Abstract
We showed previously that wild-type mice primed via intranasal inoculation with live or heat-inactivated Lactobacillus species were fully (100%) protected against the lethal sequelae of infection with the virulent pathogen, pneumonia virus of mice (PVM), a response that is associated with diminished expression of proinflammatory cytokines and diminished virus recovery. We show here that 40% of the mice primed with live Lactobacillus survived when PVM challenge was delayed for 5 months. This robust and sustained resistance to PVM infection resulting from prior interaction with an otherwise unrelated microbe is a profound example of heterologous immunity. We undertook the present study in order to understand the nature and unique features of this response. We found that intranasal inoculation with L. reuteri elicited rapid, transient neutrophil recruitment in association with proinflammatory mediators (CXCL1, CCL3, CCL2, CXCL10, TNF-alpha and IL-17A) but not Th1 cytokines. IFNγ does not contribute to survival promoted by Lactobacillus-priming. Live L. reuteri detected in lung tissue underwent rapid clearance, and was undetectable at 24 hrs after inoculation. In contrast, L. reuteri peptidoglycan (PGN) and L. reuteri genomic DNA (gDNA) were detected at 24 and 48 hours after inoculation, respectively. In contrast to live bacteria, intranasal inoculation with isolated L. reuteri gDNA elicited no neutrophil recruitment, had minimal impact on virus recovery and virus-associated production of CCL3, and provided no protection against the negative sequelae of virus infection. Isolated PGN elicited neutrophil recruitment and proinflammatory cytokines but did not promote sustained survival in response to subsequent PVM infection. Overall, further evaluation of the responses leading to Lactobacillus-mediated heterologous immunity may provide insight into novel antiviral preventive modalities.
Keywords: cytokines, neutrophils, peptidoglycan, genomic DNA, heterologous immunity
Introduction
Respiratory syncytial virus (RSV; family Paramyxoviridae; genus Pneumovirus) is the most common cause of severe lower respiratory tract disease among infants and young children and is an emerging pathogen of the elderly [Tregoning and Schwarze, 2010; Falsey and Walsh, 2005]. In most cases the disease is self -limiting but in some infants it progresses to severe bronchiolitis and respiratory compromise, resulting in more than 100,000 hospitalizations each year in the US alone. Current management for otherwise healthy infants and children hospitalized with severe RSV is supportive care only; there is no available anti-RSV vaccine, although several are under study [Wright and Piedimonte, 2011]. While monoclonal anti-RSV prophylaxis is available for identified high-risk infants [Shadman and Wald, 2011], a recent study by Hall and colleagues [Hall, et al., 2009] documented that most children hospitalized with severe RSV infection had no identified predisposing risk factors, a finding that highlights the need for more effective management strategies for this disease.
The most severe forms of RSV infection are associated with significant inflammatory pathology: this is clear from human post mortem studies, analyses of samples from the airways of mechanically ventilated infants, and from various animal infection models [Rosenberg and Domachowske, 2012; Dyer et al., 2012]). In order to explore the pathogenesis of RSV disease in vivo, we have developed a mouse model using the related pathogen, pneumonia virus of mice (PVM, family Paramyxoviridae, genus Pneumovirus). PVM is a natural rodent pneumovirus that replicates in bronchial epithelial cells and elicits severe inflammatory pathology in most inbred strains of mice [Bem, et al., 2011; Rosenberg and Domachowske, 2008]. Findings at peak morbidity include high virus titer, prominent neutrophil influx, and edema, similar to that described by Welliver and colleagues for fatal RSV infection [Welliver et al., 2008]. We have used this model to explore various immunomodulatory therapies for pneumovirus infection, including those targeting the cytokine CCL3 and its receptor, CCR1, strategies which limit neutrophil influx and result in diminished mortality [Bonville et al., 2003; Bonville et al., 2004].
In our previous work, we documented the immunomodulatory properties of Lactobacillus strains, specifically, their ability to modulate the antiviral inflammatory response to acute PVM infection [Gabryszewski et al., 2011]. Specifically, we found that wild-type mice primed via intranasal inoculation with live or heat-inactivated Lactobacillus plantarum or Lactobacillus reuteri were completely protected against lethal sequelae of this infection, with significant protection (60% survival) persisting even when virus infection was initiated three months after initial priming with live L. plantarum. Priming with live lactobacilli resulted in diminished granulocyte recruitment and diminished expression of multiple proinflammatory cytokines characteristic of acute PVM infection. These findings represent an original and robust example of heterologous immunity, or “innate imprinting,” a concept introduced by Hussell and colleagues [Goulding et al., 2007; Didierlaurent et al., 2007; Hussell and Cavanaugh, 2009] to explain the increased resistance or susceptibility to an unrelated pathogen generated upon recovery from a primary innate or inflammatory response. As some examples of this concept, Nguyen and colleagues [2008] found that primary infection with murine gammaherpesvirus infection resulted in diminished recovery of mouse adenovirus-1 upon challenge with this unrelated pathogen up to three weeks later. Similarly, Williams and colleagues [2004] elicited protection against virus and fungal pathogens by intranasal administration of an E. coli-derived heat-labile antigen. Likewise, Li and colleagues [2010] documented the protective effects of intranasal administration of live Bordetella pertussis (BPEZI, pertussis-toxin inactivated) against subsequent infection with influenza, although curiously, no protection was observed in response to inoculation with similar, toxin-gene-deleted (ΔPT B. pertussis) strain [Ayala et al., 2011]. Of note, not all of these heterologous responses result in positive outcomes or are directed toward promoting homeostasis; several molecular mechanisms have been proposed to explain the well-known increased susceptibility to bacterial pneumonia observed clinically in individuals recovering from acute severe influenza infection (reviewed in [Ballinger et al., 2010; Steinberg et al., 2010]).
In this manuscript we have characterized the innate inflammatory responses in the airways that are elicited in response to priming with live Lactobacillus species as part of an ongoing effort to elucidate the mechanisms that promote this unique and notably robust form of heterologous immunity to respiratory virus infection.
Materials and Methods
Mice
Wild-type BALB/c and C57BL/6 mice were purchased from Division of Cancer Therapeutics, Frederick, MD. IFNγ−/− mice [Line 208; Dalton, et al., 1993] are maintained by NIAID-Taconic contract. All mouse studies were carried out in accordance with Animal Study Protocol LAD-8E approved by the National Institutes of Allergy and Infectious Diseases Animal Care Committee.
Lactobacillus inoculations
Lactobacillus plantarum NCIMB 8826 (ATCC BAA-793) and Lactobacillus reuteri F275 (ATCC 23272) were grown overnight at 37°C in Difco Lactobacilli MRS Broth (BD Biosciences, Sparks, MD). Correlations between OD600 and colony forming units (cfu) were as previously described [Gabryszewski et al., 2011]. Bacteria were harvested by centrifugation (5 min, 1500 rpm Sorvall RT6000B centrifuge) washed with sterile tissue-culture grade phosphate-buffered saline (PBS) and re-suspended in PBS supplemented with 1% bovine serum albumin (PBS/BSA) at 2 × 1010 colony forming units (cfu) / mL (L. plantarum) or 2 × 109 cfu / mL (L. reuteri). Mice were inoculated intranasally with 50 µL PBS/BSA with 109 or 108 cfu live L. plantarum or L. reuteri in 50 µL PBS/BSA, respectively, at time points indicated. In some experiments, mice were inoculated intranasally with purified Gram-positive peptidoglycan (100 µg / 50 µL PBS/BSA; Invivogen, San Diego, CA).
Virus preparation and inoculations
PVM strain J3666 is maintained as an in vivo passaged stock; virus copy number per unit volume was determined by a quantitative PCR method [Gabryszewski et al., 2011] that targets the PVM small hydrophobic (SH) gene. All PVM-infected mice were inoculated intranasally with 50 µL of PVM stock (2 × 105 PVMSH/µL) diluted 1:2000 (C57BL/6 mice) or 1:3000 (BALB/c mice) in culture medium while under isoflurane anesthesia.
Leukocyte recruitment
Lung tissue was harvested and single cell suspensions prepared essentially as described in [Gabryszewski et al., 2011]; here, fresh digestion medium was added after first 45 minutes of incubation, and a 70-micron filter cell strainer was used. Live/dead stain (Invitrogen) was added to the cells and antibody binding to Fc receptors was blocked with anti-mouse CD16/CD32 (BD Biosciences, Durham, NC). Cells were then stained with anti-CD3-PE-Cy5, anti-CD3-Alexa-700, anti-CD19-Alexa-647, anti-CD49b (DX5)-PE, anti-GR1-APC, anti-GR1-V450, anti-CD11b-PE (BD Biosciences); anti-CD11c-Alexa-488, and/or anti-MHC II (I-A/I-E)-APC (eBiosciences, San Diego) in PBS/BSA at 4°C for 1hr and washed with PBS/BSA. A minimum of 100,000 events was collected on an LSRII flow cytometer (BD Biosciences) and data was analyzed in FlowJo 9.2. Analyses are presented as percentage of live cells.
