Abstract
Topoisomerase (topo) II catalyzes topological changes in DNA. Although both human isozymes, topo IIα and β are phosphorylated, site-specific phosphorylation of topo IIβ is poorly characterized. Using LC-MS/MS analysis of topo IIβ, cleaved with trypsin, Arg C or cyanogen bromide (CNBr) plus trypsin, we detected four +80-Da modified sites: tyr656, ser1395, thr1426 and ser1545. Phosphorylation at ser1395, thr1426 and ser1545 was established based on neutral loss of H3PO4 (−98 Da) in the CID spectra and on differences in 2-D-phosphopeptide maps of 32P-labeled wild-type (WT) and S1395A or T1426A/S1545A mutant topo IIβ. However, phosphorylation at tyr656 could not be verified by 2-D-phosphopeptide mapping of 32P-labeled WT and Y656F mutant protein or by Western blotting with phosphotyrosine-specific antibodies. Since the +80-Da modification on tyr656 was observed exclusively during cleavage with CNBr and trypsin, this modification likely represented bromination, which occurred during CNBr cleavage. Re-evaluation of the CID spectra identified +78/+80-Da fragment ions in CID spectra of two peptides containing tyr656 and tyr711, confirming bromination. Interestingly, mutation of only tyr656, but not ser1395, thr1326 or ser1545, decreased topo IIβ activity, suggesting a functional role for tyr656. These results, while identifying an important tyrosine in topo IIβ, underscore the importance of careful interpretation of modifications having the same nominal mass.
Keywords: Bromination, CNBr, Near-isobaric modifications, Phosphorylation, Technology, Topoisomerase IIβ
1 Introduction
DNA topoisomerases catalyze topological transformations of DNA and are involved in several different DNA metabolic processes, including DNA replication, transcription, chromosome condensation and segregation. Among the topoisomerases, the topoisomerase II (topo II) enzymes, which function as dimers, create transient double-stranded breaks in the DNA molecule and transport an intact strand of DNA through the cleaved one [1–3]. In humans, two homologous but physiologically distinct topo II isozymes, topo IIα and topo IIβ are present [2, 4–6]. The topo IIα isozyme, which is expressed in rapidly dividing cells during the S and G2+M phases of the cell cycle, is essential for cell proliferation and is involved in key cell cycle events such as DNA synthesis and chromosome condensation/segregation [7–10]. In contrast, the topo IIβ isozyme, which is expressed in all cells, is primarily involved in cell differentiation and required for ligand-induced gene transcription [10–17].
The functional differences between the structurally and enzymatically similar topo IIα and topo IIβ proteins suggest that these two isozymes are subject to distinct regulatory mechanisms [18]. Although topo IIα and topo IIβ display significant homology in the N-terminal and central catalytic core regions, the homology in the C-terminal region, which is dispensable for catalytic activity, is rather poor [19]. Thus, it has been proposed that the C-terminal domain may be important for regulating the activity of the enzyme [20]. In this regard, the C-terminal domain is subject to different post-translational modifications, including phosphorylation, sumoylation and ubiquitination, which may play important roles in regulating the functional activity of the enzymes [21–24]. Although site-specific phosphorylation of topo IIα has been extensively characterized and the biological function of some of these sites determined [9, 25–30], phosphorylation of the topo IIβ isozyme has been minimally evaluated. Recent studies employing global phosphoproteomic mass spectrometric analysis have reported several phosphorylation sites in the topo IIβ enzyme ([31–39] and www.phosphosite.org). However, characterization of specific phosphorylation sites in the purified enzyme has not been carried out.
Studies of protein phosphorylation, a key post-translational mechanism regulating several cellular processes and signaling cascades, have been greatly advanced due to recent progress in mass spectrometry. This procedure is highly sensitive and allows for high-throughput analysis of phosphorylation sites in proteins. However, the nominal +80-Da modification characteristic of phosphorylation on serine, threonine or tyrosine is also representative of other modifications, viz. sulfonation and bromination. The latter two modifications can occur physiologically or artifactually during processing of the sample [40–43]. Although phosphorylation at serine and threonine residues is characteristically identified by neutral loss of H3PO4 (−98-Da) during collision-induced dissociation in positive ion mode, identification of tyrosine phosphorylation, which is usually present in low abundance, can be challenging. Although tyrosine phosphorylated peptides can be identified by detection of the characteristic pY immonium fragment ion at m/z 216, formation of this ion is dependent on the size and sequence of the peptide and the type of mass spectrometer used in the analysis [44, 45]. Instruments with low m/z cut-offs, like ion trap instruments, may not be capable of detecting the phosphotyrosine-specific immonium ion. Thus, tyrosine phosphorylation is often identified by observing nominal +80-Da shifts in fragment ions of the modified as compared to the unmodified peptide, which is also characteristic of brominated peptides. Therefore, unambiguous identification of tyrosine phosphorylation could be difficult with low-resolution instruments. In this study, we demonstrate that of the four detected +80-Da modified sites, only three were phosphorylated; the fourth modification on tyr656, initially considered to be due to phosphorylation, was subsequently shown to be due to bromination (+78/+80-Da doublet), which occurred during the CNBr cleavage reaction. Another tyrosine residue, tyr711, was also shown to be brominated (+78/+80 Da) during the CNBr cleavage reaction. Interestingly, the reactive tyr656 residue was shown to be functionally important for the catalytic activity of the enzyme. In contrast, the three confirmed phosphorylation sites at ser1395, thr1426 and ser1545 minimally influenced the in vitro catalytic activity of topo IIβ.
2 Materials and methods
2.1 Reagents and cell culture
TPCK-treated trypsin was purchased from Worthington (Lakewood, NJ, USA). Trypsin Gold (mass spectrometry grade) was obtained from Promega (Madison, WI, USA). Endoproteinase Arg C was purchased from Roche (Indianapolis, IN, USA), and CNBr (97%, reagent grade) was obtained from Sigma (St. Louis, MO, USA). Kinetoplast DNA (kDNA) was obtained from Topogen (Columbus, OH, USA). The topo IIβ polyclonal antibody used in this study was raised in rabbits using a C-terminal peptide of topo IIβ (amino acids 1583-1601, SDFPTEPPSLPRTGRARKE) conjugated to keyhole limpet hemocyanin (generous gift of Dr. Ian Hickson, University of Oxford, Oxford, UK) as the immunogen. Cultures of HL-60 cells were maintained in RPMI-1640 supplemented with 10% FBS and 2 mM glutamine at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
2.2 Expression of wild-type (WT) or mutant human topo IIβ in Saccharomyces cerevisiae cells
The S. cerevisiae yeast strain, BJ201 (generous gift of Dr. Anni Andersen, Aarhus University, Aarhus, Denmark) was transformed with the pHT212 plasmid containing WT human topo IIβ (generous gift of Dr. Anni Andersen, Aarhus University, Aarhus, Denmark) or mutant (Y656F, S1395A, T1426A, S1545A or T1426A plus S1545A) topo IIβ (topo IIβ sequence based on isoform 1 comprising of 1621 amino acids – NCBI accession NP_001059) as described earlier [29]. The mutant topo IIβ plasmids were obtained by site-directed mutagenesis of WT topo IIβ using the Quick-change site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The primers used for mutation were Y656F mutation, 5′-CGC ATC TTG TTT AGA TTT GCT GGT CCT GAA GAT GAT GC -3′ and 5′-GC ATC ATC TTC AGG ACC AGC AAA TCT AAA CAA GAT GCG -3′; S1395A mutation 5′-GAG GAA TTG AAA GTT AAA GCA GCT CCC ATA ACA AAT GAT GGG G -3′ and 5′-C CCC ATC ATT TGT TAT GGG AGC TGC TTT AAC TTT CAA TTC CTC -3′; T1426A mutation, 5′-CCA GGC AAA TCA AAA GCC GCT CCA GAA AAA TCT TTG C -3′ and 5′-G CAA AGA TTT TTC TGG AGC GGC TTT TGA TTT GCC TGG -3′; and S1545A mutation, 5′-GCA AAG AAA AGG AAA GCA GCT GGC TCT GAA AAT GAA GG -3′ and 5′-CC TTC ATT TTC AGA GCC AGC TGC TTT CCT TTT CTT TGC C -3′. Site-directed mutageneis was confirmed by PCR and DNA sequence analysis.
