the cis-elements of these identified rhizome DEGs, and found that the sugar-repressive element TTATCC, related to axillary bud outgrowth and previously characterized as an RT- and RI-abundant cis-element [71], was significantly more abundant in the up-regulated genes (Table 4). A GA-responsive element, TATCCA, was also predominantly enriched in the rhizome up-regulated genes; this is consistent with the results of an earlier study in which the GA-responsive element TAACAA [AG] was found to be abundant in RT [72]. In addition, the CTCTT motif related to nodulin and the cytokinin responsive element TATTAG [73], [74] were abundantly represented in the up-regulated genes. In contrast, two motifs related to root CAACA and [GT] CACG [TA] [75], [76] were more abundant in the down-regulated gene sets.
Table 4. Identification of distinct cis-regulatory elements in the DEGs shown the same expression pattern in at least two different platforms.
| Cis-element | Up (%) | Down (%) | p valuea | Function |
| No. of tested genes | 40 | 53 | ||
| CTCTT | 85 | 67.9 | 0.029 | nodulin |
| TATCCA | 30 | 11.3 | 0.012 | gibberellin |
| TTATCC | 35 | 13.2 | 0.006 | axillary bud outgrowth |
| TATTAG | 27.5 | 13.2 | 0.042 | cytokinin |
| CAACA | 55 | 73.6 | 0.031 | root; leaf; shoot |
| [GT]CACG[TA] | 27.5 | 49.1 | 0.018 | root; hair |
P value represents the significance between up regulated and down regulated genes.
Discussion
Rhizomes are organs of fundamental importance to plant competitiveness and invasiveness, playing the contrasting roles of overwintering and vegetative propagation in perennial grasses such as O. longistaminata and S. propinquum. A thorough understanding of the molecular mechanisms of rhizome initiation and elongation can aid in the development of perennial rhizomatous grain crops and improve agricultural productivity. In this study, we used an Agilent rice gene expression microarray to profile the tissue-specific genome expression of S. propinquum. The goal of our study was the discovery and characterization of genes and putative pathways specifically responsible for rhizome development in sorghum. Utilizing the well-studied rice genome, we identified two distinct sets of genes enriched in RT and RI, and explored their functional annotation, regulatory motifs, and association with QTLs conferring rhizomatousness.
Several reports have demonstrated that the usefulness of heterologous microarrays in the investigation of gene expression in certain species for which unique microarrays had not yet been developed. Wang et al. [17] identified six genes related to the development of the bamboo rhizome bud based on rice cross-species microarray hybridization; Bagnaresi et al. [77] used a tomato microarray to analyse tuber transcriptome in potato. In both caes, the organ-specific gene expression was confirmed by qPCR analysis. Because the heterologous oligonucleotide microarray analysis would certainly miss some S. propinquum specific genes, a comparison of rhizome-enriched genes in O. longistaminata and S. propinquum — derived from our study as well as previous studies—was also carried out to analyze and categorize genes by function, pathway metabolism, hormone response, and regulatory motif. Although the small set of rhizome-specifically and differentially expressed genes detected in this study is incomplete, the data represent an important contribution to the determination of rhizome formation and development in S. propinquum. Detailed examination and comparative analysis of the functions of these genes should provide insights into molecular mechanisms associated with S. propinquum rhizome development and growth.
While gene expression patterns in the two studied distinct rhizome regions (RT and RI) were not very similar, expression levels in RT more closely resembled those in the above-ground plant part ST. In a previous study of S. propinquum, Jang et al. [11] also found that expression patterns in RT more closely resembled those in ST than those in RI. Because RT and ST are actively growing organs and virtually the entire rhizome tip eventually emerges from the ground to become a shoot tip, it is not surprising that RT and ST show similar expression patterns. Our observation that RT and RI have extremely different expression patterns is consistent with the fact that while RT is an actively growing organ, RI is largely a storage organ with new buds springing up from the joint of rhizome internodes only occasionally.
