Abstract
Disorders characterized by expansion of an unstable nucleotide repeat account for a number of inherited neurological diseases. Here, we review examples of unstable repeat disorders that nicely illustrate the three of the major pathogenic mechanisms associated with these diseases: loss-of-function typically by disrupting transcription of the mutated gene, RNA toxic gain-of-function, and protein toxic gain-of-function. In addition to providing insight into the mechanisms underlying these devastating neurological disorders, the study of these unstable microsatellite repeat disorders has provided insight into very basic aspects of neuroscience.
Introduction
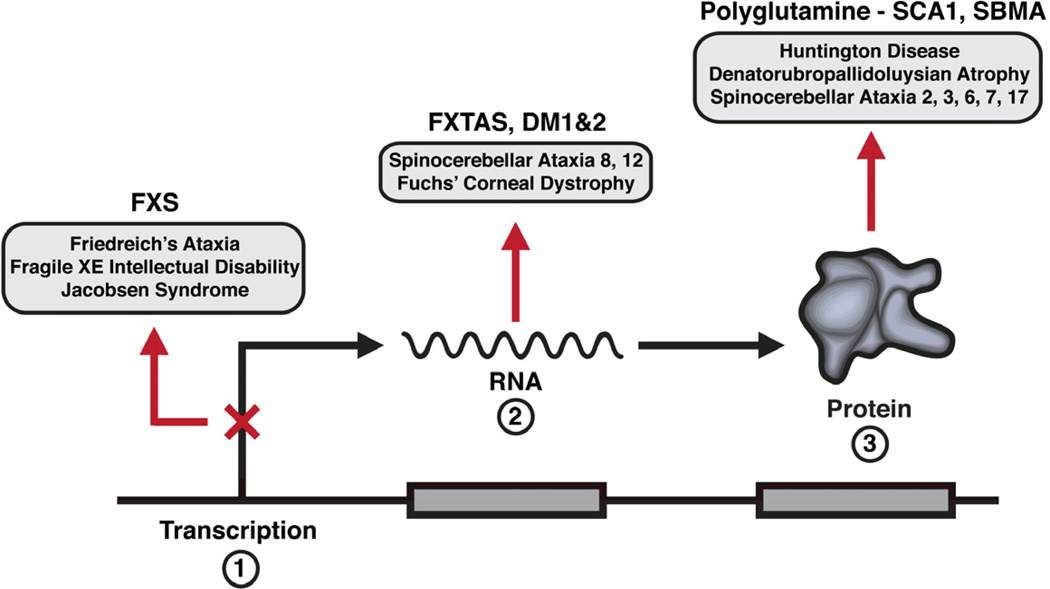
Some twenty years ago expansion of unstable nucleotide (microsatellite) repeats, notably trinucleotide repeats, were identified as a novel mutational mechanism underlying certain human diseases. Over the years other disease-associated expandable microsatellite repeats were discovered including additional trinucleotide repeats as well as unstable tetra-, penta-, hexanucleotide and longer repeats (Mirkin, 2007). Remarkably many of the unstable DNA microsatellite repeat disorders manifest with neurological and neuromuscular symptoms. These include fragile X syndrome, the most common inherited form of intellectual disability, myotonic dystrophy a common genetic muscular dystrophy, Huntington’s disease, a group of dominantly inherited ataxias, and most recently an unstable hexanucleotide repeat in the C9ORF72 gene as a frequent cause of frontotemporal dementia/amyotrophic lateral sclerosis (DeJesus-Hernandez et al., 2011; Renton et al., 2011). While the disorders highlighted in this review reflect the experiences and interests of the authors, they also illustrate three key means by which expansion of a unstable microsatellite repeat results in disease (Figure 1); loss-of-function typically by disrupting transcription of the mutated gene (fragile X syndrome - FXS), a RNA toxic gain-of-function (Fragile X tremor-ataxia syndrome - FXTAS), and a protein toxic gain-of-function in two of the polyglutamine diseases (Spinocerebellar ataxia type 1 – SCA1 and spinal bulbar muscular atrophy - SBMA). For more inclusive reviews and those presenting alternative points of view the reader is referred to several recent and excellent articles (Williams and Paulson, 2008; LaSpada and Taylor, 2010; Ross and Tabrizi, 2011; Pastore and Temussi, 2012; Renoux and Todd, 2012).
Figure 1.
Pathogenic mechanisms for Fragile X syndrome (FXS), Fragile X tremor-ataxia syndrome (FXTAS) and myotonic dystrophy (DM), and the polyglutamine diseases spinocerebellar ataxia type 1 (SCA1) and spinal bulbar muscular atrophy (SBMA).
Loss of function - Fragile X syndrome
Fragile X syndrome (FXS) is a common inherited form of intellectual disability (ID) frequently associated with autism spectrum disorder (ASD) (Bhakar et al., 2012; Santoro et al., 2011). The condition was first reported by Martin and Bell in 1943 as a then-novel example of ID segregating in an X-linked fashion (Martin and Bell, 1943). These authors noted what would later become a key feature of the disorder, but at the time seemed unusual in their pedigree: nonpenetrant carrier males. For years, other descriptions of such pedigrees were few, until Lubs described a similar pedigree that had the distinction of segregating a “Marker X chromosome” with the disorder (Lubs, 1969). This Marker X showed a fragile site, or nonstaining gaps in the metaphase chromosome, near the end of the long arm of the X chromosome (Figure 2A). Once it became clear that this fragile site required specific culture conditions to be visible (Sutherland, 1977), many other pedigrees were found (Jacobs et al., 1980). With the addition of these families, the issue of nonpenetrant carrier males was rediscovered by Sherman et al., who found that 20% of males who inherited the mutation were normal and termed them “normal transmitting males” since their daughters were found to all be carriers (Sherman et al., 1984). Using a total of 206 pedigrees, Sherman et al. made the remarkable observation that sibships containing a normal transmitting male were less likely to have an affected male, and such sibships occurred more frequently in earlier generations, which raised the portentous notion of alleles of three states: normal, premutated, and fully mutated (Sherman et al., 1985). Inexplicable in terms of conventional genetics, this phenomenon became known as the “Sherman paradox,” later linked to the equally mystifying concept of “genetic anticipation,” where age-of-onset decreases or disease severity increases as a mutation moves generationally through an affected family (Harper, 1989).
Figure 2.
Key elements of FXS
(A) The fragile X chromosome. The fragile site indicated by arrow.
(B) Alleles of FMR1.
(C) Important FMRP features. Nuclear localization signal (NLS) and nuclear export signal (NES). K-homology domain (KH) and Arginine-Glycine-Glycine box (RGG) indicated. Agent domains are part of a family of Tudor domains that interact with certain methylated amino acids. The scale is amino acid number.
Using a variety of emerging methodologies from the nascent genome project, both the mutation (Heitz et al., 1991; Kremer et al., 1991; Oberle et al., 1991) and gene (Verkerk et al., 1991) were identified in 1991. The gene, called Fragile X Mental Retardation 1, or FMR1, is located precisely at the fragile site on the X chromosome and contains a polymorphic CGG repeat in the 5’ untranslated region of exon 1. The unstable expansion of this repeat is the most common cause of FXS and is the archetypal “dynamic mutation” of triplet repeat expansion. FMR1 has four allelic classes (Figure 2B). Normal alleles contain 5 to 44 repeats, with the most common normal allele harboring 29 or 30 repeats. Alleles from patients with FXS, referred to as full mutation alleles, have over 200 repeats, often as many as several hundred. These expanded alleles are derived from premutation alleles that contain between 54 and 200 repeats. Premutation alleles are meiotically unstable, particularly when maternally transmitted. All full mutations are derived from maternal premutations, whereas male premutation carriers account for the normal transmitting males noted by Sherman et al. The resolution of the Sherman paradox came about when the length of the maternal premutation was recognized as being directly proportional to the risk of expansion into the full mutation range (Fu et al., 1991). For example, a woman with a premutation of 60–69 repeats has a 2% chance of the allele expanding beyond 200 repeats, compared to a woman with 90–99 repeats, who has a 94% risk (Nolin et al., 2011). Finally, the fourth allelic class is of the intermediate alleles, between 45 and 54 repeats. While these are essentially normal alleles in that they do not transmit directly to the full mutation, they are slightly unstable, often changing by one or two repeats (Zhong et al., 1996). The CGG repeat of FMR1 is often interrupted by one or more AGG triplets, and AGG interruptions are known to stabilize the premutation (Eichler et al., 1994; Kunst and Warren, 1994), with the interruption pattern recently found to refine the genetic risk of having an affected child (Nolin et al., 2011; Yrigollen et al., 2012).
Premutation alleles, although they carry the obvious genetic risk of having a child with FXS (in females), have recently been associated with phenotypes that do not resemble FXS. Women who carry the premutation allele are at ~25% risk of primary ovarian insufficiency, referred to as Fragile X-associated Premature Ovarian Insufficiency, or FXPOI (Conway et al., 1998; Sullivan et al., 2005). In these carriers, menopause prior to 40 years of age, and sometimes considerably earlier, has a major impact on their reproductive futures, as well as their risk for earlier health sequelae following menopause. While the mechanism behind FXPOI is poorly understood, the greatest risk appears among women with premutation alleles in the middle of the premutation range (Sullivan et al., 2005). Premutation males, on the other hand, are at substantial risk of Fragile X-associated Tremor Ataxia Syndrome, or FXTAS, covered in detail below.
When the CGG repeat in FMR1 expands into the full mutation range, an epigenetic trigger occurs, causing widespread methylation of the gene that is correlated with its loss of function (Pieretti et al., 1991). This methylation imprint occurs early in embryogenesis (Malter et al., 1997; Sutcliffe et al., 1992) and leads to histone changes that reflect the gene silencing (Coffee et al., 2002; Coffee et al., 1999). Thus the full mutation is a null mutation, and the absence of its encoded protein, FMRP, is responsible for the disorder (Meijer et al., 1994). It should be noted that conventional mutations of FMR1 might disrupt expression by deletion, nonsense, or splice site mutations, also leading to FXS (Coffee et al., 2008; Gronskov et al., 2011; Lugenbeel et al., 1995; Wang et al., 1997). There appears to be a deficit of reported missense mutations of FMR1, with only a single confirmed pathological amino acid substitution (I304N); a sequencing screen of males suspected of FXS without repeat expansion found only one additional missense mutation (R138Q) (Collins et al., 2010; Feng et al., 1997a; Reyniers et al., 1996). It is likely that the infrequent clinical sequencing of FMR1 and the possibility of a subtler phenotype with such mutations could account for the paucity of missense mutations.
Since the absence of FMRP results in ID and ASD, there has been intense research over the past two decades that has provided substantial insight into the normal function of FMRP and the molecular consequences of its absence. Indeed, FXS may well be mechanistically the best understood of all neurodevelopmental disorders today. The human FMR1 gene is composed of 17 exons with significant alternative splicing (Ashley et al., 1993b; Eichler et al., 1993); accordingly, Western blots reveal multiple FMRP isoforms between 67–80 Kd, with isoform 7 being the most prominent at 69 Kd (Devys et al., 1993; Verheij et al., 1993). FMRP is largely cytoplasmic, with perhaps 5% being nuclear (Feng et al., 1997b) and the protein is widely expressed, although most strongly in neurons and testes.
FMRP contains several domains (Figure 2C) that provide clues to its function. The protein contains two KH domains and a RGG box, all associated with RNA binding (Ashley et al., 1993a; Siomi et al., 1993). Indeed, FMRP is a selective RNA-binding protein that interacts with perhaps 4% of the mammalian brain mRNAs. In the brain, the majority of FMRP is associated with translating ribosomes (Feng et al., 1997b; Khandjian et al., 2004; Stefani et al., 2004). This association with polysomes reflects a key property of FMRP as the I304N mutation that causes FXS in human and mice (De Boulle et al., 1993; Zang et al., 2009) does not associate with polysomes (Feng et al., 1997a; Zang et al., 2009). In addition to RNA-binding domains, FMRP contains both a nuclear localization signal and a nuclear export signal, suggesting FMRP shuttles between the nucleus and the cytoplasm (Bardoni et al., 1997; Eberhart et al., 1996), perhaps binding its mRNA ligands co-transcriptionally (Kim et al., 2009). A precise nuclear function of FMRP, if any, is unclear, although in the extreme N-terminus of the protein are two Agenet domains, of the Tudor-family domains, and these are capable of binding methylated lysines and arginines (Maurer-Stroh et al., 2003). A role for FMRP binding to methyl-lysine or methyl-arginine has yet to be elucidated.