2.3 Protein isolation
Total cell lysates of HL-60 cells were prepared in radio-immunoprecipitation assay (RIPA) buffer as described earlier [29]. The lysates were then incubated with topo IIβ antibody and protein A-agarose (Bio-Rad, Hercules, CA, USA) at 4°C overnight. The antigen-antibody complex was washed with RIPA buffer, dissociated in lithium dodecyl sulfate sample buffer (Invitrogen, Carlsbad, CA, USA) and subjected to SDS-PAGE in NuPAGE® 3–8% Tris-acetate gels (Invitrogen). The separated proteins were stained with GelCode Blue (Pierce, Rockford, IL, USA) either in the gel for tryptic digestion or following transfer to 0.45-μm nitrocellulose membranes (Bio-Rad) for CNBr digestion. Recombinant human topo IIβ expressed in yeast cells was purified by Ni2+-nitrilotriacetic acid (Ni-NTA) agarose chromatography essentially as described earlier [29].
2.4 CNBr digestion
The stained topo IIβ band was excised from the nitrocellulose membrane, feathered and incubated with 160 mg/mL CNBr in 70% formic acid at 47°C for 90 min. The released peptides were concentrated by evaporation in a Savant Speed-Vac, washed twice in 150 μL of water and separated by SDS-PAGE on Tris-tricine peptide gels (Invitrogen). In vivo 32P-labeled topo IIβ was used for CNBr digestion to determine distribution of phosphorylated peptides. For this experiment, the separated 32P-labeled peptides were transferred to PVDF membrane and visualized with the Cyclone imager (Perkin Elmer Life Sciences). However, when sequential digestion of topo IIβ with CNBr and trypsin was employed for mass spectrometric analysis, label free-topo IIβ was used for proteolysis. In this case, the CNBr fragments were first separated by gel-electrophoresis and visualized by staining with GelCode Blue. Several of the stained bands were excised and used for in-gel trypsin digestion.
2.5 Proteolytic digestion with trypsin or ArgC
Stained bands of full-length or CNBr fragments of topo IIβ protein cut from the polyacrylamide gel, were cut into smaller pieces, washed and destained in two aliquots of 50% ethanol/5% acetic acid v/v for 1 h each. Following washing in 0.1 M ammonium bicarbonate and dehydration in ACN, topo IIβ was reduced with DTT and alkylated with iodoacetamide (30 min each). The gel pieces were dried in a Speed-Vac after dehydration in ACN and washing in 0.1 M ammonium bicarbonate. Trypsin was driven into the gel pieces by rehydrating with 30 μL of 20 ng/μL trypsin in 50 mM ammonium bicarbonate on ice, for 10 min. Excess trypsin solution was removed and samples were digested overnight at room temperature in 20 μL of 50 mM ammonium bicarbonate. The peptides were extracted in two aliquots of 30 μL 50% ACN/5% formic acid v/v and the extracts were combined, evaporated to approximately 5 μL and reconstituted to a total volume of 20 μL in 1% acetic acid for LC-MS/MS analysis. For 2-D phosphopeptide mapping (Supporting Information Method), the stained topo IIβ band was cut out from a nitrocellulose membrane, the membrane was first blocked with 0.5% PVP in 100 mM acetic acid at 37°C for 1 h and washed extensively with water, prior to overnight digestion with 0.1–0.5 μg of trypsin in 1% ammonium bicarbonate, pH 8.3, at 37°C. The tryptic peptides released in the supernatant solution were transferred to another tube, pooled with ACN (20%) washes of the membrane and evaporated in a Savant Speed-Vac. Following washing with reducing volumes of water, the peptides were solubilized in 10–20 μL of pH 1.9 buffer (2.2% formic acid and 7.8% acetic acid). Digestion of topo IIβ bound to nitrocellulose membrane with Arg C was carried out according to the manufacturer’s protocol following blocking of the membrane in 0.5% PVP.
2.6 LC-MS/MS analysis and data analysis
The tryptic peptides from the in-gel digestion (2 μL) were injected and separated by reversed-phase liquid chromatography in a 10 cm × 50 μm (id) Phenomenex Jupiter 10 μm C18 self-packed capillary column. Elution was achieved using a linear gradient of 2–70% ACN containing 0.05 M acetic acid in 50 min, at a constant flow rate of 0.2 μL/min. The effluent was analyzed using a Finnigan LCQ-Deca ion trap mass spectrometry system equipped with a Protana microelectrospray ion source (Thermo Fisher, San Jose, CA, USA) operated at ~3 kV. This mode of analysis produced about 2000 CID spectra, which were collected in positive ion mode over the mass range of 300–2000 amu. Initially, the digests were analyzed with data-dependent acquisition methods to map as many peptides (unphosphorylated and phosphorylated) as possible from the protein. A 4-step data-dependent acquisition cycle including a first full-scan mass spectrum for determining the molecular masses of co-eluting peptides followed by acquisition of three CID spectra (MS/MS) at 30% collision energy was used. This approach is routinely used in our laboratory and insures the sequence analysis by fragmentation of three most abundant ions collected at the respective time point. To confirm the identified phosphorylation sites, LC-MS analyses were carried out in multiple reaction monitoring (MRM) mode using immunoprecipitated full-length topo II protein purified by gel electrophoresis. MRM on the ion trap was performed by fragmenting specific m/z ratios over the entire course of the LC experiment and taking full mass scans of the product ions. The data were interrogated by plotting chromatograms looking for specific product ions. The chromatographic peaks were then manually interrogated to check whether the spectra matched the peptide of interest. This type of experiment is more sensitive and is used for qualitatively determining if specific peptides are present and can also be used for quantitative analysis. The setup for the MRM experiment was a 5-scan event analysis in which one scan event was a standard MS scan, and the other four were different MRM descriptors directed to various sets of phosphopeptides. To verify the recovery of the peptides from the digestion procedure and the mass spectrometer response, we typically monitored a trypsin autolysis peptide, VATVSLPR at m/z 422 (+2) and an unmodified native peptide NTVEITELPVR at m/z 636 (+2) in topo IIβ. Remaining MRM descriptors were directed to the examined phosphopeptides.
Peak lists were extracted using Thermo Fisher Xcalibur v2.0 subroutine extract.msn.exe. The resultant peak lists were searched with an in-house licensed Mascot search engine version 2.0 (Matrix Sciences, London, UK) against the human reference sequence database NCBI.nih.gov/genomes/H_sapiens/protein, August 24, 2006, 39 391 total entries. The search parameters were as follows: species = human; enzyme = trypsin; number of missed cleavages allowed = 2; fixed modifications = carbamidomethylated cysteine; variable modifications = oxidized methionine, phosphorylated serine, threonine and tyrosine; precursor ion mass tolerance of ±2.0 Da; fragment mass tolerance of ±1.0 Da. Individual spectra were accepted for a Mascot score of minimum 25. Since peptide mixtures in the samples analyzed by MS were not very complex each spectra was subjected to manual inspection. In addition to the Mascot searches, each analysis was also searched directly against the topo IIβ sequence (NCBI accession NP_001059, isoform 1 – 1621 amino acids or Swiss-Prot Q02880, isoform 2 – 1626 amino acids) with the program TurboSequest (Thermo Fisher) using essentially the same search parameters and an Xcorr minimum value of 2.0 for accepting individual spectra. Phosphopeptides were recognized by combinations of characteristic additions of 80 Da multiples (depending on the degree of phosphorylation) to peptide molecular weights and/or by characteristic neutral losses of 98 Da for H3PO4 for phosphoserine and phosphothreonine in the CID spectra. Again, all matching spectra were verified by manual inspection. All the major ions in the spectra were assigned to sequence specific y and b ions (see Supporting Information Table 1 for all identified ions). If all of the major ions could not be identified, we did not consider this a correct match. However, the spectrum was still considered if some low intensity peaks could not be assigned. For peptides of lower abundance, sequence specific peaks with abundance around 5% were also considered.