The rhizome tip is the most important tissue for rhizome initiation and development, with rhizome internodes playing a lesser but still important role. A small portion of these tissue-enriched genes without annotation in sorghum might result from incomplete annotation of sorghum genome or sequence diversity between S. propinquum and reference S. bicolor. RT- and RI-enriched genes, both of known (including homologs) and unknown function are therefore important candidates for further study. Unlike RI-enriched genes, the function of most RT-enriched genes, other than an AREB1 homologous gene, an REF6 homologous gene, and two sucrose transporters, is unknown. AREB1 is active in ABA signaling, suggesting that this hormone plays a part in rhizome initiation and development. Most RI-enriched genes function in major metabolic pathways, including transport, cell wall biogenesis, cell cycle, and plant growth and development, indicating the fundamental role RI plays as a storage organ.
Several RT- and RI-enriched genes, however, have no discernible function in rhizome growth and development. These include the RT-enriched gene REF6 functioning in the flowering pathway and three RI-enriched photosynthesis genes that are related to NAC1, another flowering pathway conferring gene. When RT up-regulated genes were compared for pathway analysis, five genes were identified that are involved in photorespiration: Os12g22030, Os11g26860, Os05g35440, Os03g52841, and Os01g65410 (Figure S1). Photosynthesis- and flowering-related genes, which function in the presence of light or in chloroplasts, should not be enriched in underground tissues. Other researchers [11], [31], [78] have speculated that these incongruities are due to ancient duplication of the transcriptome followed by subfunctionalization of expression patterns. However, the gene FLOWERING LOCUS T controls both flowering and storage organ formation in potato [79]. Considering that the rhizome eventually transforms into a shoot, perhaps some genes related to flowering and photosynthesis also function in rhizome initiation and development, possibly during the rhizome-to-shoot transition. This phenomenon requires further study.
Identification of cis-regulatory elements in tissue-enriched genes revealed that the ABA-responsive RY repeat CATGCA and the GA-responsive element TAACAA [AG] were abundant in RT, implying cross-talk of plant hormones during rhizome development. In addition, several other elements were significantly abundant in RT-enriched genes, i.e., sugar-repressive element TTATCC involved in axillary bud outgrowth, Myb core motif AACGG related to the M phase of the cell cycle, T-Box TT[AT]T[AT]TT[AT]TT found in the scaffold attachment region, AACCAA motif required for phytochrome regulation, RY repeat CATGCA [CT] relevant to storage protein, and amylase box TAACA [AG] A related to amylase. The presence of these elements indicates that compared with ST, RT cells are more active during developmental stages such as bud outgrowth and cell division [80], [81], [82], [83], [84]. In addition, the motif C [ACGT] GTT [AG], responsive to water stress, was also abundant in RT, whereas the W box TGAC [CT], involved in wounding, and the ABRE-like sequence ACGTG, related to early response to dehydration, were abundant in ST. This demonstrates that environmental responses of subterranean and aerial plant tissues differ in the types of signaling proteins involved [85], [86], [87]. Unlike the tip tissues, there were only three motifs significantly more abundant in RI than in SI. These were sugar-repressive element TTATCC related to axillary bud outgrowth, initiator element [CT] TCA [ACGT] T [CT] [CT], and Site II element TGGGC [CT] related to cytochrome. These also emphasize the fundamental role of RI as a storage organ.
Several lines of evidence point to plant hormones, such as ABA, GA, auxin, cytokinin, and SA, as key regulators of rhizome gene expression and development. Jang et al. [11] reported that a GA responsive cis-element was RT enriched in relatively highly expressed genes, implicating GA in regulation of many rhizome-specific genes. Hu et al. [18] found that several genes involved in GA biosynthesis were highly enriched in RT as compared to ST, while cis-elements related to auxin, JA, and ABA were abundant in rhizome-enriched genes. In our study, genes active in ABA signaling were RT enriched, and the cis-elements ABA-responsive RY repeat CATGCA and GA-responsive element TAACAA [AG] were significantly abundant in RT-enriched genes. Comparative analysis of rhizome-enriched genes revealed that hormone biosynthesis was the predominant pathway of integrated RT up-regulated genes in S. propinquum and O. longistaminata. According to HORMONOMETER analyses, the rhizome up-regulated genes were significantly correlated with transcriptional responses to ABA, GA, and SA, indicating that phytohormones may play important roles in rhizome initiation and development.