RNA selectivity requires recognition elements embedded within the messages for the protein to recognize, however the precise nature of the RNA recognition element remains controversial. Initial data on the analysis of FMRP-immunoprecipitated mRNAs and in vitro selected RNA ligands indicated RNA structure rather than precise sequence is important (Brown et al., 2001; Darnell et al., 2001). Stem-G-quartet loops appeared to interact with the RGG box domain, while a “kissing complex” RNA structure bound to the KH2 domain (Darnell et al., 2005). The later interaction seemed particularly compelling as kissing complex RNA could compete with natural RNA ligands, running FMRP off ribosomes. Further support for structural-based elements came from the identification of mRNAs containing a stem-loop motif called SoSLIP and U-rich structures interacting with FMRP (Bechara et al., 2009; Chen et al., 2003a; Fahling et al., 2009). However, with the advent of much more robust protein RNA cross-linking strategies to identify mRNA targets of FMRP, the structural hypothesis appeared too simplistic. Using HITS-CLIP, Darnell et al. identified 842 robust mRNA targets (Darnell et al., 2011). Although there was significant overlap with earlier immunoprecipitated mRNAs thought to interact via stem-G-quartets, there was no enrichment of such structures in the 842 messages compared to nontarget messages. More recently, Ascano et al., using a variation of the HITS-CLIP strategy, identified another set of mRNAs bound to FMRP with unclear overlap with earlier studies. Here a short sequence motif was identified in the targets consisting of multiple (~18 per transcript) occurrences of WGGA- and ACUK-containing elements (W = A/U and K = G/U) throughout the coding sequence and 3’ UTR (Ascano et al., 2012). The relationship, if any, between the WGGA/ACUK elements and earlier identified RNA structures remains to be clarified.
The most widely accepted function of FMRP is as a translation regulator (Bagni et al., 2012; Bhakar et al., 2012; Santoro et al., 2011), particularly at the synapse where it was shown that FMRP itself was locally synthesized following group I metabotropic glutamate receptor agonists (Greenough et al., 2001). One highly regarded mechanism is that FMRP stalls ribosomes on target messages (Ceman et al., 2003; Darnell et al., 2011). Thus, FMRP is considered a negative regulator of translation. In vitro FMRP can suppress translation of target messages in rabbit reticulolysate or Xenopus oocytes (Laggerbauer et al., 2001). Direct measurement of total protein synthesis in the Fmr1 knockout mouse shows excess translation in the brain, consistent with the loss of a translation repressor (Dolen et al., 2007; Muddashetty et al., 2007). Using 14C-leucine to determine regional rates of cerebral protein synthesis, Qin et al. demonstrated enhanced synthesis in vivo in the knockout mouse brain (Qin et al., 2005). Finally, in a key study linking FMRP loss and synaptic defects, Huber et al. showed that metabotropic glutamate receptor (mGluR)-dependent long-term depression (LTD), previously found to require local protein synthesis, was exaggerated and independent of new protein synthesis in the Fmr1 knockout mouse (Huber et al., 2002; Huber et al., 2001). Not only would this be consistent with a loss of a negative regulator of translation but also suggests an uncoupling of synaptic protein synthesis machinery from synaptic output. Similar to the exaggerated LTD in these studies, the internalization of alpha-amino-3-hydroxy-5-methyl-4-iso-oxazole propionic acid (AMPA) receptors is directly proportional to FMRP loss in rat hippocampal neurons or cultured hippocampal neurons of the Fmr1 knockout mouse (Henderson et al., 2012; Nakamoto et al., 2007). Thus FMRP loss leads to excess local translation of its target mRNAs, which in turn leads to a decrease in synaptic strength and dendritic spine dysmorphogenesis (Bagni and Greenough, 2005). It should be noted, however, that for a small minority of mRNA, FMRP appears to be required for translation (Bechara et al., 2009; Gross et al., 2011).
If FMRP normally functions as a negative regulator of translation, it must be regulated to allow translation of its targets in the appropriate context. Phosphorylation of FMRP at serine 500 (serine 499 in the mouse) is one attractive model of regulation that has been put forward (Ceman et al., 2003). Under this model, phospho-FMRP stalls ribosomes on target mRNA in or near the dendritic spine. Following mGluR stimulation, FMRP is instantly dephosphorylated by PP2A, releasing the translational repression and allowing a burst of local protein synthesis that dynamically modulates synaptic strength (Narayanan et al., 2007). Within minutes, FMRP is then found phosphorylated, with S6K1 proposed as the responsible kinase (Narayanan et al., 2008). Although it is possible the dephosphorylated FMRP is rephosphorylated by S6K1, recent evidence points to dephosphorylated FMRP being rapidly ubiquitinated and degraded by the ubiquitin proteasome system, and the emergence of phospho-FMRP reflects phosphorylation of newly synthesized FMRP (Nalavadi et al., 2012).
The mechanism by which FMRP regulates translation is somewhat controversial. As mentioned earlier, ribosomal stalling is a popular candidate, with such stalling mediated by the microRNA pathway via phosphorylation (Bassell and Warren, 2008). FMRP is found to interact with the RNA-induced silencing complex (RISC) and with microRNAs themselves, indicating FMRP may use the ability of the RISC to stall ribosomes (Bolduc et al., 2008; Caudy et al., 2002; Ishizuka et al., 2002; Jin et al., 2004). Indeed, Maddashetty et al. have shown that the 3’ UTR of PSD95, a mRNA target of FMRP, contains a G-rich sequence required for FMRP binding that is also the target of miR-125a (Muddashetty et al., 2011). Bidirectional control of PSD-95 expression depends on miR-125a and FMRP phosphorylation status; FMRP phosphorylation promotes the formation of an AGO2-miR-125a inhibitory complex on PSD-95 mRNA, whereas mGluR signaling of translation requires FMRP dephosphorylation and release of AGO2 from the mRNA. These findings reveal a mechanism whereby FMRP phosphorylation provides a reversible activity-dependent switch to selectively regulate mRNA translation at synapses. In contrast to this mechanism, it is known that nonpolysomal FMRP can repress translation by inhibiting cap-dependent initiation via interaction with the eIF4E-binding protein, CYFIP1 (Napoli et al., 2008). Moreover, haploinsufficiency of CYFIP1 in mice results in increased LTD and both biochemical and behavioral phenotypes reminiscent of the Fmr1 knockout mouse, which is consistent with a reduction in translational repression (Bozdagi et al., 2012). Though the significance of this mechanism has been questioned because the vast majority of FMRP is associated with ribosomes (Darnell et al., 2011), it may be that both mechanisms are correct, depending upon the cellular context FMRP is in. For example, FMRP may be repressing translation of target mRNA during their transport from soma to dendrites by inhibiting cap-dependent initiation. FMRP is found in a variety of RNA granules, including P-bodies and stress granules, where translational repression is preinitiation or ribosome association (Antar et al., 2004; Antar et al., 2005; De Diego Otero et al., 2002). Once localized to the synapse, the granule could release FMRP-bound messages (Krichevsky and Kosik, 2001), either switching then to ribosome stalling or by earlier conversion to high-density granules which contain stalled ribosomes (Kiebler and Bassell, 2006).
As our understanding of FMRP function has become more granular and our view of the consequences of its absence more finely detailed, new avenues for possible therapeutic interventions have emerged. Based on the observation of enhanced protein synthesis-dependent, mGluR5-stimulated LTD in the Fmr1 knockout mouse (Huber et al., 2002), the “mGluR theory of FXS” was born (Bear et al., 2004). This theory proposes that the signaling cascade that triggers local protein synthesis by mGluR stimulation is regulated by FMRP and in its absence runs unchecked. Therefore, reduction of the initial stimulation, using mGluR5 antagonists, may reduce the constitutive translation to achieve some level of correction. In the years since the theory was first published, compelling support for it has come from a remarkable series of studies that used both pharmacologic and genetic means to reduce mGluR signaling in a variety of organisms (detailed in Bhakar et al., 2012; Kruger and Bear, 2011). Biochemical and behavioral studies in the mouse have provided enough preclinical evidence that reducing mGluR5 can correct FXS-like phenotypes to launch clinical trials. Several such studies are underway with AFQ056 (Novartis), R04917523 (Hoffman-La Roche), and STX107 (Seaside), all mGluR5 antagonists (see clinicaltrials.gov for the latest). One study published thus far using AFQ056 suggests improvements in inappropriate speech, stereotypic behavior, and hyperactivity in FXS males with the full mutation (Jacquemont et al., 2011). Although this was a small study and efficacy emerged only after posthoc analysis, it is encouraging that ongoing trials with all the antagonists may produce some measure of improvement.
An alternative approach to dampen the exaggerated glutaminergic signaling, is to pharmacologically activate the ϒ-aminobutyric (GABA) receptors, which will stimulate the inhibitory pathway (Hampson et al., 2011). Support for this approach came from a small molecule screen to reverse glutamate-induced toxicity in FMRP-deficient Drosophila (Chang et al., 2008). The top three hits in the screen of 2,000 compounds all implicated the GABAergic pathway, and subsequent studies confirmed GABA could reverse biochemical, neurobiological, and behavioral abnormalities in the fly FXS model. These data validated earlier studies indicating the GABAergic pathway was downregulated in FXS (D'Hulst et al., 2006). Although the GABA(A) pathway is clearly compromised in the Fmr1 knockout mouse (Curia et al., 2009; D'Hulst et al., 2009; Gantois et al., 2006), there is evidence that stimulation of the GABA(B) receptor could reverse seizure susceptibility in the mouse model (Pacey et al., 2009). Recently, a comprehensive preclinical study in the mouse validated the GABA(B) agonist STX209 (arbaclofen, Seaside) as capable of reversing many phenotypes in the knockout mouse model (Henderson et al., 2012). In a randomized, placebo-controlled crossover trial of 63 subjects with FXS, STX209 appeared to have a significant beneficial treatment effect on measures of behavior (Berry-Kravis et al., 2012), although this too was a posthoc finding. Several ongoing trials with STX209 are active or recruiting (clinicaltrials.gov).
Because of its history, FXS has long been considered a model for biomedical investigation of idiopathic autism spectrum disorder, but there is now a growing body of evidence that FMRP may be intimately involved in the signaling pathways and circuits potentially disrupted in ASD. First, proteins involved in syndromic ASD, such as PTEN, TSC1, and TSC2, as well as proteins mutated in rare nonsyndromic ASD, such as NLGN4, NRXN1, and SHANK3, all modulate postsynaptic signaling, and the former are all key glutamatergic signaling elements with FMRP in the mTOR pathway regulating activity-dependent protein synthesis (Zoghbi and Bear, 2012). Second, although different approaches were used to identify the mRNA targets of FMRP, all the contemporary studies show a statistically significant enrichment of candidate autism susceptibility gene messages binding to FMRP (Ascano et al., 2012; Darnell et al., 2011). Finally, de novo gene disruptions found in ASD by exome sequencing reveal a disproportionate number of genes whose mRNA are known FMRP targets (Iossifov et al., 2012). This gives us hope that, while the etiology of ASD may be heterogeneous, many of the susceptibility loci could interact in a common pathway. Another hopeful possibility is that therapeutic strategies now in clinical trials for FXS may prove beneficial for idiopathic ASD.
Two Polyglutamine Diseases - SBMA and SCA1
Among the first trinucleotide microsatellite repeat disorders identified was the CAG repeat expansion underlying spinal and bulbar muscular atrophy (SBMA) or Kennedy’s disease (LaSpada et al., 1991). This discovery was soon followed by the elucidation of a similar mutation as the basis of a group of disorders now known as the polyglutamine neurodegenerative diseases. Along with SBMA, the polyglutamine diseases include Huntington disease (HD), dentatorubral-pallidoluysian atrophy (DRPLA), and six spinocerebellar ataxias (SCA) 1, 2, 3, 6, 7 and 17 (Orr & Zoghbi, 2007). As a group, these nine diseases are one of the more common forms of inherited neurodegeneration (Figure 1).
With elucidation of the polyglutamine neurodegenerative diseases, it was apparent that the only feature common among the affected proteins was the polyglutamine tract itself. Moreover, in most cases the polyglutamine tract was found in novel proteins whose function was unknown. Only in three diseases did the polyglutamine expansion occur in a previously known protein - SBMA in the androgen receptor, a ligand-dependent nuclear hormone receptor transcription factor; SCA6 and the 1A subunit, the transmembrane pore-forming subunit, of the P/Q-type or CaV2.1 voltage-gated calcium channel; SCA17 in the TATA-binding protein, a general transcription factor that plays an important role in initiation of transcription for all three eukaryotic polymerases. So how could the same mutation, a polyglutamine expansion in most cases to around 40 glutamines or more, in nine different proteins result in nine different diseases?
