This work shows that the photoreceptor phytochrome B is phosphorylated in vivo and demonstrates that this posttranslational modification inhibits red light–induced photomorphogenesis under nonsaturating light conditions by accelerating light-independent inactivation and dark reversion of the biologically active phyB conformer.
Abstract
The photoreceptor phytochrome B (phyB) interconverts between the biologically active Pfr (λmax = 730 nm) and inactive Pr (λmax = 660 nm) forms in a red/far-red–dependent fashion and regulates, as molecular switch, many aspects of light-dependent development in Arabidopsis thaliana. phyB signaling is launched by the biologically active Pfr conformer and mediated by specific protein–protein interactions between phyB Pfr and its downstream regulatory partners, whereas conversion of Pfr to Pr terminates signaling. Here, we provide evidence that phyB is phosphorylated in planta at Ser-86 located in the N-terminal domain of the photoreceptor. Analysis of phyB-9 transgenic plants expressing phospho-mimic and nonphosphorylatable phyB–yellow fluorescent protein (YFP) fusions demonstrated that phosphorylation of Ser-86 negatively regulates all physiological responses tested. The Ser86Asp and Ser86Ala substitutions do not affect stability, photoconversion, and spectral properties of the photoreceptor, but light-independent relaxation of the phyBSer86Asp Pfr into Pr, also termed dark reversion, is strongly enhanced both in vivo and in vitro. Faster dark reversion attenuates red light–induced nuclear import and interaction of phyBSer86Asp-YFP Pfr with the negative regulator PHYTOCHROME INTERACTING FACTOR3 compared with phyB–green fluorescent protein. These data suggest that accelerated inactivation of the photoreceptor phyB via phosphorylation of Ser-86 represents a new paradigm for modulating phytochrome-controlled signaling.
INTRODUCTION
The phytochromes (phyA to phyE) are photoreceptors that regulate the expression of ∼3000 genes of the Arabidopsis thaliana genome in a red/far-red (R/FR) light–dependent fashion and thus play a critical role in ensuring the optimal adaptation of plants to the rapidly changing light environment (Chen et al., 2004). phyA acts primarily as a FR sensor that regulates the transition from skotomorphogenesis to photomorphogenesis. phyB functions as a classical R/FR light–regulated molecular switch and is the major phytochrome involved in controlling the growth and development of mature plants (Nagy and Schäfer, 2002). Phytochromes are dimeric chromoproteins and cycle between their biologically inactive (Pr) and active (Pfr) forms. The interaction of the phyA and phyB Pfr forms with specific cellular factors is required for translocation into the nucleus (Hiltbrunner et al., 2005; Pfeiffer et al., 2012) and to launch the signaling cascade (Bae and Choi, 2008). It follows that phytochrome signaling is quantitatively determined (1) by the number of Pfr molecules available and (2) by the kinetics of protein–protein interactions between Pfr molecules and signal transducers.
Similarly to its evolutionary bacterial ancestors that are light-regulated His kinases (Yeh et al., 1997), oat (Avena sativa) phyA has also been suggested to act as a light-regulated enzyme. This hypothesis is based on the observation that autophosphorylation of oat phyA in vitro is chromophore regulated (Yeh and Lagarias, 1998) and by other studies that identified three phosphorylated amino acid residues, Ser-8, Ser-18, and Ser-598, in this photoreceptor (Lapko et al., 1996, 1997, 1999). Functional analysis revealed that substitutions inhibiting the phosphorylation (Ser8Ala and Ser18Ala) decreased, whereas phospho-mimicking mutations (Ser8Asp and Ser18Asp) increased the degradation rate of the Pfr conformer of phyA in planta (Han et al., 2010). Expression of rice (Oryza sativa) phyA with Ser residues of the extreme N-terminal region substituted by Ala residues also resulted in hypersensitivity to FR light in transgenic tobacco (Nicotiana tabacum; Stockhaus et al., 1992). These data were interpreted to mean that phosphorylation of Ser-8 and Ser-18 of oat phyA Pfr attenuates, whereas dephosphorylation enhances, FR light–controlled responses in transgenic Arabidopsis. Concerning the phosphorylation of oat phyA Ser-598, circumstantial evidence indicates that it is not an intramolecular process. Phosphorylation of Ser-598 is presumably mediated by an unknown kinase in planta, and it appears to inhibit protein–protein interactions between oat phyA and its signal-transducing partner proteins, including NUCLEOSIDE-DIPHOSPHATE KINASE2 (NDPK2) (Choi et al., 1999) and PHYTOCHROME INTERACTING FACTOR3 (PIF3; Kim et al., 2004) in vitro. It was also reported that (1) PHYTOCHROME-ASSOCIATED PROTEIN PHOSPHATASE5 (PAPP-5), a type-5 phosphatase, bound oat phyA in a conformation-specific fashion; (2) PAPP-5 dephosphorylated all three known phosphorylated sites; and (3) dephosphorylation of oat phyA increased its binding to NDPK2 and PIF3 in vitro. The same authors reported that overexpression of the phosphatase in Arabidopsis resulted in hypersensitivity, whereas papp-5 null mutants displayed reduced responsiveness to FR light (Ryu et al., 2005). Arabidopsis phyA has also been shown to be phosphorylated and phosphorylation modulated its association with CONSTITUTIVE PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYA-105 1 and FAR-RED ELONGATED HYPOCOTYL1 (FHY1)/FHY3 complexes in planta (Saijo et al., 2008), but the functional importance of these findings is not yet understood. Similar to oat and rice phyA, the extreme N-terminal region of Arabidopsis phyA is essential for signaling (Cherry et al., 1992; Jordan et al., 1997); yet, analysis of Arabidopsis phyA N-terminal mutants did not support the proposed mode of action by which phosphorylation modulates oat phyA signaling (Trupkin et al., 2007). It appears that further studies, including identification and subsequent mutational analysis of amino acid residues phosphorylated in Arabidopsis phyA, are required to resolve the existing controversy.
