Abstract
The skin of the nematode C. elegans is composed of a simple epidermal epithelium and overlying cuticle. The skin encloses the animal and plays central roles in body morphology and physiology; its simplicity and accessibility make it a tractable genetic model for several aspects of skin biology. Epidermal precursors are specified by a hierarchy of transcriptional regulators. Epidermal cells form on the dorsal surface of the embryo and differentiate to form the epidermal primordium, which then spreads out in a process of epiboly to enclose internal tissues. Subsequent elongation of the embryo into a vermiform larva is driven by cell shape changes and cell fusions in the epidermis. Most epidermal cells fuse in mid-embryogenesis to form a small number of multinucleate syncytia. During mid-embryogenesis the epidermis also becomes intimately associated with underlying muscles, performing a tendon-like role in transmitting muscle force. Post-embryonic development of the epidermis involves growth by addition of new cells to the syncytia from stem cell-like epidermal seam cells and by an increase in cell size driven by endoreplication of the chromosomes in epidermal nuclei.
Introduction
All animals are encased in skin layers that play critical roles in development and survival 1. The skin and its appendages (collectively the integument) form the outer protective layer of an animal, acting as a permeability and structural barrier. In addition to these well-known ‘barrier’ roles, skin layers have important physiological functions in innate immunity, endocrine and exocrine secretion, mechanosensation, and wound healing. We begin by outlining the basic structure and development of animal skin layers, emphasizing similarities among different animal groups and the main specializations specific to nematodes such as C. elegans (TABLE 1).
TABLE 1.
Comparison of epidermal layers in C. elegans, Drosophila, and mammals
| C. elegans | Drosophila | Mammal | |
|---|---|---|---|
| Epidermal epithelia | Simple epithelium; largely syncytial | Simple cuboidal cellular epithelium | Stratified epithelium |
| Dimensions, in adult | ~0.1 mm2, ~230 nuclei | 106–107 cells? | 2 m2, 1011 cells (human) |
| Embryonic origin | AB, C blastomeres | lateral ectoderm | Surface ectoderm |
| Major morphogenetic processes | ventral enclosure; elongation | Dorsal closure | ? |
| Post-embryonic growth | 4 molts; Seam cells; polyploidization | larval molts; imaginal disks | Basal epidermal stem cells |
| Apical surface matrices | Flexible collagenous cuticle; Epicuticle (lipid); Surface coat | Rigid chitinous cuticle; epicuticle; cuticulin | Stratum corneum |
| Major cytoskeletal elements | cytoplasmic IFs, actin, MTs | Microtubule bundles; actin | Keratins, hemidesmosomes |
| Junctional complexes | hemidesmosomes; CCC, DAC | Adherens, septate junctions | desmosomes, focal adhesions, hemidesmosomes |
| Permeability barrier | cuticle or epicuticle | epicuticle | stratum corneum |
| Fate specification pathways | ELT-1/GATA | Notch; grainy head | BMP signaling; p63; Grhl |
| Innate immune response pathways | TIR-1, p38 MAPK pathway, TGFβ pathway | IMD pathway | EGFR transactivation; |
| Epidermal antimicrobial factors | p38 MAPK pathway; nlp, cnc genes | Cecropin | Defensins, Cathelicidins |
| Wound closure pathways | calcium signals | grainy head, AP1, Rho, PDGFR | Re-epithelialization |
| Sensory functions | embedded touch neurons and sensilla | sensory hairs | mechanoreceptors, thermoreceptors, nociceptors |
| Appendages | lateral alae | trichomes, denticles | hair, nails, teeth, glands |
All epidermal layers are epithelial, with the apical surface of the epithelium facing the environment. In C. elegans, the epidermis (formerly termed the hypodermis; see Note 1) is a simple epithelium with an internal basal surface covered by a basal lamina and an apical surface that secretes a flexible collagenous cuticle. The form and function of the nematode epidermis and cuticle are so interdependent that they have been referred to as the ‘epidermis-cuticle complex’ 2. In contrast, the epidermis of insects such as Drosophila is a simple cuboidal epithelium that secretes a rigid, chitinous cuticle 3. Vertebrate skin layers are typically multilayered epithelia. In fish the stratified epidermis is composed entirely of living cells 4, whereas terrestrial vertebrates are covered by an outer layer of dead keratinized cells, the stratum corneum. In mammalian embryos the surface epidermis first generates a transient outer layer, the periderm. Subsequently the epidermis executes a stratification program, concomitant with development of barrier function, such that neonatal skin consists of four major cell layers 5.
The epidermis and the nervous system are derivatives of the ectoderm; a major early event in metazoan embryogenesis is the separation of epidermal and neuronal precursors in the ectoderm 6, and the subsequent internalization of neurons. C. elegans embryonic epidermal cells form on the dorsal side of the early embryo and enclose the ventral neurons by epiboly (see below). In Drosophila the epidermis forms laterally and encloses the embryo by epiboly at the dorsal midline. The mammalian epidermis begins as a single layer of multipotent cells that subsequently either stratifies or forms an appendage such as hair or nails; neurons develop at the dorsal midline and are internalized by neural tube closure. Growth of the skin is essential for animal growth. In C. elegans and Drosophila, which grow via larval molts, new epidermal cells are added by division of stem cells within the epidermis. The adult Drosophila skin forms from imaginal disks set aside in embryogenesis. After embryonic and postnatal growth, mammalian skin undergoes constant renewal from division of stem cells in the basal layer.
Although some aspects of the C. elegans epidermis and cuticle are clearly specialized aspects of the nematode body plan, others may reflect epidermal characters conserved among all metazoans 7. For example, the C. elegans epidermis contains cytoplasmic intermediate filaments (cIFs) and hemidesmosomes (HDs) that provide mechanical strength, analogous to keratin cIFs and HDs in mammalian skin. Although the molecular composition of the nematode and Drosophila cuticle and the mammalian stratum corneum are very different, all require enzymatic crosslinking of substrate proteins for formation of a mechanically strong permeability barrier. An outer lipid-based waterproof layer (the epicuticle in C. elegans) also seems to be a common feature of skin layers. Findings that conserved families of transcription factors regulate Drosophila and mammalian epidermal development 8 suggest that additional homologies or functional analogies may be found between skin layers. Here we review the developmental biology of the C. elegans epidermis, considered as an integrated organ system. In the following review, the differentiation and physiological roles of the epidermis are discussed.
SPECIFICATION AND MORPHOGENESIS OF THE EMBRYONIC EPIDERMIS
The C. elegans epidermis is a simple epithelium (FIGURE 1), within which several subtypes of epidermal cell can be defined 9. The specification of epidermal cell fates in development is highly invariant with respect to the cell lineage (FIGURE 2) 10. Most epidermal cells, including all post-embryonic epidermal blast cells, derive from the major ectodermal blastomere AB; a few are made by the C blastomere. The epidermis is generated piecemeal from multiple descendants of AB and C; exclusively epidermal precursors do not form until approximately 3 hours post fertilization.
FIGURE 1. Anatomy, topology, and genealogy of the epidermis.
(A) Anatomy of the C. elegans epidermis in L1 stage. Lateral views showing nuclei and cell boundaries, based on Sulston et al. (1983) and WormAtlas. In this and other figures, hyp7 is green, seam cells (H, V, T) are dark blue, ventral epidermal (P) cells are red; hyp4 is pink, hyp5 light blue, and hyp6 teal. (B) Cylindrical projections of the epidermis, separated at ventral midline and unrolled so that right is up and anterior to the left. The dorsal midline is indicated. Projections of the initial embryonic epithelium prior to cell fusion, L1 stage epidermis, and adult. The numbers and approximate disposition of cells are correct; the exact pattern of cell contacts is simplified. For more anatomically accurate projections, see Wormbook hypFIG2.
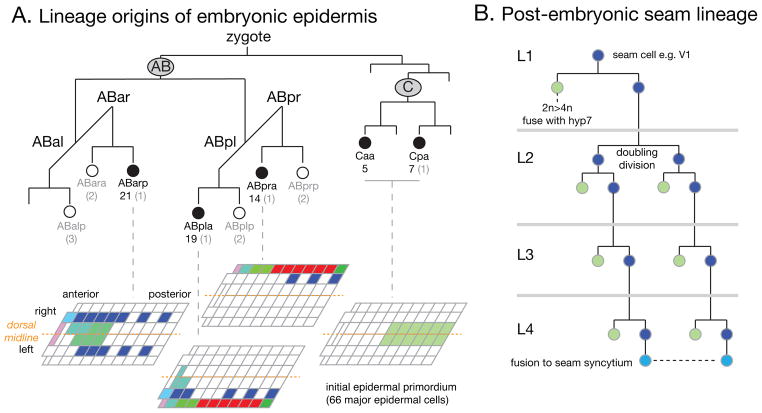
FIGURE 2. Lineage origins of the embryonic and post-embryonic epidermis.
(A) Abbreviated embryonic cell lineage showing the origin of epidermal precursors from the AB and C lineages 10. Cells are named according to standard C. elegans lineage nomenclature: AB and C are early embryonic blastomeres; a/p indicates anterior or posterior daughter and l/r indicates left or right daughter. Epidermal potential is intrinsic to AB and C and segregates at asymmetric divisions at the AB8 and C4 stages (i.e., at the division of ABar to ABara and ABarp, and so on) to five major epidermal precursors (filled circles). Epidermal cells are born in two successive rounds of cell division. 66 major epidermal cells (hyp4–7, seam, and P cells, shown in the projections) are born at ~240 minutes and comprise the initial epidermal primordium that forms dorsally and encloses the embryo. The remaining 12 minor epidermal cells of the head and tail (hyp1–3 and 8–11) are born in the next round of AB divisions (270–300 min) from ‘minor epidermal precursors’ (open circles) and are not shown in the cylindrical projections; 16 other epidermal-like cells (arcade, XXX, rectal epithelial cells, tail spike) are also born in this round of divisions and are not shown here. Numbers under cell names indicate the number of major and minor (gray) cells derived from each precursor. Projections show only the 66 major epidermal cells; note that except for V3 and V5, lineally related cells are adjacent in the epithelium. Color code as in FIGURE 1. (B) Representative lineage of a post-embryonic epidermal seam cell such as V1, illustrating asymmetric divisions in each larval stage and the L2-specific doubling division.
