The interaction between astral microtubules and the cell cortex is accompanied by constant cortical release and transport of LGN/dynein complex, which is modulated by cortical actin filaments. Regulated cortical release and transport of LGN/dynein complex along astral microtubules may contribute to spindle positioning in mammalian cells.
Abstract
Spindle positioning is believed to be governed by the interaction between astral microtubules and the cell cortex and involve cortically anchored motor protein dynein. How dynein is recruited to and regulated at the cell cortex to generate forces on astral microtubules is not clear. Here we show that mammalian homologue of Drosophila Pins (Partner of Inscuteable) (LGN), a Gαi-binding protein that is critical for spindle positioning in different systems, associates with cytoplasmic dynein heavy chain (DYNC1H1) in a Gαi-regulated manner. LGN is required for the mitotic cortical localization of DYNC1H1, which, in turn, also modulates the cortical accumulation of LGN. Using fluorescence recovery after photobleaching analysis, we show that cortical LGN is dynamic and the turnover of LGN relies, at least partially, on astral microtubules and DYNC1H1. We provide evidence for dynein- and astral microtubule–mediated transport of Gαi/LGN/nuclear mitotic apparatus (NuMA) complex from cell cortex to spindle poles and show that actin filaments counteract such transport by maintaining Gαi/LGN/NuMA and dynein at the cell cortex. Our results indicate that astral microtubules are required for establishing bipolar, symmetrical cortical LGN distribution during metaphase. We propose that regulated cortical release and transport of LGN complex along astral microtubules may contribute to spindle positioning in mammalian cells.
INTRODUCTION
Mitotic spindle orientation plays a critical role during tissue morphogenesis by regulating organ size and shape. It is also the foundation for asymmetric cell division, a key step for stem cells to function in generating cellular diversity (Gonczy, 2008; Knoblich, 2008; Siller and Doe, 2009; Gillies and Cabernard, 2011; Morin and Bellaiche, 2011).
During the asymmetric cell division of Drosophila neuroblasts and sensory organ precursor cells, the reorientation of mitotic spindle has been shown to require a protein called Partner of Inscuteable (Pins) and the Gα subunit of the heterotrimeric G proteins, which localize asymmetrically at the cell cortex during mitosis (Parmentier et al., 2000; Schaefer et al., 2000, 2001; Fuse et al., 2003; Yu et al., 2006; Chia et al., 2008). Gene products related to Pins (GPR1/2) and Gα are also essential for spindle positioning and displacement during asymmetric cell division of the Caenorhabditis elegans zygotes (Gotta and Ahringer, 2001; Gotta et al., 2003; Colombo et al., 2003; Srinivasan et al., 2003). Elegant laser cutting experiments and living-embryo time-lapse analysis suggest that GPR 1/2 and Gα subunits are required for generating unequal pulling forces on the mitotic apparatus (Grill et al., 2001, 2003; Grill and Hyman, 2005; Pecreaux et al., 2006).
We showed previously that mammalian homologue of Drosophila Pins (LGN) functions as a conformational switch that links Gαi and the nuclear mitotic apparatus (NuMA) protein and that LGN and Gαi may exert forces on mitotic spindles in cultured mammalian cells (Du et al., 2001, 2002; Du and Macara, 2004). Subsequent studies identified Mud from Drosophila and Lin5 in C. elegans as functional homologues of NuMA (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006; van der Voet et al., 2009), suggesting the existence of an evolutionarily conserved protein complex in regulating spindle positioning. The in vivo function of mammalian Gαi/LGN/NuMA complex was not clear until recently. Accumulating evidence suggest that this ternary complex plays an important role in directing spindle orientation, not only in asymmetrically dividing stem/progenitor cells during development, but also in symmetrically dividing cells during epithelial morphogenesis (Morin et al., 2007; Konno et al., 2008; Hao et al., 2010; Poulson and Lechler, 2010; Zheng et al., 2010; Ben-Yair et al., 2011; El-Hashash and Warburton, 2011; El-Hashash et al., 2011; Peyre et al., 2011; Williams et al., 2011; Zhu et al., 2011; Matsumura et al., 2012; Xiao et al., 2012).
Spindle positioning is believed to be regulated by the interaction between astral microtubules (MTs) and the cell cortex (McCarthy and Goldstein, 2006). How the cortical Gα/LGN/NuMA ternary complex is physically linked to astral MTs is not completely understood. Although NuMA can bind to MTs, it cannot do so when associated with LGN (Du et al., 2002; Gaetz and Kapoor, 2004; Kisurina-Evgenieva et al., 2004). Another plausible candidate is cytoplasmic dynein, the most abundant minus end–directed, microtubule-based motor protein complex (Kardon and Vale, 2009). Studies from different systems suggest that cytoplasmic dynein and its functionally linked dynactin complex are involved in orienting mitotic spindles (Carminati and Stearns, 1997; Adames and Cooper, 2000; O'Connell and Wang, 2000; Dujardin and Vallee, 2002; Ahringer, 2003). The relationship between dynein and the Pins family of proteins is evident in asymmetrically dividing C. elegans zygotes, in which GPR-1/2 and Gα are linked to subunits of the dynein/dynactin complex in generating pulling forces on astral MTs (Grill et al., 2001, 2003; Pecreaux et al., 2006; Couwenbergs et al., 2007; Nguyen-Ngoc et al., 2007). More recently, the link between the ternary complex and cytoplasmic dynein was also established in mitotic HeLa cells (Toyoshima et al., 2007; Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012). However, it is not clear which component of the ternary complex mediates the association with cytoplasmic dynein and how the association is regulated.
The current view of dynein-mediated force generation on astral MTs is that dynein is anchored at the cell cortex and interacts with astral MTs (Grill and Hyman, 2005; Hendricks et al., 2012; Laan et al., 2012). How dynein is anchored and regulated at the cell cortex is not known. Furthermore, dynein was also observed to localize along astral MTs (Busson et al., 1998), and in principle, dynein movement along astral MTs can also produce pulling forces (Kimura and Kimura, 2011), although such evidence is missing in mitotic mammalian cells. Recent studies also suggest that cortical actin filaments are involved in spindle positioning, either intrinsically or in response to external cues (Thery et al., 2005; Toyoshima and Nishida, 2007; Kunda and Baum, 2009; Fink et al., 2011). Whether and how actin filaments are linked to dynein-based force generator are not clear.
Here we show that LGN and cytoplasmic dynein heavy chain form a complex and regulate each other for their cortical localization and that actin filaments help to maintain LGN/dynein at the cell cortex and counteract astral microtubule–mediated cortical release and transport of LGN/dynein complex.
RESULTS
Gαi-regulated association of LGN and dynein heavy chain, cytoplasmic 1 in mammalian cells
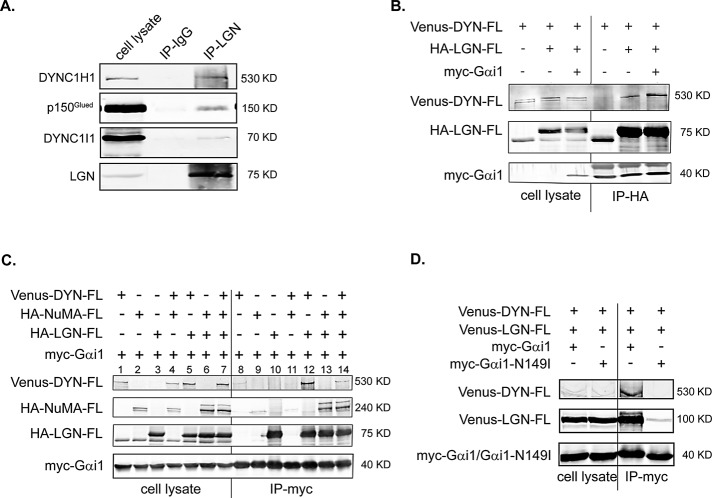
To test whether LGN associates with the dynein/dynactin complex in mammalian cells, we performed immunoprecipitation analysis using Madin–Darby canine kidney (MDCK) cells that were partially synchronized in mitosis. As shown in Figure 1A, anti-LGN antibodies, but not rabbit immunoglobulin G (IgG), could efficiently immunoprecipitate endogenous dynein heavy chain, cytoplasmic 1 (DYNC1H1). Small but specific amounts of dynein IC 1 (DYNC1I1) and dynactin subunit p150Glued were also detected in the anti-LGN immunoprecipitates, indicating that endogenous LGN and cytoplasmic dynein form a complex. Our results are consistent with a recent study showing that ectopically expressed green fluorescent protein (GFP)–LGN forms a complex with cytoplasmic dynein in HeLa cells (Kiyomitsu and Cheeseman, 2012).