2.7 Decatenation of kDNA by topo IIβ and determination of covalent topo IIβ-DNA complex
The catalytic activity of WT or mutant topo IIβ was determined by measuring decatenation of kDNA. Varying amounts of WT or mutant topo IIβ protein were incubated at 37°C with 300 ng of kDNA in 20 μL of buffer containing 50 mM Tris, pH 7.9, 88 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 10 mM ATP, 10 mM DTT and 100 μg/mL BSA. The reaction was stopped at different times by the addition of 5 μL of a stop solution containing 5% SDS, 25% Ficoll and 0.5% bromophenol blue. The decatenated and catenated DNA were separated by agarose (1%) gel electrophoresis in Tris acetate EDTA buffer and visualized by ethidium bromide staining. The relative decatenation activity was calculated as the percent ratio of the intensity of decatenated to total (catenated+decatenated) DNA.
Formation of covalent topo IIβ-DNA complex was determined by precipitation of 3′-end labeled 32P-pcDNA3 [29]. 32P-pcDNA3 (0.1 μg) was incubated with 0.1–10 pmol of purified topo IIβ protein (WT or mutant) in the presence of ATP at 37°C for 30 min. The reaction was stopped by addition of SDS and the complex precipitated with KCl. Following two washes, the precipitate was dissolved in water at 65°C and the radioactivity present in the precipitate was determined by liquid scintillation counting.
2.8 Homology modeling of the frame 449–1108 of human topo IIβ
The frame 449–1108 of topo IIβ, comprising tyrosine 656 and 711, was modeled starting from topo II from S. cerevisiae PDB code 3l4j [46]. Refined alignment and interactive threading were performed with ClustalW [47] and SLIDE [48]. Side chain modeling in sequence conserved regions and ab initio generation of variable loops were performed with the Homology module of Insight II from Accelrys. To relieve for steric conflicts, the model was further refined by local simulated annealing in nonconserved loops followed by global simulated annealing and repeated rounds of energy minimization with absolute positional harmonic restraints on backbone atoms found in definite secondary structure states (H, I, E). Model refinement was performed using the Discover module of InsightII using the cvff force field. Finally, the model quality was evaluated with PROCHECK V.3.4.4 [48] to verify if it met the crystallographic standards.
3 Results
3.1 Detection of potential phosphorylation (+80-Da modified) sites in topo IIβ by LC-MS/MS
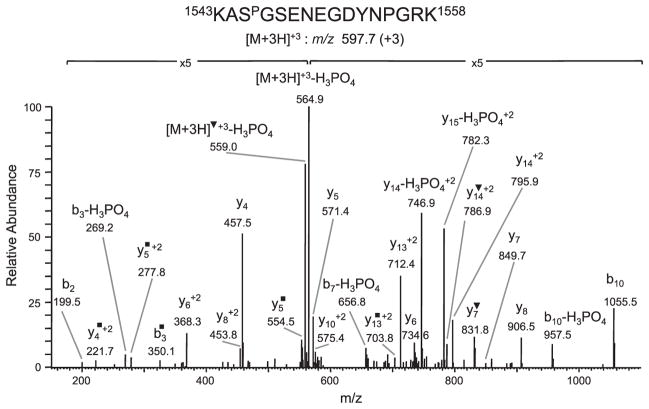
Since topo IIβ is a relatively large protein (Mr = 183 kD) and the tryptic digest is composed of a complex peptide mixture, initial attempts to identify phosphorylation sites by conventional LC-MS/MS of topo IIβ digested with trypsin yielded poor sequence coverage and led to the detection of one +80-Da modified peptide, 1543KASGSENEGDYNPGRK1558, m/z 597.7 (+3). Analysis of the CID tandem mass spectra (Fig. 1) demonstrated an abundant fragment ion corresponding to a neutral loss of H3PO4 from the precursor ion, m/z 564.9 (+3) and from the b3, b7, b10, y14 (+2) and y15 (+2) fragment ions, indicating the presence of a phosphate group. The site of phosphorylation was determined to be ser1545 based on detection of an unmodified y13 (+2) ion, m/z 712.4, but modified y14 (+2), m/z 795.9/y14 (+2)–H2O, m/z 786.9 ions and a modified b3–NH3 ion, m/z 350.1 (Fig. 1).
Figure 1.
Identification of phosphorylated ser1545 in the 1543KASGSENEGDYNPGR1558 tryptic peptide of topo IIβ. CID spectrum of the triply charged peptide at m/z 597.7 (+3) demonstrating neutral loss of H3PO4 from the precursor ion, m/z 564.9 (+3) and from the b3, b7, b10, y14 (+2) and y15 (+2). The site of phosphorylation was determined based on detection of an unmodified y13 (+2) ion, m/z 712.4, but modified y14 (+2), m/z 795.5/y14 (+2)–H2O, m/z 786.9 and modified b3-NH3 ion, m/z 350.1. ▼ denotes loss of H2O; ■ denotes loss of NH3.
To improve sequence coverage so as to detect additional phosphorylation sites, we employed two other proteolytic cleavage procedures. These included digestion with Arg C and a sequential two-step proteolytic cleavage procedure, involving initial cleavage with CNBr followed by digestion with trypsin. The latter procedure was employed since the CNBr cleavage map of topo IIβ (Supporting Information Fig. 1) predicts the generation of several reasonably sized peptides, including the large C-terminal peptide (amino acids 1250–1624, Mr = 41 kDa), wherein majority of the phosphorylation sites are thought to reside. Cleavage of immunoprecipitated topo IIβ with CNBr and separation of the resulting peptides by SDS-PAGE revealed a range of peptides with varying molecular weights (Supporting Information Fig. 2A). The two slowest migrating bands (labeled β1 and β2) that had apparent Mr of 65 and 54 kDa, respectively, corresponded to the C-terminal region (based on N-terminal sequencing and LC-MS/MS analysis of tryptic digests). These two bands incorporated most of the radioactivity when cells were labeled with 32P-inorganic phosphate (Supporting Information Fig. 2B).