Varieties of epidermal cell types
The C. elegans epidermis is made up of a small number of cells, of several types, each with multiple distinct roles in epidermal development and physiology.
Major postmitotic epidermal cells
Most of the larval and adult epidermis is composed of postmitotic syncytia, named hyp1 through hyp11, which form by the fusion of mononucleate epidermal precursors in mid-embryogenesis (see Box 1 and FIGURES 1,2). The larval C. elegans body is largely covered by a single epidermal cell, hyp7 (dark green in FIGURE 1,2). Additional cells fuse with hyp7 in each larval stage, so that in adults hyp7 contains 139 nuclei, over 80% of all epidermal nuclei. Although the 23 embryonic hyp7 precursors eventually fuse into a single syncytium, they can have distinct fates and roles: for example, fate transformations between hyp7 cells can lead to morphological defects, as in vab-7 mutants 11. The smaller epidermal cells hyp1 to hyp5 and hyp8 to hyp11 form the head and the tail respectively (FIGURE 1A,B); these are generated during embryogenesis and do not acquire additional nuclei by fusion during post-embryonic development.
BOX 1. A model for cell fusion: syncytial nature of the epidermis.
The post-embryonic C. elegans epidermis consists almost entirely of multinucleate syncytia that form by fusion of epidermal precursors 64. These epidermal syncytia range in size from 2 nuclei (e.g. hyp3) to 139 nuclei (adult hyp7). Genetic analysis led to the discovery of mutants defective in cell-cell fusion 150. EFF-1 is a transmembrane fusogen protein necessary and sufficient for epidermal and other cell types to fuse 151. Conversely, inappropriate fusions between epidermal cells are inhibited by the vacuolar ATPase 152. Fusion-defective mutants such as eff-1 are subviable and display aberrant epidermal elongation and subsequent morphogenesis, whereas mutants with excessive fusion are also inviable, suggesting that the precise pattern of syncytial fusion is critical for aspects of epidermal morphogenesis. Regulated cell fusions are also important in morphogenesis of post-embryonic epidermal structures such as the vulva 153.
Seam cells
Two chains of lateral epidermal cells known as ‘seam cells’ run the length of each side of the C. elegans body. The seam cells (H0–H2, V1–V6, and T) are born during embryogenesis (FIGURE 1B, 2A, dark blue) and, with the exception of H0, undergo stem cell-like divisions in post-embryonic development. After the seam cells divide in the L1 stage, their anterior daughters endoreduplicate their DNA then fuse with the surrounding hyp7 syncytium; their posterior daughters remain unfused (FIGURE 2B). In the second larval stage, the posterior daughters of the V cells undergo an additional ‘doubling division’ prior to the asymmetric seam division. An exception to this pattern is V5.p, whose anterior daughter is a neuroblast. In the third and fourth larval stages, seam cells again divide in a stem cell-like pattern as in the L1 stage. During the mid-L4 stage the posterior seam cells terminally differentiate and fuse to form a separate syncytium. Seam cells are responsible for the formation of cuticular specializations, the lateral alae, and play key roles in epidermal elongation and molting.
Ventral epidermal cells
Twelve ventral epidermal cells P1–12, along with adjacent hyp7 cells, form the ‘ventral pocket’ during epidermal enclosure (FIGURE 1B, C, red cells). The ventral epidermis secretes guidance cues such as netrins that play important roles in emnbryonic patterning. The P cells comprise six bilaterally symmetrical pairs (denoted P1/2, P3/4, etc) that are developmentally equivalent until they migrate and intercalate at the ventral midline in the early L1 stage. During the L1 stage each P cell divides to generate an anterior daughter neuroblast (Pn.a cell) and a posterior daughter (Pn.p cell). Pn.p cells either fuse with the hyp7 syncytium or remain unfused until the L3 stage when they take on sexually dimorphic blast cell roles in vulva or male tail sdevelopment (see below) (FIGURE 1C).
Interfacial epidermal cells
The mouth, rectum and hermaphrodite vulva are orifices lined by rings of specialized epidermal cells (FIGURE 1B). The mouth is made by three small epidermal toroids (hyp1–3) and the epidermal-like arcade. The excretory pore forms a smaller opening and is lined by a single cell, the excretory socket. Numerous sensilla are embedded within the epidermis via specialized socket cells that form unicellular toroids around neuronal endings 12. Socket cells are glial-like 13, yet partly resemble epidermal cells in that they secrete cuticle and are attached to surrounding epidermis via adherens and gap junctions 14. Indeed, the seam cell T acts as the phasmid socket cell before it divides and transfers the socket function to its descendants.
Specification of epidermal fates
Epidermal fates are specified by a hierarchy of transcriptional regulators expressed in early embryogenesis (FIGURE 3). Many of these factors remain expressed in the post-embryonic epidermis and thus both initiate and maintain epidermal fates. The GATA factor ELT-1 is necessary and sufficient for specification of most epidermal fates and can be thought of as a master regulator of epidermal ‘organ identity’ 15–17. In the C lineage ELT-1 itself is activated by blastomere-specific factors such as PAL-1 18; less is known about how ELT-1 is activated in the AB lineage.
FIGURE 3. Transcriptional hierarchy regulating epidermal identity and differentiation.
Regulatory hierarchy in embryonic epidermal specification, based on 16, 26 and 29. Lineage-specific regulators activate the ‘epidermal organ identity’ gene ELT-1 in descendants of the AB and C blastomeres. ELT-1 initiates epidermal identity by inhibiting other organ identity modules and by directly activating three epidermal ‘differentiation’ factors: LIN-26, ELT-1, and NHR-25. LIN-26 confers generic epithelial characteristics 23. ELT-3 promotes epidermal-specific aspects of terminal differentiation (e.g. cuticle collagen expression) but is not itself essential for epidermal development. The three differentiation factors appear to indirectly repress one another via negative feedback on ELT-1. NHR-23 and Grainyhead/GRH-1 promote other aspects of epidermal differentiation, but their location in the hierarchy has not been defined. In parallel, lineage-specific regulators specify subtypes of epidermal cell. Dorsal epidermal cells require the T-box genes TBX-8, 9; in C-derived hyp7, PAL-1 likely activates TBX-8/9 directly. One target of TBX-8/9 in C-derived hyp7 is the even-skipped ortholog VAB-7 27. DIE-1 is expressed in dorsal hyp7 in response to unknown signals. Lateral epidermal cells are specified by CEH-16 and ELT-5/6, which in turn repress the differentiation factor ELT-3. Post-embryonic seam cells are maintained in a stem-cell like state by the action of RNT-1/Runx and BRO-1/CBFβ; BRO-1 is directly activated by ELT-1 155. Dorsal and ventral cells (all non-seam epidermis) express ELT-3.
At least three major targets of ELT-1 execute distinct aspects of epidermal differentiation: the GATA factor ELT-3 19, the zinc finger protein LIN-26 20, and the nuclear hormone receptor NHR-25 21. LIN-26 and NHR-25 are widely expressed in the embryonic epidermis and are essential for epidermal morphogenesis, whereas ELT-3 is restricted to non-seam epidermis and is not individually essential. A second nuclear hormone receptor, NHR-23, is also widely expressed in embryonic epidermis and is required for embryonic epidermal morphogenesis and post-embryonic molting 22; NHR-23 and NHR-25 have closely related functions both in embryonic and post-embryonic epidermal development (see below); however their exact regulatory relationships have not yet been clearly elucidated. LIN-26 induces expression of genes involved in epithelial differentiation such as the junctional complex proteins AJM-1 and DLG-1 and the apical trafficking component CHE-14 (see below) 23.
Both the AB and C blastomeres have the autonomous potential to differentiate epidermal tissue 24. In both lineages, epidermal potential is segregated asymmetrically at divisions of the AB grand-daughters and C daughters. Like other embryonic asymmetries the asymmetric distribution of epidermal potential requires POP-1 25. In the C lineage POP-1 asymmetry allows ELT-1 to remain active in anterior daughters, promoting epidermal organ identity. ELT-1 activates the three ‘differentiation’ factors, which together act as an integrated module to promote epidermal specification and repress muscle specification 26. Epidermal specification in the AB lineage is more intricate, but can be viewed as a binary choice between epidermal versus neuronal potential. ELT-1 is not expressed in ‘minor’ epidermal cells, suggesting epidermal identity can be initiated independent of ELT-1.
The subtypes of epidermal cell are specified by the expression of distinct transcriptional regulators. Dorsal epidermal fates are specified by a redundant pair of T-box genes, TBX-8/9 27, which act upstream of VAB-7; in C-derived hyp7, TBX-8/9 are activated by PAL-1 18. The Zinc finger protein DIE-1 also functions to specify aspects of dorsal epidermal fates 28. The seam cell subtype is specified by the engrailed-like homeobox gene CEH-16 29, which activates two partly redundant GATA factors ELT-5 and ELT-6 30. ELT-5 and ELT-6 together repress ELT-3 in seam cells. ELT-1 itself is expressed in post-embryonic seam and is required for seam cell differentiation 31. CEH-16 also promotes the L2-specific proliferative divisions of seam cells (Huang 2009). The RUNX complex RNT-1/BRO-1 acts in parallel to CEH-16 to control seam cell proliferative divisions 32, 33. Factors specifying the ventral epidermal P cell fates have not yet been reported.