FIGURE 1:
Gαi-regulated interaction between LGN and DYNC1H1. (A) Endogenous LGN and cytoplasmic dynein form a complex. MDCK II cells were partially synchronized by treating with nocodazole (200 nM) for 12 h. After being released from the treatment for 40 min, cells were harvested and subjected to immunoprecipitation using anti-LGN antibodies or rabbit IgG. The immunoprecipitates were separated by SDS–PAGE and blotted using specific antibodies. (B) Gαi enhances the association between LGN and DYNC1H1. Cos 7 cells were transfected as indicated. Cell lysates (1/30 of total) and immunoprecipitates were separated in a 6% SDS–PAGE gel and blotted using specific antibodies. Note that in 6% gel, myc-Gαi1 comigrated with and was masked by the light chain of anti-HA antibody in the HA-LGN immunoprecipitates. (C) Gαi, LGN, and DYNC1H1 form a complex in vivo, independent of NuMA. Cos 7 cells were transfected as indicated. Cell lysates (1/30 of total) and immunoprecipitates were separated in a 7% SDS–PAGE gel and blotted. (D) The GoLoco-insensitive Gαi1 cannot associate with LGN and DYNC1H1. Cos 7 cells were transfected as indicated. Cell lysates were subjected to analysis as in C.
Because DYNC1H1 turned out to be the major coimmunoprecipitate of LGN, we further studied the interaction between LGN and DYNC1H1 by coexpressing full-length DYNC1H1 and LGN in Cos 7 cells. Immunoprecipitation of HA-LGN resulted in the coprecipitation of Venus-DYNC1H1 (Figure 1B), further suggesting that they can form a complex in vivo. The C-terminus of LGN contains four GoLoco motifs that bind to GDP-bound Gαi/o (Lanier, 2004; Willard et al., 2004). LGN is normally in its closed conformation due to intramolecular interactions, and Gαi binding could switch LGN to an open state (Du and Macara, 2004). To test the effect of Gαi on the association between LGN and DYNC1H1, we included Gαi1 in the cotransfection and immunoprecipitation experiments. Of interest, coexpression of myc-Gαi1 led to obviously enhanced coimmunoprecipitation of Venus-DYNC1H1 with HA-LGN (Figure 1B), suggesting that the association between LGN and DYNC1H1 might rely on the conformation of LGN and could be regulated by Gαi1 binding.
Next we tested whether DYNC1H1, LGN, and Gαi can form a complex in vivo by transfecting cells with different combinations of Gαi1, LGN, and DYNC1H1. As shown in Figure 1C, myc-Gαi1 could efficiently immunoprecipitate HA-LGN (lanes 3 and 10). In the absence of HA-LGN, myc-Gαi1 could immunoprecipitate little, if any, Venus-DYNC1H1 (lanes 1 and 8). However, when HA-LGN was coexpressed, myc-Gαi1 could efficiently immunoprecipitate Venus-DYNC1H1 (lanes 5 and 12), suggesting that Gαi1, LGN, and DYNC1H1 can form a ternary complex in vivo.
We showed previously that NuMA can also bind to LGN and form a complex with LGN and Gαi (Du and Macara, 2004). NuMA was also shown to associate with dynein/dynactin complex in Xenopus egg extracts (Merdes et al., 1996). It is possible that endogenous NuMA is mediating the association between Gαi/LGN and DYNC1H1. To test the effect of NuMA on the DYNC1H1/LGN/Gαi complex formation, we included NuMA in the cotransfection experiments. Whereas myc-Gαi1 can immunoprecipitate HA-NuMA in the presence of HA-LGN (Figure 1C, lanes 6 and 13), coexpression of HA-NuMA resulted in reduced Venus-DYNC1H1 in the myc-Gαi1/HA-LGN complex (lanes 7 and 14), suggesting that Gαi1/LGN/DYNC1H1 complex formation is unlikely to be mediated through NuMA.
To exclude the possibility that the association between DYNC1H1 and Gαi1 is mediated through other Gαi1-binding proteins such as Gβ/γ (Sachdev et al., 2007), we used a GoLoco-insensitive Gαi mutant (Gαi1-N149I) as a control. This mutant Gαi1 specifically lacks the ability to bind to the GoLoco motif but is otherwise equivalent to wild-type Gαi1 in terms of other functional aspects, including binding to G β/γ subunits (Willard et al., 2008). As shown in Figure 1D, whereas wild-type Gαi1 could efficiently immunoprecipitate LGN and DYNC1H1, the GoLoco-insensitive Gαi1 mutant failed to do so, suggesting that the Gαi1/DYNC1H1 complex formation is mediated through LGN.
LGN is necessary and sufficient for the cortical localization of DYNC1H1 during mitosis
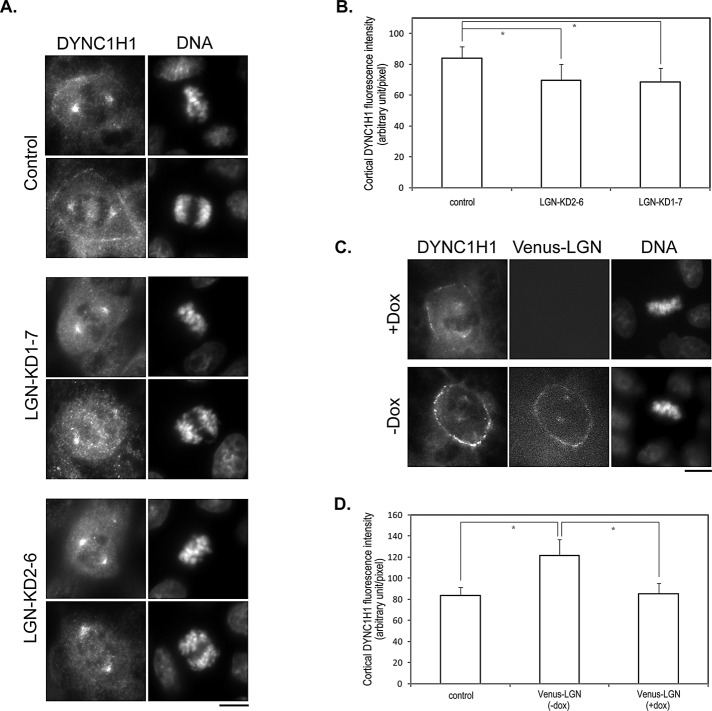
The interaction between LGN and DYNC1H1 prompted us to test whether LGN is required for the cortical localization of DYNC1H1. Cortical localization of dynactin subunits in MDCK cells had been described (Busson et al., 1998; Faulkner et al., 2000). Using an antibody against DYNC1H1, we could detect both spindle and cortical localization of DYNC1H1 (Figure 2A). The authenticity of the antibody staining was verified by using two different short hairpin RNAs (shRNAs) against canine DYNC1H1, which, upon expression, significantly eliminated the cortical and spindle staining of DYNC1H1 (Supplemental Figure S1). We then compared DYNC1H1 staining between control and stable LGN knockdown cells that we established previously (Zheng et al., 2010). Remarkably, depletion of LGN led to significantly reduced cortical localization of DYNC1H1, whereas the spindle localization of DYNC1H1 appeared normal (Figure 2, A and B). These results are consistent with recent studies in HeLa cells showing that LGN is required for mitotic cortical localization of dynein/dynactin subunits (Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012).
FIGURE 2:
LGN is required for the cortical localization of DYNC1H1 during mitosis. (A) LGN depletion results in reduced cortical localization of DYNC1H1. MDCK cells transduced by control lentivirus (control) or lentivirus expressing shRNAs targeting different regions of LGN (LGN-KD1-7 and LGN-KD2-6) were stained with anti-DYNC1H1 antibody and DNA dye. Bar, 10 μm. (B) Quantitation of the fluorescence intensity of cortical DYNC1H1 from images acquired in A. n = 50 for each set; *p < 0.01. (C) Slight overexpression of LGN leads to enhanced cortical localization of DYNC1H1. Stable Tet-Off MDCK cells expressing Venus-LGN were cultured in the presence (+Dox) or absence (–Dox) of doxycycline. At 24 h later, cells were stained as in A. Bar, 10 μm. (D) Quantitation of the fluorescence intensity of cortical DYNC1H1 from images acquired in C. n = 50 for each set; *p < 0.01.
We had established inducible MDCK cell lines that slightly overexpress Venus-tagged LGN (Du and Macara, 2004). Slight overexpression of LGN led to enhanced cortical localization of LGN and profound spindle oscillation, suggesting unbalanced forces on astral MTs (Du and Macara, 2004). We used these cell lines to test the effect of LGN overexpression on DYNC1H1 localization during mitosis. Of interest, slight overexpression of Venus-LGN resulted in significantly enhanced cortical localization of DYNC1H1 (Figure 2, C and D). These data suggest that LGN is capable of recruiting DYNC1H1 to the cell cortex during mitosis, and, most important, endogenous LGN is required for the cortical localization of DYNC1H1 during mitosis.