Histology
Lung tissue was evaluated as indicated. Lungs were inflated transtracheally with 250 µL of 10% phosphate buffered formalin; lungs and heart were removed and stored at 4°C in 10% phosphate buffered formalin. Samples were paraffin embedded, sectioned and stained with hematoxylin and eosin (Histoserv, Germantown, MD).
Cytokine and Immunoglobulin ELISAs
At selected time points, lungs of mice primed with L. reuteri (108 cfu), Gram-positive peptidoglycan, L. reuteri genomic DNA (see below) followed by PVM infection, or PBS/BSA at time points indicated were collected and blade-homogenized in 1 mL PBS/BSA. Clarified supernatants were aliquoted and stored at −80°C for later use. ELISAs were performed to evaluate cytokine responses (R&D Systems, Minneapolis, MN) as per the manufacturer’s instructions. Cytokine production was normalized to total lung protein determined by a BCA assay (Pierce, Rockford, IL). Cytokines and IgG were measured in sera from mice at selected time points after intranasal priming with L. reuteri or PBS/BSA control. Cytokine levels were measured with a mouse cytokine/chemokine Milliplex kit (Millipore, Billerica, MA) following manufacturer’s instructions. Serum IgG was measured using a designated kit from Kamiya Biomedical (Seattle, WA). Seroconversion to PVM was evaluated using the SMART-M12 ELISA as per manufacturer’s instructions (Biotech Trading Partners).
Bacterial clearance
Lung tissues from mice primed with 108 cfu L. reuteri or PBS/BSA were collected 6, 12, and 24 hrs after intranasal inoculations. Lungs were blade-homogenized in 1 mL PBS/BSA and 100 µL of undiluted, 1:50, 1:100 and 1:200 dilutions were plated on MRS agar; colonies were enumerated after overnight incubation at 37°C.
Preparation of Lactobacillus genomic DNA
L. reuteri was grown overnight at 37°C in Difco MRS Broth (BD Biosciences). Ten mL of bacteria was harvested by centrifugation for 10 min at 1500 rpm in a Sorvall RT6000B centrifuge, washed twice with sterile PBS, resuspended in 1 mL Gram-positive bacterial lysis buffer (20mM Tris-HCl pH 8.0, 2mM EDTA, 1.2% Triton X-100, 50 mg/mL lysozyme (Sigma Aldrich)), and incubated at 37°C for 1 hour. 100 mL of 10% SDS and 10uL of Purgene Proteinase K solution (Qiagen) was added followed by incubation at 37°C for 3 hours until the solution appeared clear. Remaining debris was removed by centrifugation at 25°C 13,000 RPM for 5 minutes; 750 µL supernatant harvested was extracted with 1 volume Ultrapure Phenol:Chloroform:Isoamyl Alcohol (Invitrogen). Upper aqueous phase was re-extracted twice with phenol:chloroform, followed by a final extraction with of 1/10th by volume chloroform (J.T. Baker). DNA was precipitated from the upper aqueous phase with 1/10 volume of 3M Sodium Acetate pH 5.2 and 1 volume of isopropanol followed by centrifugation at 4°C at 13,000 rpm for 10 minutes. The pellet was washed with 75% ethanol, centrifuged at 4°C at 7500 rpm for 10 minutes, the supernatant was decanted, and the remaining pellet was allowed to dry for 10 minutes. The final pellet was suspended in sterile dH2O and the concentration of DNA was measured by Nanodrop Spectrophotometer ND-1000 (Thermo Scientific). This procedure yields 300 – 500 µg genomic DNA per 10 mL Lactobacillus culture.
Detection of Lactobacillus genomic DNA in mouse lung tissue
Mice were inoculated intranasally with 108 cfu L. reuteri in 50 uL PBS/BSA and lungs were harvested at the time points indicated and stored at −80°C in RNAlater (Ambion). Approximately 7 – 10 mg of the lung tissue was washed in dH2O, minced in a petri dish using a straight razor blade, and transferred to a 15 mL tube containing 2 mL Tissue Lysis Buffer (60 mM Tris-HCl pH 8.0, 100 mM EDTA, 20 mg/mL lysozyme). Lungs were stirred for 1 hour at 37°C, then 0.5% SDS and 10uL of Purgene Proteinase K (Qiagen) were added and samples were stirred for an additional 4 hours at 55°C until the solution was clear. Samples were then spun at 25°C 4,000 RPM in a Sorvall RT6000B centrifuge for 5 minutes to remove any remaining debris and remaining supernatant was extracted and DNA precipitated as described above. This procedure yields 600 – 1500 ng genomic DNA from mouse lung tissue. L. reuteri genomic DNA was detected by quantitative PCR using primers and probe designed to detect 16s–23s intergenic region as described in [Harman and Knol, 2006], including forward primer, 5’-ACC GAG AAC ACC GCG TTA TTT-3’; reverse primer, 5’-CAT AAC TTA ACC TAA ACA ATC AAA GAT TGT CT-3’; and probe, 5’-ATC GCT AAC TCA ATT AAT-3’-MGB (minor groove binding). 300 ng of extracted DNA from L. reuteri or control-inoculated mouse lung tissue was added to TaqMan universal PCR master mix and evaluated on an ABI 7500 Real Time PCR system according to manufacturer’s instructions (Applied Biosystems). Results were calibrated via a standard curve, which included 1 to 108 copies of 1 kb LR 16–23s sequence (Genbank AF080100.1, generated by PCR using primers LR16sF (5'-CGGATCAGCATGCTGCGGTGAACTA-3') and LR16sR (5'-TGTTAGTCCCGTCCTTCATCGGCT-3') [Chagnaud et al., 2001] and cloned into the pCR 2.1 vector (Invitrogen) and confirmed by bidirectional sequencing.
Detection of peptidoglycan in the lung after L. reuteri inoculation
Lung tissue from mice receiving inoculations with 108 cfu L.reuteri or PBS/BSA (negative control) was collected 6, 24, 48, and 96 hours thereafter. Lungs were blade-homogenized in 1 mL PBS/BSA passed through a 40µm strainer and stored at −80°C. Peptidoglycan (PGN) was detected in whole lung homogenates using the silkworm larvae plasma (SLP) reagent set from Wako (Richmond, VA), which measures the formation of melanin in response to the presence of PGN in a sample [Inada et al., 2003]. Briefly lung homogenates (diluted 1:1000 in endotoxin-free water) or PGN standard dilution series (Wako) were dispensed into wells of a microplate followed by addition of an equal volume of SLP test solution. To measure the formation of melanin, a kinetic assay was run and absorbance at 650 nm was measured every 2 min for 60 minutes using a Bio-rad Benchmark plus reader. Concentrations were calculated from a standard curve generated by doing a linear regression of a logarithmic plot of concentrations of the standards vs. onset time [Suppl. Fig. 1]. Onset time was defined as the time at which the standards or samples reached A650 = 0.4, a midpoint within the linear range.
Statistical Analysis
Data were evaluated using t test, Mann-Whitney u-test or the log-rank test for survival curves as appropriate. Statistical outliers were detected using the Grubb’s test. All statistical tests used are included in the GraphPad Prism 5 Software (GraphPad Software, La Jolla, CA). Bar graphs indicate mean ± SEM.
Results
Priming of the respiratory mucosa protects mice from lethal respiratory virus infection
The standard experimental protocol and timeline is shown in Fig. 1A. Mice were inoculated intranasally on day -14 and again on day -7 with L. plantarum, L. reuteri, or PBS/BSA diluent control. On day 0 or at time points thereafter, mice were inoculated with pneumonia virus of mice (PVM). We reported previously that adult mice primed with L. plantarum or L. reuteri as per this protocol were fully protected from the lethal sequelae of subsequent PVM infection [Gabryszewski et al., 2011]). Here we extend this observation, and show that 40% of the mice inoculated with 109 cfu L. plantarum on day -14 and again on day -7 survive lethal infection when challenged with PVM at day 153, or 5 months after initial Lactobacillus priming [Fig. 1B]. Lactobacillus-priming elicited no anti-PVM IgG that could be detected in serum at day 0 [Gabryszewski et al., 2011]. Likewise, no anti-PVM IgG was detected in serum at two months after initial priming (data not shown).