To what extent are these mechanisms of epidermal fate specification conserved? Many bilaterian phyla use ELT-1-like GATA factors to specify ectodermal fates 34. Likewise, NHR-25 is related to the Ftz-F1 family of orphan nuclear receptors, implicated in epidermal cuticle formation in Drosophila 35. Grainy head family transcription factors play conserved roles in epidermal development and repair in mammals and Drosophila 8, 36, 37. The C. elegans grainy head ortholog grh-1 is required for late aspects of epidermal differentiation 38 but its position in the epidermal transcriptional hierarchy remains to be determined. Conversely, LIN-26 is a divergent C2H2 Zinc finger protein with relatives identified only in other nematodes.
Morphogenesis of the embryonic epidermis
The major epidermal cells are generated on the dorsal surface of the gastrulation stage embryo and form an epithelial sheet, here referred to as the epidermal primordium (FIGURE 2,4A). Epidermal morphogenesis includes all the processes that shape the epidermal primordium into a skin layer covering the elongated embryo. Morphogenesis occurs over a period of ~3 hours in mid-embryogenesis and can be divided into three phases: dorsal intercalation, ventral enclosure, and elongation (FIGURE 4). These movements are driven by changes in cell shape or adhesion and do not involve cell proliferation; indeed morphogenetic processes such as enclosure are resistant to decreased epidermal proliferation 39.
FIGURE 4. Embryonic epidermal morphogenesis.
Early stages of epidermal morphogenesis, illustrated as projections and in frames from movies of the epidermal junctional marker DLG-1::GFP (xnIs17). (A) Initial formation of the dorsal epidermal epithelium, ~230–250 minutes post first cleavage. The epidermis occupies the posterior 2/3 of the dorsal part of the embryo. (B) Dorsal intercalation (~250–390 minutes), early and late stages. Intercalation is not as synchronous or as consistent as in the cartoon. (C) Ventral enclosure: migrations of four leading cells to the ventral midline, 365–375 min. (D) Ventral enclosure II: closure of the ventral pocket by P cells and hyp7 ventral cells, 370–385 min. (E) Enclosure III: formation of the anterior epidermis, 380–395 min. The leading cells move anteriorly; hyp4 and hyp5 cells enclose the head in short-range movements. Dorsal hyp7 cells begin to fuse. DLG-1::GFP images in A, B are dorsal views, from Movie 2 in Chisholm and Hardin 2005, WormBook. Frames in C-E are ventral views (C.A. Giurumescu and A.D.C., unpublished).
Dorsal intercalation
The dorsal-most epidermal cells, hyp6 and hyp7 precursors, initially form two bilaterally symmetrical rows that intercalate into a single dorsal row (FIGURE 4B). Intercalation involves polarization and directed protrusions of the dorsal cells 40 and is regulated by DIE-1 28. A Wnt/β-catenin signaling pathway promotes cell fate determination of the dorsal epidermis 41, 42. Following intercalation, the nuclei of dorsal cells undergo contralateral migrations, a process dependent on a polarized MT cytoskeleton 43 and the Arp2/3 complex 44. Dorsal intercalation may play a convergent extension-like role to elongate the dorsal epidermis prior to enclosure. Intercalation may also help align the MT and actin cytoskeletons into circumferential arrays required in enclosure or elongation.
Ventral enclosure
Ventral enclosure or epidermal enclosure involves spreading of the epidermal sheet over lateral and ventral cells to meet at the ventral midline. Enclosure is an example of type of epithelial spreading movement known as epiboly; other examples include epiboly of fish embryos 45 and dorsal closure of the Drosophila epidermis 46. Mechanisms of epiboly are of broad interest as they may be related to those underlying closure of epithelial wounds 47.
At least three partly independent processes have been defined in C. elegans epidermal enclosure 48 (FIGURE 4C-E): (1) four ‘leading cells’ in the midbody initiate enclosure by sending processes to meet at the ventral midline; (2) more posterior ‘ventral pocket’ cells then enclose the posterior body, possibly via a ‘supracellular purse-string’ mechanism; (3) finally anterior epidermal cells (hyp1–5) enclose the head. Several processes contribute to epidermal motility during enclosure, including the epidermal actin cytoskeleton 49, 50 and IP3/Calcium signaling 51, 52. Cadherin-mediated adhesion is not required for motility of the spreading epidermis 53 but is necessary for stable adhesion of leading cells after they meet at the ventral midline 54.
Before enclosure the ventral surface of the embryo is covered by a set of neuroblasts and assorted non-neural cells that form the substrate for epidermal spreading. Correct development of the substrate is critical for epidermal enclosure 55. Several partly redundant pathways required for normal epidermal morphogenesis are primarily involved in migration movements within the substrate. The first such neuronal pathway to be elucidated was the Eph receptor/VAB-1 and ephrin ligand signaling pathway 56. Other pathways with partly overlapping functions include the LAR-like receptor tyrosine phosphatase PTP-3 57, semaphorin MAB-20/ephrin EFN-4 signaling 58–60, and the KAL-1 (Kallmann syndrome/Anosmin)/HSPG pathway 61, 62. Loss of function in any one of these pathways results in variable and incompletely penetrant defects in neuronal substrate formation and subsequent epidermal enclosure; loss of function in any two pathways causes highly penetrant enclosure defects. These observations do not yet distinguish whether substrate cells are permissive or instructive for epidermal enclosure. Substrate cells undergo complex dynamic rearrangements during enclosure (C.A. Giurumescu and A.D.C., unpublished results) and form specific contacts with P cells during ventral pocket closure 63, consistent iwth the neuronal substrate playing an active role in epidermal movements.
Formation of epidermal syncytia by fusion
As noted above, the epidermis of the newly hatched larva consists of multinucleate syncytia in addition to the lateral and ventral blast cells (see BOX). The cell-cell fusions that generate these syncytia commence during epidermal enclosure and proceed in a stereotyped manner 64, although the exact order of fusion within hyp7 may not be completely invariant. The first epidermal cells to fuse are the posterior pair of ventral leading hyp7 cells (18 and 19 in FIGURE 1B), followed by cells 1 and 2 in dorsal hyp7. Subsequent fusions progress from anterior to posterior and are complete by the two-fold stage. As described below, post-embryonic seam-derived cells fuse with hyp7 within hours of their birth; in late L3 stage the hyp6 syncytium as a whole fuses with hyp7 65.
Expression of the fusogen EFF-1 in many cases appears necessary and sufficient for adjacent cells to fuse; for example seam cells do not fuse with hyp7 because they express ELT-5, which transcriptionally represses EFF-1 30. These observations raise the question of how the epidermis can form multiple adjacent syncytia without the two syncytia fusing together. For example, EFF-1 is required for the fusions that form hyp6 and hyp7, yet hyp6 and hyp7 do not fuse until the L3 stage. At least two explanations may apply. First, EFF-1 expression or function in adjacent cells could be temporally separate 66. Second, distinct fusogens may be involved in distinct sets of cells. The EFF-1-related fusogen AFF-1 is required for embryonic fusion of hyp5 and the late larval fusions of seam cells 67. Alternating expression of non-homotypic fusogens might be sufficient to explain the ability of adjacent syncytia to form independently. Ephrin signaling has also been implicated in hyp6 fusion, although the mechanism remains unclear 68.
Epidermal elongation: role of the epidermal cytoskeleton
Immediately after enclosure the embryo undergoes dramatic elongation from a bean or comma-like shape into an elongated worm, folded over within the eggshell. Embryonic elongation involves rapid changes in shape of epidermal cells and of the entire embryo, ultimately driven by forces generated by the epidermal cytoskeleton. Classic embryological studies demonstrated that the epidermal actin cytoskeleton was essential for elongation 69. The epidermal MT cytoskeleton distributes forces generated by actomyosin based contraction and may be important for the epidermal response to contraction 70. Although elongation appears as a single continuous process genetic analysis has revealed that elongation beyond the two-fold stage requires the contraction of underlying body wall muscles 71. In the wild type, muscle contractions begin at the 1.75-fold stage of elongation. Muscle contractions influence epidermal cytoskeletal remodeling via a novel tension-sensing mechanism in trans-epidermal attachments (see part II). After elongation is complete the formation of the first cuticle maintains the elongated form of the larva.
Epidermal specification and morphogenesis in other nematodes
Epidermal development in C. elegans appears to be broadly representative of epidermal development in the nematode phylum. Embryonic development of a variety of nematode species has been examined, in some cases by timelapse microscopy and cell lineage analysis 72–74. Although relatively few studies explicitly describe epidermal development, all nematode embryos examined pass through ‘comma’ or ‘tadpole’ morphogenetic stages in mid-embryogenesis, corresponding to the point at which embryonic cell divisions are essentially complete and the epidermis has enclosed the embryo. Late embryonic elongation and hatching are likewise conserved, and result in similar early larval forms. Development before the comma stage appears to be evolutionarily flexible, and in many cases does not closely resemble that of C. elegans.
Species in the class Chromadoria display epidermal development closely reminiscent of C. elegans: epidermal precursors are non-clonal descendants of AB and C, and divide to form of a dorsal epidermal primordium that encloses over a neuronal substrate 75, 76. Other species, such as the Cephalobid A. nanus, display more regulative early development, yet arrive at a comma-like stage 77. Gastrulation of a blastula with a prominent blastocoel occurs in Tobrilus, yet again this leads to a comma-like stage 78.
There are fewer studies of the embryology of the two more ‘ancestral’ or ‘ancestrally diverged’ nematode classes Enoplia and Dorylaimia, also termed clades I and II 79. The Enoplid Enoplus brevis displays a variable early cell lineage in which fates may be allocated by position not ancestry 80. Epidermal cells form dorsally, and might enclose the embryo by epiboly as in C. elegans. Epidermal cells in Enoplus do not fuse, and instead form a cellular epidermis in larval stages; it has been suggested that a syncytial epidermis is a derived character in nematodes. Among the Dorylaimids, Trichinella spiralis passes through a tadpole-like stage 72. In Romanomermis culicivorax, epidermal cells originate from a single blastomere ‘S2’, possibly equivalent to the C blastomere in C. elegans. Epidermal cells form a series of rings at the posterior of the embryo before spreading anteriorly over a substrate of neurons 81.