Dynein and astral MTs regulate the cortical localization of LGN
Surprisingly, when we compared the localization of endogenous LGN in control and DYNC1H1-knockdown cells, we found that the cortical localization of LGN was obviously enhanced in DYNC1H1-knockdown cells (Figure 3, A and C), suggesting that DYNC1H1 also regulates LGN localization. As the minus end–directed motor protein, cortical dynein is believed to exert its function through astral MTs. We wondered whether disrupting astral MTs would also affect LGN distribution. We treated cells with low doses of nocodazole, which is generally used to disrupt astral MTs (Jordan et al., 1992). Indeed, treating MDCK cells with 50 nM nocodazole led to significant loss of astral MTs, whereas the spindle MTs were not affected (Supplemental Figure S2). Of interest, disruption of astral MTs also led to enhanced cortical localization of LGN (Figure 3, B and C). Thus, whereas LGN is required for the cortical recruitment of dynein, dynein also regulates cortical LGN through astral MTs.
FIGURE 3:
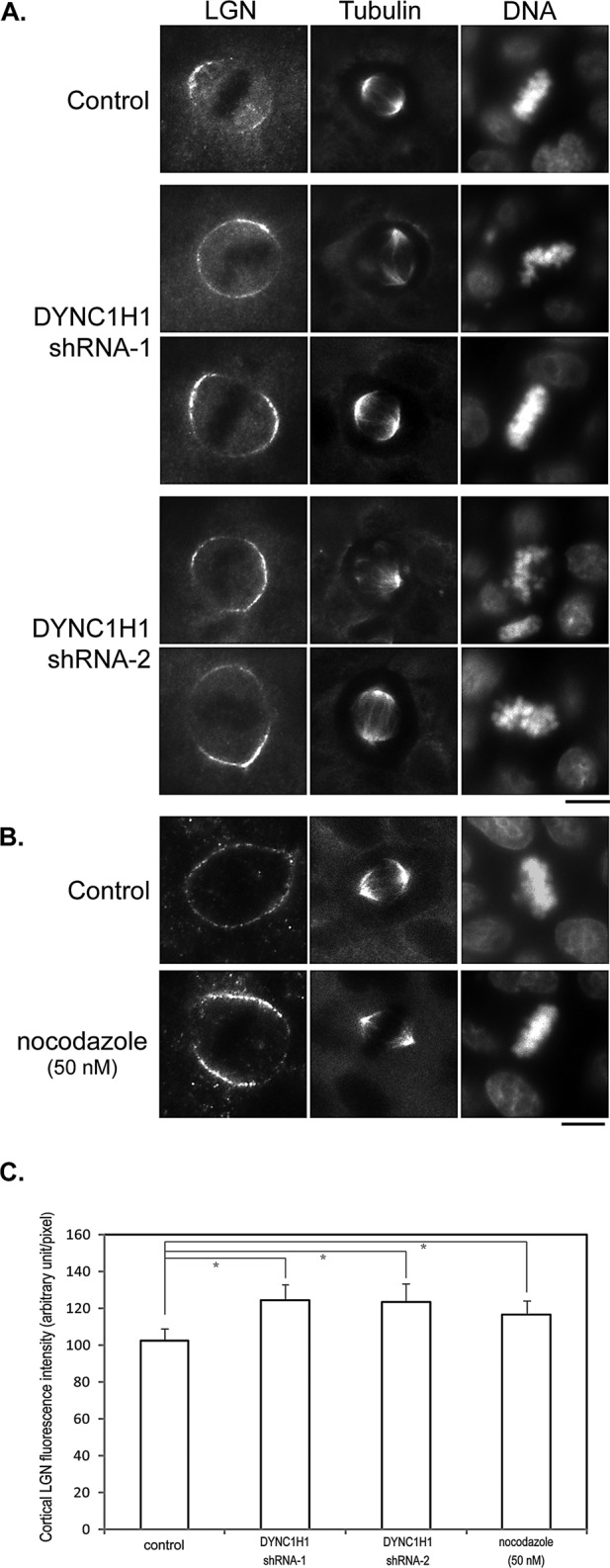
Knocking down DYNC1H1 or disrupting astral MTs leads to enhanced cortical localization of LGN. (A) MDCK cells were transfected with plasmids expressing control shRNA (control) or shRNAs targeting DYNC1H1 (shRNA1 and shRNA2). At 48 h later, cells were fixed and stained with anti-LGN, anti–α-tubulin antibodies, and DNA dye. Bar, 10 μm. (B) MDCK cells were cultured in media containing 50 nM nocodazole for 40 min. Cells were then fixed and stained as in A. Bar, 10 μm. (C) Quantitation of the fluorescence intensity of cortical LGN from images acquired in A and B. n = 50 for each set; *p < 0.01.
Dynein- and astral MT–dependent turnover of cortical LGN
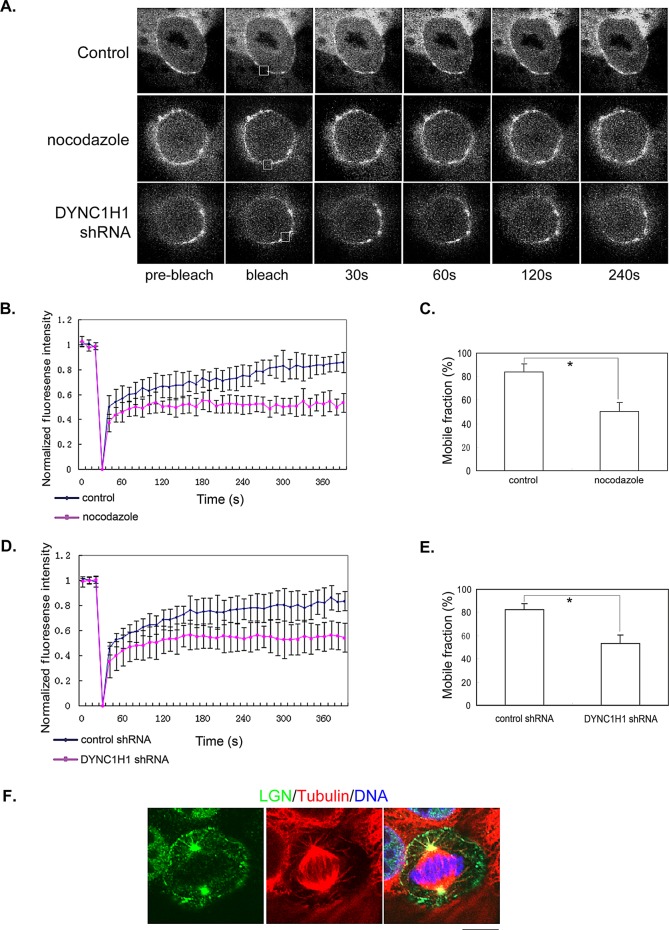
The fact that cortical LGN is modulated by dynein and astral MTs led us to speculate that it might be dynamic. We performed fluorescence recovery after photobleaching (FRAP) experiments to study the kinetics of cortical LGN using our Venus-LGN cell line. As we showed previously (Du and Macara, 2004), in live MDCK cells, cortical Venus-LGN usually exhibits a patchy distribution (Figure 4A), which is consistent with a recent report showing that GFP-DYNC1H1 also shows patchy localization at the cell cortex (Collins et al., 2012). Our FRAP analysis indicated that; indeed the fluorescence of cortical Venus-LGN recovered gradually after photobleaching, suggesting dynamic turnover (Figure 4 and Supplemental Movie S1). Next we set out to test whether astral MTs and DYNC1H1 are involved in the dynamic turnover of cortical LGN. We carried out similar FRAP experiments under conditions when astral MTs were disrupted using low doses of nocodazole or DYNC1H1 was knocked down by specific shRNA. Of interest, the fluorescence recovery of cortical Venus-LGN was significantly inhibited under both conditions (Figure 4, A–E, and Supplemental Movies S2 and S3), suggesting that astral MTs and DYNC1H1 contribute to the dynamic turnover of cortical LGN.