Independent of how phosphorylation of phyA modulates signaling, there has been considerable debate concerning whether plant phyAs phosphorylate any other proteins. With regard to oat phyA, the in vitro data demonstrating phosphorylation of CRYPTOCHROME1, PHYTOCHROME KINASE SUBSTRATE1, and auxin/indole-3-acetic acid by phyA has not been confirmed in planta (Ahmad et al., 1998; Fankhauser et al., 1999; Colón-Carmona et al., 2000). For Arabidopsis phyA, it has been recently shown that FHY1, a key mediator of the light-induced nuclear import of phyA, (1) is phosphorylated in R/FR light–reversible fashion and (2) that phosphorylation of FHY1 in R light requires the presence of phyA (Shen et al., 2009; Chen et al., 2012). These authors also demonstrated that phosphorylation of FHY1 reduces the rate of nuclear import of phyA Pfr as well as its interaction with ELONGATED HYPOCOTYL5 and PIF3 and thus results in the inactivation of transcription complexes assembled in FR (Yang et al., 2009; Chen et al., 2012). However, to show that Arabidopsis phyA indeed acts as a low-fluence response R light–activated and FR light–inactivated kinase requires additional experimentation.
Compared with phyA, scarce data have been published concerning phyB phosphorylation and its impact on phyB signaling. On the one hand, it was shown that papp-5 null mutants display hyposensitive responses to R light, whereas PAPP-5 phosphatase binds phyB in in vitro pull-down assays and these proteins colocalize in photobodies of transgenic plants (Ryu et al., 2005). On the other hand, a more recent report demonstrated that phyB also interacts with another phosphatase designated PAPP2C in vitro, and the loss-of-function mutant of papp-2c also exhibited reduced responsiveness to R light (Phee et al., 2008). Both of these phosphatases interact preferentially with the Pfr conformer of phyB, suggesting that reversible phosphorylation might play a role in modulating phyB-regulated signaling and photomorphogenesis. To determine if phyB is indeed phosphorylated in planta, we performed matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis on phyB–tandem affinity purification (TAP)–yellow fluorescent protein (YFP) fusions purified from transgenic phyB-9 plants grown under light/dark cycles. Data obtained demonstrated phosphorylation of Ser-86 in the extreme N-terminal of the photoreceptor. Next, we showed that phyB-9 plants expressing phyB–green fluorescent protein (GFP) and mutant phyB-YFP fusion proteins mimicking constant phosphorylation (Ser86Asp) or nonphosphorylation (Ser86Ala) display characteristically different responsiveness to R light. Molecular, physiological, and photobiological analysis of transgenic plants suggests that phosphorylation of phyB Ser-86 negatively regulates phyB activity by accelerating the light-independent relaxation (dark reversion) of phyB Pfr.