In summary, nematode epidermal development illustrates well the concepts of the ‘phylotypic stage’ 72, 82 and the ‘developmental hourglass’ 83. Specification of epidermal precursors, formation of the epidermal primordium, and epidermal enclosure can occur in a variety of ways. However all nematode species converge on a comma-like phylotypic stage, which then elongates into the typical nematode larva. Whether the comma and elongation stages are indeed the most conserved in nematode evolution needs to be substantiated by further comparative studies focusing on the epidermis. In subsequent larval development the epidermis and cuticle diverge in form according to the particular ecological niche of the species.
POST-EMBRYONIC GROWTH AND PATTERNING
The size and shape of the nematode body is principally determined by the post-embryonic growth of the epidermis and cuticle. The epidermis provides a model for understanding how the growth of a single tissue is integrated into organismal size control.
Growth: seam cell divisions and polyploidization
During larval development C. elegans grows six-fold in length and several-fold in diameter, an overall 32-fold increase in volume 84. This increase in size is in large part a direct result of the increase in hyp7 nuclear number and DNA content. C. elegans continue to grow over the first 4 days of adult life, approximately doubling in volume 85. Such post-L4 growth involves increased epidermal cell size, driven by polyploidization of epidermal nuclei.
Seam cells: stem cell-like epidermal blast cells
Epidermal seam cells have provided many insights into how self-renewing asymmetric stem cell-like divisions are controlled within a polarized epithelium. During each larval stage the seam cells divide asymmetrically to generate anterior daughters that fuse with the embryonically generated hyp7 syncytium; in total the seam cell divisions add a total of 98 hyp7 nuclei to the syncytium. Seam-derived hyp7 nuclei undergo round of DNA endoreduplication immediately before fusion. An additional 12 hyp7 nuclei are contributed by fusion of Pn.p cells or their daughters in the L1 and L3 stages. The hyp7 syncytium thus increases its nuclear DNA content about ten-fold from L1 to adult 86. The higher ploidy of the seam-derived epidermal nuclei presumably increases the transcriptional capacity of the epidermis, allowing tissue growth. The DNA replication factor LIN-6/MCM-4 is required for endoreduplication cycles and is required in the epidermis for post-embryonic growth 87. Nuclei that endoreduplicate in C. elegans divide in other nematode species such as Panagrellus, suggesting endoreduplication is an evolutionary abbreviation of cell division.
Wnt signaling plays multiple roles in seam development. Wnt signals directly or indirectly orient the polarity of asymmetric seam divisions. In at least some instances, such as the division of T, asymmetrically distributed Wnt signals play instructive roles in orienting the polarity of seam cells 88, 89. In animals lacking all five C. elegans Wnts, seam cells generally divide asymmetrically but their polarity becomes more random 90. These results suggest that the Wnts themselves are not essential for asymmetry per se, but function to orient asymmetry. In wild type seam cells the asymmetry of seam cell divisions is reflected in the asymmetric distribution of POP-1 activity in the anterior and posterior seam daughters: anterior daughters have high levels of nuclear POP-1 and fuse with hyp7, whereas posterior daughters have low nuclear POP-1 and remain seam cells 25, 91. The intracellular components of Wnt pathways are involved in generation of asymmetry per se. Animals with reduced function in multiple Wnt receptors (Frizzleds) 90 or in the Wnt/β-catenin asymmetry pathway 92 display global loss of asymmetry in seam divisions, and consequent under- or over-proliferation of seam cells. Finally, Wnt signals act redundantly with other cues such as cell shape to constrain the orientation of seam divisions to the long axis of the larva 93.
Several studies have focused on how the seam switches between asymmetric self-renewal divisions and the L2-specific symmetric proliferative division. L2 developmental programs in general are regulated by heterochronic genes that include the HBL-1 transcription factor and microRNAs of the let-7 family 94, 95. The RNT-1/BRO-1 complex is necessary and sufficient for the L2 seam proliferative division, as is the engrailed-like gene CEH-16 96, 97. POP-1 remains asymmetric during the symmetrical proliferative divisions of the seam in the L2 stage, suggesting that POP-1 asymmetry can be uncoupled from seam fate asymmetry.
Genetic regulation of body size: TGFβ signaling and endoreduplication
The importance of TGFβ signals in regulation of body size was discovered through genetic analysis of Small body size (sma) mutants, most of which affect genes encoding TGFβ ligands, receptors, or downstream signaling components 98. Animals lacking this pathway have normal body size and shape in the L1 stage and proceed through the normal number of molts, yet fail to grow to normal size. TGFβ signals are not required for divisions of seam cells that add nuclei to the epidermis. Animals lacking the TGFβ ligand DBL-1 display reduced endoreduplication of the seam-derived hyp7 nuclei. DBL-1 is expressed in the nervous system 99, suggesting signals from neurons might regulate epidermal polyploidization. However the precise source of DBL-1 is less critical than overall levels of signaling 100. Conversely, loss of function in the epidermal glypican-like protein LON-2, a negative regulator of DBL-1, results in animals 25% longer than wild type 101. The CRISP family protein LON-1 is a downstream target of DBL-1 that appears to repress polyploidization 102.
DBL-1 does not solely act via epidermal polyploidization. For example, overexpression of DBL-1 increases body length without increasing endoreduplication 103. The cuticle collagen LON-3 is negatively regulated by TGFβ, in parallel to LON-1 104. Thus the DBl-1 pathway acts as a coordinate regulator of epidermal growth, influencing multiple targets in the epidermis that act together to regulate epidermal size 105. Rictor signals also promote body size increase, in parallel to the DBL-1/polyploidization pathway 106.
Adult epidermal growth involves polyploidization and is sensitive to environmental conditions
Somatic cell or nuclear division has not yet been observed in adult C. elegans. Although germline proliferation continues for several days, animals lacking germlines are larger not smaller than wild type, suggesting the germline is a source of inhibitory growth signals. Instead, the post-L4 growth of the adult is at least partly due to continuing epidermal polyploidization 107. Adult growth is also reduced by dietary restriction and sensory deprivation 108. Sensory deprivation appears to act on the DBL-1 pathway via the cGMP kinase EGL-4 109, 110. Dietary restriction may also influence epidermal size independent of DBL-1, by increasing epidermal autophagy 111.
Spatial and temporal patterning of the post-embryonic epidermis
The epidermis has long been a tractable model for many aspects of spatial and temporal patterning in a single tissue. The post-embryonic seam and ventral blast cells, which show region-specific variations of serially reiterated lineages, provide excellent examples of spatial patterns. Larval stage-specific patterns of seam cell division exemplify temporal patterning of epidermal cell fates by the heterochronic gene system and are reviewed elsewhere. Finally, the epidermal components of the vulva and male tail are classic examples of inductive patterning.
Anteroposterior patterning of the epidermis: Hox clusters and Pax genes
The lateral epidermis in the L1 stage consists of the ten seam cells H0, H1, H2, V1–6, and T. Apart from V1–V4, each has its own distinct fate and lineage. Seam patterning in the body and tail is regulated by the C. elegans Hox cluster, a set of loosely clustered homeobox-containing genes whose chromosomal position is colinear with the anteroposterior location of the cells they affect (FIGURE 5) 112. The fates of V5 and V6 are patterned by the Hox genes mab-5 113 and egl-5 114. The AbdB-related posterior HOX gene NOB-1 is required for posterior epidermal morphogenesis 115 and functions with a related AbdB-like gene PHP-3 to specify the T fate 116. The labial-like Hox gene ceh-13 may be involved in specification of anterior seam cells V1 and H2 117, although lineage transformations have not yet been described. The Hox cluster has not been clearly implicated in specification of the most anterior seam cells; instead, patterning of H0 and H1 is mediated by isoforms of the VAB-3/PAX-6 paired domain protein 118, 119.
FIGURE 5. Anteroposterior pattern in the larval epidermis.
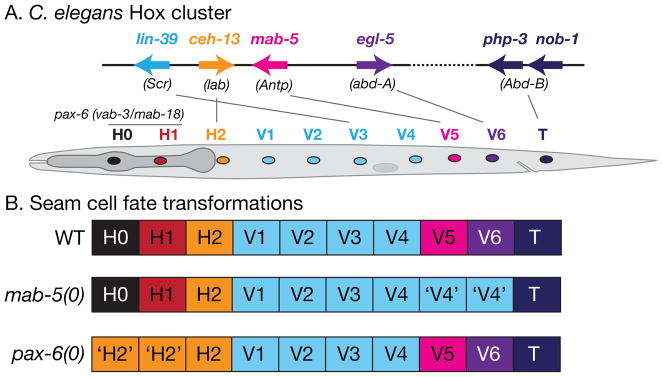
Patterning of the seam in the trunk and tail is specified by genes in the C. elegans Hox gene cluster. Schematic showing Hox cluster genes on part of chromosome III; putative Drosophila homologs are indicated 156. Examples of cell fate transformations: in mab-5 mutants the posterior seam cells V5 and V6 adopt V1–V4-like fates (‘V4’) 113. Patterning of the head seam cells is specified by the PAX-6 locus vab-3/mab-18. 118, 119. In pax-6 null mutants the anterior-most seam cells H0 and H1 are transformed to H2-like fates. Hox cluster genes also regulate anteroposterior pattern in the ventral epidermal (P) cells.