Figure 1. Priming of the respiratory mucosa with live Lactobacillus species protects mice from lethal sequelae of subsequent virus challenge.
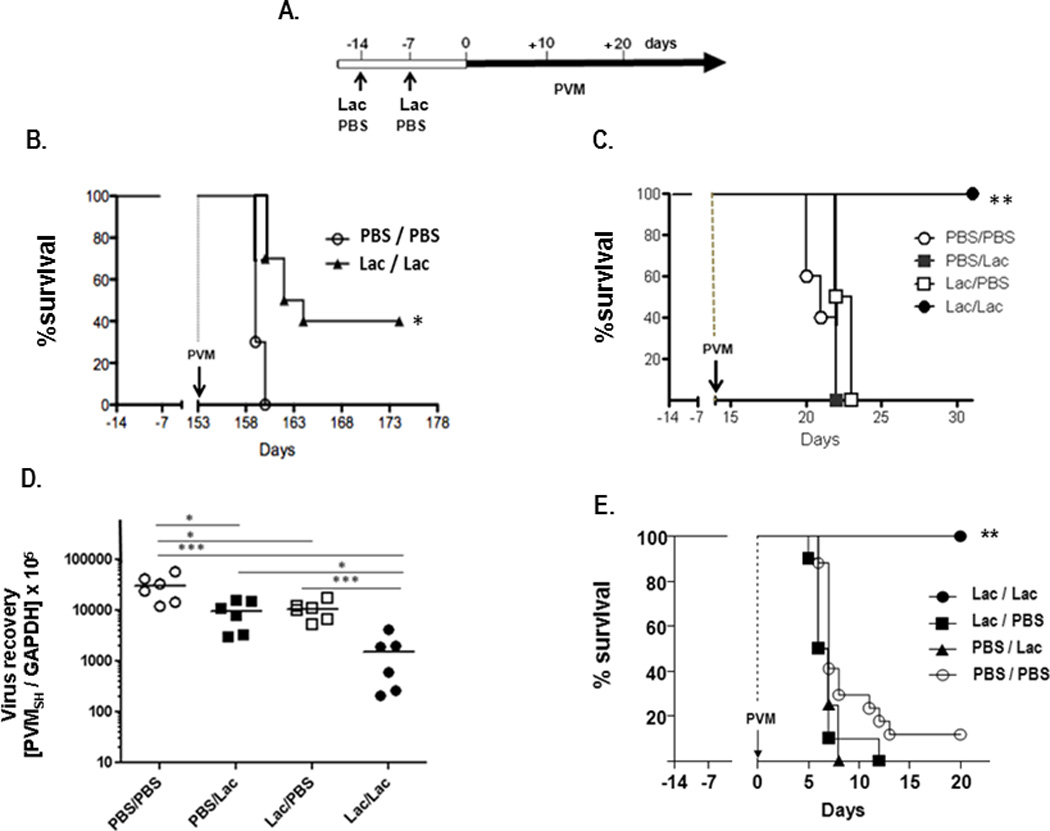
A. Standard experimental protocol and timeline. Mice are inoculated intranasally with Lactobacillus (Lac), either 109 cfu live Lactobacillus plantarum, 108 cfu live Lactobacillus reuteri, or PBS/BSA (PBS) control on day -14 and again on day -7. On day 0 or at a time point thereafter, mice are challenged with pneumonia virus of mice (PVM). B. Survival of 8 week old BALB/c mice inoculated with L. plantarum as indicated in A., and challenged with PVM on day +153 (5 months), n = 10 mice per group. C. Survival of 8 week old BALB/c mice primed on day -14 and day -7 with PBS/BSA, with PBS/BSA on day -14 and L. plantarum on day -7, with L. plantarum on day -14 and PBS/BSA on day -7, or with L. plantarum on day -7 and on day -14; all inoculated with PVM on day +14, n = 5 mice/group. D. Virus recovery on day +17 (PVMSH/GAPDH) from lung tissue of mice primed and challenged with PVM as in (C.) E. Survival of 3 week old C57BL/6 mice primed on day -14 and day -7 with PBS/BSA only, with L.reuteri on day -14 and PBS/BSA on day -7, with PBS/BSA on day -14 and L. reuteri on day -7, or L. reuteri on day -7 and day -14; n = 10 mice/group; statistical significance, *p < 0.05, **p < 0.01, ***p < 0.005.
Although our priming protocol was designed to include two intranasal inoculations with L. plantarum or L. reuteri, we had not determined previously whether one or both inoculations were necessary to elicit full protection against lethal virus challenge. As shown in Fig. 1C, we found that at least two inoculations with L. plantarum were required to elicit protection via this protocol. Interestingly, although the single inoculations with Lactobacillus at either day -14 or day -7 had no impact on survival, they did have a significant impact on virus recovery [Fig. 1D]. Virus recoveries from control-primed mice were ~3-fold higher than those from mice primed only once with L. plantarum (*p < 0.05), while virus recoveries from control-primed mice were ~20-fold higher than those from mice primed twice with Lactobacillus (*p < 0.005). Overall, these findings are consistent with our previous observations, specifically: while Lactobacillus priming has a clear impact on virus replication and clearance, virus recovery may be variable. While statistically significant, relatively small changes in virus recovery may not predict changes in survival.
Finally, similar to findings in adult mice, we show here that three-week old C57BL/6 mice receiving two intranasal inoculations, each at 108 cfu L. reuteri, were also fully protected from the lethal sequelae of subsequent PVM infection [Fig. 1E]. Mice that received only one intranasal Lactobacillus inoculation either at one week (day -7) or two weeks (day -14) prior to virus challenge were not protected, and exhibited survival rates that were indistinguishable from the controls.
Inflammatory cytokine production and leukocyte recruitment
In order to understand the nature of the antiviral state, we proceeded to examine the inflammatory responses to Lactobacillus-mediated priming, including pro-inflammatory cytokine production and leukocyte recruitment to lung tissue. Elevated levels of leukocyte chemoattractants CXCL1, CCL3, and IL-17A and the proinflammatory mediator TNF-α were detected in lung tissue within 24 hours after the first intranasal inoculation with L. reuteri (day -13, timeline as in Fig. 1A). These responses were transient; levels of these mediators fell to baseline by day -10 [Fig. 2]. In response to the second L. reuteri inoculation, cytokine production was more robust and sustained. In addition to the aforementioned mediators, we also detected significantly elevated levels of CCL2 and CXCL10 at 24 hrs after the second inoculation (day -6), with levels of all but TNF-α remaining above baseline control (PBS/PBS) levels at the day -3 time point. In parallel experiments, levels of CXCL1 and CXCL10 were elevated in serum at days -13 and -6, within 24 hrs after Lactobacillus-priming, and total IgG levels remained unchanged [Suppl. Fig. 2]. No immunoreactive IFN-α or IFN-β was detected in response to priming with L. reuteri at any time points (data not shown). Similarly, Lactobacillus-priming elicited only minimal production of IFNγ [Fig. 3A]; no biologically-active IL-12p70 was detected over baseline (data not shown). Lactobacillus-priming protects IFNγ gene-deleted mice just as effectively as wild-type mice [Fig. 3B], overall suggesting no direct role for Th1 cytokines in this response.
Figure 2. Proinflammatory cytokines detected in response to Lactobacillus priming.