In this study, 74 rhizome-enriched genes co-localized with 11 rhizome-related QTL regions of sorghum, and 26 genes co-localized with 7 rice rhizome-related QTLs. These rhizome-enriched genes identified by our cross-species microarray represent multiple, evolutionarily conserved genes between S. propinquum and O. sativa, and appear to be heavily involved in rhizome development in perennial grasses. In addition, the 104 DEGs showing the same expression pattern on at least two different platforms, especially the 43 up-regulated ones, confirmed expression levels in the same species as well as those of evolutionarily conserved genes in both rice and sorghum.
In conclusion, a whole rice genome oligonucleotide microarray was used to profile gene expression across five tissues of the perennial wild sorghum S. propinquum. Expression patterns of the five tissues were consistent with the different functions of each organ, and RT- and RI-enriched genes revealed clues to molecular mechanisms of rhizome development. Plant hormones, including ABA, GA, and SA, function as key regulators of rhizome gene expression and development. To shed further light on the identities of rhizome-specific genes, rhizome-enriched candidates were identified using QTL co-localization and comparative analysis.
Supporting Information
An overview of rhizome tip up regulated genes in Sorghum propinquum and O. longistaminata mapped to major metabolic pathways in rice (ssp. japonica ).
(PPT)
Primer list for the real-time PCR analysis.
(DOC)
Commonly and uniquely expressed genes in the five tissues.
(DOC)
A complete list of 548 differentially expressed genes in five tissues of Sorghum propinquum .
(DOC)
The list of genes enriched specifically in the young leaf relative to other tissues.
(DOC)
The list of genes enriched specifically in shoot tips relative to other tissues.
(DOC)
The list of genes enriched specifically in shoot internodes relative to other tissues.
(DOC)
The list of genes enriched specifically in rhizome internodes relative to other tissues.
(DOC)
Rhizome-enriched genes on the rhizome-related QTLs regions identified in sorghum and rice.
(DOC)
The list of down regulated genes in the comparison of the rhizome related DEGs.
(DOC)
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant No. 30760094 and U0836605) and the Key Project from MOA (Grant No. 2008ZX001–003). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smith CW, Frederiksen R, editors (2000) Sorghum: origin, history, technology, and production. New York:John Wiley and Sons.824 p. [Google Scholar]
- 2.Doggett H, editor (1988) Sorghum. Ed 2 ed.Ames, IA:Blackwell Publishing. [Google Scholar]
- 3. Zhu-Salzman K, Salzman RA, Ahn JE, H Koiwa (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134: 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Salzman RA, Brady JA, Finlayson SA, Buchanan CD, Summer EJ, et al. (2005) Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol 138: 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pratt LH, Liang C, Shah M, Sun F, Wang HM, et al. (2005) Sorghum expressed sequence tags identify signature genes for drought, pathogenesis, and skotomorphogenesis from a milestone set of 16,801 unique transcripts. Plant Physiol 139: 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchanan CD, Lim S, Salzman RA, Kagiampakis I, Morishige DT, et al. (2005) Sorghum bicolor's transcriptome response to dehydration, high salinity and ABA. Plant Mol Biol 58: 699–720. [DOI] [PubMed] [Google Scholar]
- 7. Park SJ, Huang YH, Ayoubi P (2006) Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta 223: 932–947. [DOI] [PubMed] [Google Scholar]
- 8.ClaytonWD,VorontsovaMS,HarmanKT,Williamson H(2006 onwards). GrassBase - The Online World Grass Flora. Available: http://www.kew.org/data/grasses-db.html.Accessed 2013 Feb 27.