RESULTS
Arabidopsis phyB Is Phosphorylated in Planta
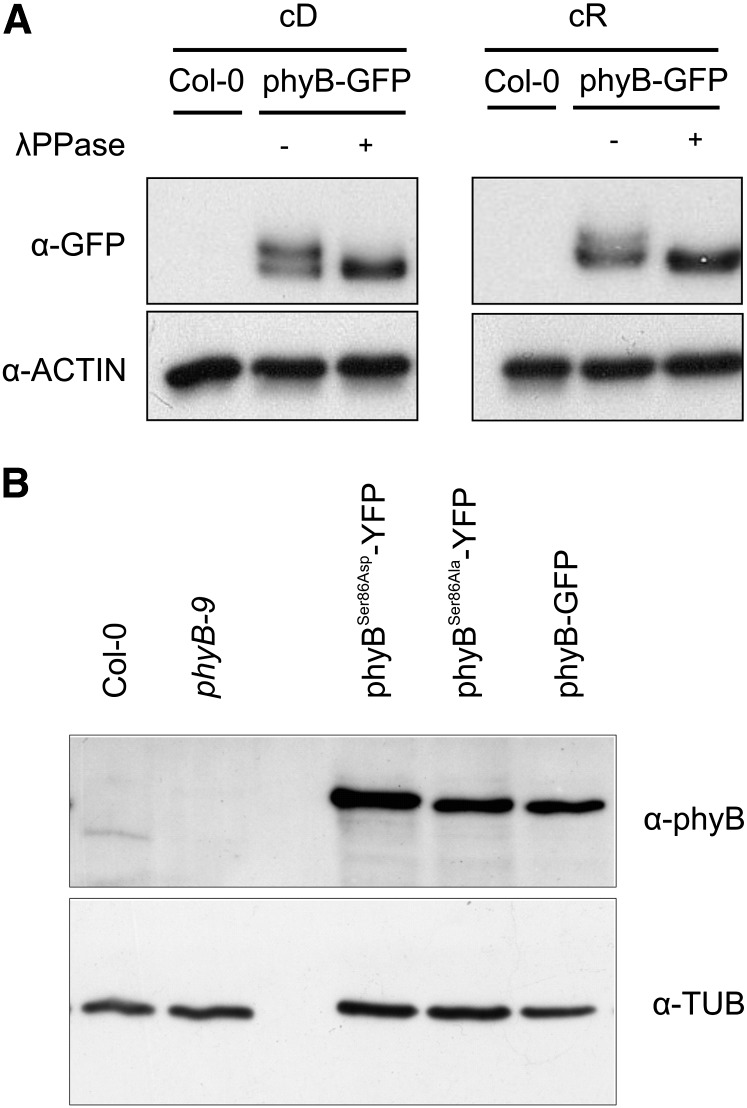
To determine whether phyB is phosphorylated in planta and if so on which amino acids, we purified the phyB-TAP fusion protein from 3-week-old plants and subjected the samples to MALDI-TOF-MS analysis. The detailed description of growth conditions, sample preparation, and MALDI-TOF-MS analysis is provided in Supplemental Methods 1 online. Repeated rounds of mass spectrometry (MS) analysis indicated phosphorylation of various amino acid residues localized mainly in the N-terminal domain of the photoreceptor. Supplemental Figure 1A online illustrates the domain structure of phyB, and Supplemental Figure 1B online shows the quantitative time-of-flight collision-induced dissociation spectrum of the ion observed at mass-to-charge ratio 577.73(2+) that revealed phosphorylation of the peptide SFDYSQSLK [80-88]. To provide an independent line of evidence for in planta phosphorylation of phyB, we analyzed phosphorylation of the phyB-YFP fusion protein with or without phosphatase treatment by Zn-Phos-Tag PAGE, a method suitable to separate phosphorylated and nonphosphorylated derivatives of the same protein (Kinoshita and Kinoshita-Kikuta, 2011). The phyB-YFP was phosphorylated in both dark- and R light–grown seedlings, although the amount of phosphorylated phyB appeared to be slightly lower in the latter sample (Figure 1A). As MALDI-TOF-MS experiments spanned a relatively long period of time, we present data obtained by characterizing the biological function of the phosphorylation of Ser-86 residue that was first unambiguously identified by MS analysis. To test the effect of Ser-86 phosphorylation on phyB-controlled responses, we replaced Ser-86 with either Asp (phyBSer86Asp-YFP) or Ala (phyBSer86Ala-YFP) and expressed these as well the wild-type phyB-GFP or phyB-YFP fusion proteins under the control of the 35S promoter (P35S) in phyB-9 mutants. Transgenic plants expressing these fusion proteins at nearly identical levels were identified and used for further studies (Figure 1B). This figure also shows that expression level of the various phyB fusion proteins was ∼25 times higher compared with that of the native phyB in Columbia-0 (Col-0).
Figure 1.
In Planta Phosphorylation and Expression Analysis of phyB-GFP.
(A) Phytochrome B is phosphorylated in vivo. Wild-type (Col-0) or transgenic plants expressing the phyB-GFP fusion protein were grown in darkness (cD) or in cR light at 40 µmol m−2 s−1 fluence rate for 4 d on wet filter papers. Total protein extracts were prepared and treated (+) or not (−) with λ phosphatase (λPPase), and the different forms of phyB-GFP protein were separated by Zn-Phos-tag PAGE and visualized using antibody-specific for GFP. The lanes contain equal amounts of total protein as shown by the comparable levels of ACTIN.