Anteroposterior pattern in the ventral epidermis is also defined by Hox gene activity 120, 121. LIN-39, in concert with the homeobox cofactor CEH-20 (orthologous to Drosophila Exd/Pbx), specifies midbody P cell fates, including the vulval equivalence group 122. LIN-39 appears to repress fusion of vulval precursor cells by activation of the seam GATA factors ELT-5 and ELT-6 123. LIN-39 and MAB-5 interact combinatorially in more posterior P cells, whereas EGL-5 acts in the most posterior P cell 114. Studies of C. elegans epidermal patterning have thus elucidated how Hox gene clusters can pattern a tissue at the level of individual cell fates. How the C. elegans Hox genes become active in an anteroposterior sequence in mid embryogenesis remains to be addressed, as equivalents of the Drosophila segmentation gene hierarchy do not appear to be involved.
Post-embryonic organogenesis and sexual dimorphism in the epidermis
The epidermis contributes to two major sexually dimorphic organs, the hermaphrodite vulva and the male tail. The hermaphrodite vulva develops in the ventral midbody whereas the male tail develops in the lateral and ventral tail region. The vulva and male tail are integrated organs comprising neurons, muscles, and epidermis; here we focus on the epidermal contribution to these organs.
The vulva develops from three epidermal precursors P(5–7).p; three additional cells (P3.p, P4.p, P8.p) do not normally form the vulva but are developmentally equivalent, thus the vulval equivalence group of the midbody consists of P(3–8).p. The formation of the vulva is a dramatic example of orifice formation in an epithelium. Vulval precursors are induced by signals from gonadal cells, but their fates are also regulated by the surrounding syncytial epidermis. Loss of function in epidermally expressed genes such as lin-15 or lin-35/Rb results in excessive vulval induction (the Multivulva or Muv phenotype). These genes act in the hyp7 syncytium to repress expression of the inductive signal LIN-3 124, 125. hyp7 is also one source of Wnt signals that act as competence factors for the vulva, preventing fusion of vulval precursors with hyp7 126. Vulval precursors interact extensively among themselves to ensure a highly invariant pattern of fates that then undergo morphogenetic movements to form the external opening to the uterus; genes such as VAB-23 act downstream of the midbody Hox gene LIN-39 to regulate vulval morphogenesis programs 127.
Male tail epidermal structures are a dramatic example of epidermal sculpting. Three posterior lateral epidermal seam cells (V5, V6, T) generate the sensory rays and acellular fan. The patterning of the male tail seam cells is under the control of Hox genes (MAB-5, EGL-5) 128 and the PAX-6 isoform MAB-18 129, which activate expression of neurogenic genes such as LIN-32 to trigger ray formation. The positions of specific rays are determined by interactions with the surrounding epidermis, and are regulated by multiple signaling pathways, including Semaphorin, Ephrin 130, 131, and TGFβ 132–134. Ray positioning is also dependent on myc family transcription factors expressed in epidermal cells 135. The final stages of male tail morphogenesis involve extensive remodeling of the tail epidermis and cuticle and require specific cuticle collagens 136 and cell surface proteins 137, 138.
The sexual dimorphism of epidermal precursors to the vulva and male tail imply that epidermal fate specification intersects with the sex determination pathway. Somatic sex determination in C. elegans is regulated by the TRA-1 GLI family transcription factor, which is expressed in XX (hermaphrodite) animals and represses male developmental programs 139. Few direct targets of TRA-1 in the sexually dimorphic epidermis have been characterized. Although vulval fates are hermaphrodite-specific, TRA-1 appears to inhibit rather than promote vulval differentiation, acting as a repressor of LIN-39 140. In the male tail, the DM domain transcription factor MAB-3 acts downstream of MAB-5 in specification of male V5 and V6 fates 141, 142. However MAB-3 is expressed throughout the lateral epidermis in both sexes 143, implying that sexual dimorphism results from regulation at other levels in the pathway.
Plasticity and regulation in the epidermis
Despite the high degree of invariance of wild type epidermal development, laser ablation experiments have revealed a limited capacity for regulation in the lateral epidermis of the male tail 144, suggesting cell signals can contribute to patterning of the lateral epidermis. Direct contact between seam cells is important in specifying lateral epidermal cell fates 145, 146. The nature of the contact-dependent signal between seam cells is not yet known; it appears to counteract Wnt signals 147. Semaphorin/Plexin signals are known to regulate seam cell contacts and may contribute to the ‘stop’ signal that prevents seam cells from extending past one another 132.
Conclusions and Future Directions
The C. elegans epidermis is a simple model for many aspects of skin developmental biology. Analysis of embryonic morphogenesis has illuminated our understanding of processes such as epiboly, hemidesmosome biogenesis, and tension-induced cell shape change. An important goal in the future will be to take a more quantitative approach to morphogenetic movements in the developing epidermis. Improvements in live imaging should allow the dynamics of epidermal cell shape changes to be described more accurately, permitting mathematical and physical models of force generation. The ability of the C. elegans embryo to execute complex morphogenetic movements with such high reproducibility suggests the existence of buffering systems that reduce variation; the variable phenotypes of morphogenesis mutants could reflect increased sensitivity to stochastic effects in these mutants.
Acknowledgments
The DLG-1::GFP panels in Figure 4 are taken from a 4D two-photon movie made by Matthias Köppen at the Laboratory for Optical and Computational Instrumentation (Madison, WI) and from a laser scanning confocal 4D movie made by Claudiu Giurumescu. We thank Yishi Jin and the anonymous reviewers for constructive comments. Our work on C. elegans epidermal development is supported by NIH R01 GM54657.
Footnotes
The external epithelium of nematodes was historically known as the hypodermis or hypoderm 148. This term persists in the literature (e.g. Worm Atlas) and in the nomenclature for C. elegans cells (hyp7, etc). However it has long been generally accepted, based on developmental and now molecular evidence, that the hypodermal epithelium is homologous to the epidermal epithelia of other animals. We follow Wright’s view 149, namely that “The concept of phylogenetic origin of cuticles indicates that the term epidermis is more appropriate than hypodermis for the cell layer that forms the nematode cuticle”. The term hypodermis is accurately applied to the sub-dermal connective tissue of vertebrates, or the subepidermal cell layers of plants.
Further Reading/Resources
For a complete account of adult epidermal anatomy, see the WormAtlas web site (www.wormatlas.org) or the C. elegans Atlas 9, Chapters 2 (epithelial system), 9 (cuticle), and 10 (pericellular structures). For general reviews of nematode epidermis and cuticle see The Biology of Nematodes 157, chapters 2 and 7.
Contributor Information
Andrew D. Chisholm, Email: chisholm@ucsd.edu, Division of Biological Sciences, Section of Cell and Developmental Biology, University of California San Diego, La Jolla, CA 92093
Tiffany I. Hsiao, Division of Biological Sciences, Section of Cell and Developmental Biology, University of California San Diego, La Jolla, CA 92093
References
- 1.Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson DP, Geary TG. Excretion/Secretion, Ionic and Osmotic Regulation. In: Lee DL, editor. The Biology of Nematodes. Boca Raton, FL: CRC Press; 2002. pp. 291–320. [Google Scholar]
- 3.Payre F. Genetic control of epidermis differentiation in Drosophila. Int J Dev Biol. 2004;48:207–215. [PubMed] [Google Scholar]
- 4.Le Guellec D, Morvan-Dubois G, Sire JY. Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio) The International journal of developmental biology. 2004;48:217–231. doi: 10.1387/ijdb.15272388. [DOI] [PubMed] [Google Scholar]
- 5.Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Ann Rev Cell Dev Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- 6.Sasai Y, De Robertis EM. Ectodermal patterning in vertebrate embryos. Developmental biology. 1997;182:5–20. doi: 10.1006/dbio.1996.8445. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt-Rhaesa A. The Evolution of Organ Systems. New York: Oxford University Press; 2007. [Google Scholar]
- 8.Moussian B, Uv AE. An ancient control of epithelial barrier formation and wound healing. Bioessays. 2005;27:987–990. doi: 10.1002/bies.20308. [DOI] [PubMed] [Google Scholar]
- 9.Hall DH, Altun ZF. C. elegans Atlas. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- 10.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 11.Ahringer J. Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev. 1996;10:1120–1130. doi: 10.1101/gad.10.9.1120. [DOI] [PubMed] [Google Scholar]
- 12.Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- 13.Shaham S. Glia-neuron interactions in the nervous system of Caenorhabditis elegans. Curr Opin Neurobiol. 2006;16:522–528. doi: 10.1016/j.conb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Altun ZF, Chen B, Wang ZW, Hall DH. High resolution map of Caenorhabditis elegans gap junction proteins. Dev Dyn. 2009;238:1936–1950. doi: 10.1002/dvdy.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labouesse M, Mango SE. Patterning the C. elegans embryo: moving beyond the cell lineage. Trends Genet. 1999;15:307–313. doi: 10.1016/s0168-9525(99)01750-3. [DOI] [PubMed] [Google Scholar]