FIGURE 4:
Dynamic turnover of cortical LGN relies on astral MTs and DYNC1H1. (A) FRAP analysis of cortical LGN. Representative images from live-cell time-lapse series were shown. The photobleaching areas were marked by squares. Stable MDCK cells expressing Venus-LGN were either untreated (top), treated with 50 nM nocodazole for 1 h (middle), or transfected with DYNC1H1 shRNA for 48 h (bottom) before being subjected to FRAP analysis. (B–E) Quantitative analysis of FRAP experiments. (B, D) Plots of normalized fluorescence intensity of cortical Venus-LGN in cells treated with DMSO (B, blue diamonds), 50 nM nocodazole (B, pink squares), or transfected with control shRNA (D, blue diamonds) or DYNC1H1 shRNA (D, pink squares) vs. time (in seconds) after photobleaching. Data are expressed as mean ±SEM (n = 8 for each set). (C, E) The mobile fractions of cortical Venus-LGN in control, nocodazole-treated, or DYNC1H1-knockdown cells were calculated from the fluorescence recovery curves shown in B and D. Data are expressed as mean ±SEM. *p < 0.01. (F) Association of Venus-LGN with astral MTs. MDCK cells expressing Venus-LGN were preextracted with microtubule stabilization buffer containing 0.2% Triton X-100 and then fixed with 4% PFA. Fixed cells were stained with anti-LGN (green), anti–α-tubulin (red) antibodies, and DNA dye (blue). Bar, 10 μm.
It is possible that astral MTs and dynein-dependent turnover of cortical LGN are mediated through the dissociation of LGN from the dynein complex into cytosol when astral MTs reach the cell cortex. Alternatively, LGN might be transported by dynein along astral MTs. To distinguish these possibilities, we asked whether we could detect LGN on astral MTs. Indeed, in Venus-LGN cells, we observed LGN localization along astral MTs by using either the anti-LGN antibody (Figure 4F) or the anti-GFP antibody (unpublished data). Of note, Venus-LGN was not observed to localize on spindle MTs.
Dynein and astral MTs mediate transport of LGN from cell cortex to spindle poles
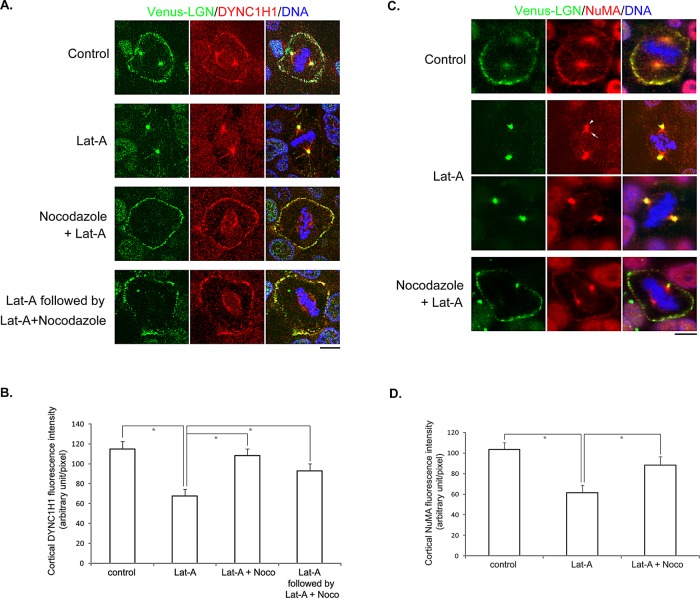
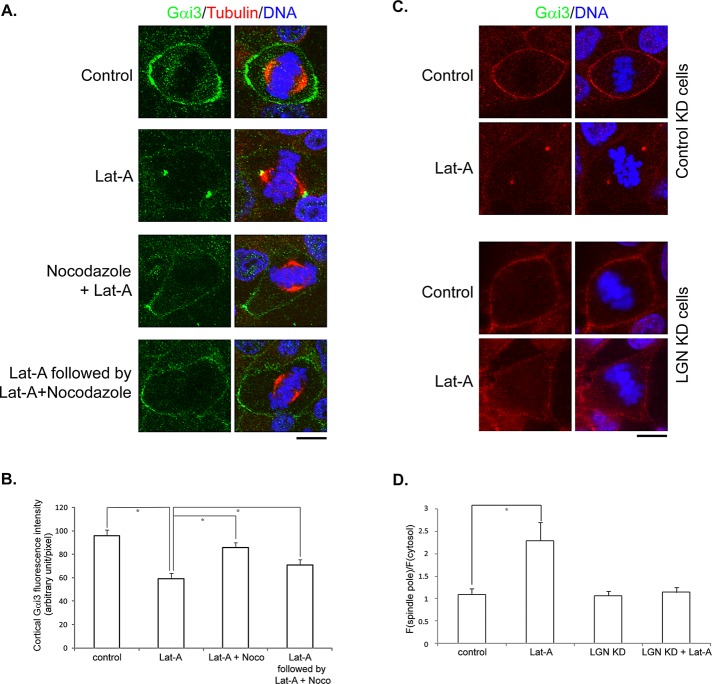
The cortical localization of LGN and dynein appears to rely on actin filaments (Busson et al., 1998; Kaushik et al., 2003; Toyoshima et al., 2007), although the underlying mechanism is not known. Indeed, LGN dissociated from the cell cortex during mitosis when MDCK cells were treated with latrunculin A (LatA; Figure 5A), a drug that depolymerizes actin filaments (Spector et al., 1983; Supplemental Figure S3). Consistent with previous studies in HeLa cells (Toyoshima and Nishida, 2007), disruption of actin filaments led to spindle misorientation in MDCK cells, although the overall spindle organization appeared normal (unpublished data). Of interest, although we could hardly detect spindle pole localization of endogenous LGN in untreated MDCK cells, we observed an obvious accumulation of LGN at the spindle poles in cells treated with LatA (Figure 5, A and B, and Supplemental Figure S3A). Of importance, the spindle pole accumulation of the anti-LGN staining was not observed in LGN-stable knockdown cells treated with LatA (Supplemental Figure S3B), indicating that it was endogenous LGN that translocated to the spindle poles when actin filaments were disrupted. Furthermore, after we washed out LatA from the cells, LGN regained cortical localization and disappeared from the spindle poles (Figure 5A). Similar results were observed when cells were treated with cytochalasin D, another inhibitor of actin filaments (unpublished data).
FIGURE 5:
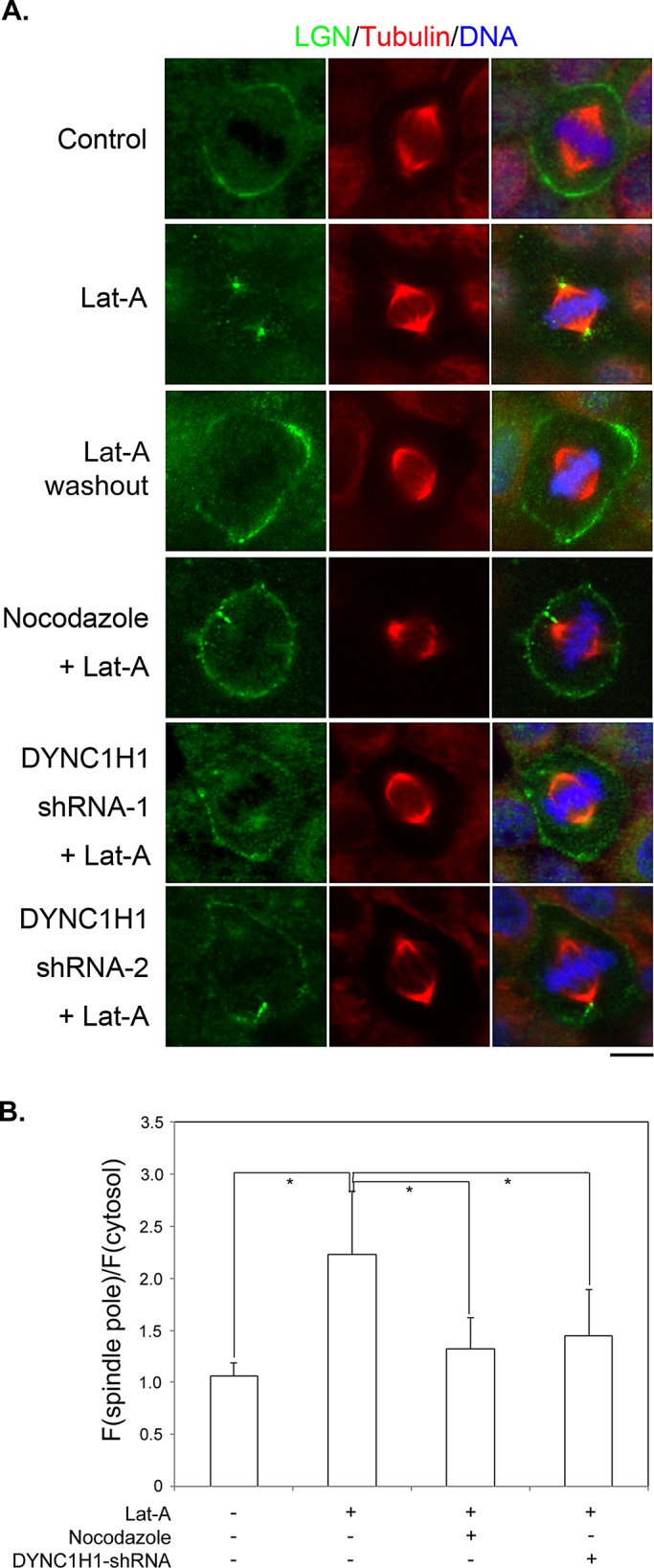
Disruption of actin filaments leads to astral microtubule– and dynein-dependent cortical dissociation and spindle pole accumulation of LGN. (A) MDCK II cells were either untreated (Control) or treated as labeled. LatA: 1 μM of LatA for 45 min; Nocodazole + LatA: 50 nM of nocodazole plus 1 μM of LatA for 45 min; DYNC1H1 shRNA-1, -2 + LatA: transfected with DYNC1H1 shRNA-1 or -2 for 48 h and then treated with 1 μM LatA for 45 min. MG132, 5 μM, was added 1 h before treatments and maintained during treatments. Cells were fixed after treatments and stained with anti-LGN (green), anti–α-tubulin (red) antibodies, and DNA dye (blue). (B) Quantitation of LGN signals at spindle poles as described in Materials and Methods. n = 50 for each set; *p < 0.01.