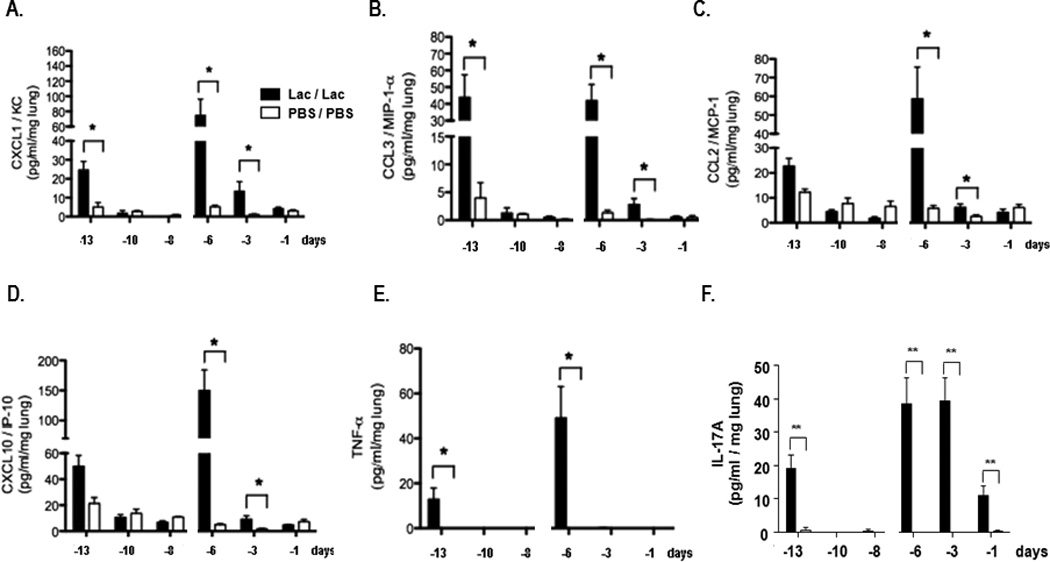
A. CXCL1, B. CCL3, C. CCL2, D. CXCL10, E. TNF-α and F. IL-17A detected in whole lung homogenates (pg / mL / mg lung tissue) at three time points after the first (day -14) and second (day -7) intranasal inoculation with L. reuteri (Lac, filled bars) or PBS/BSA control (PBS, open bars); n = 4 – 8 mice/time point, *p < 0.05, **p < 0.01, ***p < 0.005. No type I interferons were detected at any time point. Data compiled from 2 independent experiments.
Figure 3. Lactobacillus-priming and interferon-γ.
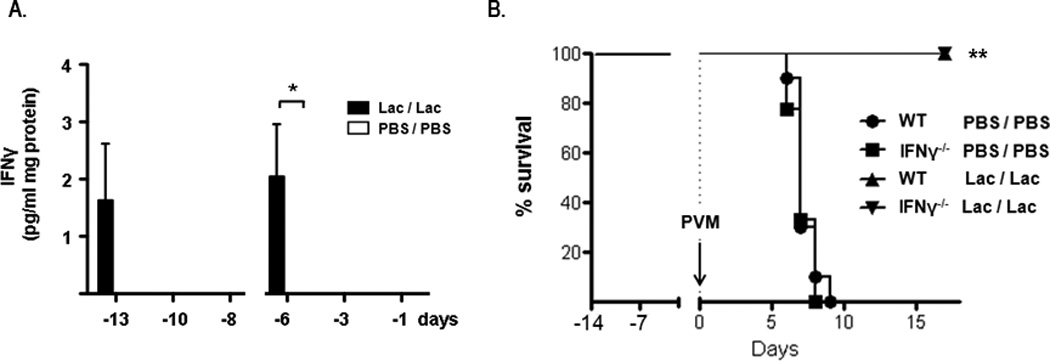
A. IFNγ detected in whole lung homogenates (pg/mL /mg lung tissue) at three time points after first (day -14) and second (day -7) intranasal inoculation with L. reuteri (Lac, filled bars) or PBS/BSA control (PBS, open bars); n = 4 – 8 mice / time point. B. Survival of IFNγ−/− and wild-type mice inoculated with L. plantarum as per standard protocol (Fig. 1A) and challenged at day 0 with PVM; *p < 0.05; **p < 0.01.
Similar to the pattern of cytokine production, total cell counts in single cell lung suspensions rose within 24 hrs of each intranasal inoculation with L. reuteri and returned to baseline levels immediately thereafter [Fig. 4A]. Neutrophils comprised ~60 – 70% of total cells detected at these time points [Fig. 4B]. Neutrophils were prominent in lung tissue sections [Fig. 4C and 4D], and were detected throughout the parenchyma, within the alveoli and were dispersed throughout the surrounding tissue. There were no localized inflammatory infiltrates and no evidence of edema.
Figure 4. Neutrophils are recruited to the lungs in response to Lactobacillus priming.
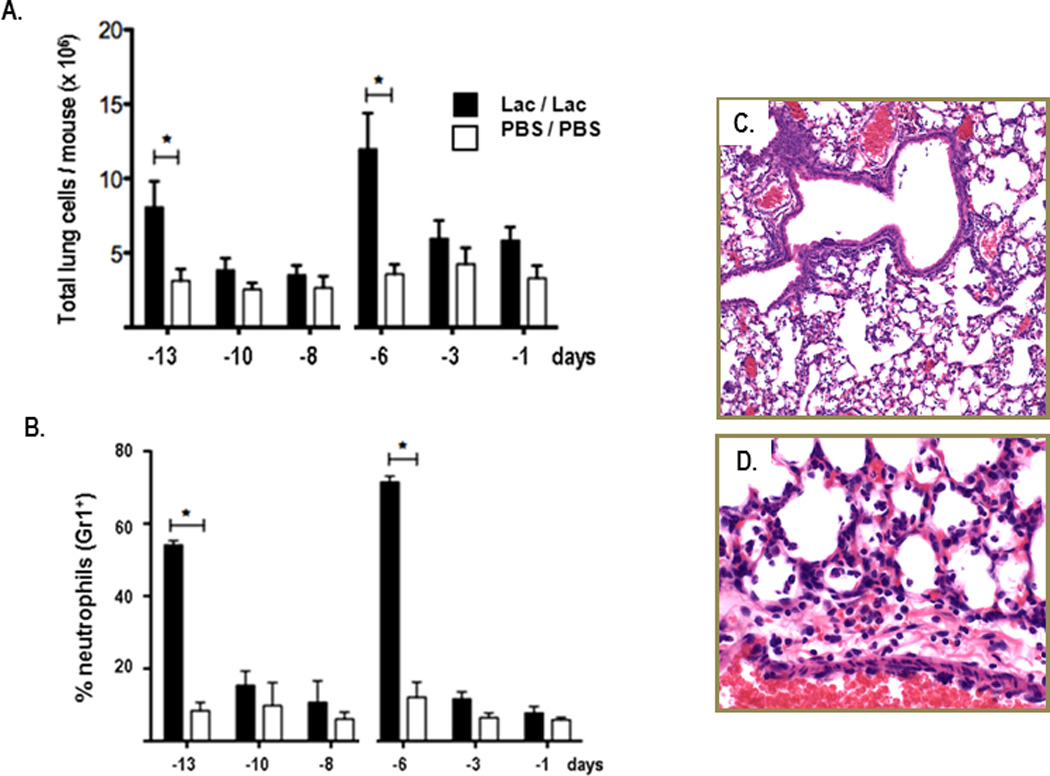
Cells from whole lung tissue evaluated by flow cytometry at three time points after the first (at day -14) and second (at day -7) inoculations with L. reuteri (Lac; filled bars) or PBS/BSA (PBS; open bars); A. Total lung cells B. Percent Gr1+ (neutrophils); n = 3 mice pooled for each time point, data compiled from 3 – 5 independent experiments; *p < 0.05; C. & D. Formalin-fixed, hematoxylin and eosin (H&E)-stained lung tissue from day -6, after second L. reuteri inoculation (see Fig. 1A); original magnifications, 10X and 40X, respectively.
Neither T lymphocyte [Fig. 5A], B lymphocyte, NK cell [Suppl. Fig. 3] or regulatory T cell (data not shown) recruitment was detected at any of the time points indicated. Recruitment of CD11c+ dendritic cells was first detected on day -8, nearly a full week after the first Lactobacillus inoculation [Fig. 5B]. In contrast to neutrophil recruitment which was profound but transient, dendritic cell recruitment was less prominent but persistent, notably after the second Lactobacillus inoculation.
Figure 5. Differential leukocyte recruitment in response to Lactobacillus priming.
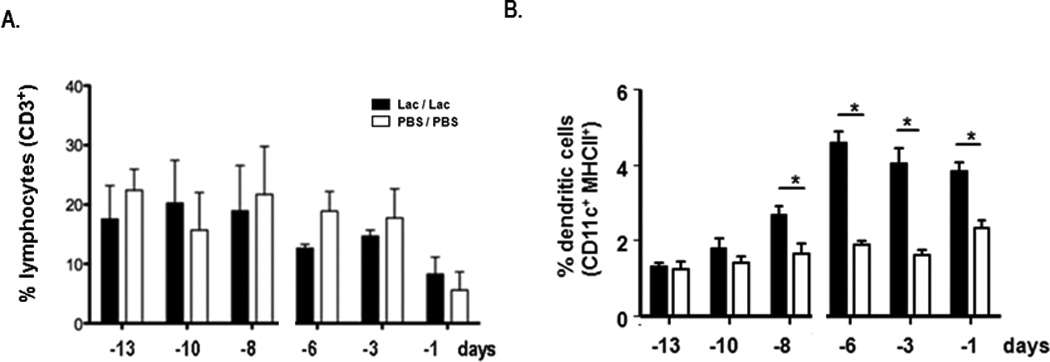
Cells from whole lung tissue evaluated by flow cytometry as in Fig. 4. A. Percent CD3+ lymphocytes B. percent CD11c+MHCIIhi dendritic cells; p < 0.05.