- 9. Chittenden L, Schertz K, Lin Y, Wing R, Paterson A (1994) A detailed RFLP map of Sorghum bicolor x S. propinquum, suitable for high-density mapping, suggests ancestral duplication of sorghum chromosomes or chromosomal segments. Theor Appl Genet 87: 925–933. [DOI] [PubMed] [Google Scholar]
- 10. Paterson AH, Schertz KF, Lin YR, Liu SC, Chang YL (1995) The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proc Natl Acad Sci U S A 92: 6127–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jang CS, Kamps TL, Skinner DN, Schulze SR, Vencill WK, et al. (2006) Functional classification, genomic organization, putatively cis-acting regulatory elements, and relationship to quantitative trait loci, of sorghum genes with rhizome-enriched expression. Plant Physiol 142: 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jang CS, Kamps TL, Tang H, Bowers JE, Lemke C, et al. (2009) Evolutionary fate of rhizome-specific genes in a non-rhizomatous sorghum genotype. Heredity 102: 266–273. [DOI] [PubMed] [Google Scholar]
- 13.Ghesquiere A, editor (1991) Re-examination of genetic control of the reproductive barrier between Oryza longistaminata and O. sativa and relationship with rhizome expression. Philippines: International Rice Research Institute (IRRI).729–730 p .
- 14. Ghesquiere A, Causse M (1992) Linkage study between molecular markers and genes controlling the reproductive barrier in interspecific backcross between O. sativa and O. longistaminata . RGN 9: 28–31. [Google Scholar]
- 15. Maekawa M, Inukai T, Rikiishi K, Matsuura T, Govidaraj K (1998) Inheritance of the rhizomatous traits in hybrid of Oryza longistaminata Chev. et Roehr. and O. sativa L. SABRAO. J Breeding Genet 30: 69–72. [Google Scholar]
- 16. Hu FY, Tao DY, Sacks E, Fu BY, Xu P, et al. (2003) Convergent evolution of perenniality in rice and sorghum. Proc Natl Acad Sci U S A 100: 4050–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang K, Peng H, Lin E, Jin Q, Hua X, et al. (2009) Identification of genes related to the development of bamboo rhizome bud. J Exp Bot 61: 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu FY, Wang D, Zhao XQ, Zhang T, Sun HX, et al. (2011) Identification of rhizome-specific genes by genome-wide differential expression analysis in Oryza longistaminata . BMC Plant Biol 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao X, Zhang T, Huang L, Wu H, Hu F, et al. (2011) Comparative metabolite profiling and hormone analysis of perennial and annual rice. J Plant Biol 55: 73–80. [Google Scholar]
- 20. Zhang T, Li L, Hu F, Zhao X, Fu B, et al. (2011) Analysis of ESTs from a normalized cDNA library of the rhizome tip of Oryza longistaminata . J Plant Biol 55: 33–42. [Google Scholar]
- 21. Horvath DP, Schaffer R, West M, Wisman E (2003) Arabidopsis microarrays identify conserved and differentially expressed genes involved in shoot growth and development from distantly related plant species. Plant J 34: 125–134. [DOI] [PubMed] [Google Scholar]
- 22. Renn SCP, Aubin-Horth N, Hofmann HA (2004) Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray. BMC Genomics 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kassahn KS, Caley MJ, Ward AC, Connolly AR, Stone G, et al. (2007) Heterologous microarray experiments used to identify the early gene response to heat stress in a coral reef fish. Mol Ecol 16: 1749–1763. [DOI] [PubMed] [Google Scholar]
- 24. Schreiber AW, Sutton T, Caldo RA, Kalashyan E, Lovell B, et al. (2009) Comparative transcriptomics in the Triticeae. BMC Genomics 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davey MW, Graham NS, Vanholme B, Swennen R, May ST, et al. (2009) Heterologous oligonucleotide microarrays for transcriptomics in a non-model species; a proof-of-concept study of drought stress in Musa. BMC Genomics 10: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang SS, Valdes-Lopez O, Xu WW, Bucciarelli B, Gronwald JW, et al. (2010) Transcript profiling of common bean (Phaseolus vulgaris L.) using the GeneChip soybean genome array: optimizing analysis by masking biased probes. BMC Plant Biol 10: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park ES, Kim SJ, Kim SW, Yoon SL, Leem SH, et al. (2011) Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis. Proc Natl Acad Sci U S A 108: 17456–17461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker SJ, Wang YL, Grant KA, Chan F, Hellmann GM (2006) Long versus short oligonucleotide microarrays for the study of gene expression in nonhuman primates. J Neurosci Methods 152: 179–189. [DOI] [PubMed] [Google Scholar]
- 29. Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- 30. Kellogg EA (1998) Relationships of cereal crops and other grasses. Proc Natl Acad Sci U S A 95: 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paterson AH (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A 101: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu J, Hu S, Wang J, Wong GK, Li S, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]
- 33. Matsumoto T, Wu JZ, Kanamori H, Katayose Y, Fujisawa M, et al. (2005) The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- 34. Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, et al. (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35: D883–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, et al. (2008) The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res 36: 1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kikuchi S (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379. [DOI] [PubMed] [Google Scholar]
- 37. Guo Y, Guo HY, Zhang L, Xie HY, Zhao X, et al. (2005) Genomic analysis of anti-Hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J Virol 79: 14392–14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, et al. (2006) Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nature Biotechnol 24: 1140–1150. [DOI] [PubMed] [Google Scholar]
- 39. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, et al. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, et al. (2010) Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform 11: 40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson D, editor (1991) In-situ hybridization in plants. Oxford,UK:Oxford University Press.163–174 p .