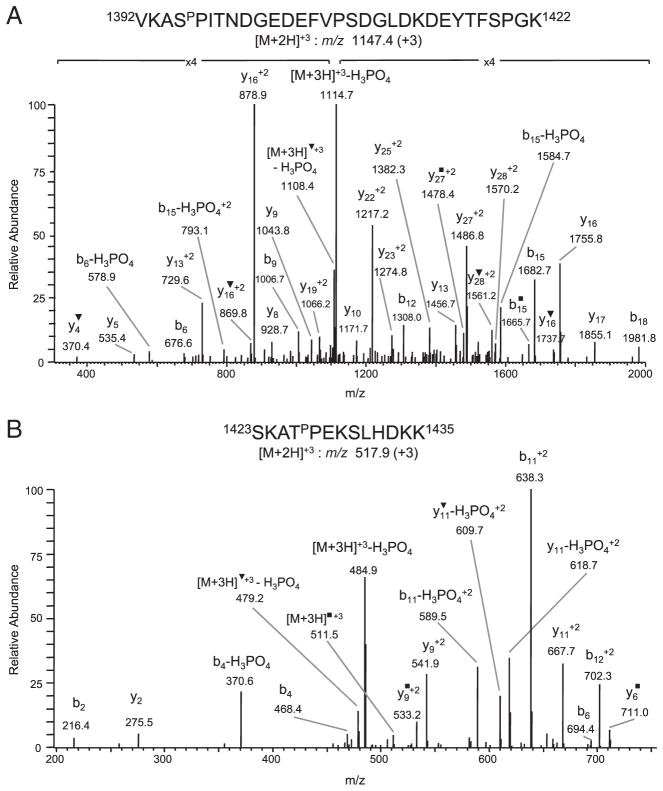
LC-MS/MS analysis of tryptic peptides of the β1 and β2 CNBr bands revealed the presence of two additional +80-Da modified peptides, 1392VKASPITNDGEDEFVPSDGL-DKDEYTFSPGK1422 m/z 1147.4 (+3) and 1423SKATPEKSLHDKK1435, m/z 517.9 (+3), which were detected in both CNBr bands. The CID spectrum of the 1392VKASPITNDGEDEFVPSDGLDKDEYTFSPGK1422peptide contained several ions undergoing neutral loss of H3PO4 [precursor ion (+3), m/z 1114.7, precursor ion–H2O (+3), m/z 1108.4, b6 and b15] (Fig. 2A). The site of phosphorylation was determined to be ser1395 based on the mass difference between the y27 (+2), m/z 1486.8 and y28 (+2), m/z 1570.2 ions, which corresponds to phosphorylated serine, and on the presence of a +80-Da modified b6 ion, m/z 676.6. Absence of modified y ions, with the exception of y28 (+2), indicated that the other four potential phosphorylation sites in this peptide were not phosphorylated. The +80-Da modification in the 1423SKATPEKSLHDKK1435 peptide was also confirmed to be due to phosphorylation, since abundant peaks corresponding to neutral loss of H3PO4 from the precursor ion (+3) m/z 484.9, precursor ion–H2O (+3) m/z 479.2, b4, b11 (+2), y11 (+2) and y11–H2O (+2), were observed (Fig. 2B). The site of phosphorylation was identified to be thr1426, based on the mass difference between the b2, m/z 216.4 and b4, m/z 468.4 ions and between the y9 (+2), m/z 541.9 and y11, m/z 667.7 (+2) ions, which corresponds to alanine-phosphorylated threonine (Fig 2B). The presence of an unmodified b2 ion, m/z 216.4 and modified b6 ion, m/z 694.4 also indicated that thr1426 was phosphorylated. In addition to detecting phosphorylation at ser1395 and thr1426 in the CNBr β1 and β2 bands, phosphorylation of ser1545 was also detected in the β1, but not in the β2, fragment. We were also able to confirm, in subsequent experiments, phosphorylation at ser1395 and thr1426 by LC-MS/MS analyses of tryptic digests of full-length topo IIβ protein and of thr1426 and ser1545 in Arg C digests.
Figure 2.
Identification of phosphorylated ser1395 and thr1426. (A) CID spectrum of triply charged ion 1392VKASPITNDGEDEFVPSDGLDKDEYTFSPGK1422 m/z 1147.4 demonstrating neutral loss of H3PO4 from the precursor ion (+3), m/z 1114.7, precursor ion–H2O (+3), m/z 1108.4, b6 ion and b15 ion. The site of phosphorylation was determined to be ser1395 based on the detection of an unmodified y27 (+2) and modified y28 (+2) ion and a modified b6 ion. (B) CID spectrum of the triply charged peptide 1423SKATPEKSLHDKK1435, m/z 517.9 demonstrating neutral loss of H3PO4 for the precursor ion (+3), m/z 484.9, precursor ion–H2O (+3) m/z 479.2, b4, b11 (+2), b12 (+2), y11 (+2) and y11–H2O (+2). The site of phosphorylation was identified to be thr1426, based on detection of an unmodified b2 ion and modified b4 ion, and an unmodified y9 (+2) ion but modified y11 (+2) ion. ▼ denotes loss of H2O; ■ denotes loss of NH3.
Phosphorylation at ser1395, thr1426 and ser1545 was confirmed by comparing 2-D-phosphopeptide maps of in vivo 32P-labeled WT topo IIβ with that of mutant topo IIβ, in which the phosphoserine or threonine residue was mutated to alanine (Supporting Information Fig. 3A and B). Comparison of the 2-D-phosphopeptide of 32P-labeled WT with S1395A mutant topo IIβ demonstrated the absence of a single phosphorylated peptide in the mutant protein (Supporting Information Fig. 3A) and comparison of the 2-D-phosphopeptide maps of WT with T1426A plus S1545A mutant topo IIβ demonstrated the absence of two phosphorylated peptides in the 2-D map of the double mutant protein (Supporting Information Fig. 3B). The phosphorylated amino acid in the phosphopeptide that was deficient in the 2-D phosphopeptide map of S1395A mutant topo IIβ was identified to be serine. The phosphorylated amino acids in the two peptides that were deficient in the 2-D phosphopeptide maps of T1395A/S1545A double mutant topo IIβ protein were identified to be threonine (indicated as thr1426 in the map of the WT protein) and serine (indicated as ser1545 in the map of the WT protein).
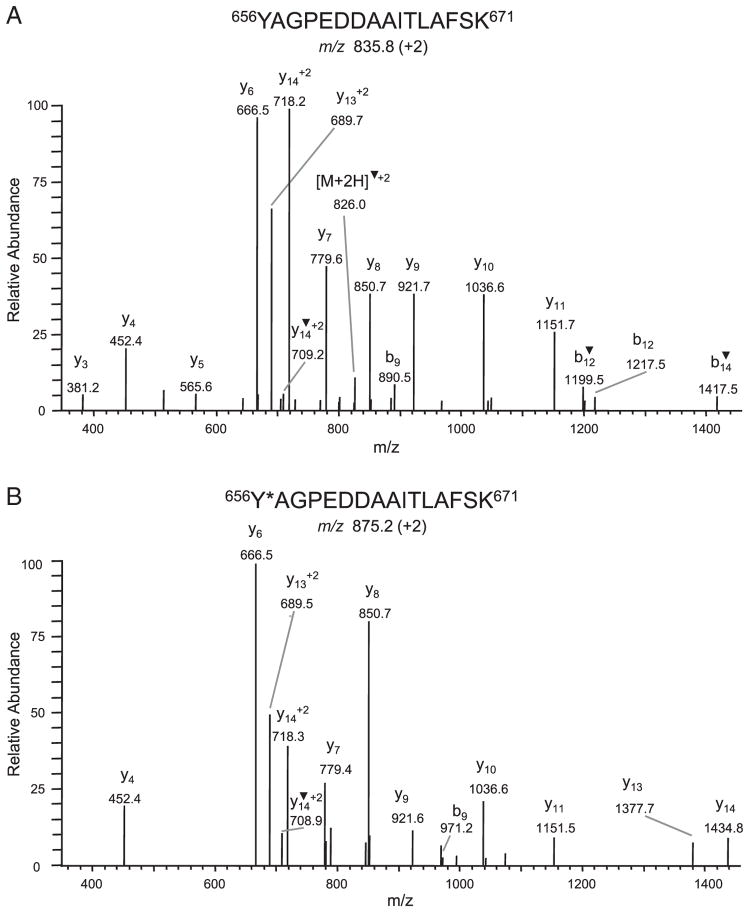
In addition to the three serine or threonine phosphorylation sites, a fourth +80-Da modified peptide was detected by LC-MS/MS analysis in the tryptic digest of a CNBr band migrating at ~15 kDa (labeled β5 in Supporting Information Fig. 2A). This peptide, 656YAGPEDDAAITLAFSK671, was detected in both the unmodified m/z 835.8 (+2) and modified m/z 875.2 (+2) form (Fig. 3), with the modified form being present in low abundance. Of the three possible sites, tyr656, thr666 and ser670, that could have a +80-Da modification, tyr656 was identified as the modified site based on absence of neutral loss of H3PO4 (−98), no change in the mass of y ions in the CID spectrum of the modified as compared to the unmodified peptide ion and a shift of +80 Da for the b9 ion, m/z 971.2 (present in low abundance). Unlike detection of the +80-Da modification at ser1395, thr1426 and ser1545 in tryptic digests of either full-length topo IIβ or CNBr fragments of topoIIβ, we were unable to detect the +80-modification on tyr656 in MRM analysis of tryptic digests of full-length topo IIβ. Further, comparison of 2-D phosphopeptide maps of 32P-labeled WT topo IIβ with that of mutant topo IIβ, in which tyrosine 656 was mutated to phenylalanine (Y656F), did not reveal any significant difference (Supporting Information Fig. 3C). This result suggested that the +80-Da modification on tyr656 may not have resulted from in vivo phosphorylation of tyr656.