Lactobacillus clearance from lung tissue
Consistent with the aforementioned pattern of neutrophil recruitment and production of proinflammatory mediators, we found that live L. reuteri were cleared rapidly from lung tissue; there was no evidence of bacterial replication or colonization. Live L. reuteri were isolated from lung tissue homogenates at 6 and 12 hours after inoculation, but no live L. reuteri was detected in lung tissue at 24 hours after the first [Fig. 6A] or second (data not shown) L. reuteri inoculations. Peak levels of L. reuteri genomic DNA (gDNA) were similarly detected at the first time point (4 hours) after inoculation; prominent detection continued 24 and 48 hours after inoculation [Fig. 6B]. Similarly, L. reuteri peptidoglycan was detected in lung tissue for 1 – 3 days after complete clearance of live L. reuteri [Fig. 6C].
Figure 6. Lactobacillus clearance from lung tissue.
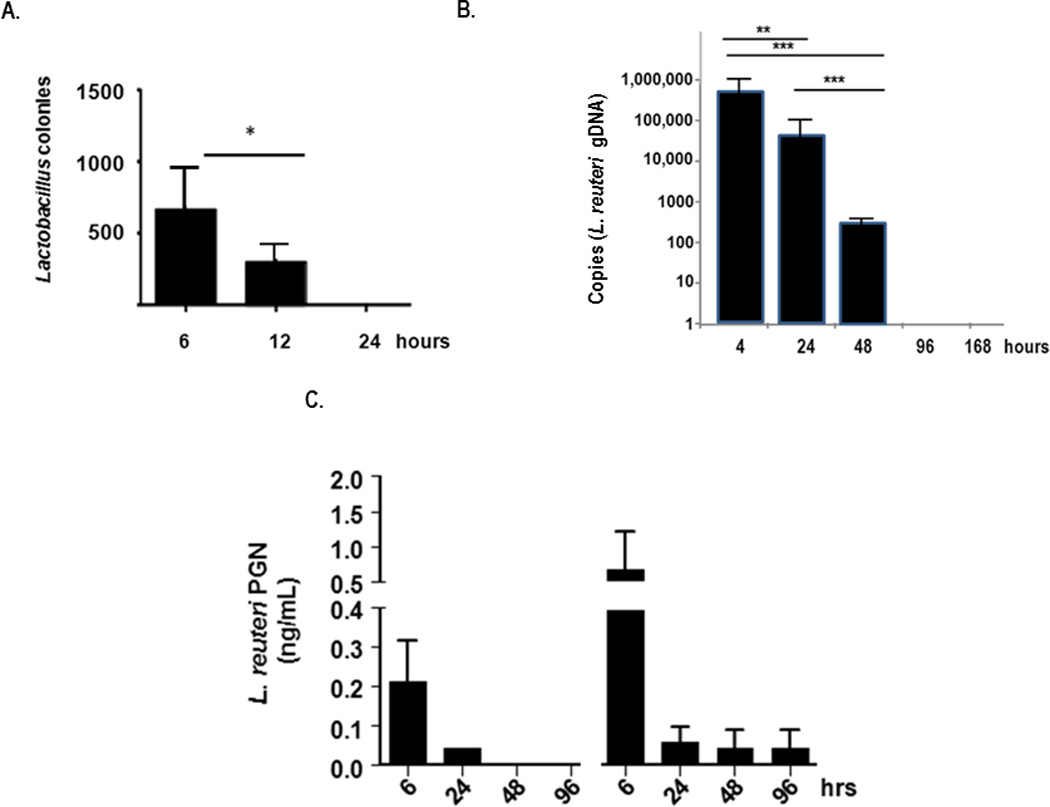
A. Live L. reuteri detected in lung tissue at time points after inoculation with 108 cfu; n = 3 mice/time point. B. L. reuteri genomic DNA (gDNA) detected by qPCR targeting 16S–23S intergenic region, normalized to a standard curve. C. L. reuteri PGN detected by kinetic melanin-generating spectrophotometric assay (see Suppl. Fig. 1) normalized to S. aureus standards; *p < 0.05; **p < 0.01; ***p < 0.005.
The impact of genomic DNA on inflammation and survival in response to PVM infection
In contrast to results obtained upon priming with live L. reuteri, priming with L. reuteri genomic DNA (gDNA) in amounts ~100–1000 fold greater than found in the bacterial inoculum did not promote neutrophil recruitment to lung tissue [Fig. 7A]. Likewise, priming with L. reuteri gDNA had only minimal and transient impact on virus recovery [Fig. 7B]. L. reuteri gDNA also had no impact on induction of the proinflammatory mediator, CCL3 [Fig. 7C], or on survival in response to PVM infection [Fig. 7D].
Figure 7. Priming with L. reuteri genomic DNA does not elicit protection from the lethal sequelae of PVM infection.
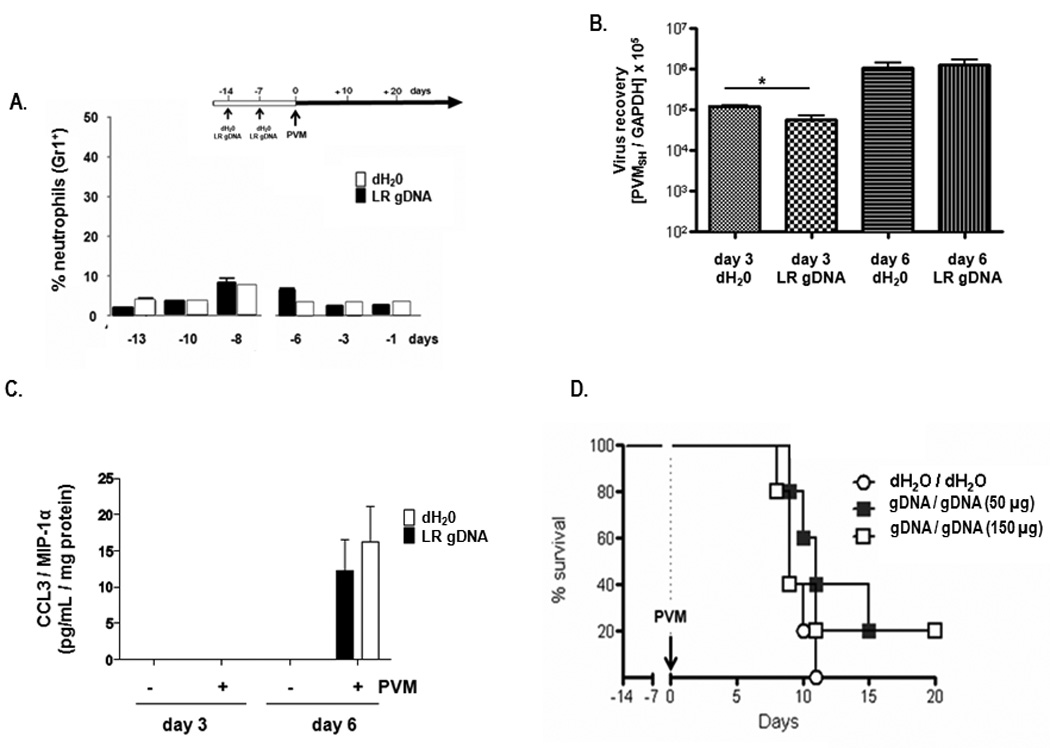
A. Cells from whole lung tissue evaluated by flow cytometry after a first (day -14) and second (day -7) inoculation with 150 µg L. reuteri gDNA or diluent (dH2O). Percent neutrophils (Gr1+ cells) at each time point are as shown; n = 3 mice per time point per condition, data compiled from 3 experiments. B. Virus recovery (PVMSH /GAPDH) from mice primed with 50 µg L. reuteri genomic DNA or diluent (dH20) at days +3 and +6 after PVM challenge on day 0 C. Detection of CCL3 / MIP-1α in whole lung homogenates from mice primed and PVM-challenged as in B. D. Survival of mice inoculated with L. reuteri gDNA (50 or 150 µg at days -7 and -14) or dH2O diluent control, and challenged with PVM on day 0, n = 5 mice per group;
The impact of peptidoglycan on inflammation and survival in response to PVM infection
In our previous study [Gabryszewski et al., 2011], we found that priming with both live and heat-inactivated L. plantarum and L. reuteri, as well as an unrelated live and heat-inactivated Gram-positive Listeria innocua, elicited full protection against a subsequent lethal PVM infection. Prominent neutrophil recruitment was observed in response to priming with Gram-positive PGN (50 µg / mouse), at levels indistinguishable from those observed in response to live L. reuteri [Fig. 8A]. The cytokine expression patterns detected in response to PGN alone were also similar to those observed in response to priming with L. reuteri [Fig. 8B – F]. However, although priming with a large inoculum of PGN (100 µg / mouse / inoculation, roughly equivalent to a PGN inoculum from 109 Gram-positive bacteria) resulted in delayed mortality (median survival, t½ = 9.0 vs.10.5 days, *p < 0.05 [Fig. 8G]), it did not confer sustained survival such as that observed in response to priming with live L. reuteri. No protection against lethal PVM challenge was observed in response to priming with 10 or 50 µg PGN / mouse / inoculation (data not shown).