- 16.Page BD, Zhang W, Steward K, Blumenthal T, Priess JR. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 1997;11:1651–1661. doi: 10.1101/gad.11.13.1651. [DOI] [PubMed] [Google Scholar]
- 17.Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol. 2001;21:2533–2544. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, Brown EL, Hunter CP. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development. 2005;132:1843–1854. doi: 10.1242/dev.01782. [DOI] [PubMed] [Google Scholar]
- 19.Gilleard JS, Shafi Y, Barry JD, McGhee JD. ELT-3: A Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Dev Biol. 1999;208:265–280. doi: 10.1006/dbio.1999.9202. [DOI] [PubMed] [Google Scholar]
- 20.Labouesse M, Sookhareea S, Horvitz HR. The Caenorhabditis elegans gene lin-26 is required to specify the fates of hypodermal cells and encodes a presumptive zinc-finger transcription factor. Development. 1994;120:2359–2368. doi: 10.1242/dev.120.9.2359. [DOI] [PubMed] [Google Scholar]
- 21.Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- 22.Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintin S, Michaux G, McMahon L, Gansmuller A, Labouesse M. The Caenorhabditis elegans gene lin-26 can trigger epithelial differentiation without conferring tissue specificity. Dev Biol. 2001;235:410–421. doi: 10.1006/dbio.2001.0294. [DOI] [PubMed] [Google Scholar]
- 24.Gendreau SB, Moskowitz IP, Terns RM, Rothman JH. The potential to differentiate epidermis is unequally distributed in the AB lineage during early embryonic development in C. elegans. Dev Biol. 1994;166:770–781. doi: 10.1006/dbio.1994.1355. [DOI] [PubMed] [Google Scholar]
- 25.Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- 26.Yanai I, Baugh LR, Smith JJ, Roehrig C, Shen-Orr SS, Claggett JM, Hill AA, Slonim DK, Hunter CP. Pairing of competitive and topologically distinct regulatory modules enhances patterned gene expression. Mol Syst Biol. 2008;4:163. doi: 10.1038/msb.2008.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pocock R, Ahringer J, Mitsch M, Maxwell S, Woollard A. A regulatory network of T-box genes and the even-skipped homologue vab-7 controls patterning and morphogenesis in C. elegans. Development. 2004;131:2373–2385. doi: 10.1242/dev.01110. [DOI] [PubMed] [Google Scholar]
- 28.Heid PJ, Raich WB, Smith R, Mohler WA, Simokat K, Gendreau SB, Rothman JH, Hardin J. The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev Biol. 2001;236:165–180. doi: 10.1006/dbio.2001.0315. [DOI] [PubMed] [Google Scholar]
- 29.Cassata G, Shemer G, Morandi P, Donhauser R, Podbilewicz B, Baumeister R. ceh-16/engrailed patterns the embryonic epidermis of Caenorhabditis elegans. Development. 2005;132:739–749. doi: 10.1242/dev.01638. [DOI] [PubMed] [Google Scholar]
- 30.Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–2880. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- 31.Smith JA, McGarr P, Gilleard JS. The Caenorhabditis elegans GATA factor elt-1 is essential for differentiation and maintenance of hypodermal seam cells and for normal locomotion. J Cell Sci. 2005;118:5709–5719. doi: 10.1242/jcs.02678. [DOI] [PubMed] [Google Scholar]
- 32.Kagoshima H, Nimmo R, Saad N, Tanaka J, Miwa Y, Mitani S, Kohara Y, Woollard A. The C. elegans CBFβ homologue BRO-1 interacts with the Runx factor, RNT-1, to promote stem cell proliferation and self-renewal. Development. 2007;134:3905–3915. doi: 10.1242/dev.008276. [DOI] [PubMed] [Google Scholar]
- 33.Xia D, Zhang Y, Huang X, Sun Y, Zhang H. The C. elegans CBFβ homolog, BRO-1, regulates the proliferation, differentiation and specification of the stem cell-like seam cell lineages. Dev Biol. 2007;309:259–272. doi: 10.1016/j.ydbio.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Gillis WJ, Bowerman B, Schneider SQ. Ectoderm- and endomesoderm-specific GATA transcription factors in the marine annelid Platynereis dumerilli. Evol Dev. 2007;9:39–50. doi: 10.1111/j.1525-142X.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 35.Yamada M, Murata T, Hirose S, Lavorgna G, Suzuki E, Ueda H. Temporally restricted expression of transcription factor betaFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development. 2000;127:5083–5092. doi: 10.1242/dev.127.23.5083. [DOI] [PubMed] [Google Scholar]
- 36.Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308:381–385. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- 37.Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- 38.Venkatesan K, McManus HR, Mello CC, Smith TF, Hansen U. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 2003;31:4304–4316. doi: 10.1093/nar/gkg644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardin J, King R, Thomas-Virnig C, Raich WB. Zygotic loss of ZEN-4/MKLP1 results in disruption of epidermal morphogenesis in the C. elegans embryo. Dev Dyn. 2008;237:830–836. doi: 10.1002/dvdy.21455. [DOI] [PubMed] [Google Scholar]
- 40.Williams-Masson EM, Heid PJ, Lavin CA, Hardin J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol. 1998;204:263–276. doi: 10.1006/dbio.1998.9048. [DOI] [PubMed] [Google Scholar]
- 41.Walston T, Guo C, Proenca R, Wu M, Herman M, Hardin J, Hedgecock E. mig-5/Dsh controls cell fate determination and cell migration in C. elegans. Dev Biol. 2006;298:485–497. doi: 10.1016/j.ydbio.2006.06.053. [DOI] [PubMed] [Google Scholar]
- 42.King RS, Maiden SL, Hawkins NC, Kidd AR, 3rd, Kimble J, Hardin J, Walston TD. The N- or C-terminal domains of DSH-2 can activate the C. elegans Wnt/β-catenin asymmetry pathway. Dev Biol. 2009;328:234–244. doi: 10.1016/j.ydbio.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong H, Mohler WA, Soto MC. The branched actin nucleator Arp2/3 promotes nuclear migrations and cell polarity in the C. elegans zygote. Dev Biol. 2011;357:356–369. doi: 10.1016/j.ydbio.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lepage SE, Bruce AE. Zebrafish epiboly: mechanics and mechanisms. The International journal of developmental biology. 2010;54:1213–1228. doi: 10.1387/ijdb.093028sl. [DOI] [PubMed] [Google Scholar]
- 46.Jacinto A, Woolner S, Martin P. Dynamic analysis of dorsal closure in Drosophila: from genetics to cell biology. Dev Cell. 2002;3:9–19. doi: 10.1016/s1534-5807(02)00208-3. [DOI] [PubMed] [Google Scholar]
- 47.Martin P, Parkhurst SM. Parallels between tissue repair and embryo morphogenesis. Development. 2004;131:3021–3034. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- 48.Williams-Masson EM, Malik AN, Hardin J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 1997;124:2889–2901. doi: 10.1242/dev.124.15.2889. [DOI] [PubMed] [Google Scholar]
- 49.Sawa M, Suetsugu S, Sugimoto A, Miki H, Yamamoto M, Takenawa T. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J Cell Sci. 2003;116:1505–1518. doi: 10.1242/jcs.00362. [DOI] [PubMed] [Google Scholar]
- 50.Patel FB, Bernadskaya YY, Chen E, Jobanputra A, Pooladi Z, Freeman KL, Gally C, Mohler WA, Soto MC. The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Dev Biol. 2008;324:297–309. doi: 10.1016/j.ydbio.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas-Virnig CL, Sims PA, Simske JS, Hardin J. The inositol 1,4,5-trisphosphate receptor regulates epidermal cell migration in Caenorhabditis elegans. Curr Biol. 2004;14:1882–1887. doi: 10.1016/j.cub.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Vazquez-Manrique RP, Nagy AI, Legg JC, Bales OA, Ly S, Baylis HA. Phospholipase C-epsilon regulates epidermal morphogenesis in Caenorhabditis elegans. PLoS Genet. 2008;4:e1000043. doi: 10.1371/journal.pgen.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raich WB, Agbunag C, Hardin J. Rapid epithelial-sheet sealing in the Caenorhabditis elegans embryo requires cadherin-dependent filopodial priming. Curr Biol. 1999;9:1139–1146. doi: 10.1016/S0960-9822(00)80015-9. [DOI] [PubMed] [Google Scholar]
- 55.George SE, Simokat K, Hardin J, Chisholm AD. The VAB-1 Eph receptor tyrosine kinase functions in neural and epithelial morphogenesis in C. elegans. Cell. 1998;92:633–643. doi: 10.1016/s0092-8674(00)81131-9. [DOI] [PubMed] [Google Scholar]
- 56.Chin-Sang ID, George SE, Ding M, Moseley SL, Lynch AS, Chisholm AD. The ephrin VAB-2/EFN-1 functions in neuronal signaling to regulate epidermal morphogenesis in C. elegans. Cell. 1999;99:781–790. doi: 10.1016/s0092-8674(00)81675-x. [DOI] [PubMed] [Google Scholar]
- 57.Harrington RJ, Gutch MJ, Hengartner MO, Tonks NK, Chisholm AD. The C. elegans LAR-like receptor tyrosine phosphatase PTP-3 and the VAB-1 Eph receptor tyrosine kinase have partly redundant functions in morphogenesis. Development. 2002;129:2141–2153. doi: 10.1242/dev.129.9.2141. [DOI] [PubMed] [Google Scholar]
- 58.Chin-Sang ID, Moseley SL, Ding M, Harrington RJ, George SE, Chisholm AD. The divergent C. elegans ephrin EFN-4 functions in embryonic morphogenesis in a pathway independent of the VAB-1 Eph receptor. Development. 2002;129:5499–5510. doi: 10.1242/dev.00122. [DOI] [PubMed] [Google Scholar]
- 59.Roy PJ, Zheng H, Warren CE, Culotti JG. mab-20 encodes Semaphorin-2a and is required to prevent ectopic cell contacts during epidermal morphogenesis in Caenorhabditis elegans. Development. 2000;127:755–767. doi: 10.1242/dev.127.4.755. [DOI] [PubMed] [Google Scholar]
- 60.Nakao F, Hudson ML, Suzuki M, Peckler Z, Kurokawa R, Liu Z, Gengyo-Ando K, Nukazuka A, Fujii T, Suto F, et al. The PLEXIN PLX-2 and the ephrin EFN-4 have distinct roles in MAB-20/Semaphorin 2A signaling in Caenorhabditis elegans morphogenesis. Genetics. 2007;176:1591–1607. doi: 10.1534/genetics.106.067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rugarli EI, Di Schiavi E, Hilliard MA, Arbucci S, Ghezzi C, Facciolli A, Coppola G, Ballabio A, Bazzicalupo P. The Kallmann syndrome gene homolog in C. elegans is involved in epidermal morphogenesis and neurite branching. Development. 2002;129:1283–1294. doi: 10.1242/dev.129.5.1283. [DOI] [PubMed] [Google Scholar]