Figure 3.
Identification of a +80-Da modification on tyr656. CID spectra of the doubly charged peptide 656YAGPEDDAAITLAFSK671 detected in both the unmodified m/z 835.8 (A) and modified m/z 875.2 (B) forms. Tyr656 was identified as the modified residue based on a shift of +80 Da for the b9 ion in the spectrum of the modified peptide. ▼ denotes loss of H2O. *corresponds to a modification.
3.2 The +80-Da modification at tyr656 observed in LC-MS/MS is not due to in vivo phosphorylation but due to in vitro oxidative bromination
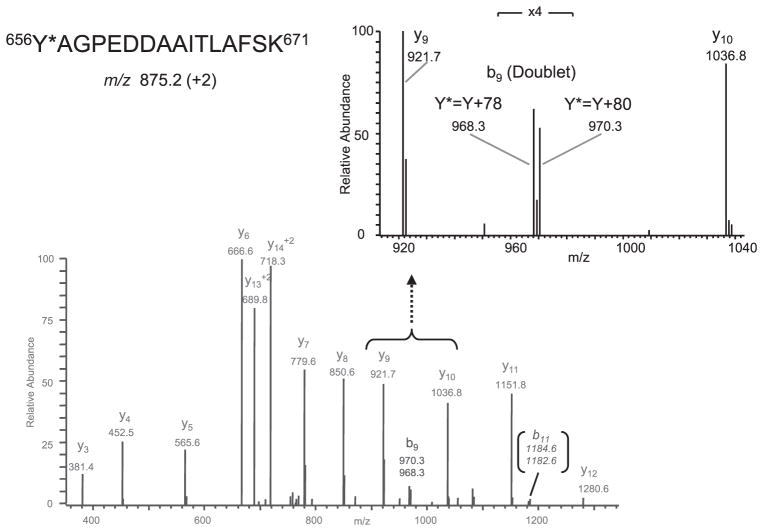
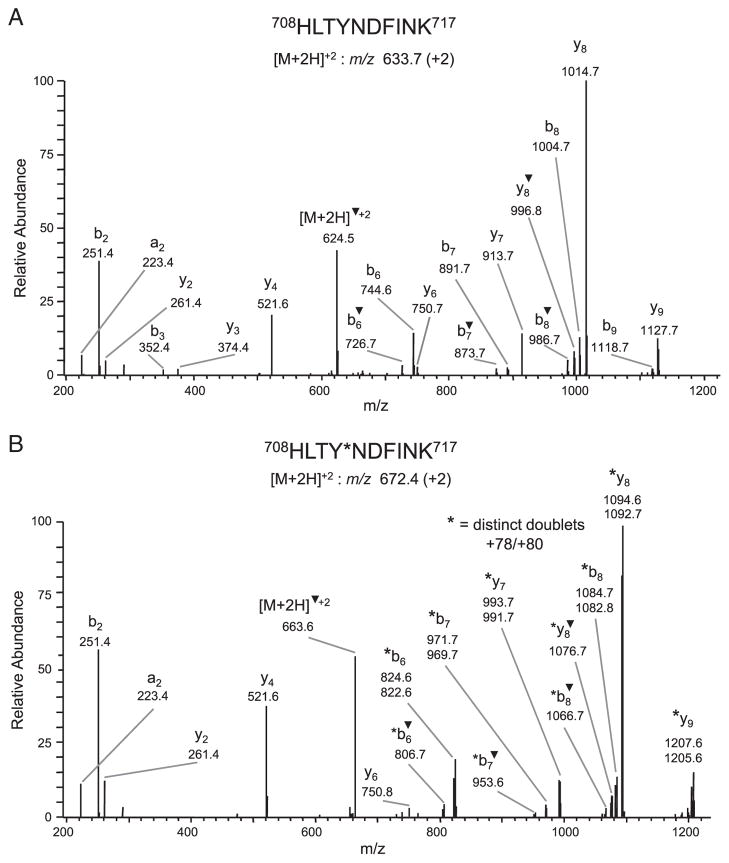
Since the 2-D phosphopeptide maps of WT and Y656F mutant protein were similar and the +80-Da modification on tyr656 was not observed by LC-MS/MS analysis following tryptic digestion of the full-length protein, we performed Western blot analysis of immunoprecipitated WT and mutant Y656F topo IIβ as well as CNBr-digested topo IIβ with a phosphotyrosine-specific antibody to determine whether topo IIβ was phosphorylated at tyrosine. However, no tyrosine phosphorylation was detected in full-length or CNBr-digested topo IIβ (data not shown), further suggesting that the modification at tyr656 was likely not due to phosphorylation. Because a nominal mass increase of 80 Da could represent three near-isobaric modifications, viz., phosphorylation, sulfonation or bromination, we re-examined the CID spectra to consider alternative possibilities, i.e. sulfonation or bromination. Of these two possibilities, we first considered the latter one, primarily because modification at tyr656 was observed only when topo IIβ was cleaved with CNBr and trypsin. In our studies, bromination would be expected to result as a side-reaction of CNBr cleavage, rather than due to an in vivo oxidative reaction. Since elemental bromine comprises two stable isotopes (atomic mass of 79 and 81 Da) that are present in equimolar proportions, the CID spectrum for a brominated tyr656 peptide would comprise a doublet corresponding to +78- and +80-Da mass shifts. Indeed, re-analysis of the fragmentation spectra revealed the presence of +78/+80-Da doublets in the CID spectrum of two modified peptides, 656YAGPEDDAAITLAFSK671 m/z 875.2 (+2) (Fig. 4) and 708HLTYNDFINK717 m/z 672.4 (+2) (Fig. 5B), respectively. These modified peptides along with the unmodified forms, 656YAGPEDDAAITLAFSK671 m/z 875.2 (+2) (data not shown) and 708HLTYNDFINK717 m/z 633.7 (+2), (Fig. 5A) were detected in the tryptic digest of the CNBr band β6, located adjacent to β5 band (Supporting Information Fig. 2A) in which the +80-Da modification on tyr656 was first identified. Although the fragmentation spectrum of the 656YAGPEDDAAITLAFSK671 peptide contained only one distinct +78/+80-Da doublet corresponding to modified b9 ions and a low intensity (<5%) doublet corresponding to modified b11 ion (Fig. 4), several distinct +78/+80-Da modified ions (b6, b6–H2O, b7, b7–H2O, b8, b8–H2O, y7, y8, y8–H2O and y9) were observed in the CID spectrum of the 708HLTYNDFINK717 peptide. The site of modification in the 708HLTYNDFINK717 peptide was determined to be tyr711 based on detection of unmodified y6 ion, m/z 750.8, but +78/+80-Da modified y7 ions, m/z 991.7/993.7, respectively. This result strongly suggested that tyr656, as well as tyr711, were brominated during cleavage with CNBr.
Figure 4.
Identification of bromination at tyr656 in the modified doubly charged peptide, 656Y*AGPEDDAAITLAFSK671, m/z 875.2 (+2). CID spectrum of the modified peptide displaying two +78/+80-Da doublets corresponding to a modified b9 ion (also see inset showing the region of the b9 ion scaled-up 4 ×) and b11 ion (low intensity), which are characteristic of bromination.* corresponds to a modification.
Figure 5.