Figure 8. Priming with peptidoglycan results in neutrophil recruitment and proinflammatory cytokine production but does not protect mice against the lethal sequelae of PVM infection.
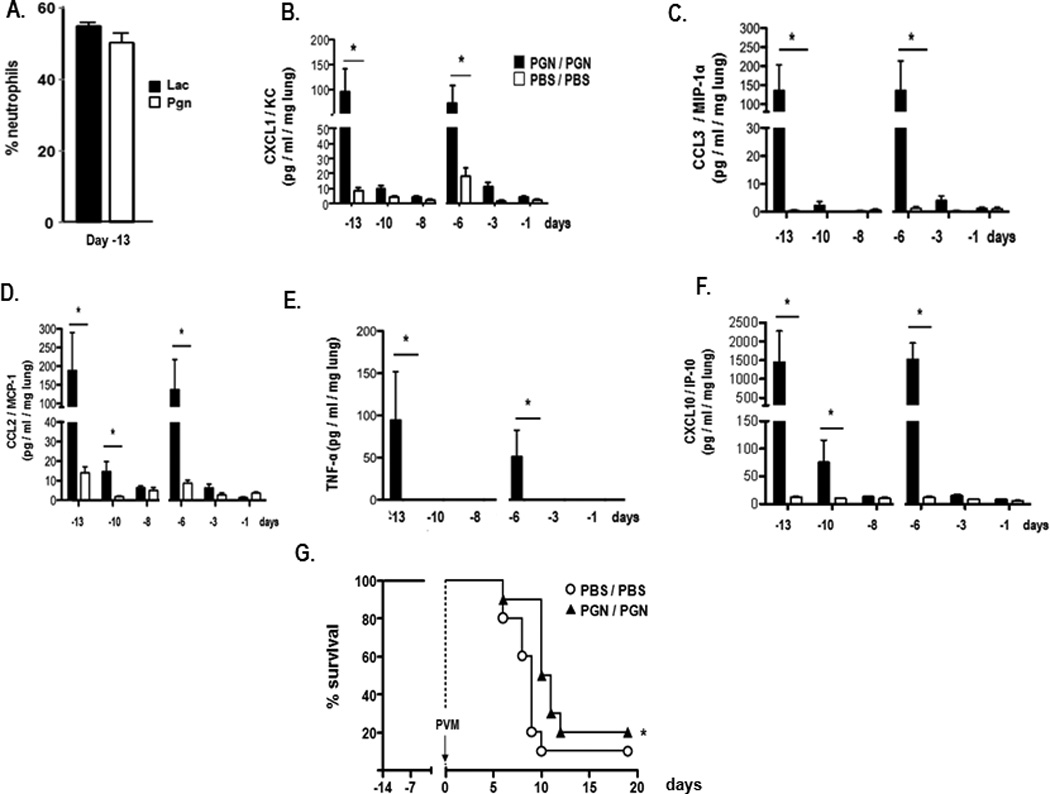
A. Percentage of GR1+ neutrophils detected in single cell suspensions from whole lung from mice inoculated with 100 µg PGN compared to 108 cfu L. reuteri (Lac). B. – F. Proinflammatory cytokines CXCL1, CCL3, CCL2, TNF-α, and CXCL10 detected in whole lung homogenates after first (day -14) and second (day - 7) intranasal inoculations with 100 µg PGN (filled symbols) or PBS/BSA control (open symbols); G. Survival of mice primed with PGN (100 µg / mouse; filled symbols) or PBS/BSA (PBS; open symbols) prior to inoculation with PVM on day 0; n = 10 mice/group; statistical significance, *p < 0.05.
Discussion
In our previous work, we showed that lactobacilli targeted to the respiratory tract were highly effective at suppressing virus-induced inflammation and protecting against lethal disease. Specifically, we found that wild-type adult mice primed via intranasal inoculation with live or heat-inactivated Lactobacillus strains were completely protected against lethal infection with pneumonia virus of mice (PVM), and that significant protection against subsequent virus challenge persisted for at least three months after initial priming [Gabryszewski et al., 2011]. Several other groups have reported similar results with influenza A, albeit findings limited to short-term protection only in response to intranasal inoculation with various Lactobacillus species [Harata et al., 2010; Izumo et al., 2010; Hori et al., 2001; Young et al., 2012]
Here, we have extended these findings and found that Lactobacillus-mediated protection can be sustained for as long as five months, can be elicited in younger mice, and that at least two intranasal inoculations with L. plantarum or L. reuteri were required to elicit protection against a lethal virus challenge. Although priming with L. reuteri results in suppression of inflammation upon virus challenge [Gabryszewski et al., 2011], we show here that priming itself elicits a robust but transient inflammatory response, including production of proinflammatory cytokines and leukocyte chemoattractants but interestingly, no type I interferons. Parker and Prince [2011] recently reviewed the literature on type I interferons and extracellular bacteria in the airways, which include specific responses to Gram-positive Staphylococcus aureus and Streptococcus pneumoniae and Gram-negative Pseudomonas aeruginosa. It is not immediately clear why these microoganisms (and others, as discussed below) elicit prominent type I interferon responses in lung tissue while inoculation with L. reuteri does not. It is also not immediately clear what the impact of this finding is with respect to the outcome of a subsequent infection, particularly with an unrelated virus pathogen.
Although lactobacilli are cleared rapidly, both peptidoglycan and genomic DNA from L. reuteri are detected for several days after live bacteria have cleared. Several groups have explored the immunomodulatory potential of Lactobacillus genomic DNA. Kim and colleagues [2012] reported that pretreatment of monocytic THP-1 cell line with L. plantarum gDNA inhibited production of TNFα in response to LPS. Likewise, Bouladoux and colleauges [2012] have identified unique immunosuppressive motifs enriched in the DNA of Lactobacillus species that inhibit activation of dendritic cells in the gastrointestinal tract. Despite prominent recruitment of dendritic cells to the respiratory tract in response to whole Lactobacillus-priming, we find that L. reuteri genomic DNA administered in amounts far exceeding that received with bacterial inoculation had no impact on any parameters associated with protection against lethal virus infection.
Pattern recognition receptors (PRRs) that recognize gram-positive bacteria and their components are among the factors that might be considered important in Lactobacillus-mediated priming in the lung. The nucleotide-binding oligomerization (NOD) protein, NOD2 is a cytosolic PRR expressed in epithelial and antigen presenting cells (dendritic cells and macrophages) that detects muramyl-dipeptides from both gram-positive and gram-negative bacteria and activates inflammatory gene transcription via NF-kB activation pathways (reviewed in [Strober et al., 2006; Sorbara & Philpott, 2011; Biswas et al., 2012]). At present, there is a small literature on the interactions between Lactobacillus strains and the NOD2 receptor. Among the most interesting, Macho Fernandez and colleagues (2011) found that peptidoglycan from the specific L. salivarius strain Ls33 protected mice from chemical colitis in a NOD2-dependent, MyD88-independent fashion. Although the main focus has been the gastrointestinal mucosa, NOD2 is expressed in cells recruited to lung tissue, and NOD2 gene-deleted mice display aberrant inflammatory responses to both bacterial and viral respiratory pathogens as well as to pathogen-derived antigens [Theivanthiran et al., 2012; Coulombe et al., 2012; Divangahi et al., 2008; Frutuoso et al., 2010; Davis et al. 2011].