- 62.Hudson ML, Kinnunen T, Cinar HN, Chisholm AD. C. elegans Kallmann syndrome protein KAL-1 interacts with syndecan and glypican to regulate neuronal cell migrations. Dev Biol. 2006;294:352–365. doi: 10.1016/j.ydbio.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 63.Ikegami R, Simokat K, Zheng H, Brown L, Garriga G, Hardin J, Culotti J. Semaphorin and Eph Receptor Signaling Guide a Series of Cell Movements for Ventral Enclosure in C. elegans. Current biology: CB. 2012;22:1–11. doi: 10.1016/j.cub.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Podbilewicz B, White JG. Cell fusions in the developing epithelia of C. elegans. Dev Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- 65.Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.del Campo JJ, Opoku-Serebuoh E, Isaacson AB, Scranton VL, Tucker M, Han M, Mohler WA. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Current biology: CB. 2005;15:413–423. doi: 10.1016/j.cub.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 67.Sapir A, Choi J, Leikina E, Avinoam O, Valansi C, Chernomordik LV, Newman AP, Podbilewicz B. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell. 2007;12:683–698. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Roy PJ, Holland SJ, Zhang LW, Culotti JG, Pawson T. Multiple ephrins control cell organization in C. elegans using kinase-dependent and -independent functions of the VAB-1 Eph receptor. Mol Cell. 1999;4:903–913. doi: 10.1016/s1097-2765(00)80220-8. [DOI] [PubMed] [Google Scholar]
- 69.Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- 70.Ciarletta P, Ben Amar M, Labouesse M. Continuum model of epithelial morphogenesis during Caenorhabditis elegans embryonic elongation. Philos Transact A Math Phys Eng Sci. 2009;367:3379–3400. doi: 10.1098/rsta.2009.0088. [DOI] [PubMed] [Google Scholar]
- 71.Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hope IA. Embryology, Developmental Biology, and the Genome. In: Lee DL, editor. The Biology Of Nematodes. Boca Raton, FL: CRC Press; 2002. pp. 121–146. [Google Scholar]
- 73.Schierenberg E. Embryological variation during nematode development. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.55.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulze J, Schierenberg E. Evolution of embryonic development in nematodes. EvoDevo. 2011;2:18. doi: 10.1186/2041-9139-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Houthoofd W, Jacobsen K, Mertens C, Vangestel S, Coomans A, Borgonie G. Embryonic cell lineage of the marine nematode Pellioditis marina. Dev Biol. 2003;258:57–69. doi: 10.1016/s0012-1606(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 76.Lahl V, Halama C, Schierenberg E. Comparative and experimental embryogenesis of Plectidae (Nematoda) Dev Genes Evol. 2003;213:18–27. doi: 10.1007/s00427-002-0289-1. [DOI] [PubMed] [Google Scholar]
- 77.Wiegner O, Schierenberg E. Specification of gut cell fate differs significantly between the nematodes Acrobeloides nanus and Caenorhabditis elegans. Dev Biol. 1998;204:3–14. doi: 10.1006/dbio.1998.9054. [DOI] [PubMed] [Google Scholar]
- 78.Schierenberg E. Unusual cleavage and gastrulation in a freshwater nematode: developmental and phylogenetic implications. Dev Genes Evol. 2005;215:103–108. doi: 10.1007/s00427-004-0454-9. [DOI] [PubMed] [Google Scholar]
- 79.Blaxter M. Nematodes: the worm and its relatives. PLoS Biol. 2011;9:e1001050. doi: 10.1371/journal.pbio.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Voronov DA, Panchin YV. Cell lineage in marine nematode Enoplus brevis. Development. 1998;125:143–150. doi: 10.1242/dev.125.1.143. [DOI] [PubMed] [Google Scholar]
- 81.Schulze J, Schierenberg E. Embryogenesis of Romanomermis culicivorax: an alternative way to construct a nematode. Dev Biol. 2009;334:10–21. doi: 10.1016/j.ydbio.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 82.Slack JM, Holland PW, Graham CF. The zootype and the phylotypic stage. Nature. 1993;361:490–492. doi: 10.1038/361490a0. [DOI] [PubMed] [Google Scholar]
- 83.Schierenberg E. Three sons of fortune: early embryogenesis, evolution and ecology of nematodes. BioEssays. 2001;23:841–847. doi: 10.1002/bies.1119. [DOI] [PubMed] [Google Scholar]
- 84.Knight CG, Patel MN, Azevedo RB, Leroi AM. A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol Dev. 2002;4:16–27. doi: 10.1046/j.1525-142x.2002.01058.x. [DOI] [PubMed] [Google Scholar]
- 85.Flemming AJ, Shen ZZ, Cunha A, Emmons SW, Leroi AM. Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc Natl Acad Sci U S A. 2000;97:5285–5290. doi: 10.1073/pnas.97.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 1985;107:128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- 87.Korzelius J, The I, Ruijtenberg S, Portegijs V, Xu H, Horvitz HR, van den Heuvel S. C. elegans MCM-4 is a general DNA replication and checkpoint component with an epidermis-specific requirement for growth and viability. Developmental biology. 2011;350:358–369. doi: 10.1016/j.ydbio.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goldstein B, Takeshita H, Mizumoto K, Sawa H. Wnt signals can function as positional cues in establishing cell polarity. Developmental cell. 2006;10:391–396. doi: 10.1016/j.devcel.2005.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizumoto K, Sawa H. Two βs or not two βs: regulation of asymmetric division by β-catenin. Trends Cell Biol. 2007;17:465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 90.Yamamoto Y, Takeshita H, Sawa H. Multiple Wnts redundantly control polarity orientation in Caenorhabditis elegans epithelial stem cells. PLoS Genet. 2011;7:e1002308. doi: 10.1371/journal.pgen.1002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herman M. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development. 2001;128:581–590. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- 92.Gleason JE, Eisenmann DM. Wnt signaling controls the stem cell-like asymmetric division of the epithelial seam cells during C. elegans larval development. Dev Biol. 2010;348:58–66. doi: 10.1016/j.ydbio.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wildwater M, Sander N, de Vreede G, van den Heuvel S. Cell shape and Wnt signaling redundantly control the division axis ofC. elegans epithelial stem cells. Development. 2011;138:4375–4385. doi: 10.1242/dev.066431. [DOI] [PubMed] [Google Scholar]
- 94.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 96.Nimmo R, Antebi A, Woollard A. mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development. 2005;132:5043–5054. doi: 10.1242/dev.02102. [DOI] [PubMed] [Google Scholar]
- 97.Huang X, Tian E, Xu Y, Zhang H. The C. elegans engrailed homolog ceh-16 regulates the self-renewal expansion division of stem cell-like seam cells. Dev Biol. 2009;333:337–347. doi: 10.1016/j.ydbio.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 98.Wang J, Tokarz R, Savage-Dunn C. The expression of TGFβ signal transducers in the hypodermis regulates body size in C. elegans. Development. 2002;129:4989–4998. doi: 10.1242/dev.129.21.4989. [DOI] [PubMed] [Google Scholar]
- 99.Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, Padgett RW, Wood WB. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- 100.Savage-Dunn C, Yu L, Gill K, Awan M, Fernando T. Non-stringent tissue-source requirements for BMP ligand expression in regulation of body size in Caenorhabditis elegans. Genet Res. 2011;93:427–432. doi: 10.1017/S0016672311000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gumienny TL, MacNeil LT, Wang H, de Bono M, Wrana JL, Padgett RW. Glypican LON-2 is a conserved negative regulator of BMP-like signaling in Caenorhabditis elegans. Curr Biol. 2007;17:159–164. doi: 10.1016/j.cub.2006.11.065. [DOI] [PubMed] [Google Scholar]
- 102.Maduzia LL, Gumienny TL, Zimmerman CM, Wang H, Shetgiri P, Krishna S, Roberts AF, Padgett RW. lon-1 regulates Caenorhabditis elegans body size downstream of the dbl-1 TGFβ signaling pathway. Dev Biol. 2002;246:418–428. doi: 10.1006/dbio.2002.0662. [DOI] [PubMed] [Google Scholar]
- 103.Nystrom J, Shen ZZ, Aili M, Flemming AJ, Leroi A, Tuck S. Increased or decreased levels of Caenorhabditis elegans lon-3, a gene encoding a collagen, cause reciprocal changes in body length. Genetics. 2002;161:83–97. doi: 10.1093/genetics/161.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki Y, Morris GA, Han M, Wood WB. A cuticle collagen encoded by the lon-3 gene may be a target of TGF-β signaling in determining Caenorhabditis elegans body shape. Genetics. 2002;162:1631–1639. doi: 10.1093/genetics/162.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roberts AF, Gumienny TL, Gleason RJ, Wang H, Padgett RW. Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans. BMC Dev Biol. 2010;10:61. doi: 10.1186/1471-213X-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lozano E, Saez AG, Flemming AJ, Cunha A, Leroi AM. Regulation of growth by ploidy in Caenorhabditis elegans. Curr Biol. 2006;16:493–498. doi: 10.1016/j.cub.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 108.Tain LS, Lozano E, Saez AG, Leroi AM. Dietary regulation of hypodermal polyploidization in C. elegans. BMC Dev Biol. 2008;8:28. doi: 10.1186/1471-213X-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hirose T, Nakano Y, Nagamatsu Y, Misumi T, Ohta H, Ohshima Y. Cyclic GMP-dependent protein kinase EGL-4 controls body size and lifespan in C elegans. Development. 2003;130:1089–1099. doi: 10.1242/dev.00330. [DOI] [PubMed] [Google Scholar]