Identification of bromination at tyr711. CID spectra of the doubly charged peptide 708HLTYNDFINK717 detected in both the unmodified m/z 633.7 (A) and modified m/z 672.4 (B) forms. The site of bromination was identified as tyr711 based on detection of unmodified y4, m/z 521.6 and y6 m/z 750.8 ions, but +78/+80-Da modified y7 ions, m/z 991.7/993.7, respectively, in the CID spectrum of the modified peptide. ▼ denotes loss of H2O. *corresponds to a modification.
3.3 Phosphorylation at ser1395, thr1426 and ser1545 does not significantly affect the activity of topo IIβ, whereas mutation of tyr656 leads to a significant decrease in the catalytic activity of topo IIβ
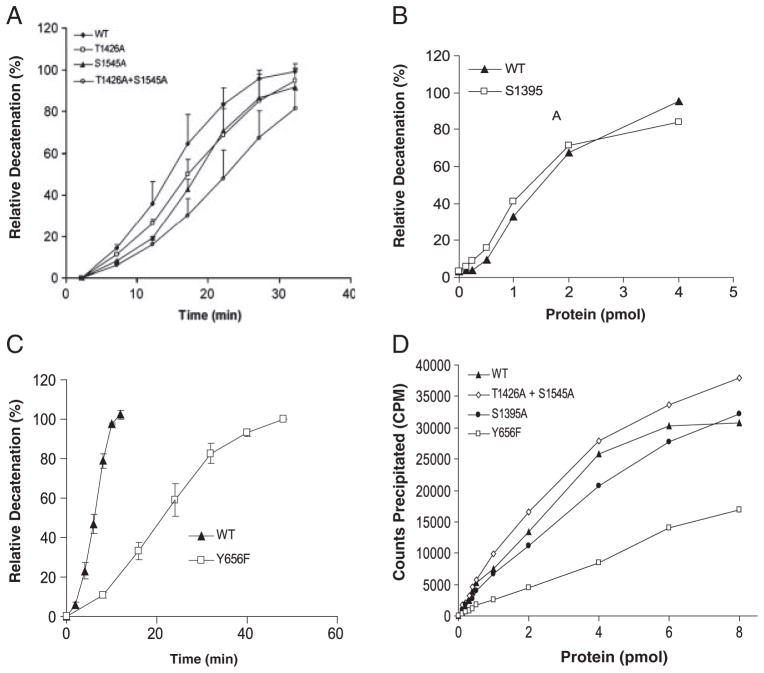
To determine whether phosphorylation at ser1395, thr1426 and ser1545 affects enzyme activity, we compared the ability of WT topo IIβ to decatenate kDNA and to form protein–DNA cleavable complex with that of mutant topo IIβ proteins, S1395A, T1426A, S1545A and T1426A plus S1545A. No significant difference in the decatenation activity of WT topo IIβ and mutant topo IIβ proteins, S1395A, T1426A and S1545A, mutated at a single amino acid residue, was observed (Fig. 6A, B). Similarly, no difference was observed for the amount of topo IIβ DNA cleavable complex formed by the WT, S1395A or T1426A plus S1545A mutant topo IIβ (Fig. 6D). However, mutation at both thr1426 and ser1545 led to a modest decrease (twofold) in the decatenation activity (Fig. 6A).
Figure 6.
Comparison of the decatenation activity and DNA cleavable complex formation of WT and mutant forms of topo IIβ protein. Recombinant human topo IIβ protein expressed in yeast cells was purified by Ni2+-nitrilotriacetic acid agarose chromatography and incubated with 300 ng kDNA (A–C) at 37°C. An aliquot of the reaction mixture was electrophoresed on 1% agarose gel to separate the decatenated DNA (mini circles) from the kDNA substrate. Percent decatenation was calculated as the relative intensity of the decatenated DNA to that of the substrate DNA plus decatenated DNA. (A) Time course of percent relative decatenation/ng of WT, T1426A, S1545A or T1426A plus S1545A topo IIβ. (B) Percent relative decatenation catalyzed by varying concentrations of WT or S1395A mutant topo IIβ protein in 30 min. (C) Time course of percent relative decatenation/ng of WT or tyr656F mutant topo IIβ protein. (D) Varying concentrations of purified WT, Y656F, S1395A or T1426A plus S1550A topo IIβ protein were incubated with 32P-labeled pcDNA3 as described in Section 2. The protein-DNA complex was precipitated following addition of KCl and the radioactivity in the precipitate determined by liquid scintillation counting.
Since, detection of a +80-Da modification on tyr656 was initially interpreted by us to result from in vivo phosphorylation at this site; we mutated this tyrosine residue to simultaneously confirm in vivo phosphorylation and to determine whether phosphorylation, if present, was essential for regulating enzyme activity. Although we were unable to confirm in vivo phosphorylation at tyr656, the decatention activity of the Y656F mutant protein was significantly less than that of WT topo IIβ (Fig. 6C), indicating that this amino acid was important for enzyme activity. The rate of decatenation, based on the initial slope of the reaction curve, by the Y656F mutant protein was five to six-fold less as compared to the rate of decatenation by WT topo IIβ (Fig. 6C). Further, formation of the topo IIβ DNA cleavable complex was significantly less in the Y656F mutant protein as compared to WT topo IIβ (Fig. 6D). Thus, detection of an artifactual modification on tyr656 led to the identification of an amino acid residue that was important for the catalytic activity of the topo IIβ enzyme.
3.4 Homology modeling of the frame 449–1108 of human topo IIβ comprising tyr656 and tyr711
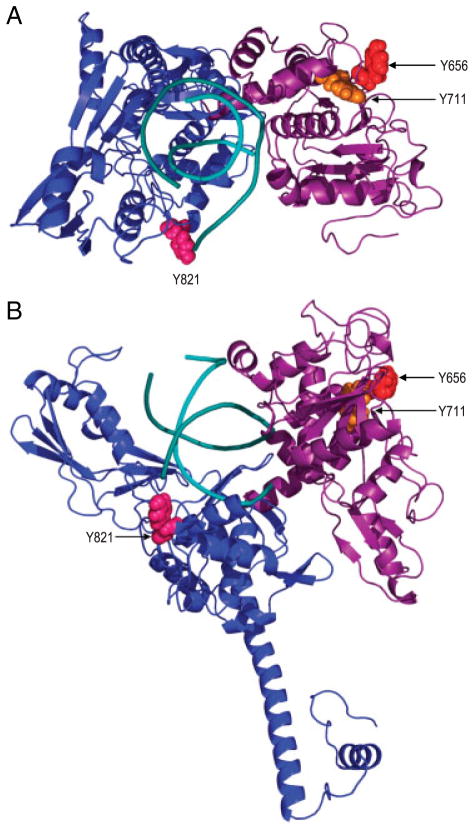
Since mutation of tyr656 significantly altered the catalytic activity of topo IIβ and preliminary studies of mutation of tyr711 revealed that this residue may also be important for topo IIβ function (data not shown), we modeled the region of topo IIβ (amino acids 449–1108) containing these two reactive residues to determine whether the structural location of these two amino acids could potentially influence functioning of the topo IIβ enzyme. Modeling was based on the crystal structure of the S. cerevisiae topo II enzyme that was co-crystallized with a 15-bp polydeoxyribonucleotide. The crystal structure was determined at 2.4 Å resolution with no missing coordinates over a frame corresponding to amino acids 449–1108 of human topo IIβ [45]. The level of homology (over 48% identity and 75% similarity) between human topo IIβ and the yeast homologue made comparative modeling highly reliable, in close limit to the crystal structure resolution, especially in the sequence conserved regions of the structure which comprise also the amino acids of interest. Based on the derived model (Fig. 7), it can be seen that tyr656 and tyr711, which are separated in sequence by over 50 amino acids, are in close proximity to each other (dCα-Cα ~ 7.6Å), within the B′ domain of topo IIβ. Although tyr711 is buried, tyr656 is highly exposed on the surface of B′ in a region opposite to the groove hosting the DNA. Functional and coarse grained models of topo II [50] indicate that this region is within or very close to the dimerization interface, which suggests that these amino acids may be critical for topo IIβ dimer formation. Thus, mutation or possible oxidation of these residues might interfere directly or indirectly with the formation of topo IIβ dimer.