The aforementioned observations—cortical LGN underwent astral MTs and dynein-dependent turnover, and LGN was detected along astral MTs (Figure 4)—led us to wonder whether LatA-induced cortical dissociation and spindle pole accumulation of LGN are mediated through an active transport process that involved astral MTs and dynein. If so, we would expect that blocking such transport might attenuate the effect of LatA on LGN. To test this hypothesis, we included low doses of nocodazole in the LatA treatment. Strikingly, the presence of nocodazole significantly preserved the cortical localization of LGN and inhibited the spindle pole accumulation of LGN (Figure 5, A and B). It is noteworthy that nocodazole-mediated preservation of cortical LGN was not due to incomplete disruption of actin filaments (Supplemental Figure S3A). Similar results were obtained when DYNC1H1-knockdown cells were treated with LatA (Figure 5, A and B), suggesting that the translocation of LGN after LatA treatment requires astral MTs and cytoplasmic dynein. To explore whether such phenomena are specific to adherent, polarized epithelial cells, we performed identical experiments in MDA-231 and HeLa cells and observed similar results (Supplemental Figure S4 and unpublished data).
Astral MTs mediate release and transport of cortical dynein and NuMA when actin filaments are disrupted
We showed that LGN forms a complex with DYNC1H1 and is required for the cortical localization of DYNC1H1 (Figures 1 and 2). If LGN is transported by cortical dynein, we would expect dissociation of DYNC1H1 from the cortex when cells were treated with LatA. We used our Venus-LGN cell line to investigate the distribution of DYNC1H1, as more robust cortical staining of DYNC1H1 was observed in these cells (Figure 2C). As shown in Figure 6A (second from top), LatA treatment led to diminished cortical localization of Venus-LGN and endogenous DYNC1H1. We also observed more Venus-LGN and DYNC1H1 accumulating at the spindle poles (Figure 6A, second from top). Intriguingly, including low doses of nocodazole prevented LatA-induced cortical dissociation and spindle pole accumulation of Venus-LGN and DYNC1H1 (Figure 6, A, second from bottom, and B), further suggesting that dynein-mediated transport of LGN underlies the observed translocation of LGN upon disruption of actin filaments. These results also suggest that the cortical targeting of LGN/DYNC1H1 complex may not require actin filaments. It is still possible, however, that actin filaments are required for the initial cortical targeting of LGN/DYNC1H1 complex before the LatA plus nocodazole treatment. To clarify this issue, we pretreated cells with LatA to dissociate LGN/DYNC1H1 complex from the cell cortex and then added low doses of nocodazole while maintaining same amount of LatA in the medium. Such treatments resulted in significant relocalization of Venus-LGN and DYNC1H1 to the cell cortex (Figure 6, A, bottom, and B). Live-cell analysis also revealed LatA-induced cortical dissociation and spindle pole accumulation of Venus-LGN and the rescue effect of nocodazole in the presence of LatA (Supplemental Figure S5). These results indicate that LGN-mediated cortical targeting of dynein does not require actin filaments, and actin filaments may serve to maintain LGN/dynein at the cell cortex during mitosis.
FIGURE 6:
Disruption of actin filaments leads to astral microtubule–dependent cortical dissociation and spindle pole accumulation of DYNC1H1 and NuMA. (A) Venus-LGN–expressing cells were treated as in Figure 5A except for the results at the bottom, for which cells were first treated with 1 μM LatA for 45 min and then treated with 1 μM LatA plus 50 nM nocodazole for another 45 min. Cells were stained with anti-DYNC1H1 antibody (red) and DNA dye (blue). (B) Quantitation of cortical DYNC1H1 fluorescence intensity as described in Materials and Methods. n = 50 for each set; *p < 0.01. (C) Venus-LGN–expressing cells were treated as in A. Cells were stained with anti-NuMA antibody (red) and DNA dye (blue). Second from top, arrow points to the original crescent-shaped NuMA, and arrowhead points to NuMA accumulated at the spindle pole. (D) Quantitation of cortical NuMA fluorescence intensity. n = 50 for each set; *p < 0.01. Bars, 10 μm.
Next we tested whether NuMA also follows a similar pattern as LGN and dynein. In untreated Venus-LGN cells, in addition to the well-known crescent-shaped structure near the spindle poles, endogenous NuMA was also observed at the cell cortex and along astral MTs (Figure 6C, top). After LatA treatment, NuMA dissociated from the cell cortex and was often seen to accumulate and colocalize with Venus-LGN at the spindle poles (Figure 6, C, middle, arrowhead, and D) adjacent to the original crescent (Figure 6C, middle, arrow). Disrupting astral MTs blocked LatA-induced dissociation of NuMA from the cell cortex (Figure 6, C, bottom, and D). Similar results were observed in MDA-231 cells (Supplemental Figure S4).
Astral microtubule– and LGN-mediated transport of Gαi
Because Gαi binds to LGN and regulates the association between LGN and DYNC1H1, we studied the effect of LatA treatment on the distribution of endogenous Gαi. In untreated MDCK cells, Gαi3 localized at the cell cortex in a similar pattern as LGN (Figure 7A). Of interest, LatA treatment resulted in significant cortical reduction and spindle pole accumulation of Gαi3 (Figure 7, A and B), which is reminiscent of the effects of LatA on the distribution of LGN, DYNC1H1, and NuMA. Furthermore, the effect of LatA on the localization of Gαi3 could be inhibited by low concentrations of nocodazole (Figure 7, A and B), suggesting that it was mediated by astral MTs. To test whether LatA-induced translocation of Gαi3 was mediated through LGN, we compared control and LGN-knockdown cells. Consistent with previous work (Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012), knockdown of LGN did not obviously affect the membrane association of Gαi3 in untreated cells (Figure 7, C and D). However, in the absence of LGN, LatA treatment did not lead to spindle pole accumulation of Gαi3 (Figure 7, C and D). These results suggest that when actin filaments were disrupted, cortical Gαi3 was also transported to the spindle poles along astral MTs through the LGN/dynein complex.
FIGURE 7:
Disruption of actin filaments leads to astral microtubule– and LGN-dependent cortical release and spindle pole accumulation of Gαi3. (A) MDCK II cells were treated as in Figure 5A. Cells were fixed after treatments and stained with anti-Gαi3 (green), anti–α-tubulin (red) antibodies, and DNA dye (blue). (B) Quantitation of cortical Gαi3 fluorescence intensity. n = 50 for each set; *p < 0.01. (C) Stable MDCK cells expressing control shRNA (top two) or shRNA against LGN (bottom two) were either untreated (control) or treated with 1 μM of LatA for 45 min (LatA). Cells were fixed and stained with anti-Gαi3 (red) and DNA dye (blue). (D) Quantitation of Gαi3 signals at spindle poles as described in Materials and Methods. n = 50 for each set; *p < 0.01.