- 44. Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun QW, Zhou DX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105: 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koch M, Vorwerk S, Masur C, Sharifi-Sirchi G, Olivieri N, et al. (2006) A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J 47: 629–639. [DOI] [PubMed] [Google Scholar]
- 47. Kang XJ, Zhou Y, Sun XD, Ni M (2007) Hypersensitive to Red and Blue 1 and its C-terminal regulatory function control FLOWERING LOCUS T expression. Plant J 52: 937–948. [DOI] [PubMed] [Google Scholar]
- 48. Martin V, Villarreal F, Miras I, Navaza A, Haouz A, et al. (2009) Recombinant plant gamma carbonic anhydrase homotrimers bind inorganic carbon. FEBS Lett 583: 3425–3430. [DOI] [PubMed] [Google Scholar]
- 49. Tanaka R, Rothbart M, Oka S, Takabayashi A, Takahashi K, et al. (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc Natl Acad Sci U S A 107: 16721–16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vainonen JP, Sakuragi Y, Stael S, Tikkanen M, Allahverdiyeva Y, et al. (2008) Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana . FEBS J 275: 1767–1777. [DOI] [PubMed] [Google Scholar]
- 52. Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, et al. (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci U S A 101: 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, et al. (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8 ′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, et al. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655. [DOI] [PubMed] [Google Scholar]
- 55. Yoo SY, Kim Y, Kim SY, Lee JS, Ahn JH (2007) Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS One 2: e642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Noh B, Lee SH, Kim HJ, Yi G, Shin EA, et al. (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu FL, Cui X, Zhang SB, Jenuwein T, Cao XF (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nature Genetics 43: 715–U144. [DOI] [PubMed] [Google Scholar]
- 58. Yamaguchi-Shinozaki K, Fujita Y, Fujita M, Satoh R, Maruyama K, et al. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scofield GN, Hirose T, Aoki N, Furbank RT (2007) Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J Exp Bot 58: 3155–3169. [DOI] [PubMed] [Google Scholar]
- 60. Scofield GN, Aoki N, Hirose T, Takano M, Jenkins CLD, et al. (2007) The role of the sucrose transporter, OsSUT1, in germination and early seedling growth and development of rice plants. J Exp Bot 58: 483–495. [DOI] [PubMed] [Google Scholar]
- 61. Ezcurra I, Ellerstrom M, Wycliffe P, Stalberg K, Rask L (1999) Interaction between composite elements in the napA promoter: both the B-box ABA responsive complex and the RY/G complex are necessary for seed specific expression. Plant Mol Biol 40: 699–709. [DOI] [PubMed] [Google Scholar]
- 62. Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, et al. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Welchen E, Gonzalez D (2005) Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol 39: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, et al. (2008) Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yamaji N, Ma JF (2009) A transporter at the node responsible for intervascular transfer of silicon in rice. Plant Cell 21: 2878–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Murcha MW, Elhafez D, Millar AH, Whelan J (2005) The C-terminal region of TIM17 links the outer and inner mitochondrial membranes in Arabidopsis and is essential for protein import. J Biol Chem 280: 16476–16483. [DOI] [PubMed] [Google Scholar]
- 69. Ouyang J, Cai ZY, Xia KF, Wang YQ, Duan J, et al. (2010) Identification and analysis of eight peptide transporter homologs in rice. Plant Sci 179: 374–382. [Google Scholar]