Although it is generally regarded that a major aspect of polyglutamine pathogenesis is a polyglutamine-induced toxic gain-of-function activity on host proteins, the more detailed molecular basis of these disorders remains controversial. Perhaps the most seminal unresolved issue is the extent to which the pathogenic mechanism is due a direct toxicity of an expanded polyglutamine tract and, thus, common among the diseases. One at least initially widely held hypothesis whereby pathogenesis is driven by the polyglutamine tract, is one where disease is caused by the accumulation of aggregates containing a misfolded form of the affected protein (Pastore and Temussi, 2012). This hypothesis has its roots in the observation that a pathological hallmark of many of these diseases is the presence of intracellular inclusions containing the mutant polyglutamine protein. The ability of an expanded polyglutamine stretch to promote protein aggregation and subsequent disruption of cellular protein homeostasis in some model systems provides experimental evidence in support of a misfolded protein/aggregation hypothesis (Powers et al., 2009). At the core of this mechanism is the concept that it is a misfolded, nonnative conformation of the polyglutamine protein, induced by an expanded polyglutamine tract, which initiates the pathogenic process. However, there is evidence questioning whether the large aggregates/inclusions of mutant protein are directly pathogenic (Klement et al., 1998; Saudou et al. 1998; Cummings et al., 1999; Arasate et al., 2004). In fact some studies suggest these inclusions likely have, at least early in disease progression, a protective role perhaps by sequestering mutant protein (Saudou et al., 1998; Watase et al., 2002). In the case of Huntington’s disease, considerable evidence using a variety of experimental approaches supports a model of pathogenesis where release by proteolytic cleavage of a toxic polyglutamine fragment from Huntingtin is key (reviewed by Ross and Tabrizi, 2011).
An additional hypothesis poses that a normal function and, thereby, native conformation/interactions underlie the mechanism of pathogenesis (Riley and Orr, 2006). Key in the development of this idea were findings that residues/regions outside of the polyglutamine tract have a crucial role in pathogenesis and in some instances the demonstration that polyglutamine expansion in itself is not sufficient to cause disease (Emamian et al., 2003; Gu et al., 2009; Humbert et al, 2002; Katsuno et al., 2002; Klement et al. 1998; Takeyama et al., 2002). These residues function to modulate the normal biology of the polyglutamine protein and presumably pathogenesis by impacting its normal subcellular location, degradation, and interaction with other cellular molecules. Of the polyglutamine disorders, spinobulbar muscular atrophy (SBMA) and spinocerebellar ataxia type 1 (SCA1) are two where the concept of normal function and conformation being tied to pathogenesis is well demonstrated (Kratter and Finkbeiner, 2010).
Amongst the polyglutamine disorders, SBMA in many ways is the prototype in linking normal function to pathogenesis. SBMA, also known as Kennedy’s disease, is a progressive late-onset disease that consists of motor neuron degeneration in the brainstem and spinal cord (Kennedy et al., 1968). As mentioned above, SBMA is due to expansion of a CAG repeat in the androgen receptor (AR) gene located on the X chromosome (LaSpada et al., 1991). Importantly, SBMA affects only males. Even females homozygous for ARs with an expanded polyglutamine fail to develop the neurodegeneration and neurological symptoms seen in affected males, suggesting that perhaps high levels of circulating androgen have a role in pathogenesis (Schmidt et al., 2002). Direct evidence in support of this concept was obtained with the development of transgenic mouse and Drosophila models in which only males with an expanded polyglutamine AR gene developed disease (Katsuno et al., 2002: Takeyama et al., 2002). Removal of testosterone from young mutant transgenic male mice blocked development of disease (Katsuno et al., 2002), while in older mutant males castration restored normal function (Chevalier-Larsen et al., 2004). Correspondingly, female mice carrying a mutant AR gene developed disease upon administration of testosterone (Katsuno et al., 2002). This same androgen-dependent neurodegeneration is seen in Drosophila expressing a polyglutamine expanded AR (Takeyama et al., 2002). Although Drosophila lacks an ortholog to human AR, the nuclear hormone transactivation pathways are well conserved (King-Jones and Thummel, 2005; Takeyama et al., 2004).
Unlike the polyglutamine diseases where little was known about the function of the affected protein, the normal structure/function of AR is well characterized. The AR consists of several functional elements/domains (Figure 3). The AR amino-terminal region contains, along with the polyglutamine tract, two so-called activation functions AF-1 and AF-5. In absence of ligand, androgen receptor resides in the cytoplasm in an inactive form in a complex with a chaperone. Upon binding androgen, the hormone-receptor complex translocates to the nucleus, binds to DNA at specific recognition sites upstream of its target genes. Expression or repression of these genes occurs through the interaction of two co-regulator regions of AR, AF-1 and AF-2 (Figure 3), with other transcription factors. In a recent and elegant study, Nedelsky and colleagues also showed that ligand binding was necessary for expanded polyglutamine AR to cause degeneration of Drosophila adult photoreceptors and larval motor neurons (Nedelsky et al., 2010). This study further demonstrated that in order for mutant AR to cause degeneration, it had to translocate to the nucleus and bind to DNA. It was shown that one of the interaction regions of AR, the AF-2 transactivation domain, is required for toxicity. Moreover, a genetic screen revealed that several known AF-2 interacting proteins are modifiers of mutant AR induced degeneration. These results show that the mutant AR pathogenic pathway induced by expansion of the polyglutamine tract overlaps considerably with the normal AR-ligand binding pathway. Nedelsky et al. speculated that is an enhancement of a yet to be identified aspect of normal AR function, involving the AF-2 transduction domain, that causes neurodegeneration. The observation that Drosophila overexpressing wild type AR show signs of disease provides additional support to a native AR function being critical for pathogenesis (Nedelsky et al., 2010).
Figure 3.
Key structural features of the androgen receptor (SBMA) and ataxin-1 (SCA1). (A) Schematic depiction of the androgen receptor protein. Features depicted are: Q)n, polymorphic polyglutamine tract; NTD, N-terminal domain; DBD, DNA binding domain; NLS, nuclear location signal; LBD, ligand binding domain; AF1, transcription activation function 1 region; AF2, transcription activation function 2 region. (B) Diagram of the Ataxin-1 protein. Features depicted are: Q)n, polymorphic polyglutamine tract; AXH, ataxin-1/HBP1 homology domain; NLS, nuclear location signal (expanded to indicate location of S776, serine 776, 14-3-3 LM, 14-3-3 binding site; ULM, binding site for RBM17 and U2AF65); SAR, self-association region; RNA binding region; CIC, binding region for Capicua.
Recently, expression of polyglutamine expanded AR in a mouse model of SBMA was shown to upregulate the gene encoding calcitonin gene-related peptide a (CGRP1) (Minamiyama et al., 2012). CGRP1 was found to be upregulated in motor neurons of patients with SBMA. Importantly, mutant AR increased transcription at the CGRP1 promoter consistent with altered regulation of transcription being key to the pathogenic actions of mutant AR. Minamiyama et al. further reported that the toxic effects of increased CGRP1 expression were linked to the activation of c-Jun N-terminal kinase (JNK). Moreover, pharmacological and genetic inactivation of CGRP1 reduced neurodegeneration in a mouse model of SBMA. Together these results provide compelling evidence that stimulation of CGRP1 transcription and subsequently activation of the JNK pathway by mutant AR is crucial to SBMA pathogenesis.
Linkage of SCA1 to the HLA complex on the shortarm of chromosome 6 was seminal to isolating the gene (Yakura et al., 1974; Nino et al., 1980; Morton et al., 1980). An equally critical finding was the report in 1950 of genetic anticipation (Schut, 1950) that subsequently was shown to have a form of SCA in linkage with the HLA complex (Haines et al., 1984). In contrast to SBMA, spinocerebellar ataxia type 1 (SCA1) is an example of a disorder where the host protein, Ataxin-1 (ATXN1), is novel and whose function was unknown when the disease-causing mutation was identified. Subsequent to cloning the SCA1 gene (Orr et al., 1993), studies identified several functional/structural motifs in ATXN1 (Figure 3).
These include the AXH domain that folds into an OB-fold forming both a putative RNA-binding and a protein-protein interaction surface (Chen et al., 2004), a nuclear localization signal (NLS) (Klement et al., 1998) that overlaps with or is immediately adjacent to a 14-3-3 binding site (Chen et al., 2003b) and a UHM ligand motif (ULM) present in proteins associated with RNA splicing (de Chiara et al., 2009). Moreover, ATXN1 interacts with a variety of nuclear components including RNA (Yue et al., 2001; Irwin et al., 2005), several regulators of transcription, SMRT (Tsai et al., 2004), Capicua (Lam et al., 2006), Gfi-1 (Tsuda et al., 2005), and the Rora/Tip60 complex (Serra et al., 2006; Gehrking et al., 2011) and proteins involved in RNA processing such as the RNA splicing factors RBM17 (Lim et al., 2008) and U2AF65 (de Chiara et al., 2009), and p80 coilin involved in the assembly of U snRNPs (Hong et al. 2003).
The ATXN1/RORα/Tip60 interaction may have implications for there being a relationship between Purkinje cell development and SCA1-mediated degeneration later on in adults. First of all, conditional SCA1 mice were used to demonstrate that a compromise in early Purkinje cell development impacts severity of mutant ATXN1-mediated Purkinje cell degeneration in adult mice (Serra et al., 2006). SCA1 mice that express mutant ATXN1 protein after the first three weeks appeared normal with respect to behavior and cerebellar morphology out past one year of age. In contrast, SCA1 mice with the transgene on during early postnatal development developed ataxia and Purkinje cell degeneration as adults. Mechanistic insight for this finding came by comparing altered cerebellar gene expression in SCA1 mice with the mouse cerebellar mutant known staggerer, having an intragenic deletion in the retinoic acid-related orphan receptor α (RORα) gene (Hamilton et al., 1996). This comparison yielded an extensive overlap of RORα-regulated target genes. In addition, there is a reduction in the amount of RORα protein in Purkinje cells of SCA1 mice. As assessed by coimmunoprecipitation from cerebellar lysates, RORα and ATXN1 interact. Yet, these proteins do not interact directly in vitro. Rather ATXN1 seems to interact directly the RORα coactivator Tip60 (Serra et al., 2006; Gehrking et al., 2011).
In agreement with an ATXN1 nuclear function being critical for pathogenesis is the finding that a single amino acid substitution in the NLS of ATXN1 that dramatically reduces translocation of the protein to the nucleus of murine cerebellar Purkinje cells prevents ATXN1 with an expanded polyglutamine tract from causing disease (Klement et al., 1998). While wild-type ATXN1 shuttles between the cytoplasm and nucleus, ATXN1 with an expanded polyglutamine tract is unable to be exported from the nucleus and, thus, is retained in the nucleus (Irwin et al., 2005). Therefore, pathogenesis of both SBMA and SCA1 stem from altered events in the nucleus. What about the other polyglutamine disorders? Might alterations in the regulation of gene expression underlie pathogenesis of these diseases as a group? There are three points that argue in favor of such an idea. First, many regulators of transcription contain glutamine-rich activation domains that are typical of an extensive family of highly conserved transcriptional activators. These glutamine-rich activation domains are an important class of protein-protein interacting motifs that enable transcription factors to interact with one another and thus regulate gene expression (Tanese and Tjian, 1993). Second, are the observations that transcriptional alterations appear early in disease progression for the various polyglutamine diseases and that mutant polyglutamine proteins interfere with a large number of transcription factors (Sugars and Rubinsztein, 2003). The one outlier in a nuclear-mediated mechanism for polyglutamine pathogenesis is SCA6 where the mutation occurs in the CACNA1A gene that encodes the α1 subunit of a voltage-gated calcium channel (Zhuchenko et al., 1997), indicating that an alteration in ion channel function underlies SCA6. However, Christopher Gomez and colleagues recently obtained compelling evidence that this is not the case (Du et al., 2013). They found that CACNA1A produces a bicistronic mRNA bearing a cryptic internal ribosomal entry site with the first cistron encoding the α1A subunit and the second cistron encoding a novel transcription factor, designated α1ACT. α1ACT contains the polyglutamine tract and regulates expression of a set of genes involved in neural and Purkinje cell development. α1ACT carrying an expanded polyglutamine tract, lacks transcription factor function, causes cell death in culture, and leads to ataxia and cerebellar atrophy in transgenic mice.