Figure 7.
Spatial location of tyr654 and tyr711 with respect to the active site tyrosine, tyr821. The A′ domain is shown in blue, the B′ domain is shown in purple while the DNA is shown in cyan.
4 Discussion
In this study, we identified four +80-Da modified sites, tyr656, ser1395, thr1426 and ser1545 in LC-MS/MS analysis of in vivo phosphorylation sites in the human topo IIβ enzyme. Three of the +80-Da modified sites at ser1395, thr1426 and ser1545 were confirmed to be phosphorylated in vivo. However, the modification at tyr656, which was initially thought to be due to in vivo phosphorylation, was subsequently shown to result from artifactual bromination that occurred during the CNBr cleavage reaction. Bromination was also shown to occur on another tyrosine residue, tyr711. Although phosphorylation at ser1395, thr1426 and ser1545 did not significantly influence the in vitro decatenation activity of the enzyme, the reactive tyr656 residue was shown to be functionally involved in topo IIβ enzyme activity. These results while enhancing our knowledge of the biochemical characteristics of the topo IIβ enzyme, underscore the importance of accurately identifying modifications of same nominal mass, especially when using low-mass accuracy instruments.
Of the three phosphorylation sites, ser1395, thr1426 and ser1545, identified in this study, phosphorylation of ser1395 and ser1545 has been reported previously in experiments employing global proteomic LC-MS/MS analysis of phosphorylated peptides [[31–39] and www.phosphosite.org]. However, this is the first report identifying phosphorylation at thr1426. All of the identified phosphorylation sites were found to reside in the C-terminal region of topo IIβ, which is consistent with previous studies demonstrating predominant phosphorylation of topo IIβ in the C-terminal region and our observation that most of the radioactivity is incorporated in the C-terminal CNBr peptides of in vivo 32P-labeled topo IIβ. Since most of the phosphorylation sites in the topo IIα isozyme are also located in the C-terminal domain, phosphorylation within this region may represent an important mechanism by which the two isozymes are regulated. Indeed, phosphorylation of some of the sites in the C-terminal region of topo IIα has been shown to be functionally important [9]. However, in this study, we were unable to demonstrate a significant effect of phosphorylation at the three individual sites in the C-terminal domain on the in vitro catalytic activity of topo IIβ. Although phosphorylation at individual sites did not affect the in vitro decatenation activity of topo IIβ, phosphorylation at both thr1426 and ser1545 modestly enhanced (two-fold) the decatenation activity. Thus, it is possible that phosphorylation at multiple sites may be important for regulating the enzymatic activity of topo IIβ. Alternatively, since topo IIβ has been shown to associate with protein complexes involved in hormone- or ligand-induced gene transcription [16, 17], phosphorylation at different sites may regulate the ability of topo IIβ to associate with different DNA binding-protein complexes and thereby influence the physiologic activity of the enzyme.
Tandem mass spectrometry has become the method of choice for identifying in vivo phosphorylation of proteins. Although a nominal mass increase of 80 Da is associated with protein phosphorylation, this shift is also observed for other modifications, viz. sulfonation and bromination. Although it is possible to distinguish phosphorylation from the other two modifications, based on accurate mass measurements (<5ppm) with high-resolution mass spectrometers or by distinct characteristics of the CID spectra, instrument limitations could result in misinterpretation of the +80-Da modifications as phosphorylation, especially when the modified peptide is present in low abundance. This problem may be particularly important for modifications on tyrosine residues, since phosphorylated tyrosine is present in low abundance and fragmentation in positive ion mode of phosphotyrosine containing peptides does not lead to the characteristic neutral loss of H3PO4 (−98 Da). Although, a characteristic feature of the fragmentation pattern of tyrosine phosphorylated peptides is formation of the immonium ion of pY at m/z 216, efficiency of generation of this species is dependent on collisional offset and on the size and sequence of the peptide [44]. Further, because of the low m/z value of this species, it may not be possible to detect it with instruments that have low mass cut-offs, such as the ion trap instrument used in this study. Typically, tyrosine phosphorylation is identified by detecting +80-Da shifts in fragment y or b ions in the CID spectrum of the modified peptide as compared to that of the unmodified ion. This pattern is distinct from that observed for tyrosine sulfonated peptides, since the CID spectrum of sulfonated peptides do not contain +80-Da fragment y or b ions due to rapid elimination of the labile SO3 group, which occurs prior to fragmentation of the peptide backbone. However, the CID spectrum of brominated peptides also exhibit +80-Da mass shifts in fragment ions and in addition displays +78-Da shifts due to the presence of two stable isotopes of atomic mass 79 and 81 Da for elemental bromine. Nevertheless, if the brominated sequence specific ions are present in low abundance, it may not always be possible to detect the characteristic doublet (separated by + 2 Da) in the CID spectrum. This could lead to misidentification of bromination for phosphorylation. This scenario was indeed encountered by us in this study when initial data-dependent analysis followed by TurboSequest searches for +80-Da shifts in fragment ions of tryptic peptides of CNBr-digested topo IIβ led to the detection of a fragment ion at m/z 971.2, which was assigned as a +80-Da modified b9 ion (theoretical m/z value 970.3). Since low intensity signals detected by ion trap instruments are often observed at higher m/z values and we were not searching for +78-Da shifts, the low intensity fragment ion at m/z 968.1 (b9+78), which was separated from the fragment ion at m/z 971.1 by +3 Da was not considered at that time to represent the characteristic bromination doublet. Only upon subsequent re-analysis of the CID spectra, which was carried out when we were unable to conclusively demonstrate phosphorylation on tyr656 by other independent methods, were we able to detect a distinct doublet in another CID spectrum of the tyr656 containing peptide and confer that the modification on tyrosine was due to bromination. Interestingly, this re-analysis also led to the detection of +78/+80-Da shifts in several fragment ions of another peptide 708HLTYNDFINK717, in which tyr711 was identified as the modified residue. These spectra, which were not analyzed in our initial data-dependent analysis, indicated that CNBr cleavage of topo IIβ led to artifactual bromination on, at least, two tyrosine residues.
The rationale for using CNBr as a proteolytic cleavage reagent in this study was to improve sequence coverage of the large topo IIβ protein for detecting phosphorylated amino acids. However, this procedure led to the bromination of at least two tyrosine residues, tyr656 and tyr711, which enabled us to identify functionally important residues in topo IIβ. Homology modeling of the region containing these reactive tyrosine residues revealed that tyr656 and tyr711, which are separated in sequence by over 50 amino, are located in close proximity to each other and are within, or very close to the dimerization interface. This suggests that these amino acids may be critical for topo IIβ dimer formation.
Supplementary Material
Acknowledgments
This work was supported by United States Public Health Service Grant RO1 CA 117928. M. A. M. and A.-J. P. were funded by CNCSIS ID-249 168/2007 and POSDRU/89/1.5/S/60746.
Abbreviations
- kDNA
kinetoplast DNA
- MRM
multiple reaction monitoring
- RIPA
radioimmunoprecipitation assay
- WT
wild-type
Footnotes
The authors have declared no conflict of interest.