Astral microtubule–mediated transport is important for the establishment of bipolar, symmetrical cortical LGN distribution during metaphase
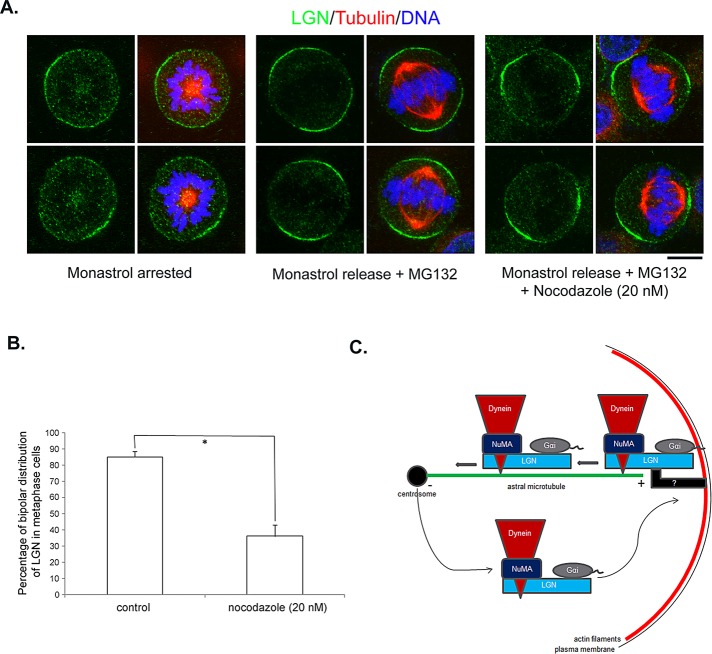
To maintain spindle positioning within a cell, cortical cues that exert forces on astral MTs need to be placed at confined regions of the cell cortex. Indeed, in metaphase HeLa cells, LGN is enriched at regions of the cell cortex that are close to the spindle poles, correlating perfectly with the spindle axis (Kiyomitsu and Cheeseman, 2012; Matsumura et al., 2012). Recently a chromosome-associated Ran-GTP mechanism was proposed for excluding LGN from cortical regions near the spindle midzone (Kiyomitsu and Cheeseman, 2012). However, whether this mechanism can wholly account for the establishment of LGN distribution during metaphase is not clear. Our finding of dynein- and astral microtubule–mediated dynamic turnover of cortical LGN led us to test whether it is also involved in installing LGN at the cell cortex. We synchronized HeLa cells using thymidine block and arrested cells in mitosis with monastrol, an Eg5 kinesin inhibitor that prevents centrosome separation (Mayer et al., 1999). In monastrol-arrested cells, LGN exhibited either diffused cortical localization or slight enrichment at one side of the cell cortex (Figure 8A, left). We then washed out monastrol and incubated cells in medium containing MG132, a proteasome inhibitor that prevents anaphase onset (Kisselev and Goldberg, 2001). One hour after monastrol washout, most of the mitotic cells had established bipolar spindles attached to properly aligned chromosomes (Figure 8A, middle). In these cells, LGN showed confined, bipolar and symmetrical distribution at cortical regions facing the spindle poles and was excluded from the central cell cortex (Figure 8A, middle), identical to its localization in untreated metaphase cells. Thus the monastrol washout experiments allowed us to study how the spatial arrangement of cortical LGN was achieved. To test whether astral MTs were involved, we included 20 nM nocodazole in the medium after monastrol washout. Of interest, such low concentration of nocodazole treatment led to LGN mislocalization in a large portion of cells with apparently normal chromosome alignment and bipolar spindle formation (Figure 8, A, right, and B), although the chromosome-mediated exclusion mechanism appeared to be working in most of the cases. These results suggest that astral MTs, and probably astral microtubule-mediated shuttling and subsequent recycling of cortical LGN, are required for establishing proper LGN distribution at the cell cortex.
FIGURE 8:
Astral MTs are required for establishing bipolar, symmetrical cortical LGN distribution during metaphase. (A) HeLa cells were partially synchronized by treating with 2 mM thymidine for 16 h. After release in normal medium for 8 h, cells were treated with 50 mM monastrol for 3 h. Monastrol-arrested cells were either directly fixed (left) or washed and released in normal medium containing 5 μM MG132 (middle) or 5 μM MG132 plus 20 nM nocodazole (right) for 1 h and fixed. Fixed cells were stained with anti-LGN (green), anti–α-tubulin (red) antibodies, and DNA dye (blue). Bar, 10 μm. (B) Quantitation of bipolar, symmetrical cortical LGN localization. Only cells with normal chromosome condensation and alignment were included in the quantification. n = 100 from three independent experiments; *p < 0.001. (C) Schematic working model illustrating astral microtubule–mediated transport and recycling of the Gα/LGN/dynein/NuMA complex during mitosis.
DISCUSSION
Proper spindle positioning is believed to be achieved when astral MTs receive confined cortical forces (Grill and Hyman, 2005). The evolutionarily conserved Gα/LGN/NuMA ternary complex has emerged as an important cortical force generator (Gonczy, 2008; Knoblich, 2008; Siller and Doe, 2009), but how this complex exerts forces on astral MTs is not clear.
Here we show that Gαi, LGN, and DYNC1H1 form a complex in mammalian cells. Of interest, Gαi binding enhances the association between LGN and DYNC1H1, further supporting the conformation switch hypothesis for the Pins family of proteins (Du and Macara, 2004; Nipper et al., 2007). It also suggests that regulators of Gαi may affect the interaction between LGN and DYNC1H1. Our results, together with recent studies in HeLa cells (Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012), indicate that LGN serves as a major cortical factor that recruits cytoplasmic dynein to the cell cortex during mitosis, probably through membrane-bound Gα. It is surprising that NuMA, the other LGN-binding partner, has an inhibitory effect on the association between Gαi/LGN and DYNC1H1. NuMA was shown to associate with cytoplasmic dynein in Xenopus egg extracts (Merdes et al., 1996). While the manuscript was under revision, Kotak et al. (2012) reported that the N-terminal of NuMA associates with cytoplasmic dynein, although a direct physical link with a specific dynein subunit is still missing. Our results suggest that Gαi/LGN and DYNC1H1 might form a complex in a NuMA-independent manner. It is possible, however, that in our immunoprecipitation analysis, the overexpressed NuMA might sequester DYNC1H1 to a cellular compartment that is not accessible for the Gαi/LGN complex. Nevertheless, we showed that NuMA also localizes to astral MTs and is transported to the spindle poles when actin filaments are disrupted, suggesting that it is in a complex with LGN and dynein. We propose that NuMA may associate with other component of the dynein/dynactin complex and function with LGN in recruiting or modulating dynein at the cell cortex during mitosis. Further studies are needed to determine whether the association between LGN and DYNC1H1 is direct or indirect and how the interaction is regulated by related proteins.
A prevalent view of dynein function in spindle positioning is that dynein is anchored at the cell cortex and exerts forces on astral MTs either by controlling microtubule dynamics or by driving microtubule sliding along the cell cortex (Grill and Hyman, 2005; Hendricks et al., 2012; Laan et al., 2012). We now provide evidence that cortical dynein may not be static; instead, it can be released and move, in complex with LGN and NuMA, along astral MTs. The movement of the dynein complex along astral MTs may generate pulling forces, as astral MTs could also be captured by other cortical microtubule-binding proteins rather than dynein (Samora et al., 2011). Alternatively, the release and transport of dynein complex may provide a novel mechanism for mammalian cells to limit, balance, or terminate cortical dynein-generated pulling forces. Our results are consistent with a recent study in C. elegans suggesting that MTs regulate cortical localization of GPR-1/2 (Werts et al., 2011). It would be interesting to test whether dynein-mediated transport of LGN could be applied to other systems.
An interesting phenomenon that has been observed in multiple mammalian cell lines is that, during metaphase, LGN exhibits bipolar, symmetrical cortical localization, which correlates nicely with spindle orientation, suggesting an intrinsic mechanism for spindle positioning. How is the confined LGN distribution established? Recently an elegant chromosome-mediated, cortical exclusion model was proposed (Kiyomitsu and Cheeseman, 2012), although the underlying mechanism is not clear. Here we show that astral MTs are also required for proper cortical LGN distribution, suggesting the involvement of multiple mechanisms. We propose that astral MT–mediated transport and recycling may help to sort and enrich LGN at cortical regions facing the spindle poles, where the interaction between astral MTs and the cell cortex should be more intensive.
It is intriguing to observe that when actin filaments were disrupted, endogenous Gαi also dissociated from the cell cortex and accumulate at the spindle poles in an astral microtubule– and LGN-dependent manner. The membrane localization of Gαi is mediated through cotranslational or posttranslational lipid modification (Chen and Manning, 2001) and, in principle, should not be affected by cortical actin filaments. We propose that during mitosis, a large portion of Gαi is associated with LGN at the cell cortex in MDCK cells. Dynein-generated force could overcome the membrane attachment of Gαi. Alternatively, other mitotic-specific mechanisms, such as depalmitoylation, may exist. In line with our hypothesis, a recent study in C. elegans revealed that the Gβ subunit GBP-1, a negative regulator of Gα/GPR1,2 complex, has the lowest membrane level during mitosis (Thyagarajan et al., 2011). We are in the process of testing whether Gβ/γ and other Gαi regulators are involved in the transport of LGN/Gαi.