Studies reveal that the majority of soluble ATXN1 in the nucleus is normally associated with other proteins in large (approx. 1.8MDa) multimeric complexes (Lam et al., 2006; Lim et al, 2008). In regards to pathogenesis, are the complexes ATXN1 forms with the transcription factor capicua (Cic) and the splicing factor RBM17. Along with ATXN1, both Cic and RBM17 are highly expressed in the nuclei of Purkinje cells where polyglutamine ATXN1 is believed to do the most harm. The effect polyglutamine expansion has on ATXN1’s interaction with and alteration of Cic function is multifaceted. Cic is transcriptional repressor that contains a Sox-like high mobility group (HMG) box. Cic binds directly to the AXH domain of ATXN1 (Lam et al., 2006). In a transfected cell-based transcriptional assay, wild-type ATXN1 synergizes with Cic to repress transcription. The ability of ATXN1 to synergize with Cic in repression of transcription is compromised with polyglutamine-expanded ATXN1, suggesting that polyglutamine-expanded ATXN1 might cause a loss of Cic transcriptional function (i.e. derepression). A more recent study indicates that for some Cic gene targets expanded ATXN1 causes a loss of function while at other targets polyglutamine-expanded ATXN1 enhances Cic binding inducing a state of hyper-repression (Fryer et al., 2011). This study postulated that it is the hyper-repressive effects of expanded polyglutamine ATXN1 that are toxic. In support of this idea is the demonstration that in a mouse model of SCA1 a reduction in Cic either genetically or, intriguingly, by exercise dampened toxic gain of function of polyglutamine expanded ATXN1.
In another study, genes with altered expression due to ATXN1 and corepressors with which it binds were identified using chromatin immunopreciptiation along with functional transcriptional read-out assays (Cvetanovic et al., 2011). One such gene encoding the angiogenic/trophic factor VEGF was identified as a gene repressed by mutant ATXN1. Importantly, restoration of VEGF expression either genetically or pharmacologically dampens mutant ATXN1-induced pathology. These results support the notion that transcriptional deficits play a role in SCA1 pathogenesis.
In the case of the interaction of ATXN1 with the splicing factor RBM17, polyglutamine expansion and the amino acid at position 776 affect complex formation (Lam et al., 2008). Expansion of the polyglutamine tract in ATXN1 enhances its interaction with RBM17. Normally ATXN1 has a serine at position 776, which is one of seven endogenous sites of phosphorylation in ATXN1 (Emamian et al., 2003; Huttlin et al., 2010). Evidence suggests that cyclic AMP-dependent protein kinase (PKA) is an active ATXN1-S776 kinase in the cerebellum and Purkinje cells (Jorgensen et al., 2009; Hearst et al., 2010). Placing a phosphorylation-resistant alanine at position 776 dramatically decreases the ability of wild type or polyglutamine-expanded ATXN1 to interact with RBM17 (Lam et al., 2008). Conversely, replacing serine 776 with a potentially phospho-mimicking aspartic acid increased the ability of wild type ATXN1 to interact with RBM17 to a level similar to that seen with ATXN1[82Q]-S776. Importantly, the extent to which a form of ATXN1 interacts with RBM17 correlates directly with its ability to cause Purkinje cell dysfunction and ataxia in vivo. Polyglutamine expanded ATXN1 with an alanine at position 776 is no longer pathogenic in vivo (Emamian et al., 2003). On the other hand an aspartic acid at residue 776, enhances ATXN1 induced pathogenesis such that even ATXN1 with a wild type, unexpanded polyglutamine becomes pathogenic (Duvick et al., 2010). These results indicate that the interaction with RBM17 also has a critical role in pathogenesis.
Phosphorylation of serine 776 (S776) stabilizes ATXN1 and forms a binding site for the 14-3-3 family of proteins (Chen et al., 2003b; de Chiara et al., 2009). 14-3-3 proteins are known to have a critical role in many cell signaling events (Morrison, 2008). In the case of ATXN1, data indicate that upon serine 776 phosphorylation 14-3-3 binds to ATXN1 in the cytoplasm (Lai et al., 2011). While an aspartic acid at residue 776 stabilizes ATXN1 and enhances ATXN1’s interaction with RBM17, this amino acid substitution does not enhance binding of ATXN1 with 14-3-3 (de Chiara et al., 2009; Lai et al., 2011; Menson et al., 2012). Thus, an aspartic acid at position 776 fails to mimic all of the effects of phosphorylating serine 776.
Binding of 14-3-3 to ATXN1 was shown to have several effects. First 14-3-3 binding is associated with a stabilization of ATXN1 and in the case of polyglutamine expanded ATXN1 an increase in the formation of nuclear inclusions (Chen et al., 2003b). A critical question is whether 14-3-3 binding directly or indirectly stabilizes ATXN1. Recent data suggest that perhaps the stabilization effect of 14-3-3 binding to ATXN1 is an indirect effect. 14-3-3 binds to ATXN1 in the cytoplasm blocking the dephosphorylation of serine 776 (Lai et al., 2011). Since it is the unphosphorylated form of ATXN1 that seems to be relatively unstable and more readily degraded, blocking dephosphorylation of ATXN1 at serine 766 would indirectly stabilize the protein. Second is the observation that 14-3-3 binding per se is not required for stabilization of ATXN1. ATXN1-D776, which does not bind 14-3-3, is as stable in vivo as is ATXN1-S776 (Lai et al., 2011). In addition to having a role in regulating the dephosphorylation of ATXN1 at serine 776 and, thus, the degradation of ATXN1, 14-3-3 binding also regulates the transport of ATXN1 into the nucleus. Since the 14-3-3 binding site is immediately adjacent to the NLS in ATXN1, 14-3-3 binding masks the NLS and blocks entry of ATXN1 into the nucleus (Lai et al., 2011). Thus, 14-3-3 must be released from ATXN1, presumably by some regulated process, for ATXN1 to be transported into the nucleus.
This model places the binding of 14-3-3 to ATXN1 at two steps that are crucial for polyglutamine expanded ATXN1 to cause disease – regulation of the level of mutant ATXN1 that directly correlates with disease severity (Burright et al., 1995; Lorenzetti et al., 2000; Watase et al., 2002) and entry of ATXN1 into the nucleus that is required for pathogenicity (Klement et al., 1998). This raises the question of whether a reduction in 14-3-3 binding would make disease worse or better. To examine how reducing 14-3-3 levels and thereby its interaction with ATXN1 impacts disease, knockin Sca1154Q/+ mice were crossed with mice lacking one allele of 14-3-3ε. 14-3-3ε mice were selected since 14-3-3ε interacts with ATXN1, is highly conserved, and expressed highly in the cerebellum. Sca1154Q/+ animals have an insertion of a CAG expansion of 154 in the endogenous Sca1 gene, display many features of human SCA1 including cerebellar phenotypes such as ataxia, progressive Purkinje cell degeneration, and non-cerebellar phenotypes like weight loss, and premature death (Watase et al., 2002). Consistent with 14-3-3 having a role in regulating ATXN1 protein levels, a partial reduction of 14-3-3ε reduced the amount of ATXN1 in the cerebellum (Jafar-Nejad et al., 2011). In contrast, in mice with a partial loss of 14-3-3ε in the brainstem were identical to those in wild type mice. Similarly, 14-3-3ε haploinsufficiency rescued the cerebellar pathology but had no effect on non-cerebellar phenotypes in Sca1154Q/+, mice. These data argue that 14-3-3 regulates ATXN1 levels in some regions of the brain, e.g. cerebellar Purkinje cells, but not in other regions, e.g. the brainstem. Thus, pathogenic pathways may vary among the different susceptible brain regions, adding another layer of complexity to SCA1 pathogenesis.
As one contemplates a treatment for any neurodegenerative disease including SCA1, the extent to which disease is reversible after onset is of importance. To assess reversibility of SCA1, a conditional mouse model of SCA1 was developed (Zu et al., 2004). The approach was to use the Pcp2/L7 promoter to drive a tetracycline-transactivator (Tta) transgene specifically in Purkinje cells and cross these mice with mice carrying a TRE-ATXN11[82Q] transgene. Administration of doxycycine (dox), a tetracycline derivative able to cross the blood brain barrier, abolishes the Tta/TRE interaction turning off expression of ATXN1. At the early stage of disease, i.e. 6-week-old mice, the rotating-rod deficit was reversed completely after 6 weeks of dox treatment. In addition, Purkinje cell pathology improved by assessment of molecular layer thickness and dendritic arborization, although heterotopic Purkinje cells were still present. When ATXN1 expression is stopped at 12 weeks of age, a mid-stage disease, in animals with the gene off for 4 weeks continued to have rotating-rod deficits similar to 12 and 16 week-on animals. After longer periods of dox administration, 8 weeks and 12 weeks, rotating-rod performance improved, but only partially. Interestingly, this 8-week time point coincided with the return of mGluR1α glutamate receptors at the Purkinje cell-parallel fiber synapses, albeit at a lower expression level. There was recovery of molecular layer thickness and improved arborization of Purkinje cell dendrites demonstrating that halting ATXN1[82Q] expression at the time of typical onset of ataxia prevents the progression of and partially reverses degenerative changes in Purkinje cells. When dox was given to animals at 32 weeks of age, there was no significant improvement of ataxia, but there was some improvement of Purkinje cell pathology. At all disease stages, Purkinje cells rapidly eliminated ATXN1[82Q]-containing nuclear inclusions upon administration of dox and cessation of ATXN1 expression. Thus, even at a late stage of disease Purkinje cells are able to clear ATXN1 protein with an expanded polyglutamine. Overall, these studies revealed that Purkinje cell pathology induced by ATXN1 with an expanded polyglutamine could be reversed with cessation of ATXN1 protein synthesis. Interestingly, the capability of Purkinje cells to recover decreases with increasing age. It remains unclear whether the age effect on recovery is due to prolonged exposure to mutant ATXN1 and/or that older neurons in general are less able to repair damage.
All of the evidence using SCA1 experimental models shows a direct relationship between disease severity and level of mutant ATXN1 expression. Identifying pathways or strategies by which the levels of the disease-causing protein could be decreased would serve as potential drug targets and or therapeutic approaches. An attractiveness of such an approach is its potential effectiveness irrespective of a lack of clarity regarding the mechanisms that underlie pathogenesis. Participation of S776 and its phosphorylation in regulating degradation of ATXN1 provides one potential therapeutic target for SCA1. Much effort has gone into and is going into understanding signaling pathways regulated by protein kinases. Protein kinases are one of the most active groups under investigation as drug target owing to their involvement in many pathological conditions. Yet there are considerable challenges facing the therapeutic targeting of protein kinases in disorders of the brain like SCA1 (Chico et al., 2009). Not the least of which is the penetration of the blood-brain barrier and selectively targeting a multi-functional kinase like PKA. Regardless, identification of the kinase(s) responsible for ATXN1-S776 phosphorylation in affected neurons affords an opportunity to elucidate processes/signals upstream to the kinase that regulate its action on ATXN1 as wells as elucidation of downstream events triggered by ATXN1-S776 phosphorylation. Either of which could result in identification of viable drug targets.
The ability to achieve gene silencing by selectively modulating RNA function is a genetic approach that offers another potential means for treating dominantly inherited neurodegenerative diseases. In the case of SCA1, proof-of-concept for RNA interference as a possible treatment approach was demonstrated by Xia et al. (Xia et al., 2004). AAVs expressing shRNAs targeting the ATXN1[82Q] RNA were injected into the cerebellum of SCA1 mice and found to improve molecular and motor phenotypes. Since artificial miRNAs are reported to be less toxic than shRNAs, current thought would be to use artificial miRNAs for gene silencing (Boudreau et al. 2009). siRNAs can be delivered directly to achieve knockdown of a gene of interest. To block their digestion by nucleases the siRNAs need to be chemically modified. Allele-specific silencing of mutant Htt in Huntington disease (HD) mouse models was recently achieved using chemically modified single stranded siRNAs (Yu et al., 2012; Lima et al., 2012).
An alternative approach for reducing mutant protein synthesis in the brain is infusion of single-stranded antisense oligonucleotides (ASOs) that guide degradation of target RNAs. Recently, infusion into the CSF of ASOs to the another polyglutamine protein, huntingtin, was shown to provide a reversal of disease phenotypes in a Huntington’s disease mouse model (Kordasiewicz et al., 2012). An exciting finding of this study was that a transient ASO infusion and reduction of huntingtin yielded a sustained reversal of disease symptoms, suggesting that a sustained therapeutic efficacy may not require continuous treatment.