References
- 1.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Kellner U, Sehested M, Jensen PB, Gieseler F, Rudolph P. Culprit and victim–DNA topoisomerase II. Lancet Oncol. 2002;3:235–243. doi: 10.1016/s1470-2045(02)00715-5. [DOI] [PubMed] [Google Scholar]
- 4.Tan KB, Dorman TE, Falls KM, Chung TD, et al. Topoisomerase IIα and topoisomerase IIβ genes: characterization and mapping to human chromosomes 17 and 3, respectively. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 5.Lang AJ, Mirski SE, Cummings HJ, Yu Q, et al. Structural organization of the human TOP2A and TOP2B genes. Gene. 1998;221:255–266. doi: 10.1016/s0378-1119(98)00468-5. [DOI] [PubMed] [Google Scholar]
- 6.Drake FH, Hofmann GA, Bartus HF, Mattern MR, et al. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 7.Heck MM, Earnshaw WC. Topoisomerase II: A specific marker for cell proliferation. J Cell Biol. 1986;103:2569–2581. doi: 10.1083/jcb.103.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 9.Li H, Wang Y, Liu X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J Biol Chem. 2008;283:6209–6221. doi: 10.1074/jbc.M709007200. [DOI] [PubMed] [Google Scholar]
- 10.Turley H, Comley M, Houlbrook S, Nozaki N, et al. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br J Cancer. 1997;75:1340–1346. doi: 10.1038/bjc.1997.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim Biophys Acta. 1992;1132:43–48. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIβ and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 13.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIβ. Proc Natl Acad Sci USA. 2003;100:7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chikamori K, Hill JE, Grabowski DR, Zarkhin E, et al. Downregulation of topoisomerase IIβ in myeloid leukemia cell lines leads to activation of apoptosis following all-trans retinoic acid-induced differentiation/growth arrest. Leukemia. 2006;20:1809–1818. doi: 10.1038/sj.leu.2404351. [DOI] [PubMed] [Google Scholar]
- 15.Lyu YL, Lin CP, Azarova AM, Cai L, et al. Role of topoisomerase IIβ in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 17.Perillo B, Ombra MN, Bertoni A, Cuozzo C, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 18.Isaacs RJ, Davies SL, Sandri MI, Redwood C, et al. Physiological regulation of eukaryotic topoisomerase II. Biochim Biophys Acta. 1998;1400:121–137. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 19.Austin CA, Sng JH, Patel S, Fisher LM. Novel HeLa topoisomerase II is the IIβ isoform: complete coding sequence and homology with other type II topoisomerases. Biochim Biophys Acta. 1993;1172:283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 20.Linka RM, Porter AC, Volkov A, Mielke C, et al. C-Terminal regions of topoisomerase IIα and IIβ determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007;35:3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heck MM, Hittelman WN, Earnshaw WC. In vivo phosphorylation of the 170-kDa form of eukaryotic DNA topoisomerase II. Cell cycle analysis. J Biol Chem. 1989;264:15161–15164. [PubMed] [Google Scholar]
- 22.Taagepera S, Rao PN, Drake FH, Gorbsky GJ. DNA topoisomerase IIα is the major chromosome protein recognized by the mitotic phosphoprotein antibody bMPM-2. Proc Natl Acad Sci USA. 1993;90:8407–8411. doi: 10.1073/pnas.90.18.8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le MT, Bachant J. SUMO modification of DNA topoisomerase II: Trying to get a CENse of it all. DNA Repair. 2009;8:557–568. doi: 10.1016/j.dnarep.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 25.Wells NJ, Addison CM, Fry AM, Ganapathi R, Hickson ID. Serine 1524 is a major site of phosphorylation on human topoiosmerase IIα protein in vivo and is a substrate for casein kinase II in vitro. J Biol Chem. 1994;269:29746–29751. [PubMed] [Google Scholar]
- 26.Ishida R, Iwai M, Marsh KL, Austin CA, et al. Threonine 1342 in human topoisomerase IIα is phosphorylated throughout the cell cycle. J Biol Chem. 1996;271:30077–30082. doi: 10.1074/jbc.271.47.30077. [DOI] [PubMed] [Google Scholar]
- 27.Escargueil AE, Plisov SY, Filhol O, Cochet C, Larsen AK. Mitotic phosphorylation of DNA topoisomerase IIα by protein kinase CK2 creates the MPM-2 phosphoepitope on ser-1469. J Biol Chem. 2000;275:34710–34718. doi: 10.1074/jbc.M005179200. [DOI] [PubMed] [Google Scholar]
- 28.Iida M, Matsuda M, Komatani H. Plk3 phosphorylates topoisomerase IItopo IIα at Thr1342; a site that is not recognized by Plk1. Biochem J. 2008;411:27–32. doi: 10.1042/BJ20071394. [DOI] [PubMed] [Google Scholar]
- 29.Chikamori K, Grabowski DR, Kinter M, Willard BB, et al. Phosphorylation of serine 1106 in the catalytic domain of topoisomerase IIα regulates enzymatic activity and drug sensitivity. J Biol Chem. 2003;278:12696–12702. doi: 10.1074/jbc.M300837200. [DOI] [PubMed] [Google Scholar]
- 30.Grozav AG, Chikamori K, Kozuki T, Grabowski DR, et al. Casein kinase I δ/ε phosphorylates topoisomerase IIα at serine 1106 and modulates DNA cleavage activity. Nucleic Acids Res. 2009;37:382–392. doi: 10.1093/nar/gkn934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen JV, Blagoev B, Gnad F, Macek B, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Nousiainen M, Sillje HHW, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giorgianni F, Zhao Y, Desiderio DM, Beranova-Giorgianni S. Toward a global characterization of the phosphoproteome in prostate cancer cells: identification of phosphoproteins in the LNCaP cell line. Electrophoresis. 2007;28:2027–2034. doi: 10.1002/elps.200600782. [DOI] [PubMed] [Google Scholar]
- 35.Yu LR, Zhu Z, Chan KC, Issaq HJ, et al. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J Proteome Res. 2007;6:4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 37.Cantin GT, Yi W, Lu B, Park SK, et al. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res. 2008;7:1346–1351. doi: 10.1021/pr0705441. [DOI] [PubMed] [Google Scholar]
- 38.Dephoure N, Zhou C, Villén J, Beausoleil SA, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen RQ, Yang QK, Lu BW, Yi W, et al. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 2009;69:2663–2668. doi: 10.1158/0008-5472.CAN-08-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medzihradszky KF, Darula Z, Perlson E, Fainzilber M, et al. O-Sulfonation of serine and threonine: mass spectrometric detection and characterization of a new post-translational modification in diverse proteins throughout the eukaryotes. Mol Cell Proteomics. 2004;3:429–443. doi: 10.1074/mcp.M300140-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Wu W, Chen Y, d’Avignon A, Hazen SL. 3-Bromotyrosine and 3, 5-dibromotyrosine are major products of protein oxidation by eosinophil peroxidase: potential markers for eosinophil-dependent tissue injury in vivo. Biochemistry. 1999;38:3538–3548. doi: 10.1021/bi982401l. [DOI] [PubMed] [Google Scholar]
- 42.Gharib M, Marcantonio M, Lehmann SG, Courcelles M, et al. Artifactual sulfation of silver-stained proteins: implications for the assignment of phosphorylation and sulfation sites. Mol Cell Proteomics. 2009;8:506–518. doi: 10.1074/mcp.M800327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahali V, Gueguen J. Chemical cleavage of bovine β-lactoglobulin by BNPS-Skatole for preparative purposes: comparative study of hydrolytic procedures peptide characterization. J Protein Chem. 1999;18:1–11. doi: 10.1023/a:1020635130077. [DOI] [PubMed] [Google Scholar]
- 44.Salek M, Alonso A, Pipkorn R, Lehmann WD. Analysis of protein tyrosine phosphorylation by nanoelectrospray ionization high-resolution tandem mass spectrometry and tyrosine-targeted product ion scanning. Anal Chem. 2003;75:2724–2729. doi: 10.1021/ac020657y. [DOI] [PubMed] [Google Scholar]
- 45.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanganu A, Micluta MA, Popa BA, Spiridon LN, Tacutu R. SLIDE: An interactive threading refinement tool for homology modeling. Rom J Biochem. 1009;46:123–127. [Google Scholar]
- 49.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK:a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 50.Christmann-Franck S, Bertrand HO, Goupil-Lamy A, der Garabedian PA, et al. Structure-based virtual screening: an application to human topoisomerase II alpha. J Med Chem. 2004;47:6840–6853. doi: 10.1021/jm049745w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.