(B) Levels of the phyB-GFP, phyBSer86Ala-YFP, and phyBSer86Asp-YFP fusion proteins are identical in the selected transgenic phyB-9 lines. Transgenic phyB-9 seedlings were grown in darkness for 4 d on wet filter paper, and the expression levels of the various fusion proteins were determined by immunoblot analysis using anti-GFP antibody. The lanes contain equal amounts of total protein as shown by the comparable levels of tubulin.
Light-Induced Growth Responses of the Phospho-Mimic P35S:PHYBSer86Asp-YFP Seedlings Are Hyposensitive
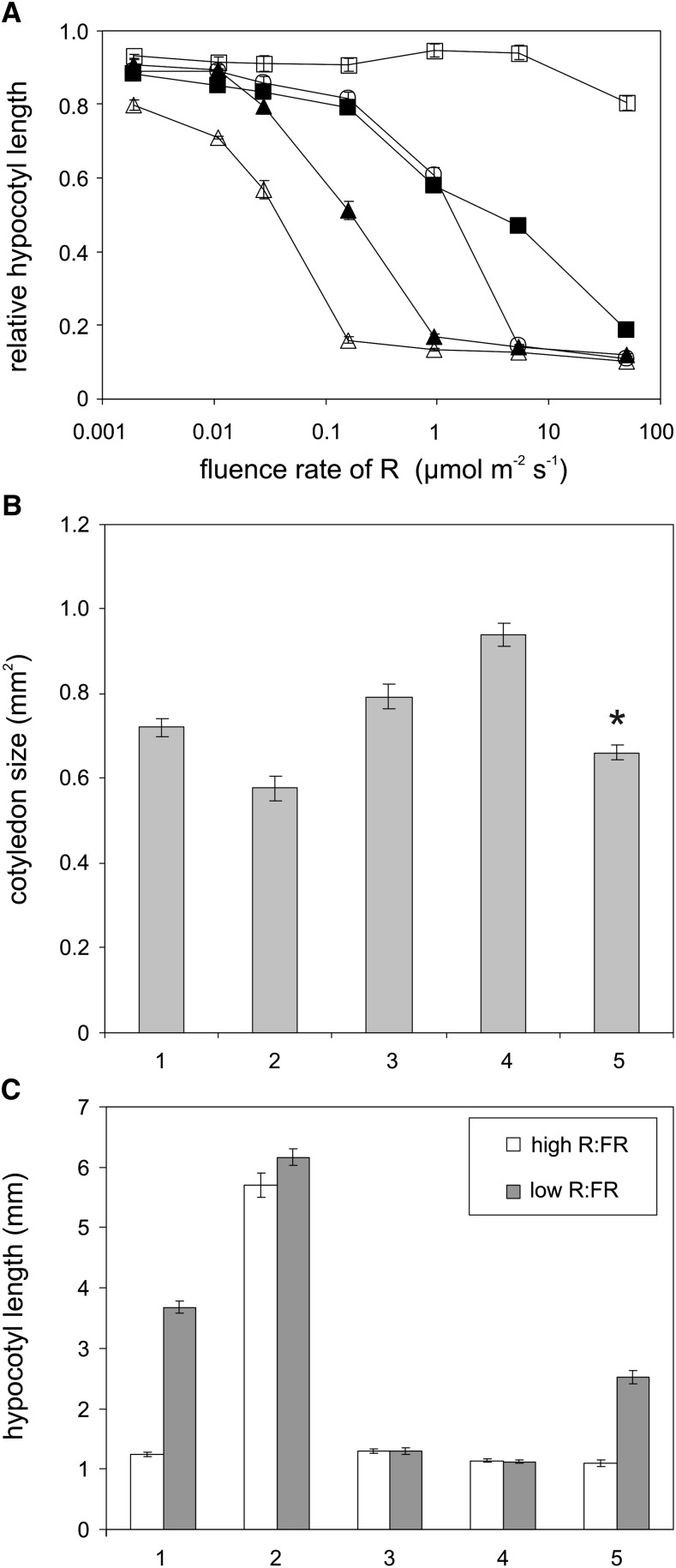
We determined to what extent the Ser86Ala and Ser86Asp substitutions affect light-induced phyB-controlled physiological responses. Analysis of fluence rate–dependent growth inhibition of hypocotyls in constant R (cR) light demonstrated that P35S:PHYBSer86Ala-YFP seedlings displayed a strong hypersensitivity, whereas P35S:PHYBSer86Asp-YFP seedlings exhibited a pronounced hyposensitivity, especially at lower fluence rates compared with P35S:PHYB-GFP or Col-0 (Figure 2A). Seedlings expressing the mutant photoreceptor that was not phosphorylated at Ser-86 were by two orders of magnitude more sensitive to cR than those expressing the phospho-mimic phyBSer86Asp-YFP fusion protein. At a fluence rate of ∼10 µmol m−2 s−1, the growth inhibition response displayed by the mutants and P35S:PHYB-GFP seedlings was saturated and uniformly hypersensitive when compared with nontransgenic Col-0 seedlings. R light–regulated cotyledon expansion at a low fluence rate (1 µmol m−2 s−1) again indicated that signaling by the phyBSer86Asp-YFP photoreceptor was significantly attenuated. The response displayed was not only hyposensitive when compared with P35S:PHYB-GFP and especially P35S:PHYBSer86Ala-YFP lines, but it was even slightly below the level of nontransgenic Col-0 (Figure 2B). At higher fluence rates (≥25 µmol m−2 s−1), all transgenic lines exhibited similar, hypersensitive cotyledon expansion responses compared with Col-0 (see Supplemental Figure 2 online).