- 110.Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- 111.Morck C, Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aboobaker A, Blaxter M. Hox gene evolution in nematodes: novelty conserved. Current opinion in genetics & development. 2003;13:593–598. doi: 10.1016/j.gde.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 113.Kenyon C. A gene involved in the development of the posterior body region of C. elegans. Cell. 1986;46:477–487. doi: 10.1016/0092-8674(86)90668-9. [DOI] [PubMed] [Google Scholar]
- 114.Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. doi: 10.1242/dev.111.4.921. [DOI] [PubMed] [Google Scholar]
- 115.Van Auken K, Weaver DC, Edgar LG, Wood WB. Caenorhabditis elegans embryonic axial patterning requires two recently discovered posterior-group Hox genes. Proc Natl Acad Sci U S A. 2000;97:4499–4503. doi: 10.1073/pnas.97.9.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Arata Y, Kouike H, Zhang Y, Herman MA, Okano H, Sawa H. Wnt signaling and a Hox protein cooperatively regulate psa-3/Meis to determine daughter cell fate after asymmetric cell division in C. elegans. Dev Cell. 2006;11:105–115. doi: 10.1016/j.devcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 117.Brunschwig K, Wittmann C, Schnabel R, Burglin TR, Tobler H, Muller F. Anterior organization of the Caenorhabditis elegans embryo by the labial-like Hox gene ceh-13. Development. 1999;126:1537–1546. doi: 10.1242/dev.126.7.1537. [DOI] [PubMed] [Google Scholar]
- 118.Chisholm AD, Horvitz HR. Patterning of the Caenorhabditis elegans head region by the Pax-6 family member vab-3. Nature. 1995;377:52–55. doi: 10.1038/377052a0. [DOI] [PubMed] [Google Scholar]
- 119.Cinar HN, Chisholm AD. Genetic analysis of the Caenorhabditis elegans pax-6 locus: roles of paired domain-containing and nonpaired domain-containing isoforms. Genetics. 2004;168:1307–1322. doi: 10.1534/genetics.104.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang BB, Muller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- 121.Clark SG, Chisholm AD, Horvitz HR. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74:43–55. doi: 10.1016/0092-8674(93)90293-y. [DOI] [PubMed] [Google Scholar]
- 122.Shemer G, Podbilewicz B. LIN-39/Hox triggers cell division and represses EFF-1/fusogen-dependent vulval cell fusion. Genes Dev. 2002;16:3136–3141. doi: 10.1101/gad.251202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Koh K, Peyrot SM, Wood CG, Wagmaister JA, Maduro MF, Eisenmann DM, Rothman JH. Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors -- apparent direct targets of the LIN-39 Hox protein. Development. 2002;129:5171–5180. doi: 10.1242/dev.129.22.5171. [DOI] [PubMed] [Google Scholar]
- 124.Cui M, Chen J, Myers TR, Hwang BJ, Sternberg PW, Greenwald I, Han M. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev Cell. 2006;10:667–672. doi: 10.1016/j.devcel.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 125.Myers TR, Greenwald I. lin-35 Rb acts in the major hypodermis to oppose ras-mediated vulval induction in C. elegans. Dev Cell. 2005;8:117–123. doi: 10.1016/j.devcel.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 126.Myers TR, Greenwald I. Wnt signal from multiple tissues and lin-3/EGF signal from the gonad maintain vulval precursor cell competence in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:20368–20373. doi: 10.1073/pnas.0709989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pellegrino MW, Farooqui S, Frohli E, Rehrauer H, Kaeser-Pebernard S, Muller F, Gasser RB, Hajnal A. LIN-39 and the EGFR/RAS/MAPK pathway regulate C. elegans vulval morphogenesis via the VAB-23 zinc finger protein. Development. 2011;138:4649–4660. doi: 10.1242/dev.071951. [DOI] [PubMed] [Google Scholar]
- 128.Chow KL, Emmons SW. HOM-C/Hox genes and four interacting loci determine the morphogenetic properties of single cells in the nematode male tail. Development. 1994;120:2579–2592. doi: 10.1242/dev.120.9.2579. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y, Emmons SW. Specification of sense-organ identity by a Caenorhabditis elegans Pax-6 homologue. Nature. 1995;377:55–59. doi: 10.1038/377055a0. [DOI] [PubMed] [Google Scholar]
- 130.Hahn AC, Emmons SW. The roles of an ephrin and a semaphorin in patterning cell-cell contacts in C. elegans sensory organ development. Dev Biol. 2003;256:379–388. doi: 10.1016/s0012-1606(02)00129-x. [DOI] [PubMed] [Google Scholar]
- 131.Ikegami R, Zheng H, Ong SH, Culotti J. Integration of semaphorin-2A/MAB-20, ephrin-4, and UNC-129 TGF-β signaling pathways regulates sorting of distinct sensory rays in C. elegans. Dev Cell. 2004;6:383–395. doi: 10.1016/s1534-5807(04)00057-7. [DOI] [PubMed] [Google Scholar]
- 132.Fujii T, Nakao F, Shibata Y, Shioi G, Kodama E, Fujisawa H, Takagi S. Caenorhabditis elegans PlexinA, PLX-1, interacts with transmembrane semaphorins and regulates epidermal morphogenesis. Development. 2002;129:2053–2063. doi: 10.1242/dev.129.9.2053. [DOI] [PubMed] [Google Scholar]
- 133.Ginzburg VE, Roy PJ, Culotti JG. Semaphorin 1a and semaphorin 1b are required for correct epidermal cell positioning and adhesion during morphogenesis in C. elegans. Development. 2002;129:2065–2078. doi: 10.1242/dev.129.9.2065. [DOI] [PubMed] [Google Scholar]
- 134.Nukazuka A, Fujisawa H, Inada T, Oda Y, Takagi S. Semaphorin controls epidermal morphogenesis by stimulating mRNA translation via eIF2α in Caenorhabditis elegans. Genes Dev. 2008;22:1025–1036. doi: 10.1101/gad.1644008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pickett CL, Breen KT, Ayer DE. A C. elegans Myc-like network cooperates with semaphorin and Wnt signaling pathways to control cell migration. Dev Biol. 2007;310:226–239. doi: 10.1016/j.ydbio.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ko FC, Chow KL. A mutation at the start codon defines the differential requirement of dpy-11 in Caenorhabditis elegans body hypodermis and male tail. Biochem Biophys Res Commun. 2003;309:201–208. doi: 10.1016/s0006-291x(03)01545-6. [DOI] [PubMed] [Google Scholar]
- 137.Tsang SW, Nguyen CQ, Hall DH, Chow KL. mab-7 encodes a novel transmembrane protein that orchestrates sensory ray morphogenesis in C. elegans. Dev Biol. 2007;312:353–366. doi: 10.1016/j.ydbio.2007.09.037. [DOI] [PubMed] [Google Scholar]
- 138.Yu RY, Nguyen CQ, Hall DH, Chow KL. Expression of ram-5 in the structural cell is required for sensory ray morphogenesis in Caenorhabditis elegans male tail. EMBO J. 2000;19:3542–3555. doi: 10.1093/emboj/19.14.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zarkower D. Somatic sex determination. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.84.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Szabo E, Hargitai B, Regos A, Tihanyi B, Barna J, Borsos E, Takacs-Vellai K, Vellai T. TRA-1/GLI controls the expression of the Hox gene lin-39 during C. elegans vulval development. Dev Biol. 2009;330:339–348. doi: 10.1016/j.ydbio.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 141.Shen MM, Hodgkin J. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell. 1988;54:1019–1031. doi: 10.1016/0092-8674(88)90117-1. [DOI] [PubMed] [Google Scholar]
- 142.Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 143.Yi W, Ross JM, Zarkower D. mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- 144.Sulston JE, White JG. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev Biol. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- 145.Waring DA, Wrischnik L, Kenyon C. Cell signals allow the expression of a pre-existent neural pattern in C. elegans. Development. 1992;116:457–466. doi: 10.1242/dev.116.2.457. [DOI] [PubMed] [Google Scholar]
- 146.Austin J, Kenyon C. Cell contact regulates neuroblast formation in the Caenorhabditis elegans lateral epidermis. Development. 1994;120:313–323. doi: 10.1242/dev.120.2.313. [DOI] [PubMed] [Google Scholar]
- 147.Hunter CP, Harris JM, Maloof JN, Kenyon C. Hox gene expression in a single Caenorhabditis elegans cell is regulated by a caudal homolog and intercellular signals that inhibit wnt signaling. Development. 1999;126:805–814. doi: 10.1242/dev.126.4.805. [DOI] [PubMed] [Google Scholar]
- 148.Chitwood BG, Chitwood MB. An introduction to nematology (reprinted) Baltimore, MD: University Park Press; 1974. [Google Scholar]
- 149.Wright KA. The nematode’s cuticle--its surface and the epidermis: function, homology, analogy--a current consensus. J Parasitol. 1987;73:1077–1083. [PubMed] [Google Scholar]
- 150.Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, Assaf N, White JG, Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 151.Podbilewicz B. WormBook. 2006. Cell fusion; pp. 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kontani K, Moskowitz IP, Rothman JH. Repression of cell-cell fusion by components of the C. elegans vacuolar ATPase complex. Dev Cell. 2005;8:787–794. doi: 10.1016/j.devcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 153.Shemer G, Podbilewicz B. Fusomorphogenesis: cell fusion in organ formation. Dev Dyn. 2000;218:30–51. doi: 10.1002/(SICI)1097-0177(200005)218:1<30::AID-DVDY4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 154.Davis MM, Engstrom Y. Immune Response in the Barrier Epithelia: Lessons from the Fruit FlyDrosophila melanogaster. Journal of innate immunity. 2012 doi: 10.1159/000332947. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brabin C, Appleford PJ, Woollard A. The Caenorhabditis elegans GATA factor ELT-1 works through the cell proliferation regulator BRO-1 and the Fusogen EFF-1 to maintain the seam stem-like fate. PLoS Genet. 2011;7:e1002200. doi: 10.1371/journal.pgen.1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hueber SD, Weiller GF, Djordjevic MA, Frickey T. Improving Hox protein classification across the major model organisms. PLoS One. 2010;5:e10820. doi: 10.1371/journal.pone.0010820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee DL, editor. The Biology of Nematodes. Boca Raton, FL: CRC Press; 2002. [Google Scholar]