Toxic RNA - Fragile X tremor/ataxia and myotonic muscular dystrophy
It was ten years after the discovery of the expanded CGG triplet repeat that caused fragile X syndrome before it became clear that the “normal transmitting males,” long known to geneticists as non-penetrant male carriers of the Fragile X mutation, were subject to a late age of onset neurodegenerative condition (Hagerman et al., 2001). Repeated questions from mothers of fragile X sons regarding their fathers’ symptoms, lead to the appreciation of a constellation of symptoms that we now refer to as fragile X tremor ataxia syndrome, or FXTAS (Greco et al., 2002; Jacquemont et al., 2003). FXTAS can affect both men and women, but male carriers of the FMR1 premutation are at significantly greater risk, with more than half exhibiting symptoms by age 70 (Jacquemont et al., 2004). Women who carry the premutation have a much lower risk (less than 20%) (Rodriguez-Revenga et al., 2009). While symptoms are variable, a Parkinsonian tremor and ataxia are common, along with cognitive decline, and mood disorders. Significantly, males with FXS, who can live to old age, do not commonly develop FXTAS. Features of FXTAS patients also include unusual findings in magnetic resonance imaging studies (middle cerebellar peduncles have increased T2-weighted density) (Jacquemont et al., 2003), and at autopsy, widespread, large eosinophilic nuclear inclusions that stain with ubiquitin and proteins involved in proteasome degradation are found in both neurons and glia (Greco et al., 2006). While the inclusions are reminiscent of findings in disorders involving polyglutamine expansions, they do not stain with antibodies that can detect polyglutamine. FXTAS patient’s brains typically demonstrate mild-tomoderate global atrophy, most common in the frontal and parietal regions, as well as in the pons and cerebellum including Purkinje neuron loss, Bergmann gliosis, spongiosis of the deep cerebellar white matter and swollen axons (Greco et al., 2006). Ubiquitin staining nuclear inclusions and neuronal loss are not commonly found in the brains of individuals with Fragile X syndrome.
Finding nuclear inclusions at autopsy and specific MRI anomalies helped to distinguish FXTAS as a distinct clinical entity, different from other neurological conditions of old age. Importantly, the differences in pathology between FXS and FXTAS demanded new hypotheses for the contribution of the FMR1 CGG repeat expansion to the disorder. FXS clearly results from loss of FMR1 function due to the large expansion mutation’s effect of shutting off transcription of the FMR1 gene. Premutation alleles were found to express somewhat more FMR1 RNA (Kenneson et al., 2001; Tassone et al., 2000), and since the repeat expansion was confined to the 5’ untranslated region of the gene, were not expected to qualitatively alter the FMR1 protein. Fortunately, progress in characterizing another trinucleotide repeat expansion disorder, myotonic muscular dystrophy (DM), provided a road map for investigations into FXTAS.
With the identification of expanded and unstable repeats in FXS and SBMA, several groups hypothesized a similar mutational mechanism for DM, since repeat instability provided a molecular mechanism to explain the long-debated genetic peculiarities of this disorder (Sutherland et al., 1991). Myotonic dystrophy is a common (~1/8000) typically adult-onset muscle disease involving myotonia and muscle wasting that includes skeletal, cardiac and smooth muscle. Numerous other symptoms are more variable, including cataracts and central nervous system involvement. DM pedigrees often show more severe disease in the younger members of the family. This includes a severe form of “congenital” DM in children born to affected mothers that can be fatal (Udd and Krahe, 2012). The term anticipation was coined a century ago to describe the observation of worsening symptoms in subsequent generations of this dominantly inherited disease. Some geneticists were reluctant to accept the phenomenon, questioning whether ascertainment bias in selection of patients was the explanation, since mutations were not known to be variable between generations (Harper et al., 1992). The examples of FXS and SBMA led to discovery by several groups of a highly unstable CTG repeat in the 3’ untranslated region of the myotonic dystrophy protein kinase (DMPK) gene (Brook et al., 1992; Fu et al., 1992; Mahadevan et al., 1992). This repeat showed instability and size ranges that nicely explained the anticipation in pedigrees; all affected members had repeat lengths longer than the range in the general population, and longer repeats led to more severe disease, with congenital cases exhibiting very large expansions. Most significantly, the repeat length tended to increase when transmitted from parent to child, providing a mechanism for anticipation: directional hypermutability (Shelbourne and Johnson, 1992).
The mechanism by which the DMPK expanded CTG repeat led to disease pathology was much less clear in 1992, since the location of the repeat did not predict an altered protein (as in SBMA) or simple loss of function of DMPK (as in FXS). Several hypotheses were proposed, including effects of the repeat expansion on neighboring genes and reduced DMPK levels due to RNA processing and transport effects. The observations in 1995 of nuclear foci containing the DMPK transcript in mutant cells (Taneja et al., 1995), with the subsequent identification of RNA-binding proteins present in nuclear foci (Mankodi et al., 2001; Miller et al., 2000) and interacting with the expanded repeat spurred a third hypothesis where repeat expansion leads to a toxic gain of function mediated through RNA. Significant support for this idea stemmed from mouse models, where CTG expressed in RNA as a transgene outside of the DMPK context recapitulated many DM features (Mankodi et al., 2000), while heterozygous loss of DMPK in the mouse showed little pathology (Reddy et al., 1996). The discovery that DM2, a very similar autosomal dominant human myotonic dystrophy, resulted from a tetranucleotide (CCTG) repeat expansion in an intronic location of a separate gene (CNBP) (Liquori et al., 2001), added strong support to the toxic gain of function hypothesis, although there remain aspects of the DM1 phenotype that may result from other effects on the DMPK locus. For example, knockout of DMPK in the mouse can cause cardiac abnormalities that are reminiscent of those in DM patients (Berul et al., 1999). Importantly, repeat sequence-mediated RNA toxicity is especially attractive for explaining the correlation of repeat length with disease severity. Longer repeats might be expected to have a more pronounced effect on cellular metabolism, with the possibility of differential susceptibility among tissues altering the onset and constellation of symptoms. Mouse and Drosophila models expressing RNAs containing the expanded CUG repeats in untranslated regions unrelated to DMPK have also provided demonstration of toxicity (de Haro et al., 2006; Mankodi et al., 2000)
The toxic gain of function that results from the DM1 and DM2 repeat expansions appears to be mediated in part through abnormal interactions of the respective RNAs with proteins controlling RNA metabolism. Skeletal and cardiac muscle have been the primary sites of investigation due to the presence of myotonia in most patients. RNA binding proteins with an affinity for CUG repeats (CELF1 and 2) (Timchenko et al., 1996) and human muscleblind proteins (MBNL1-3) (Wang et al., 2012) have received considerable attention; their typical developmental changes in abundance are modified in the presence of the expanded CUG repeat containing RNA (Fardaei et al., 2001; Lin et al., 2006; Mankodi et al., 2001; Timchenko et al., 2001). This in turn results in changes to the developmentally regulated patterns of alternative splicing of numerous RNA targets in muscle (Kalsotra et al., 2008). Modulating the levels of these RNA binding proteins in mouse models can mimic these changes, and result in similar pathology (Kanadia et al., 2003) or rescue (Kanadia et al., 2006; Charizanis et al., 2012). Echeverria and Cooper (Echeverria and Cooper, 2012) recently published an excellent review of the effects of the CUG and CCUG expansions on RNA binding proteins. Some downstream effects of the alterations in mRNA metabolism have been described. A particularly striking example is altered splicing resulting from reduced MBNL1 levels that leads to reduced levels of the chloride channel CLCN1 in both mouse models and skeletal muscle of patients. The characteristic myotonia of DM results from CLCN1 reduction, and can be rescued by overexpression of MBNL1 (Kanadia et al., 2003; Kanadia et al., 2006).
Many challenges remain for understanding the various effects of the expanded CUG sequences expressed in patients with myotonic dystrophy. The behavioral and cognitive aspects of the disorder have received significant attention, and there has been some exciting progress showing alterations in differentiated embryonic stem cells demonstrating neurite path finding defects (Marteyn et al., 2011). It is clear that many transcripts are affected at a variety of levels (Du et al., 2010), and that a large fraction of the observed effects on protein and RNA abundance may be secondary to the primary disruption by the repeat-containing RNA.
The findings that the expanded CUG repeat expressed in RNA is likely to be the principal agent of cellular damage in myotonic dystrophy and that haploinsufficiency of DMPK is not highly pathogenic allows consideration of therapeutic strategies that target the RNA directly. Mahadevan et al. demonstrated in an inducible DMPK overexpression model in the mouse that DM muscle phenotypes were reversed after shutting off expression, suggesting that damage in muscle was not permanent (Mahadevan et al., 2006). A number of recent studies have targeted the DMPK mRNA expanded CUG repeat with the aim of disassociating interacting proteins. Antisense oligonucleotides (Mulders et al., 2009; Wheeler et al., 2009) (Francois et al., 2011; Lee et al., 2012), small peptides (Garcia-Lopez et al., 2011) or small molecules (Childs-Disney et al., 2012a; Childs-Disney et al., 2012b; Parkesh et al., 2012; Warf et al., 2009) have targeted the CUG sequence to disrupt binding or destroy the mutant RNA and have shown significant efficacy in modifying downstream effects. Mahavedan argues that it might be best to destroy the expanded RNA (Mahadevan, 2012), since displacing its protein partners and eliminating RNA foci still leaves the RNA available for other possible mischief. The Cooper and Thornton groups recently describe methods to utilize gapmer oligonucleotides to induce RNaseH cleavage in the nucleus of muscle of a transgenic DM1 mouse model (Wheeler et al., 2012; Lee et al., 2012). Numerous features of the disease were corrected long term.
While evidence for a significant role for the expanded repeat carrying RNA in DM is quite persuasive, recent observations regarding the potential for expanded repeats leading to unusual protein translation offers another hypothesis regarding toxic agents. Ranum and colleagues have proposed the term Repeat-associated non-ATG (RAN) translation to describe the process encoding homopolymeric polyproteins from RNAs containing triplet repeats (Zu et al., 2011). These appear to play a role in the pathology in SCA8 (Moseley et al., 2006), and there may be polyglutamine peptides produced from the expanded DMPK allele. This phenomenon deserves attention for all of the repeat-associated disorders that involve toxic RNA.
The effort to understand the toxic nature of the non-coding CTG repeat expansion in DM1 and the intronic CCTG repeat expansion in DM2 provided considerable guidance for studying the potential pathogenic mechanisms in FXTAS. The expanded CGG repeat transcribed into mRNA in premutation carriers likely caused pathology, especially since it was not expressed in individuals with FXS, who showed no FXTAS symptoms. While the higher than normal levels of FMR1 mRNA (Kenneson et al., 2001; Tassone et al., 2000) could play a role, the expanded repeat’s enhanced ability to interact with RNA-binding proteins similarly with the DM1 mRNA was the main suspect. Jin and colleagues provided evidence that the expanded CGG in a transcript unrelated to FMR1 in a Drosophila model could cause a similar neurodegeneration to that seen in patients, including the presence of ubiquitin-positive nuclear inclusions (Jin et al., 2003). This observation strongly implicated the repeats, and subsequent work using the same model provided evidence for a role for RNA-binding proteins in modulating the phenotype (Jin et al., 2007; Sofola et al., 2007). Interestingly, one of these proteins, CELF1, also played a role in DM1 pathology, while others, PURA and HNRNPA2B1 have not been implicated. Both PURA and HNRNPA2B1 were found to bind CGG repeats in RNA, while CELF1 binds in conjunction with HNRNPA2B1.
Efforts to model the premutation associated disorder in the mouse were accelerated by existing knockin models for CGG repeat expansion that had been developed to characterize the instability of the repeat (Bontekoe et al., 1997; Lavedan et al., 1998). The model developed by Oostra and colleagues accumulated nuclear inclusions with age similarly with FXTAS (Willemsen et al., 2003), as well as behavioral and other features that correlate with the human syndrome (Brouwer et al., 2008; Van Dam et al., 2005). A limitation with the model is that animals begin to show phenotypes after six months, increasing analysis times. Hashem and colleagues developed transgenic mice that express mRNAs with 90 untranslated CGG repeats in both FMR1 and enhanced green fluorescent protein contexts specifically in Purkinje neurons, observing nuclear inclusions, cell death and behavioral deficits (Hashem et al., 2009). These mice provided evidence in a mammalian model that expressed CGG repeats were both necessary and sufficient to cause in neurodegeneration. This model also develops phenotypes more rapidly, providing for faster analyses and improved surveys of modifying genes and treatments.