Figure 2.
Phenotypic Characterization of Seedlings Expressing phyBSer86Ala-YFP and phyBSer86Asp-YFP.
(A) Hypocotyl growth inhibition of phyBSer86Ala-YFP seedlings exhibits extreme hypersensitivity to R light. Nontransgenic wild-type (Col-0) (closed square), and phyB-9 (cross) as well as transgenic phyB-9 seedlings expressing the phyB-GFP (closed triangle), phyBSer86Asp-YFP (open circle), and phyBSer86Ala-YFP (open triangle) fusion proteins were grown for 4 d at 22°C in darkness or at the indicated fluence rates of cR light on wet filter paper. Hypocotyl length was determined using the MetaMorph image analysis software, and the fluence rate response is shown as relative hypocotyl length to dark-grown samples (n = 50). Error bars indicate se.
(B) Cotyledon expansion is impaired in transgenic seedlings expressing the phospho-mimic phyBSer86Asp-YFP grown under low-intensity R light. Nontransgenic wild-type (Col-0; 1) and phyB-9 (2) as well as transgenic phyB-9 seedlings expressing the phyB-GFP (3), phyBSer86Ala-YFP (4), and phyBSer86Asp-YFP (5) fusion proteins were grown for 4 d on Murashige and Skoog medium without sugar at 1 µmol m−2 s−1 R light. The cotyledon surface area was measured using MetaMorph image analysis software (n = 40, error bars represent se). Asterisks indicate significant difference from the wild type as determined by Student’s t test (P < 0.001).
(C) Transgenic seedlings expressing the phospho-mimic phyBSer86Asp-YFP display hyposensitivity to shade. Nontransgenic wild-type (1) and phyB-9 (2) as well as transgenic P35S:PHYB-GFP (3), P35S:PHYBSer86Ala-YFP (4), and P35S:PHYBSer86Asp-YFP (5) expressing phyB-9 seedlings were grown for 7 d in high R/FR or for 3 d in high R/FR followed by 4 d in low R/FR at 20°C. The experiment was performed under constant light conditions (PAR = 130 µmol m−2 s−1). Hypocotyl lengths shown were measured using ImageJ software (n = 31 to 40). Error bars indicate se.
Plants also respond to changes in the R/FR ratio of incipient white light. This response, termed as shade avoidance, is regulated by the concerted action of phytochromes (Morelli and Ruberti, 2002), and phyB is a major regulator of growth under shade. For these experiments, seedlings were grown 3 d in constant high R/FR conditions before being transferred for four more days to low R/FR conditions. As expected, wild-type seedlings responded to this shade-mimicking treatment (low R/FR) with hypocotyl elongation, whereas the phyB-9 mutant did not show any response (i.e., it displayed a constitutive shade-mimicking, long hypocotyl phenotype, even under high R/FR conditions). Overexpression of phyB, either as phyB-GFP or phyBSer86Ala-YFP, completely inhibited hypocotyl elongation in response to low R/FR, indicating that despite the low R/FR conditions there was enough phyB Pfr to inhibit hypocotyl elongation (Figure 2C). However hypocotyl elongation of P35S:PHYBSer86Asp-YFP was partially restored in low R/FR, suggesting that the Ser86Asp mutation inhibits phyB signaling. Taken together, these data above convincingly demonstrate that R light–induced growth responses displayed by the P35S:PHYBSer86Asp-YFP and P35S:PHYBSer86Ala-YFP lines are markedly different and that the phospho-mimic Ser86Asp substitution inhibits, whereas the nonphosphorylatable Ser86Ala substitution enhances, phyB activity.