- 70. Volodarsky D, Leviatan N, Otcheretianski A, Fluhr R (2009) HORMONOMETER: A tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol 150: 1796–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lu C, Ho T, Ho S, Yu S (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14: 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sandal N, Bojsen K, Marcker K (1987) A small family of nodule specific genes from soybean. Nucleic Acids Res 15: 1507–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fusada N, Masuda T, Kuroda H, Shimada H, Ohta H, et al. (2005) Identification of a novel cis-element exhibiting cytokinin-dependent protein binding in vitro in the 5 ′-region of NADPH-protochlorophyllide oxidoreductase gene in cucumber. Plant Mol Biol 59: 631–645. [DOI] [PubMed] [Google Scholar]
- 75. Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim DW, Lee SH, Choi SB, Won SK, Heo YK, et al. (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bagnaresi P, Moschella A, Beretta O, Vitulli F, Ranalli P, et al. (2008) Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening. BMC Genomics 9: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Adams K, Cronn R, Percifield R, Wendel J (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organspecific reciprocal silencing. Proc Natl Acad Sci USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gasser S, Amati B, Cardenas M, Hofmann J (1989) Studies on scaffold attachment sites and their relation to genome function. Intnatl Rev Cyto 119: 57–96. [DOI] [PubMed] [Google Scholar]
- 81. Huang N, Sutliff TD, Litts JC, Rodriguez RL (1990) Classification and characterization of the rice alpha-amylase multigene family. Plant Mol Biol 14: 655–668. [DOI] [PubMed] [Google Scholar]
- 82. Fujiwara T, Beachy RN (1994) Tissue-specific and temporal regulation of a beta-conglycinin gene: roles of the RY repeat and other cis-acting elements. Plant Mol Biol 24: 261–272. [DOI] [PubMed] [Google Scholar]
- 83. Degenhardt J, Tobin EM (1996) A DNA binding activity for one of two closely defined phytochrome regulatory elements in an Lhcb promoter is more abundant in etiolated than in green plants. Plant Cell 8: 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Planchais S, Perennes C, Glab N, Mironov V, Inze D, et al. (2002) Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath;CycB1;1 transcription and identification of putative regulatory proteins. Plant Mol Biol 50: 111–127. [DOI] [PubMed] [Google Scholar]
- 85. Solano R, Nieto C, Avila J, Canas L, Diaz I, et al. (1995) Dual DNA-binding specificity of a petal epidermis-specific myb transcription factor (Myb.Ph 3) from Petunia hybrida . EMBO J 14: 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nishiuchi T, Shinshi H, Suzuki K (2004) Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves - possible involvement of NtWRKYs and autorepression. J Biol Chem 279: 55355–55361. [DOI] [PubMed] [Google Scholar]
- 87. Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, et al. (2006) Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60: 51–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An overview of rhizome tip up regulated genes in Sorghum propinquum and O. longistaminata mapped to major metabolic pathways in rice (ssp. japonica ).
(PPT)
Primer list for the real-time PCR analysis.
(DOC)
Commonly and uniquely expressed genes in the five tissues.
(DOC)
A complete list of 548 differentially expressed genes in five tissues of Sorghum propinquum .
(DOC)
The list of genes enriched specifically in the young leaf relative to other tissues.
(DOC)
The list of genes enriched specifically in shoot tips relative to other tissues.
(DOC)
The list of genes enriched specifically in shoot internodes relative to other tissues.
(DOC)
The list of genes enriched specifically in rhizome internodes relative to other tissues.
(DOC)
Rhizome-enriched genes on the rhizome-related QTLs regions identified in sorghum and rice.
(DOC)
The list of down regulated genes in the comparison of the rhizome related DEGs.
(DOC)