A hallmark feature of FXTAS is the presence of ubiquitin-stained nuclear inclusions. These are also found in fly and mouse models. They are large and typically only one is found per nucleus. They stain with antibodies to a variety of proteins with chaperone and proteolysis functions (e.g. HSP70, 20s subunit of the proteosome) (Iwahashi et al., 2006), and resemble inclusions found in polyglutamine disorders, but do not stain with anti-polyglutamine antibodies. As in DM1, CGG-repeat containing RNA can be found in the inclusions (Sellier et al., 2010; Tassone et al., 2004), and it is likely that as the RNA seeds accumulation of proteins into the inclusion, it alters these proteins’ abundance or localization, leading to events that can be toxic to the cell. Unlike DM1, the bulk of the mutant RNA is not located in the inclusion, since FMR1 mRNA is translated and there is little or no reduction of FMRP detected in patient lymphoblasts (Kenneson et al., 2001; Tassone et al., 2000). Interestingly, reduced FMRP is found in brains of mice with large CGG repeat expansions (Iliff et al, 2013). While evidence for similar premutation-associated changes in brains of individuals with FXTAS is lacking, these men do not exhibit cognitive difficulties prior to onset of the disorder later in life, suggesting that any reduction in protein levels that may be present does not affect intelligence. It remains unclear whether the inclusions represent a toxic entity or clearance of the toxic RNA. Interest in the components of the inclusion led to their purification from human autopsy material with subsequent identification of proteolysis products with mass spectroscopy (Iwahashi et al., 2006). A large number of additional proteins were found, including histones, intermediate filaments, microtubules, myelin associated proteins and RNA binding proteins HNRNPA2B1 and MBNL1.
Sellier and colleagues described sequestration of SAM68 into nuclear inclusions in cells transfected with CGG expressing transgenes and subsequently in a mouse FXTAS model and in human samples (Sellier et al., 2010). They propose that SAM68 is required for MBNL1 and HNRNPG association in the inclusions. SAM68 is an RNA-binding protein with a role in control of alternative splicing, and they show changes to SAM68 targets’ splicing patterns in patients. PURA is one of the RNA-binding proteins that interact with expanded CGGs in RNA, can be found in the inclusions in patients, and modifies phenotypes in the Drosophila model (Jin et al., 2007). Mice lacking PURA are born without symptoms but rapidly develop severe tremor and seizures (Hokkanen et al., 2012; Khalili et al., 2003). Qurashi and colleagues defined more than 100 proteins that interact with PURA in Drosophila, including FMRP (Qurashi et al., 2011). Among these is Rm62, which shows reduced levels in the presence of expanded CGG transcripts, with subsequent nuclear accumulation of mRNA encoding Hsp70 and other stress response transcripts. It appears likely that FXTAS, like DM1 and DM2, results at least partially from changes in patterns of alternative RNA splicing, transport and translation driven by quantitative changes in RNA-binding proteins.
Therapeutic intervention strategies for FXTAS are following the paths defined for DM1, although with the much larger challenge of targeting neurons instead of muscle. Already, small molecules that disrupt CGG structure are in development (Disney et al., 2012), and show efficacy in cell culture models. A drug screen utilizing the Drosophila model has provided small molecules that reduce the neurodegeneration phenotype (Qurashi et al., 2012). Among these are phospholipase A(2) inhibitors, which may have promise for treatment. FXTAS mouse models will provide data on the ability of these compounds to alter the course of FXTAS phenotypes.
The myotonic dystrophies and FXTAS demonstrate that expanded microsatellites expressed in RNA can have toxic effects. Several other neurodegenerative disorders resulting from non-coding trinucleotide repeat expansions appear to have similar mechanisms, including SCA8 and HDL2. Antisense transcripts in many of the disease loci provide additional possibilities, and it remains possible that aspects of the coding-sequence repeat expansion disorders result from toxic RNA (Tan et al., 2012). Additional enlarged microsatellites expressed in RNA appear capable of inducing neurodegeneration. Examples include ATTCT repeat expansion in SCA10 (Matsuura et al., 2000), GGCCTG repeat expansion in SCA36 (Kobayashi et al., 2011) and GGGGCC repeat expansion in a common dominant familial form of amyotrophic lateral sclerosis and frontotemporal dementia (DeJesus-Hernandez et al., 2011; Renton et al., 2011). As more examples of transcribed polymorphic microsatellites conferring pathology are uncovered, and as it becomes clear that a large fraction of the genome is transcribed into RNA, it might be of interest to investigate role of this mechanism in variable susceptibility to more common disorders. For example, is there a liability from numerous loci with small expansions that is similar to a single locus with a large expansion? Further advances in genome sequencing will be required to capture long repeated sequences, which have been more difficult to accurately determine using current high-throughput methods.
Closing Comments
Clearly one of the seminal developments in human genetics is the identification the expansion of unstable nucleotide repeats as a mutational mechanism. In addition to providing the molecular basis for several devastating neurological disorders, study of the unstable microsatellite repeat disorders described in this review also provided insight into very basic aspects of neuroscience. For example, understanding the function of FMRP was key in appreciating the importance local mRNA translation plays in regulating synaptic function. Both FXS and FXTAS/DMs also played significant roles in understanding pathways by which RNA-binding proteins regulate RNA processing, and thereby neuronal function and development. Work on SCA1 indicates there is an overlap between development and degeneration. The eventual timing of the onset of degeneration at later ages may actually be determined during early development. Thus, we should be cautious in thinking of neural development and age-dependent degeneration as discrete entities. Progress made over the first two decades of research on these unstable microsatellite disorders provides solid platforms from which hopefully effective treatments will emanate over the next several years.
We can draw important inferences from the disorders described here, largely due to expansions of trinucleotide repeats, for elucidating the mechanisms by which expansions in other recently identified dynamic mutations lead to disease pathology, e.g. the C9ORF92 hexanucleotide repeat in ALS and FTLD, and pentanucleotide repeats associated with SCA10 and SCA31. First, a detailed characterization of the disease in patients is critical. Is it inherited in a dominant or recessive fashion? If recessive, do mutation carriers have an altered phenotype? Understanding where the repeat is located relative to surrounding genes is vital for appreciating whether expansion leads to a simple loss of function as in FXS, or a toxic RNA as in FXTAS and DM. Thus, the intronic location of the unstable microsatellite repeats in C9ORF92, SCA10 and SCA31 are suggestive of either a toxic RNA as in FXTAS and DM or a loss of expression as seen in Friedreich’s ataxia. The recent report of non-ATG-mediated translation from repeat containing RNA raises the possibility that toxic peptides also contribute to disease and recent evidence from study of C9ORF72 showing that dipeptide-repeat proteins are produced from the expanded hexanucleotide repeat and aggregate (Mori et al, 2013; Ash et al., 2013) suggests there may be a role for protein toxicity in this disorder. While an expanded microsatellite can have many molecular outcomes, the key is to find those that have a substantive contribution to disease. Research highlighted here demonstrates the importance of applying a cross-species spectrum of animal models from invertebrates to mammals has in revealing and testing pathways fundamental to pathogenesis. Identification of mechanisms that are fundamental for dysfunction and degeneration of affected neural systems maximizes the likelihood of developing effective therapeutic approaches.
ACKNOWLEDGMENTS
The authors’ work in this area was supported by grants from the N.I.H. (NS022920 & NS045667, H.T.O.; HD020521 & HD024064, S.T.W.; NS051630 & HD024064, D.L.N.) Figures were drawn with assistance of J. Frisch.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tusch lT. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013 doi: 10.1016/j.neuron.2013.02.004. Published online 12 February 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993a;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Ashley CT, Sutcliffe JS, Kunst CB, Leiner HA, Eichler EE, Nelson DL, Warren ST. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat. Genet. 1993b;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat. Rev. Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J. Clin. Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni B, Sittler A, Shen Y, Mandel JL. Analysis of domains affecting intracellular localization of the FMRP protein. Neurobiol. Dis. 1997;4:329–336. doi: 10.1006/nbdi.1997.0142. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, Bardoni B. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis EM, Hessl D, Rathmell B, Zarevics P, Cherubini M, Walton-Bowen K, Mu Y, Nguyen DV, Gonzalez-Heydrich J, Wang PP, Carpenter RL, Bear MF, Hagerman RJ. Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: a randomized, controlled, phase 2 trial. Sci. Transl. Med. 2012;4:152ra127. doi: 10.1126/scitranslmed.3004214. [DOI] [PubMed] [Google Scholar]
- Berul CI, Maguire CT, Aronovitz MJ, Greenwood J, Miller C, Gehrmann J, Housman D, Mendelsohn ME, Reddy S. DMPK dosage alterations result in atrioventricular conduction abnormalities in a mouse myotonic dystrophy model. The Journal of clinical investigation. 1999;103:R1–R7. doi: 10.1172/JCI5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Ann. Rev. Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat. Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontekoe CJ, de Graaff E, Nieuwenhuizen IM, Willemsen R, Oostra BA. FMR1 premutation allele (CGG)81 is stable in mice. European journal of human genetics : EJHG. 1997;5:293–298. [PubMed] [Google Scholar]
- Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Therapy. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Dorr N, Pilorge M, Takahashi N, Buxbaum JD. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One. 2012;7:e42422. doi: 10.1371/journal.pone.0042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Brouwer JR, Severijnen E, de Jong FH, Hessl D, Hagerman RJ, Oostra BA, Willemsen R. Altered hypothalamus-pituitary-adrenal gland axis regulation in the expanded CGG-repeat mouse model for fragile Xassociated tremor/ataxia syndrome. Psychoneuroendocrinology. 2008;33:863–873. doi: 10.1016/j.psyneuen.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Chang S, Bray SM, Li Z, Zarnescu DC, He C, Jin P, Warren ST. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat. Chem. Biol. 2008;4:256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, Kumar A, Ashizawa T, Clark HB, Kimura T, Takahashi MP, Fujimura H, Jinnai K, Yoshikawa H, Gomes-Pereira M, Gourdon G, Sakai N, Nishino S, Foster TC, Ares M, Jr, Darnell RB, Swanson MS. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, Zoghbi HY. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- Chen YW, Allen MD, Veprintsev DB, Lowe J, Bycroft M. The Structure of the AXH Domain of Spinocerebellar Ataxin-1JBiol. Chem. 2004;279:3758–3765. doi: 10.1074/jbc.M309817200. [DOI] [PubMed] [Google Scholar]
- Chevalier-Larsen ES, O'Brien CJ, Wang H, Jenkins SC, Holder L, Lieberman AP, Merry DE. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J. Neurosci. 2004;24:4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico LK, Van Eldik LJ, Watterson DM. Targeting protein kinases in central nervous system disorders. Nat. Rev. Drug Discov. 2009;8:892–909. doi: 10.1038/nrd2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Hoskins J, Rzuczek SG, Thornton CA, Disney MD. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS chemical biology. 2012a;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Parkesh R, Nakamori M, Thornton CA, Disney MD. Rational Design of Bioactive, Modularly Assembled Aminoglycosides Targeting the RNA that Causes Myotonic Dystrophy Type 1. ACS chemical biology. 2012b;7:1984–1993. doi: 10.1021/cb3001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST. Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. Am. J. Med. Genet. A. 2008;146A:1358–1367. doi: 10.1002/ajmg.a.32261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am. J. Hum. Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- Collins SC, Bray SM, Suhl JA, Cutler DJ, Coffee B, Zwick ME, Warren ST. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am. J. Med. Genet. A. 2010;152A:2512–2520. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway GS, Payne NN, Webb J, Murray A, Jacobs PA. Fragile X premutation screening in women with premature ovarian failure. Hum. Reprod. 1998;13:1184–1187. doi: 10.1093/humrep/13.5.1184. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 transgenic mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- Curia G, Papouin T, Seguela P, Avoli M. Downregulation of tonic GABAergic inhibition in a mouse model of fragile X syndrome. Cereb. Cortex. 2009;19:1515–1520. doi: 10.1093/cercor/bhn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetanovic M, Patel J, Kini A, Hugo V, Opal P. VEGF ameliorates the ataxic phenotype in spinocerebellar ataxia 1 (SCA1) mice. Nat. Med. 2011;17:1445–1447. doi: 10.1038/nm.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hulst C, De Geest N, Reeve SP, Van Dam D, De Deyn PP, Hassan BA, Kooy RF. Decreased expression of the GABAA receptor in fragile X syndrome. Brain Res. 2006;1121:238–245. doi: 10.1016/j.brainres.2006.08.115. [DOI] [PubMed] [Google Scholar]