The Phospho-Mimic phyBSer86Asp-YFP Photoreceptor Shows Reduced Light-Induced Nuclear Accumulation and Association with Photobodies
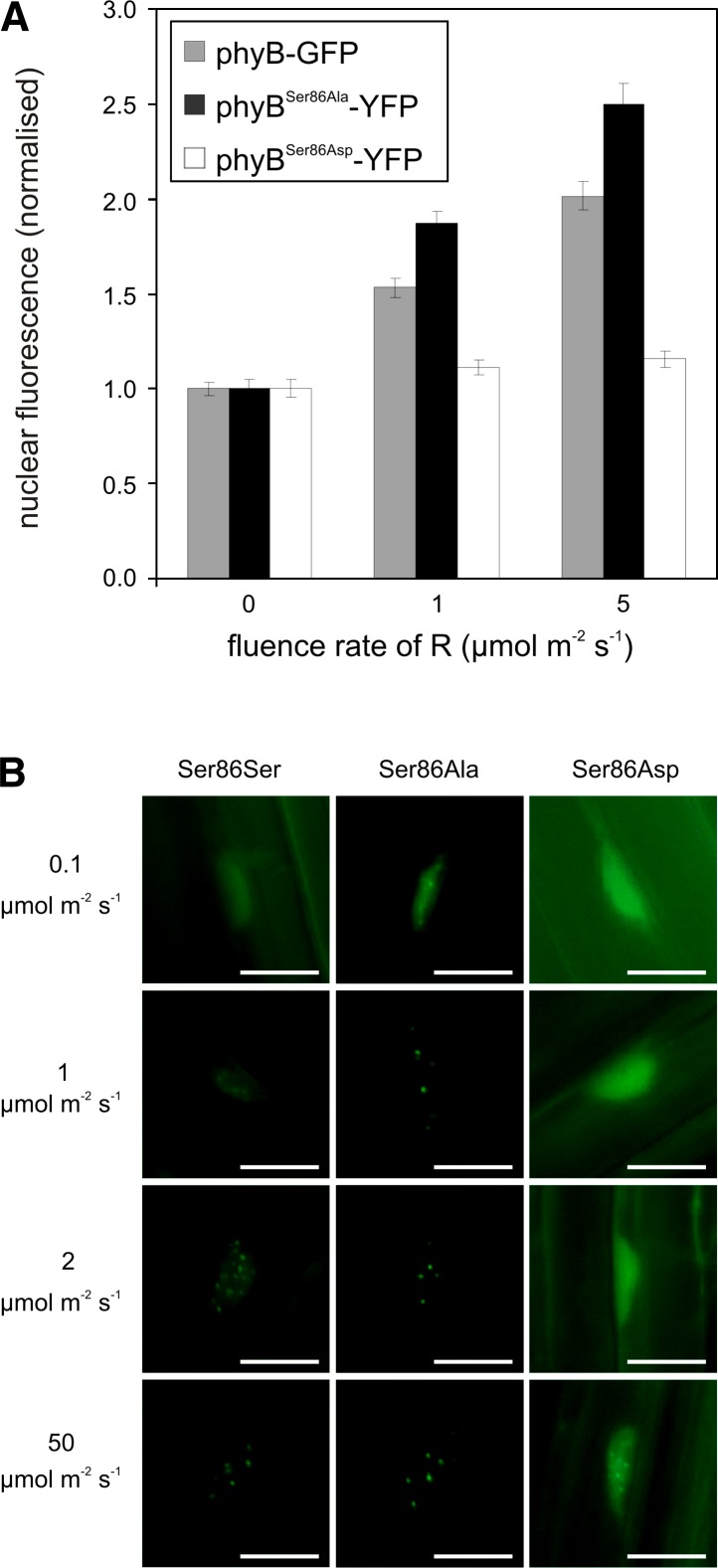
The molecular machinery mediating light quality and quantity-dependent nuclear import of phyB is only partially known. However, it has been shown that phyB does not possess an intrinsic nuclear localization signal, and its import into the nucleus can be mediated by interaction of phyB Pfr with nuclear localization signal–bearing proteins, including the PIF3 (Pfeiffer et al., 2012). To determine if substitution of Ser-86 affects nucleo/cytoplasmic partitioning, we monitored the velocity of light-induced nuclear accumulation of the various chimeric photoreceptors. Densitometric, quantitative analysis of fluorescence demonstrated that the rate of light-induced nuclear import of the various fusion proteins differed at low fluences of R. PhyBSer86Ala-YFP was imported into the nucleus upon irradiation with 1 and 5 µmol m−2 s−1 of R light, whereas the amount of nuclear phyBSer86Asp-YFP remained at the dark level (Figure 3A). By contrast, saturating R light (2 h irradiation with 43 µmol m−2 s−1) uniformly elevated the level of nuclear fluorescence, suggesting no significant difference between the nuclear accumulations of the various fusion proteins (see Supplemental Figure 3 online). These observations suggest that nuclear import of the phospho-mimic phyBSer86Asp-YFP is severely reduced under conditions when the amount of Pfr is limited.