Further experiments were designed to explore the relationship between transient neutrophil recruitment and protection against lethal virus infection. Recruitment paralleled Lactobacillus-mediated induction of numerous neutrophil chemoattractants, including CXCL1 and IL-17A [Pappu et al., 2012]. Various strategies that result in depletion of neutrophils from blood and bronchoalveolar lavage (BAL) fluid [Tate et al., 2011; Daley et al., 2008; McFarlane et al., 2008; Hahn et al., 2011] yielded only partial (~50%) depletion of neutrophils from whole lung tissue (Garcia-Crespo et al., unpublished results). Interestingly, these results (clearance from airways and blood, but not lung tissue) are similar to those observed clinically in response to anti-IL-5 monoclonal antibody therapy (mepolizumab) designed to eliminate eosinophils from the human asthmatic lung [Leckie et al., 2000]. However, to address this point from another perspective, we found that priming with PGN elicited neutrophil recruitment but not full protection against respiratory virus infection. Thus, we conclude that neutrophil recruitment, while a prominent response to Lactobacillus-mediated priming, may not be sufficient to elicit protection against a subsequent virus challenge.
Lactobacillus-mediated protection against respiratory virus infection via the protocol featured here is a unique and particularly robust form of heterologous immunity. As such, our findings can be compared and contrasted with findings reported by Tuvim, Evans, Dickey and colleagues [Evans et al., 2010; Evans et al., 2011; Tuvim et al., 2009; Clement et al., 2008; Duggan et al., 2011] who likewise examined questions related to this issue. Their model focuses specifically on pathogen resistance in lung tissue in response to multiple infusions of a sterile lysate from the Gram-negative microorganism, non-typeable Haemophilus influenza (NTHi; reviewed in [Erwin and Smith, 2007; Foxwell et al., 1998]). Among the specific similarities, these researchers have found that one or more intranasal infusions with NTHi result in protection from the lethal sequelae of infection with mouse-passaged H3N2 influenza A; likewise, NTHi infusion elicits production of inflammatory cytokines (including TNF-α) and neutrophil recruitment. However, among the marked differences between our datasets, protection elicited by the sterile NTHi infusion is short-lived. In experiments examining protection against a heterologous pathogen, the protective impact of NTHi infusion peaked within 4 hrs of the intranasal infusion, with effectiveness waning at 48 hrs [Clement et al., 2008]. In contrast, Lactobacillus-mediated priming elicits full protection against PVM when the virus pathogen is introduced one to three weeks after the final Lactobacillus inoculation, with significant protection persisting for up to 5 months. Furthermore, NTHi-mediated protection is dependent on MyD88-signaling [Duggan et al., 2011], while Lactobacillus-mediated protection against PVM can be elicited in MyD88 gene-deleted mice [Gabryszewski et al., 2011]. Another significant difference, sterile NTHi infusion results in profound and immediate induction of IFN-α, IFN-αβR, and type I interferon-related transcripts [Tuvim et al., 2009]; in contrast, no immunoreactive IFN-α or IFN-β was detected in whole lung lysates in response to Lactobacillus-priming. Thus, although both NTHi and Lactobacillus-priming may proceed to a shared outcome - protection against respiratory pathogens - the cellular and biochemical mechanisms promoting these responses are clearly unique and stimulus-specific.
Among our immediate goals is the ongoing exploration of the mechanisms promoting protection in response to Lactobacillus priming. These findings will ultimately lead us in new directions and toward novel immunomodulatory modalities.
Supplementary Material
Wild-type mice inoculated with Lactobacillus were fully protected against the lethal sequelae of subsequent PVM infection
Inflammation associated with Lactobacillus exposure is robust but transient Lactobacillus clearance is rapid, but gDNA and PGN persist
Administration of Lactobacillus gDNA did not elicit inflammation or protect mice against PVM infection.
Acknowledgments
The authors are grateful for the assistance of Dr. Alfonso Gonzalo and the staff of National Institute of Allergy and Infectious Diseases 14BS animal facility for care of mice used in these studies, Mr. Ricardo Dreyfuss of Medical Arts who prepared the photomicrographs for publication, and Dr. Stephanie Glineur for performing the studies to detect serum antibodies to PVM. HFR would also like to thank Dr. Peter J. Openshaw who brought the concept of innate imprinting to our attention. This work is funded by NIAID Division of Intramural Research (DIR) AI000943 to HFR and Children’s Miracle Network of New York to JBD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayala VI, Teijaro JR, Farber DL, Dorsey SG, Carbonetti NH. Bordetella pertussis infection exacerbates influenza virus infection through pertussis toxin-mediated suppression of innate immunity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019016. 4e10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger MN, Standiford TJ. Postinfluenza bacterial pneumonia: host defenses gone awry. J Interferon Cytokine Res. 2010;30:643–652. doi: 10.1089/jir.2010.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bem RA, J. B. Domachowske JB, Rosenberg HF. Animal models of human respiratory syncytial virus disease. Am. J. Physiol. Lung Cell Mol. Physiol. 2011;201:L148–L156. doi: 10.1152/ajplung.00065.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A, Petnicki-Ocwieja T, Kobayashi KS. Nod2: a key regulator linking microbiota to intestinal mucosal immunity. J. Mol. Med. 2012;90:15–24. doi: 10.1007/s00109-011-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonville CA, Easton AJ, Rosenberg HF, Domachowske JB. Altered pathogenesis of severe pneumovirus infection in response to combined antiviral and specific immunomodulatory agents. J. Virol. 2003;77:1237–1244. doi: 10.1128/JVI.77.2.1237-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonville CA, Lau VK, DeLeon JM, Gao JL, Easton AJ, Rosenberg HF, Domachowske JB. Functional antagonism of chemokine receptor CCR1 reduces mortality in acute pneumovirus infection in vivo. J. Virol. 2004;78:7984–7989. doi: 10.1128/JVI.78.15.7984-7989.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouladoux N, Hall JA, Grainger JR, Dos Santos LM, Kann MG, Nagarajan V, Verthelyi D, Belkaid Y. Regulatory role of suppressive motifs from commensal DNA. Mucosal Immunol. 2012 doi: 10.1038/mi.2012.36. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnaud P, Machinis K, Coutte LA, Marecat A, Mercenier A. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. J. Micro. Met. 2001;44:139–148. doi: 10.1016/s0167-7012(00)00244-x. [DOI] [PubMed] [Google Scholar]
- Clement CG, Evans SE, Evans CM, Hawke D, Kobayashi R, Reynolds PR, Moghaddam SJ, Scott BL, Melicoff E, Adachi R, Dickey BF, Tuvim MJ. Am. J. Respir. Crit. Care Med. 2008;177:1322–1330. doi: 10.1164/rccm.200607-1038OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe F, Fiola S, Akira S, Cormier Y, Gosselin J. Muramyl-dipeptide induces NOD2-dependent Ly6C(high) monocyte recruitment to the lungs and protects against influenza virus infection. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036734. e36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J. Leukoc. Biol. 2008;83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- Davis KM, Nakamura S, Weiser JN. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of Spneumonia colonization in mice. J. Clin. Invest. 2011;121:3666–3676. doi: 10.1172/JCI57761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didierlaurent A, Goulding J, Hussell T. The impact of successive infections on the lung microenvironment. Immunology. 2007;122:457–465. doi: 10.1111/j.1365-2567.2007.02729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divangahi M, Mostowy S, Coulombe F, Kozak R, Guillot L, Veyrier F, Kobayashi KS, Flavell RA, Gros P, Behr MA. NOD2-deficient mice have impaired resistance to Mycobacterium tuberculosis infection through defective innate and adaptive immunity. J. Immunol. 2008;181:7157–7165. doi: 10.4049/jimmunol.181.10.7157. [DOI] [PubMed] [Google Scholar]