- D'Hulst C, Heulens I, Brouwer JR, Willemsen R, De Geest N, Reeve SP, De Deyn PP, Hassan BA, Kooy RF. Expression of the GABAergic system in animal models for fragile X syndrome and fragile X associated tremor/ataxia syndrome (FXTAS) Brain Res. 2009;1253:176–183. doi: 10.1016/j.brainres.2008.11.075. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- de Chiara C, Menon RP, Strom M, Gibson TJ, Pastore A. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One. 2009;4:e8372. doi: 10.1371/journal.pone.0008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego Otero Y, Severijnen LA, van Cappellen G, Schrier M, Oostra B, Willemsen R. Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol. Cell. Biol. 2002;22:8332–8341. doi: 10.1128/MCB.22.23.8332-8341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haro M, Al-Ramahi I, De Gouyon B, Ukani L, Rosa A, Faustino NA, Ashizawa T, Cooper TA, Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Gilmer HF, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung G-YR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M., Jr Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Bio. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Wang J, Zhu H, Rinaldo L, Hu Y, Lamar K-M, Palmenberg AC, Hansel C, Gomez CM. A second cistron in the CACNA1A gene encodes a transcription factor that mediates cerebellar development and SCA6. Cell. 2013 doi: 10.1016/j.cell.2013.05.059. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick L, Barnes J, Ebner B, Agrawal S, Andresen JM, Lim J, Giesler GJ, Zoghbi HY, Orr HT. SCA1-like disease in mice expressing wild type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Echeverria GV, Cooper TA. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain research. 2012;1462:100–111. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nuc. Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Holden JJ, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, Ward PA, Nelson DL. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Richards S, Gibbs RA, Nelson DL. Fine structure of the human FMR1 gene. Hum. Mol. Genet. 1993;2:1147–1153. doi: 10.1093/hmg/2.8.1147. [DOI] [PubMed] [Google Scholar]
- Emamian ES, Kaytor MD, Duvick LA, Zu T, Susan KTouseyS.K, Zoghbi HY, Clark HB, Orr HT. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- Fahling M, Mrowka R, Steege A, Kirschner KM, Benko E, Forstera B, Persson PB, Thiele BJ, Meier JC, Scholz H. Translational regulation of the human achaete-scute homologue-1 by fragile X mental retardation protein. J. Biol. Chem. 2009;284:4255–4266. doi: 10.1074/jbc.M807354200. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997a;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 1997b;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois V, Klein AF, Beley C, Jollet A, Lemercier C, Garcia L, Furling D. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nature Struc. Mol. Bio. 2011;18:85–87. doi: 10.1038/nsmb.1958. [DOI] [PubMed] [Google Scholar]
- Fryer JD, Yu P, Kang H, Mandel-Brehm C, Carter A, Crespo-Barreto J, Gao Y, Flora A, Shaw C, Orr HT, Zoghbi HY. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334:690–693. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Jr, Warren ST, Oostra BA, Nelson DL, Caskey CT. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Fu YH, Pizzuti A, Fenwick RG, Jr, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P, Wieringa B, Korneluk R, Perryman MB, Epstein HF, Caskey CT. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Gantois I, Vandesompele J, Speleman F, Reyniers E, D'Hooge R, Severijnen LA, Willemsen R, Tassone F, Kooy RF. Expression profiling suggests underexpression of the GABA(A) receptor subunit delta in the fragile X knockout mouse model. Neurobiol. Dis. 2006;21:346–357. doi: 10.1016/j.nbd.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez A, Llamusi B, Orzaez M, Perez-Paya E, Artero RD. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc. Natl. Acad. Sci. USA. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrking KM, Andresen JM, Duvick L, Lough J, Zoghbi HY, Orr HT. Partial Loss of Tip60 Slows Midstage Neurodegeneration in a Spinocerebellar Ataxia Type 1 (SCA1) Mouse Model. Hum. Mol. Genet. 2011;20:2204–2212. doi: 10.1093/hmg/ddr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, Hessl D, Becker EJ, Papazian J, Leehey MA, Hagerman RJ, Hagerman PJ. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- Greenough WT, Klintsova AY, Irwin SA, Galvez R, Bates KE, Weiler IJ. Synaptic regulation of protein synthesis and the fragile X protein. Proc. Natl. Acad. Sci. USA. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronskov K, Brondum-Nielsen K, Dedic A, Hjalgrim H. A nonsense mutation in FMR1 causing fragile X syndrome. Eur. J. Hum. Genet. 2011;19:489–491. doi: 10.1038/ejhg.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Yao X, Pong DL, Jeromin A, Bassell GJ. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J. Neurosci. 2011;31:5693–5698. doi: 10.1523/JNEUROSCI.6661-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Misha R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Serines 13 and 16 are critical determinant of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- Haines JL, Schut LJ, Weitkamp LR, Thayer M, Anderson VE. Spinocerebellar ataxia in a large kindred: age at onset, reproduction, and genetic linkage studies. Neurology (Cleveland) 1984;43:1542–1548. doi: 10.1212/wnl.34.12.1542. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, Fitz-Hugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren B, Kruglyak L, Lander ES. Disruption of the nuclear hormone receptor RORα in staggerer, mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- Hampson DR, Adusei DC, Pacey LK. The neurochemical basis for the treatment of autism spectrum disorders and Fragile X Syndrome. Biochem. Pharmacol. 2011;81:1078–1086. doi: 10.1016/j.bcp.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Harper PS. Myotonic dystrophy. 2 edn. Philadelphia: WB Saunders; 1989. [Google Scholar]
- Harper PS, Harley HG, Reardon W, Shaw DJ. Anticipation in myotonic dystrophy: new light on an old problem. Amer. J. Hum. Genet. 1992;51:10–16. [PMC free article] [PubMed] [Google Scholar]
- Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, Nelson DL. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum. Mol. Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz D, Rousseau F, Devys D, Saccone S, Abderrahim H, Le Paslier D, Cohen D, Vincent A, Toniolo D, Della Valle G, Johnson S, Schlessinger D, Oberle I, Mandel JL. Isolation of sequences that span the fragile X and identification of a fragile X-related CpG island. Science. 1991;251:1236–1239. doi: 10.1126/science.2006411. [DOI] [PubMed] [Google Scholar]
- Hearst SM, Lopez ME, Shao Q, Liu Y, Vig PJ. Dopamine D2 receptor signaling modulates mutant ataxin-1 S776 phosphorylation and aggregation. J. Neurochem. 2010;114:706–716. doi: 10.1111/j.1471-4159.2010.06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C, Wijetunge L, Kinoshita MN, Shumway M, Hammond RS, Postma FR, Brynczka C, Rush R, Thomas A, Paylor R, Warren ST, Vanderklish PW, Kind PC, Carpenter RL, Bear MF, Healy AM. Reversal of disease-related pathologies in the fragile X mouse model by selective activation of GABA(B) receptors with arbaclofen. Sci. Transl. Med. 2012;4:152ra128. doi: 10.1126/scitranslmed.3004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokkanen S, Feldmann HM, Ding H, Jung CK, Bojarski L, Renner-Muller I, Schuller U, Kretzschmar H, Wolf E, Herms J. Lack of Puralpha alters postnatal brain development and causes megalencephaly. Hum. Mol. Genet. 2012;21:473–484. doi: 10.1093/hmg/ddr476. [DOI] [PubMed] [Google Scholar]
- Hong S, Ka S, Kim S, Park Y, Kang S. p80 coilin, a coiled body-specific protein, interacts with ataxin-1, the SCA1 gene product. Biochim. Biophys. Acta. 2003;1638:35–42. doi: 10.1016/s0925-4439(03)00038-3. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl. Acad. Sci. U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis--dependent long-term depression in hippocampal area CA1. J. Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff AJ, Renoux AJ, Krans A, Usdin K, Sutton MA, Todd PK. Impaired activity-dependent FMRP translation and enhanced mGluR-dependent LTD in Fragile X premutation mice. Hum. Mol. Genet. 2013 doi: 10.1093/hmg/dds525. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB, Mardis ER, Wilson RK, Schatz MC, McCombie WR, Wigler M. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin S, Vandelft M, Howell JL, Graczyk J, Orr HT, Truant R. RNA association and nucleocytoplasmic shuttling by ataxin-1. J. Cell Sci. 2005;118:233–242. doi: 10.1242/jcs.01611. [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Glover TW, Mayer M, Fox P, Gerrard JW, Dunn HG, Herbst DS. X-linked mental retardation: a study of 7 families. Am. J. Med. Genet. 1980;7:471–489. doi: 10.1002/ajmg.1320070408. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Leehey M, Grigsby J, Zhang L, Brunberg JA, Greco C, Des Portes V, Jardini T, Levine R, Berry-Kravis E, Brown WT, Schaeffer S, Kissel J, Tassone F, Hagerman PJ. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am. J. Hum. Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Leehey MA, Hall DA, Levine RA, Brunberg JA, Zhang L, Jardini T, Gane LW, Harris SW, Herman K, Grigsby J, Greco CM, Berry-Kravis E, Tassone F, Hagerman PJ. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Curie A, des Portes V, Torrioli MG, Berry-Kravis E, Hagerman RJ, Ramos FJ, Cornish K, He Y, Paulding C, Neri G, Chen F, Hadjikhani N, Martinet D, Meyer J, Beckmann JS, Delange K, Brun A, Bussy G, Gasparini F, Hilse T, Floesser A, Branson J, Bilbe G, Johns D, Gomez-Mancilla B. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci. Transl. Med. 2011;3:64ra61. doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen ND, Andresen JM, Lagalwar S, Armstrong B, Stevens S, Byam CE, Duvick LA, Lai S, Zoghbi HY, Clark HB, Orr HT. Phosphorylation of ATXN1 at Ser776 in the Cerebellum. J. Neurochem. 2009;110:675–686. doi: 10.1111/j.1471-4159.2009.06164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. oNatl. Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- Kennedy WR, Alter M, Sung JH. Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology. 1968;18:671–680. doi: 10.1212/wnl.18.7.671. [DOI] [PubMed] [Google Scholar]
- Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, Amini S, Gordon J. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol. Cell. Bio. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandjian EW, Huot ME, Tremblay S, Davidovic L, Mazroui R, Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl. Acad. Sci. U S A. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol. Cell Biol. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors—a perspective from Drosophila. Nat. Rev. Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Abe K, Matsuura T, Ikeda Y, Hitomi T, Akechi Y, Habu T, Liu W, Okuda H, Koizumi A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, Shihabuddin LS, et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratter IH, Finkbeiner S. PolyQ disease: too many Qs, too much function? Neuron. 2010;67:897–899. doi: 10.1016/j.neuron.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, Richards RI. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Ann. Rev. Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst CB, Warren ST. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lai S, O'Callaghan B, Zoghbi HY, Orr HT. 14-3-3 binding to ataxin-1(ATXN1) regulates its dephosphorylation at S776 and transport to the nucleus. J. Biol. Chem. 2011;286:34606–34616. doi: 10.1074/jbc.M111.238527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun E, Duvick LA, Orr HT, Botas J, Zoghbi HY. Mutant ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repear expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavedan C, Grabczyk E, Usdin K, Nussbaum RL. Long uninterrupted CGG repeats within the first exon of the human FMR1 gene are not intrinsically unstable in transgenic mice. Genomics. 1998;50:229–240. doi: 10.1006/geno.1998.5299. [DOI] [PubMed] [Google Scholar]
- Lee JE, Bennett CF, Cooper TA. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc. Natl. Acad. Sci. USA. 2012;109:4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–719. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, Seth PP, Swayze EE, Crooke ST. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Lorenzetti D, Watase K, Xu B, Antalffy B, Matzuk MM, Guos Q, Wang P, Orr HT, Zoghbi HT. Repeat instability and motor incoordination in mice with a targeted expanded CAG repeat in the Sca1 locus. Hum. Mol. Genet. 2000;9:779–785. doi: 10.1093/hmg/9.5.779. [DOI] [PubMed] [Google Scholar]
- Lubs HA. A Marker X-Chromosome. Am. J. Hum. Genet. 1969;21:231–244. [PMC free article] [PubMed] [Google Scholar]
- Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat. Genet. 1995;10:483–485. doi: 10.1038/ng0895-483. [DOI] [PubMed] [Google Scholar]
- Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O'Hoy K, Leblond S, Earle-Macdonald J, De Jong PJ, Wieringa B, Robert Korneluk RG. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Mahadevan MS. Myotonic dystrophy: is a narrow focus obscuring the rest of the field? Curr. Opin. Neurol. 2012;25:609–613. doi: 10.1097/WCO.0b013e328357b0d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malter HE, Iber JC, Willemsen R, de Graaff E, Tarleton JC, Leisti J, Warren ST, Oostra BA. Characterization of the full fragile X syndrome mutation in fetal gametes. Nat. Genet. 1997;15:165–169. doi: 10.1038/ng0297-165. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Marteyn A, Maury Y, Gauthier MM, Lecuyer C, Vernet R, Denis JA, Pietu G, Peschanski M, Martinat C. Mutant human embryonic stem cells reveal neurite and synapse formation defects in type 1 myotonic dystrophy. Cell Stem Cell. 2011;8:434–444. doi: 10.1016/j.stem.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Martin JP, Bell J. A Pedigree of Mental Defect Showing Sex-Linkage. J. Neurol. Psychiatry. 1943;6:154–157. doi: 10.1136/jnnp.6.3-4.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Yamagata T, Burgess DL, Rasmussen A, Grewal RP, Watase K, Khajavi M, McCall AE, Davis CF, Zu L, Achari M, Pulst SM, Alonso E, Noebels JL, Nelson DL, Zoghbi HY, Ashizawa T. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Meijer H, de Graaff E, Merckx DM, Jongbloed RJ, de Die-Smulders CE, Engelen JJ, Fryns JP, Curfs PM, Oostra BA. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum. Mol. Genet. 1994;3:615–620. doi: 10.1093/hmg/3.4.615. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamiyama M, Katsuno M, Adachi H, Doi H, Knodo N, Iida M, Ishigaki, Fujioka Y, Matsumoto S, Miyazaki Y, Tanaka F, Kurihara H, Sobue G. Naratriptan mitigates CGRP1-associated motor neuron degeneration caused by an expanded polyglutamine repeat tract. Nat. Med. 2012;18:1531–1538. doi: 10.1038/nm.2932. [DOI] [PubMed] [Google Scholar]
- Monson RP, Soong D, de Chiara C, Holt MR, Anilkumar N, Pastore A. The importance of serine 776 in Atxain-1 partner selection: a FRET analysis. Scientific Reports. 2012;2:919. doi: 10.1038/srep00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Dieter Edbauer D. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science. 2013 doi: 10.1126/science.1232927. Published online 7 February 2013. [DOI] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Bio. 2008;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton NE, Lalouel JM, Jackson JF, Currier RD, Yee S. Linkage studies in spinocerebellar ataxia (SCA) Am. J. Med. Genet. 1980;6:251–257. doi: 10.1002/ajmg.1320060309. [DOI] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, Day JW, Ranum LP. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, Wieringa B, Wansink DG. Triplet-repeat oligonucleotidemediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M, Nalavadi V, Epstein MP, Narayanan U, Bassell GJ, Warren ST. Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc. Natl. Acad. Sci. U S A. 2007;104:15537–15542. doi: 10.1073/pnas.0707484104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J. Neurosci. 2012;32:2582–2587. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J. Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J. Biol. Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino HE, Noreen HJ, Dubey DP. A family with hereditary ataxia: HLA typing. Neurology (NY) 1980;30:12–20. doi: 10.1212/wnl.30.1.12. [DOI] [PubMed] [Google Scholar]
- Nolin SL, Glicksman A, Ding X, Ersalesi N, Brown WT, Sherman SL, Dobkin C. Fragile X analysis of 1112 prenatal samples from 1991 to 2010. Prenat. Diagn. 2011;31:925–931. doi: 10.1002/pd.2815. [DOI] [PubMed] [Google Scholar]
- Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas M, Mandel J. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Orr HT, Chung MY, Banfi S, Kwiatkowski TJ, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LP, Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide Repeat Disorders. Ann. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pacey LK, Heximer SP, Hampson DR. Increased GABA(B) receptor-mediated signaling reduces the susceptibility of fragile X knockout mice to audiogenic seizures. Mol. Pharmacol. 2009;76:18–24. doi: 10.1124/mol.109.056127. [DOI] [PubMed] [Google Scholar]
- Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman D, Thornton CA, Disney MD. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J. Am. Chem. Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A, Temussi PA. The two faces of Janus: functional interactions and protein aggregation. Curr. Opin. Struct. Bio. 2012;22:30–37. doi: 10.1016/j.sbi.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Ann. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J. Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qurashi A, Li W, Zhou JY, Peng J, Jin P. Nuclear accumulation of stress response mRNAs contributes to the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2011;7:e1002102. doi: 10.1371/journal.pgen.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qurashi A, Liu H, Ray L, Nelson DL, Duan R, Jin P. Chemical screen reveals small molecules suppressing fragile X premutation rCGG repeat-mediated neurodegeneration in Drosophila. Hum. Mol. Genet. 2012;21:2068–2075. doi: 10.1093/hmg/dds024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S, Smith DB, Rich MM, Leferovich JM, Reilly P, Davis BM, Tran K, Rayburn H, Bronson R, Cros D, Balice-Gordon RJ, Housman D. Mice lacking the myotonic dystrophy protein kinase develop a late onset progressive myopathy. Nat. Genet. 1996;13:325–335. doi: 10.1038/ng0796-325. [DOI] [PubMed] [Google Scholar]
- Renoux AJ, Todd PK. Neurodegeneration the RNA way. Prog. Neurobiol. 2012;97:173–189. doi: 10.1016/j.pneurobio.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Jamie Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister J, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita V-M, Kaivorinne A-L, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, The ITALSGEN Consortium, Heckerman D, Rogaeva E, Zinman L, Rothstein J, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyniers E, Wolff G, Tariverdian G, De Boulle K, Storm K, Kooy RF, Willems PJ. Severe mental retardation and macroorchidism without mutation in the FMR1 gene. Am. J. Med. Genet. 1996;64:408–412. doi: 10.1002/(SICI)1096-8628(19960809)64:2<408::AID-AJMG35>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Revenga L, Madrigal I, Pagonabarraga J, Xuncla M, Badenas C, Kulisevsky J, Gomez B, Mila M. Penetrance of FMR1 premutation associated pathologies in fragile X syndrome families. Eur. J. Hum. Genet. 2009;17:1359–1362. doi: 10.1038/ejhg.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Schmidt BJ, Greenberg CR, Allingham-Hawkins DJ, Spriggs EL. Expression of X-linked bulbospinal muscular atrophy (Kennedy disease) in two homozygous women. Neurology. 2002;59:770–772. doi: 10.1212/wnl.59.5.770. [DOI] [PubMed] [Google Scholar]
- Schut JW. Hereditary ataxia. Clinical Study Through Six Generations. Arch. Neurol. Psych. 1950;63:535–568. [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular Mechanisms of Fragile X Syndrome: A Twenty-Year Perspective. Ann. Rev. Path. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, Lysholm A, Burright E, Zoghbi HY, Clark HB, Andresen JM, Orr HT. RORα-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Shelbourne P, Johnson K. Myotonic dystrophy: another case of too many repeats? Hum. Mut. 1992;1:183–189. doi: 10.1002/humu.1380010302. [DOI] [PubMed] [Google Scholar]
- Sherman SL, Jacobs PA, Morton NE, Froster-Iskenius U, Howard-Peebles PN, Nielsen KB, Partington MW, Sutherland GR, Turner G, Watson M. Further segregation analysis of the fragile X syndrome with special reference to transmitting males. Hum. Genet. 1985;69:289–299. doi: 10.1007/BF00291644. [DOI] [PubMed] [Google Scholar]
- Sherman SL, Morton NE, Jacobs PA, Turner G. The marker (X) syndrome: a cytogenetic and genetic analysis. Ann. Hum. Genet. 1984;48:21–37. doi: 10.1111/j.1469-1809.1984.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trend Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- Sullivan AK, Marcus M, Epstein MP, Allen EG, Anido AE, Paquin JJ, Yadav-Shah M, Sherman SL. Association of FMR1 repeat size with ovarian dysfunction. Hum. Reprod. 2005;20:402–412. doi: 10.1093/humrep/deh635. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Sutherland GR. Fragile sites on human chromosomes: demonstration of their dependence on the type of tissue culture medium. Science. 1977;197:265–266. doi: 10.1126/science.877551. [DOI] [PubMed] [Google Scholar]
- Sutherland GR, Haan EA, Kremer E, Lynch M, Pritchard M, Yu S, Richards RI. Hereditary unstable DNA: a new explanation for some old genetic questions? Lancet. 1991;338:289–292. doi: 10.1016/0140-6736(91)90426-p. [DOI] [PubMed] [Google Scholar]
- Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, Kanuka H, Miura M, Tabata T, Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor inDrosophila. Neuron. 2002;35:855–864. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Takeyama K, Ito S, Sawatsubashi S, Shirode Y, Yamamoto A, Suzuki E, Maki A, Yamagata K, Zhao Y, Kouzmenko A, Tabata T, Kato S. A novel genetic system for analysis of co-activators for the N-terminal transactivation function domain of the human androgen receptor. Biosci. Biotechnol. Biochem. 2004;68:1209–1215. doi: 10.1271/bbb.68.1209. [DOI] [PubMed] [Google Scholar]
- Tan H, Xu Z, Jin P. Role of noncoding RNAs in trinucleotide repeat neurodegenerative disorders. Experimental neurology. 2012;235:469–475. doi: 10.1016/j.expneurol.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Bio. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Bio. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nuc. Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Tsai C-C, Kao H-Y, Mitzutani A, Banayo E, Rajan H, McKeown M, Evans RM. Ataxin-1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc. Natl. Acad. Sci. USA. 2004;101:4047–4052. doi: 10.1073/pnas.0400615101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Jafar-Nejad H, Patel AJ, Sun Y, Chen HK, Rose MF, Venken KJ, Botas J, Orr HT, Bellen HJ, Zoghbi HY. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–644. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Udd B, Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. doi: 10.1016/S1474-4422(12)70204-1. [DOI] [PubMed] [Google Scholar]
- Van Dam D, Errijgers V, Kooy RF, Willemsen R, Mientjes E, Oostra BA, De Deyn PP. Cognitive decline, neuromotor and behavioural disturbances in a mouse model for fragile-X-associated tremor/ataxia syndrome (FXTAS) Behav. Brain Res. 2005;162:233–239. doi: 10.1016/j.bbr.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Verheij C, Bakker CE, de Graaff E, Keulemans J, Willemsen R, Verkerk AJ, Galjaard H, Reuser AJ, Hoogeveen AT, Oostra BA. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature. 1993;363:722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, Eussen BE, van Ommen G-JB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Caskey CT, Nelson DL, Oostra BA, Warren ST. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wang YC, Lin ML, Lin SJ, Li YC, Li SY. Novel point mutation within intron 10 of FMR-1 gene causing fragile X syndrome. Hum. Mutat. 1997;10:393–399. doi: 10.1002/(SICI)1098-1004(1997)10:5<393::AID-HUMU10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, Lécuyer E, Burge CB. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watase K, Weeber EJ, Xu B, Antalffy B, Yuva-Paylor L, Hashimoto K, Kano M, Atkinson DL, Sweatt JD, Orr HT, Paylor R, Zoghbi HY. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–919. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, van Unen L, Tassone F, Hoogeveen AT, Hagerman PJ, Mientjes EJ, Oostra BA. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum. Mol. Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SI, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BI. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Yakura H, Wakisaka A, Fujimoto S, Itakura K. Hereditary ataxia and HLA genotypes. N. Engl. J. Med. 1974;291:154–155. doi: 10.1056/NEJM197407182910314. [DOI] [PubMed] [Google Scholar]
- Yrigollen CM, Durbin-Johnson B, Gane L, Nelson DL, Hagerman R, Hagerman PJ, Tassone F. AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet. Med. 2012;14:729–736. doi: 10.1038/gim.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Pendergraff H, Liu J, Kordasiewicz H, Cleveland DW, Swayze EE, Lima WF, Crooke SY, Prakash TP, Corey DR. Single-Stranded RNAs Use RNAi to Potently and Allele-Selectively Inhibit Mutant Huntingtin Expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S, Serra H, Zoghbi HY, Orr HT. The SCA1 protein, ataxin-1, has RNA-binding activity that is inversely affected by the length of its polyglutamine tract. Hum. Mol. Genet. 2001;10:25–30. doi: 10.1093/hmg/10.1.25. [DOI] [PubMed] [Google Scholar]
- Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, Darnell JC, Darnell RB. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N, Ju W, Pietrofesa J, Wang D, Dobkin C, Brown WT. Fragile X "gray zone" alleles: AGG patterns, expansion risks, and associated haplotypes. Am. J. Med. Genet. 1996;64:261–265. doi: 10.1002/(SICI)1096-8628(19960809)64:2<261::AID-AJMG5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, Dobyns WB, Subramony SH, Zoghbi HY, Lee CC. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat. Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harbor Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Duvick LA, Kaytor MD, Berlinger M, Zoghbi HY, Clark HB, Orr HT. Recovery from polyglutamine-Induced neurodegeneration in conditionalSCA1, transgenic mice. J. Neurosci. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, Nan Z, Forster C, Low WC, Schoser B, Somia NV, Clark HB, Schmechel S, Bitterman PB, Gourdon G, Swanson MS, Moseley M, Ranum LP. Non-ATGinitiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]