Figure 3.
Phosphorylation of phyB Ser-86 Affects the Nuclear Translocation of the Photoreceptor.
(A) Initial rate of light-induced accumulation of the phospho-mimic phyBSer86Asp-YFP fusion protein in the nucleus is reduced. phyB-9 seedlings expressing the phyB-GFP, phyBSer86Asp-YFP, or phyBSer86Ala-YFP fusion proteins were grown for 5 d in darkness and subsequently exposed for 2 h to the indicated fluences of R light. Quantification of the nuclear accumulation of the various fusion proteins was performed as described (Pfeiffer et al., 2012). Nuclear fluorescence normalized to dark levels is shown (n = 50). Error bars indicate se.
(B) PhyBSer86Asp-YFP photobodies are detected only after irradiation with high intensity R light. phyB-9 seedlings expressing the phyB-GFP, phyBSer86Asp-YFP, and phyBSer86Ala-YFP fusion proteins were grown for 5 d in darkness and subsequently exposed for 6 h to the indicated fluences of R light. Representative pictures are shown (n = 50). Bars = 10 µm.
Nuclear-localized phyB is associated with subnuclear protein complexes, termed speckles, nuclear bodies, or photobodies (Kircher et al., 1999, Van Buskirk et al., 2012), and formation of these structures occurs in a fluence rate–dependent fashion (Chen et al., 2003). Here, we show that phyBSer86Ala-YFP photobodies are already formed after 6 h of irradiation with 0.1 µmol m−2 s−1 R light, whereas detection of phyB-YFP photobodies requires irradiation with an order of magnitude higher fluence rate of R light (Figure 3B). More strikingly, our data indicate the phyBSer86Asp-YFP photobodies were not detectable at fluences up to 8 µmol m−2 s−1. Under saturating light conditions (50 µmol m−2 s−1) all three phyB fusion proteins were associated with photobodies, although the number and brightness of phyBSer86Asp-YFP photobodies appeared to be somewhat reduced (Figure 3B).
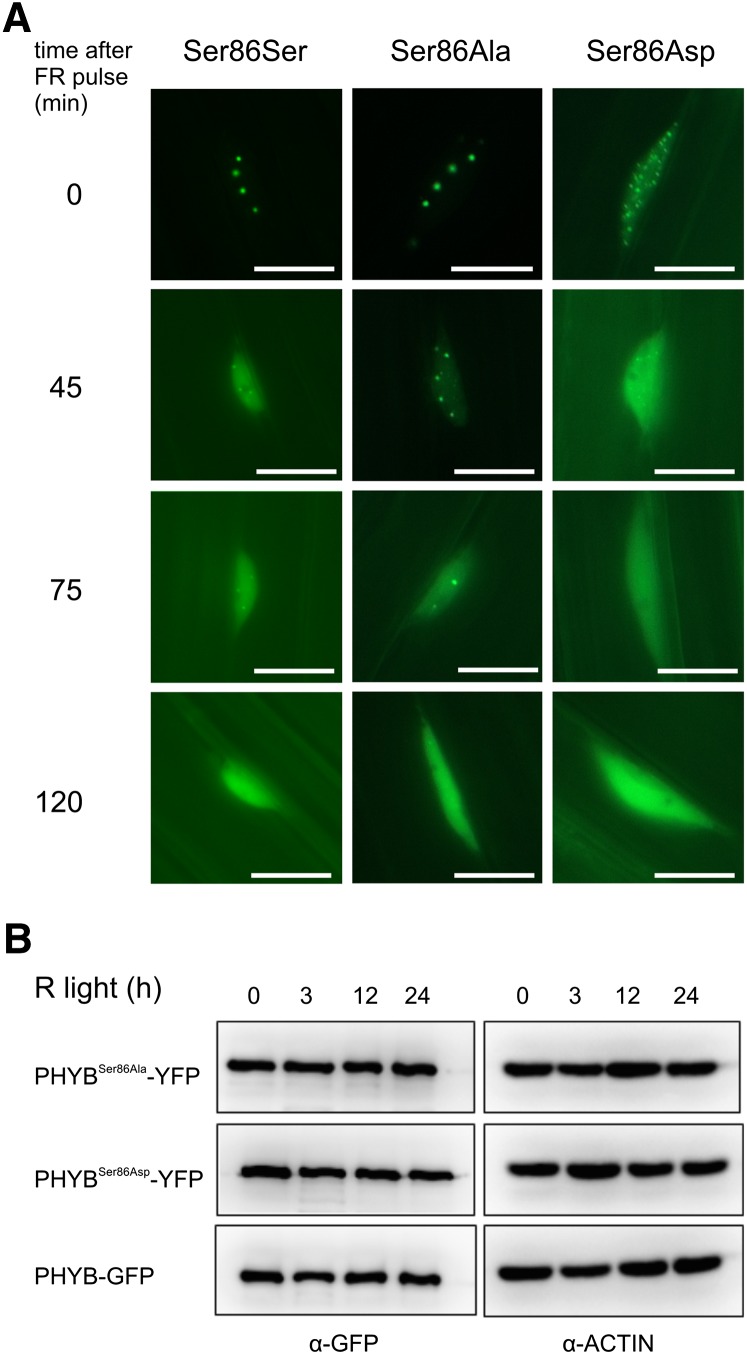
phyB photobodies dissociate in extended darkness, and this process is significantly accelerated by preirradiation with FR light, thus indicating that these subnuclear structures contain phyB Pfr (Nagy and Schäfer, 2002). Here, we show that phyBSer86Asp-YFP photobodies display an increased sensitivity to FR light. After receiving a short pulse of