The relationship between cortical actin filaments and dynein has long been an enigma. Previous studies suggest that actin filaments are required for the cortical localization of LGN and dynein subunits (Busson et al., 1998; Kaushik et al., 2003; Toyoshima et al., 2007). Through careful analysis, we found that cortical dissociation of LGN and dynein upon disruption of actin filaments is not a simple event of anchorage detachment but involves astral MT–mediated transportation. We now provide a more definitive link between cortical actin filaments and the LGN/dynein complex. Our results suggest that actin filaments are not required for the cortical targeting of LGN/dynein; instead, they serve as a stabilizing structure to maintain the LGN/dynein complex at the cell cortex. We propose the existence of actin filament–associated or regulated protein(s) that links the LGN complex to cortical actin filaments and counteracts astral MT–mediated release of LGN/dynein complex. Such actin filament–mediated maintenance of cortical LGN/dynein may sustain dynein-generated forces or serve as a rate-limiting factor—by regulating the release and transport of the LGN/dynein complex—in modulating pulling forces on astral MTs under different physiological conditions.
Taking our results together, we propose the following working hypothesis (Figure 8C): membrane-associated Gα recruits LGN to the cell cortex, which in turn recruits NuMA and dynein. Cortical Gα/LGN/dynein/NuMA complex is stabilized and maintained by actin filaments through unknown factor(s). The interaction between astral MTs and the cell cortex is accompanied by constant release and transport of a portion of the Gα/LGN/dynein/NuMA complex from the cell cortex to spindle poles. Once reaching the spindle poles, the Gα/LGN/dynein/NuMA complex leaves the spindle poles and recycles back to the cell cortex. When dynein is knocked down or in the absence of astral MTs, the cortical release and transport of LGN will be compromised, and more LGN will be trapped at the cell cortex. On the other hand, when cortical actin filaments are disrupted, astral MTs mediate rapid release and transport of the Gα/LGN/dynein/NuMA complex, leading to its spindle pole accumulation. The cortical release, transport, and recycling of LGN is important for establishing the bipolar, symmetric cortical distribution of LGN during metaphase. The transport of LGN/dynein complex may also generate pulling forces on astral MTs or serve as a regulatory mechanism in balancing the pulling forces on astral MTs. Further studies are needed to elucidate how the dynamic behavior of cortical LGN complex is regulated at the molecular level and, of importance, to what extent the regulated transport of LGN/dynein complex contributes to spindle positioning.
MATERIALS AND METHODS
Cell lines and reagents
Cos 7 and MDCK cells were maintained in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum and antibiotics at 37°C in a humidified 5% CO2 atmosphere. Control, LGN-stable knockdown MDCK cell lines and Tet-Off MDCK cell line expressing Venus-LGN were described previously (Du and Macara, 2004; Zheng et al., 2010).
Rabbit anti-LGN antibodies were described previously (Du and Macara, 2004). Rabbit anti-NuMA antibody was a kind gift from Duane Compton (Dartmouth Medical School, Hanover, NH). The following antibodies were also used: primary monoclonal 9E10 anti-myc, 12CA5 anti-hemagglutinin (HA; Santa Cruz Biotechnology, Santa Cruz, CA), anti-dynein (IC, DYNC1I1; clone 70.1; Sigma-Aldrich, St. Louis, MO), anti-p150Glued (Cell Signaling Technology, Danvers, MA), polyclonal rabbit anti-DYNC1H1, anti-Gαi3 (Santa Cruz Biotechnology), and anti-GFP (Torry Pines Biolabs, Secaucus, NJ); secondary Alexa 488, Alexa 594, Alexa 660, and Alexa 680 (Invitrogen, Carlsbad, CA) and IRDye800-conjugated (Rockland ImmunoChemicals, Gilbertsville, PA) goat anti-mouse or rabbit antibodies. Rhodamine–phalloidin was from Invitrogen; nocodazole, latrunculin A, monastrol, and MG132 were from Sigma-Aldrich.
Plasmids
Full-length human DYNC1H1 cDNA was transferred from KIAA cDNA clone (0325; Kazusa DNA Research Institute, Kisarazu, Japan) by PCR and restriction enzyme digestion and cloned in pK-VENUS (Du et al., 2001). Human NuMAcDNA was PCR amplified and cloned into pKHA3 to generate pKHA3-NuMA. Wild-type Gαi1 constructs were described previously (Du and Macara, 2004). Mutant Gαi1 (N149I) was PCR amplified and cloned into pKmyc. For knocking down DYNC1H1 in MDCK cells, we used the pRNAi-neo plasmid (Biosettia, San Diego, CA) and the following target sequences: 5′-GCTGAAATCTGAAGCACTTAA-3′ (DYNC1H1 shRNA-1) and 5′-GCACTTAAAGATCGCCATTGG-3′ (DYNC1H1 shRNA-2).
Immunoprecipitation and Western blotting
Cos 7 cells were electroporated with mammalian expression vectors using an Amaxa Nucleofection device (Lonza, Basel, Switzerland) following the manufacturer's instructions. Cells were collected in lysis buffer (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 0.5 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml leupeptin, 20 μg/ml aprotinin, and 20 μg/ml pepstatin). Equal amounts of cell lysate were incubated with 2 μg of anti-myc, anti-HA, anti-LGN antibodies or rabbit IgG at 4°C for 2 h or overnight. GammaBind-Plus Sepharose beads (GE Healthcare, Pittsburgh, PA) were added, and the mixture was incubated for 1 h at 4°C. Beads were washed four times with cell lysis buffer, and bound proteins were separated by SDS–PAGE. Proteins were transferred to nitrocellulose membrane and detected using anti-myc (1:1000), anti-HA (1:2000), anti-LGN (1:500), anti-GFP (1:2000), anti-DYNC1H1 (1:200), anti-DYNC1I1 (1:1000), or anti-p150Glued (1:1000) antibodies.
Immunofluorescence microscopy
Cells were grown on poly-lysine–coated coverglass and fixed using either 4% paraformaldehyde (PFA) or 4% PFA/0.25% Triton X-100 in phosphate-buffered saline (PBS) as indicated. For detecting astral MT localization of LGN or cortical localization of DYNC1H1 and NuMA, cells were preextracted using PHEM buffer (25 mM HEPES, 60 mM 1,4-piperazinediethanesulfonic acid, 10 mM ethylene glycol tetraacetic acid, 2 mM MgCl2, pH 6.9) containing 0.2% Triton X-100 and fixed in 4% PFA. Fixed cells were blocked with 1% bovine serum albumin/10% normal goat serum in PBS for 1 h and incubated in primary antibodies for 1 h at room temperature or overnight at 4°C. Cells were then washed and incubated for 1 h with the DNA dye Hoechst 33342 and secondary antibodies coupled with Alexa 488 or Alexa 594 (Invitrogen). A SlowFade Gold AntiFade kit (Invitrogen) was used to reduce photobleaching. Cells were imaged using either a 60×/numerical aperture (NA) 1.2 oil-immersion objective on a Nikon TE2000 inverted microscope (Nikon Instruments, Melville, NY) or a 63×/NA 1.4 objective on a Zeiss 510 LSM confocal microscope (Carl Zeiss, Oberkochen, Germany).
Measurement of the relative fluorescence intensity of cortical and spindle pole LGN and Gαi3 and of cortical NuMA and DYNC1H1
Measurements of the relative fluorescence intensity of cortical LGN or DYNC1H1 were performed as described (Wan et al., 2012). Briefly, cells were stained with identical procedure, and images were taken with identical microscopic settings. Fifty metaphase cells in each group were randomly selected, and the mean fluorescence intensity of cortical LGN or DYNC1H1 was measured using ImageJ software (National Institutes of Health, Bethesda, MD). The standard deviation was calculated, and statistical significance was determined by Student's t test.
Similarly, for the measurement of the relative fluorescence intensity of spindle pole LGN, a 60-pixel circle was drawn around each spindle pole and in areas 10 pixels away using ImageJ software. Fluorescence intensities at the spindle poles and the cytosol were referred to as F(spindle pole) and F(cytosol), respectively. The ratio F(spindle pole)/F(cytosol) was collected for each group of cells and analyzed.