- Duggan JM, You D, Cleaver JO, Larson DT, Garza RJ, Guzman Pruneda FA, Tuvim MJ, Zhang J, Dickey BF, Evans SE. Synergistic interactions of TLR2/6 and TLR9 induce a high level of resistance to lung infection in mice. J Immunol. 2011;186:5916–5926. doi: 10.4049/jimmunol.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KD, Garcia-Crespo KE, Glineur S, Domachowske JB, Rosenberg HF. The pneumonia virus of mice (PVM) model of acute respiratory infection. Viruses. 2012 doi: 10.3390/v4123494. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin AL, Smith AL. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Micrbiol. 2007;15:355–362. doi: 10.1016/j.tim.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Evans SE, Tuvim MJ, Fox CJ, Sachdev N, Gibiansky L, Dickey BF. Inhaled innate immune ligands to prevent pneumonia. Brit. J. Pharmacol. 2011;163:195–206. doi: 10.1111/j.1476-5381.2011.01237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SE, Xu Y, Tuvim MJ, Dickey BF. Inducible innate resistance of lung epithelium to infection. Ann. Rev. Physiol. 2010;72:413–435. doi: 10.1146/annurev-physiol-021909-135909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey AR, Walsh EE. Respiratory syncytial virus infection in elderly adults. Drugs Aging. 2005;22:577–587. doi: 10.2165/00002512-200522070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxwell AR, Kyd JM, Cripps AW. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Micro. Mol. Biol. Revs. 1998;62:294–308. doi: 10.1128/mmbr.62.2.294-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frutuoso MS, Hori JI, Pereira MS, Junior DS, Sonego F, Kobayashi KS, Flavell RS, Cunha FQ, Zamboni DS. The pattern recognition receptors Nod1 and Nod2 account for neutrophil recruitment to the lungs of mice infected with Legionella pneumophila. Microbes Infect. 2010;12:819–827. doi: 10.1016/j.micinf.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Gabryszewski SJ, Bachar O, Dyer KD, Percopo CM, Killoran KE, Domachowske JB, Rosenberg HF. Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J. Immunol. 2011;186:1151–1161. doi: 10.4049/jimmunol.1001751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding J, Snelgrove R, Saldana J, Didierlaurent A, Cavanagh M, Gwyer E, Wales J, Wissinger EL, Hussell T. Respiratory virus infections: do we ever recover? Proc. Am. Thorac. Soc. 2007;4:618–625. doi: 10.1513/pats.200706-066TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. App. Env. Micro. 2006;72:2359–2365. doi: 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn I, Klaus A, Janze AK, Steinwede K, Ding N, Bohling J, Brumshagen C, Serrano H, Gauthier F, Paton JC, Welte T, Maus UA. Cathepsin G and neutrophil elastase play critical and nonredundant roles in lung-protective immunity against Streptococcus pneumoniae in mice. Inf. Immun. 2011;79:4893–4901. doi: 10.1128/IAI.05593-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 2009;360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harata G, He F, Hiruta N, Kawase M, Kubota A, Hiramatsu M, Yausi H. Intranasal administration of Lactobacillus rhamnosus GG protects mice from H1N1 influenza virus infection by regulating respiratory immune responses. Lett Appl Microbiol. 2010;50:597–602. doi: 10.1111/j.1472-765X.2010.02844.x. [DOI] [PubMed] [Google Scholar]
- Hori T, Kiyoshima J, Shida K, Yasui H. Effect of intranasal administration of Lactobacillus casei Shirota on influenza infection of the upper respiratory tract in mice. Clin. Diag. Lab. Immunol. 2001;8:593–597. doi: 10.1128/CDLI.8.3.593-597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T, Cavanagh MM. The innate immune rheostat: influence on lung inflammatory disease and secondary bacterial pneumonia. Biochem. Soc. Trans. 2009;37:811–813. doi: 10.1042/BST0370811. [DOI] [PubMed] [Google Scholar]
- Inada K, Takahashi K, Ichinohe S, Suda H, Tsuchiya M, Takahashi J, Matsuura S, Kasai T, Yoshida M, Endo S, Sato S. A silkworm larvae plasma test for detecting peptidoglycan in cerebrospinal fluid is useful for the diagnosis of bacterial meningitis. Microbiol Immunol. 2003;47:701–707. doi: 10.1111/j.1348-0421.2003.tb03439.x. [DOI] [PubMed] [Google Scholar]
- Izumo T, Maekawa T, Ida M, Noguchi A, Kitagawa Y, Shibata H, Yasui H, Kiso Y. Effect of intranasal administration of Lactobacillus pentosus S-PT84 on influenza virus infection in mice. Int Immunopharmacol. 2010;10:1101–1106. doi: 10.1016/j.intimp.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Kim CH, Kim HG, Kim JY, Kim NF, Jung BJ, Jeong JH, Chung DK. Probiotic genomic DNA reduces the production of pro-inflammatory cytokine tumor necrosis factor-alpha. FEMS Microbiol Lett. 2012;328:13–19. doi: 10.1111/j.1574-6968.2011.02470.x. [DOI] [PubMed] [Google Scholar]
- Leckie MJ, et al. Effects of an interleukin-5 blocking monoclonal antibody on eosinophils, airway hyper-responsiveness, and the late asthmatic response. Lancet. 2000;356:2144–2148. doi: 10.1016/s0140-6736(00)03496-6. [DOI] [PubMed] [Google Scholar]
- Li R, Lim A, Phoon MC, Narasaraju T, Ng JK, Poh WP, Sim MK, Chow VT, Locht C, Alonso S. Attenuated Bordetella pertussis protects against highly pathogenic influenza A viruses by dampening the cytokine storm. J Virol. 2010;84:7105–7113. doi: 10.1128/JVI.02542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho Fernandez E, Valenti V, Rockel C, Hermann C, Pot B, Boneca IG, Grangette C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60:1050–1059. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- McFarlane E, Perez C, Charmoy M, Allenbach C, Carter KC, Alexander J, Tacchini-Cottier F. Neutrophils contribute to development of a protective immune response during onset of infection with Leishmania donovani. Inf. Immun. 2008;76:532–541. doi: 10.1128/IAI.01388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Y, McGuffie BA, Anderson VE, Weinberg JB. Gammaherpesvirus modulation of mouse adenovirus type 1 pathogenesis. Virology. 2008;380:182–190. doi: 10.1016/j.virol.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu R, Rutz S, Ouyang W. Regulation of epithelial immunity by IL-17 family cytokines. Trends Immunol. 2012;33:343–349. doi: 10.1016/j.it.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Parker D, Prince A. Type I interferon response to extracellular bacteria in the airway epithelium. Trends Immunol. 2011;32:582–588. doi: 10.1016/j.it.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg HF, Domachowske JB. Pneumonia virus of mice: severe respiratory infection in a natural host. Immunol Lett. 2008;118:6–12. doi: 10.1016/j.imlet.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg HF, Domachowske JB. Inflammatory responses to Respiratory Syncytial Virus (RSV) infection and the development of immunomodulatory pharmacotherapeutics. Curr. Med. Chem. 2012;19:1424–1431. doi: 10.2174/092986712799828346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadman KA, Wald ER. A review of paluvizumab and emerging therapies for respiratory syncytial virus. Expert. Opin. Biol. Ther. 2011;11:1455–1467. doi: 10.1517/14712598.2011.608062. [DOI] [PubMed] [Google Scholar]
- Sorbara MT, Philpott DJ. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol. Revs. 2011;243:40–60. doi: 10.1111/j.1600-065X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- Steinberg BE, Goldenberg NM, Lee WL. Do viral infections mimic bacterial sepsis? The role of microvascular permeability: a review of mechanisms and methods. Antiviral Res. 2012;93:2–15. doi: 10.1016/j.antiviral.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nature Revs. Immunol. 2006;6:9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- Tate MD, Ioannidis LJ, Croker B, Brown LE, Brooks AG, Reading PC. The role of neutrophils during mild and severe influenza infections of mice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017618. 3e17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theivanthiran B, Batra S, Balamayooran G, Cai S, Kobayashi K, Flavell RA, Jeyaseelan S. NOD2 signaling contributes to host defense in the lungs against Escherichia coli infection. Infect. Immun. 2012;80:2558–2569. doi: 10.1128/IAI.06230-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tuvim MJ, Evans SE, Clement CG, Dickey BF, Gilbert BE. Augmented lung inflammation protects against influenza pneumonia. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004176. 1e4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welliver TP, Reed JL, Welliver RC., Sr Respiratory syncytial virus and influenza infections. Observations from tissues of fatal infant cases. Pediatr. Infect. Dis. J. 2008;27:S92–S96. doi: 10.1097/INF.0b013e318168b706. [DOI] [PubMed] [Google Scholar]
- Williams AE, Edwards L, Humphreys IR, Snelgrove R, Rae A, Rappuoli R, Hussell T. Innate imprinting by the modified heat-labile toxin of Escherichia coli (LTK63) provides generic protection against lung infectious disease. J. Immunol. 2004;173:7435–7443. doi: 10.4049/jimmunol.173.12.7435. [DOI] [PubMed] [Google Scholar]
- Wright M, Piedimonte G. Respiratory syncytial virus prevention and therapy: past, present, and future. Pediatr Pulmonol. 2011;46:324–347. doi: 10.1002/ppul.21377. [DOI] [PubMed] [Google Scholar]
- Young HN, Lee DH, Lee YN, Park JK, Yuk SS, Yang SY, Lee HJ, Woo SH, Kim HM, Lee JB, Park SY, Choi IS, Song CS. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antiviral Res. 2012;93:138–143. doi: 10.1016/j.antiviral.2011.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