FRAP analysis
Stable Tet-Off MDCK cells expressing Venus-LGN were cultured in the absence (–Dox) of doxycycline to allow expression of low levels of Venus-LGN. Untreated cells or cells transfected with either control shRNA or DYNC1H1 shRNA were grown on Delta T dishes (Bioptechs, Butler, PA) in DMEM supplemented with 10% fetal calf serum (FCS) and antibiotics. At 1 h before FRAP analysis, the medium was replaced with F10 medium containing 10% FCS, and dimethyl sulfoxide (DMSO; 0.5%) or 50 nM nocodazole was added in the medium as needed. The dish was then placed in a temperature control system (Bioptechs) that maintained a temperature of 37°C during the FRAP experiment. FRAP experiments were performed on a LSM 7 live laser-scanning confocal microscope (Carl Zeiss) with a plan-Apochromat 63×/NA 1.4 oil objective. Because LGN accumulates at the cell cortex only during mitosis, it is easy to identify mitotic cells showing cortical Venus-LGN signal. We specifically chose those cortical areas where Venus-LGN fluorescence was relatively bright and smooth as the regions of interest (ROIs), which were 4 μm2 in size and centered on the cortical Venus-LGN fluorescence. ROIs were first scanned with a 488-nm diode laser at 1.1% power for three cycles (10 s each cycle) to determine a measurement of initial fluorescence intensity in the ROI. Next ROIs were subjected to 12 iterations with 488-nm diode laser at 100% power to photobleach the ROI. The fluorescent images were then acquired every 10 s at 1.1% laser power for another 37 cycles using ZEN 2009 software (Carl Zeiss). The fluorescence recovery in the ROI at every time point was normalized according to 100(It − I0)/(Ic − I0), where It represents fluorescence intensity in the ROI at the given time point, I0 represents the intensity of fluorescence in ROI after photobleaching, and Ic represents the average value of three measurements of the fluorescence intensity in the ROI before photobleaching. Recovery measurements were quantified by fitting normalized fluorescence intensities of bleached areas to a one-phase exponential association by using ZEN 2009 software (Carl Zeiss). Standard SEM was calculated, and statistical significance was determined by Student's t test.
Supplementary Material
Acknowledgments
We thank Kazusa DNA Research Institute in Japan for kindly providing the KIAA clone. We are grateful to Duane Compton for providing the anti-NuMA antibody. This work was supported by grants from American Cancer Society (RSG0717601CSM) and the National Institutes of Health (GM079506) to Q.D.
Abbreviations used:
- DYNC1H1
dynein heavy chain, cytoplasmic 1
- DYNC1I1
dynein IC 1
- FRAP
fluorescence recovery after photobleaching
- LatA
latrunculin A
- LGN
mammalian homologue of Drosophila Partner of Inscuteable
- MT
microtubule
- NuMA
nuclear mitotic apparatus
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-06-0458) on February 6, 2013.
REFERENCES
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahringer J. Control of cell polarity and mitotic spindle positioning in animal cells. Curr Opin Cell Biol. 2003;15:73–81. doi: 10.1016/s0955-0674(02)00018-2. [DOI] [PubMed] [Google Scholar]
- Ben-Yair R, Kahane N, Kalcheim C. LGN-dependent orientation of cell divisions in the dermomyotome controls lineage segregation into muscle and dermis. Development. 2011;138:4155–4166. doi: 10.1242/dev.065169. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CA, Manning DR. Regulation of G proteins by covalent modification. Oncogene. 2001;20:1643–1652. doi: 10.1038/sj.onc.1204185. [DOI] [PubMed] [Google Scholar]
- Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell. 2012;23:3380–3390. doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol. 2007;179:15–22. doi: 10.1083/jcb.200707085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–49. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Du Q, Stukenberg PT, Macara IG. A mammalian Partner of Inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- Du Q, Taylor L, Compton DA, Macara IG. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- El-Hashash AH, Turcatel G, Al Alam D, Buckley S, Tokumitsu H, Bellusci S, Warburton D. Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development. 2011;138:1395–1407. doi: 10.1242/dev.058479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- El-Hashash AH, Warburton D. Cell polarity and spindle orientation in the distal epithelium of embryonic lung. Dev Dynam. 2011;240:441–445. doi: 10.1002/dvdy.22551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O'Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Fink J, et al. External forces control mitotic spindle positioning. Nat Cell Biol. 2011;13:771-U401. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- Fuse N, Hisata K, Katzen AL, Matsuzaki F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr Biol. 2003;13:947–954. doi: 10.1016/s0960-9822(03)00334-8. [DOI] [PubMed] [Google Scholar]
- Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies TE, Cabernard C. Cell division orientation in animals. Curr Biol. 2011;21:R599–R609. doi: 10.1016/j.cub.2011.06.055. [DOI] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Gotta M, Ahringer J. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol. 2003;13:1029–1037. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Hao Y, Du QS, Chen XY, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur ELF. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr Biol. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Jordan MA, Thrower D, Wilson L. Effects of vinblastine, podophyllotoxin and nocodazole on mitotic spindles. Implications for the role of microtubule dynamics in mitosis. J Cell Sci. 1992;102:401–416. doi: 10.1242/jcs.102.3.401. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik R, Yu F, Chia W, Yang X, Bahri S. Subcellular localization of LGN during mitosis: evidence for its cortical localization in mitotic cell culture systems and its requirement for normal cell cycle progression. Mol Biol Cell. 2003;14:3144–3155. doi: 10.1091/mbc.E03-04-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Kimura A. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc Natl Acad Sci USA. 2011;108:137–142. doi: 10.1073/pnas.1013275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- Kisurina-Evgenieva O, Mack G, Du Q, Macara I, Khodjakov A, Compton DA. Multiple mechanisms regulate NuMA dynamics at spindle poles. J Cell Sci. 2004;117:6391–6400. doi: 10.1242/jcs.01568. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009;19:174–179. doi: 10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier SM. AGS proteins, GPR motifs and the signals processed by heterotrimeric G proteins. Biol Cell. 2004;96:369–372. doi: 10.1016/j.biolcel.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Matsumura S, Hamasaki M, Yamamoto T, Ebisuya M, Sato M, Nishida E, Toyoshima F. ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat Commun. 2012;3:626. doi: 10.1038/ncomms1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- McCarthy EK, Goldstein B. Asymmetric spindle positioning. Curr Opin Cell Biol. 2006;18:79–85. doi: 10.1016/j.ceb.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Morin X, Jaouen F, Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci USA. 2007;104:14306–14311. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier ML, Woods D, Greig S, Phan PG, Radovic A, Bryant P, O'Kane CJ. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J Neurosci. 2000;20:RC84. doi: 10.1523/JNEUROSCI.20-14-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecreaux J, Roper JC, Kruse K, Julicher F, Hyman AA, Grill SW, Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Peyre E, Jaouen F, Saadaoui M, Haren L, Merdes A, Durbec P, Morin X. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J Cell Biol. 2011;193:141–154. doi: 10.1083/jcb.201101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson ND, Lechler T. Robust control of mitotic spindle orientation in the developing epidermis. J Cell Biol. 2010;191:915–922. doi: 10.1083/jcb.201008001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev P, Menon S, Kastner DB, Chuang JZ, Yeh TY, Conde C, Caceres A, Sung CH, Sakmar TP. G protein beta gamma subunit interaction with the dynein light-chain component Tctex-1 regulates neurite outgrowth. EMBO J. 2007;26:2621–2632. doi: 10.1038/sj.emboj.7601716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:U1040–U1059. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- Thyagarajan K, Afshar K, Gonczy P. Polarity mediates asymmetric trafficking of the G beta heterotrimeric G-protein subunit GPB-1 in C. elegans embryos. Development. 2011;138:2773–2782. doi: 10.1242/dev.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F, Matsumura S, Morimoto H, Mitsushima M, Nishida E. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev Cell. 2007;13:796–811. doi: 10.1016/j.devcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, Boxem M, van den Heuvel S. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- Wan QW, Liu J, Zheng Z, Zhu HB, Chu XG, Dong Z, Huang S, Du QS. Regulation of myosin activation during cell-cell contact formation by Par3-Lgl antagonism: entosis without matrix detachment. Mol Biol Cell. 2012;23:2076–2091. doi: 10.1091/mbc.E11-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts AD, Roh-Johnson M, Goldstein B. Dynamic localization of C. elegans TPR-GoLoco proteins mediates mitotic spindle orientation by extrinsic signaling. Development. 2011;138:4411–4422. doi: 10.1242/dev.070979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard FS, et al. A point mutation to Galphai selectively blocks GoLoco motif binding: direct evidence for Galpha. GoLoco complexes in mitotic spindle dynamics. J Biol Chem. 2008;283:36698–36710. doi: 10.1074/jbc.M804936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol. 2010;30:3519–3530. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZN, Wan QW, Du QS, Zheng Z. Galpha/LGN-mediated asymmetric spindle positioning does not lead to unequal cleavage of the mother cell in 3-D cultured MDCK cells. Biochem Biophys Res Commun. 2012;420:888–894. doi: 10.1016/j.bbrc.2012.03.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Kuo CT, Jan YN. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Zhu HB, Wan QW, Liu J, Xiao ZN, Siderovski DP, Du QS. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J Cell Biol. 2010;189:275–288. doi: 10.1083/jcb.200910021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JW, Wen WY, Zheng Z, Shang Y, Wei ZY, Xiao ZN, Pan Z, Du QS, Wang WN, Zhang MJ. LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G alpha i/LGN/NuMA pathways. Mol Cell. 2011;43:418–431. doi: 10.1016/j.molcel.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.