FR light, phyBSer86Asp-YFP photobodies dissociated much faster in subsequent darkness when compared with phyBSer86Ala-YFP (Figure 4A). It has been shown that degradation of phyB is induced by R light and that this process requires interaction of phyB Pfr with PIF proteins (Leivar et al., 2008). A significantly altered degradation rate of the phyB Pfr at lower fluence rates could explain the observed hyposensitivity of phyBSer86Asp-YFP controlled responses. Therefore, we determined the stability of the various phyB fusion proteins in seedlings exposed to weak cR light. Data shown in Figure 4B clearly indicate that phosphorylation of Ser-86 did not significantly alter R light–promoted degradation of the various fusion proteins at the fluence rate of 0.1 µmol m−2 s−1.
Figure 4.
Phosphorylation of Ser-86 Affects Photobody Dissociation but Not the Abundance of phyB.
(A) phyBSer86Ala-YFP photobodies exhibit increased stability under FR light irradiation. Transgenic phyB-9 seedlings expressing the phyB-GFP (S86S), phyBSer86Ala-YFP (S86A), and phyBSer86Asp-YFP (S86D) fusion proteins were grown for 5 d in darkness and then exposed to 24 h of R light (22 µmol m−2 s−1) followed by irradiation with FR light for 5 min (20 µmol m−2 s−1). phyB-associated photobodies were monitored as described by Pfeiffer et al. (2012). The experiments were repeated three times (n = 50), and representative pictures are shown. Bars = 10 µm.
(B) Substitutions of Ser-86 do not alter cR-induced degradation of the various phyB fusion proteins. Transgenic phyB-9 seedlings were grown in darkness for 4 d on wet filter paper and exposed to cR (0.1 µmol m−2 s−1) as indicated. The levels of the various fusion proteins were determined by immunoblot analysis using anti-GFP antibody. The lanes contain equal amounts of total protein as shown by the comparable levels of actin.
The Capacity of the Phospho-Mimic phyBSer86Asp-YFP Photoreceptor to Bind PIF3 Is Significantly Reduced under Nonsaturating Light Conditions in Vitro
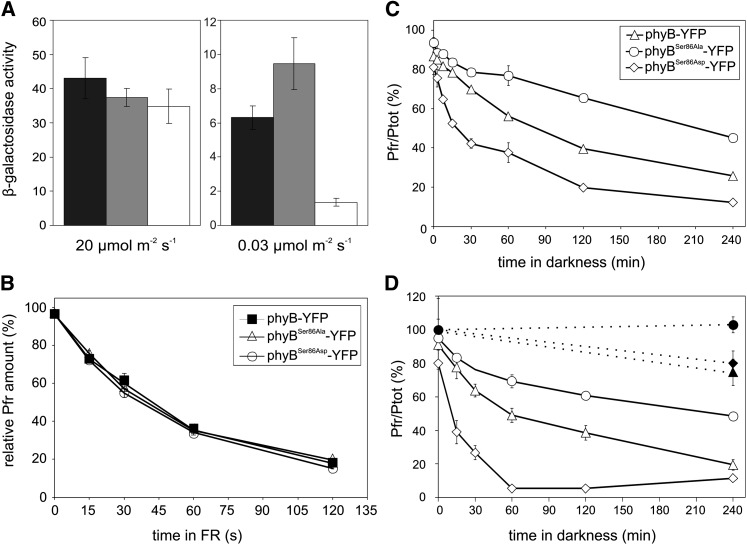
phyB interacts in a conformation-specific reversible fashion with a battery of downstream signaling components, including the well-characterized PIFs (Ni et al., 1998; Al-Sady et al., 2006; Bae and Choi, 2008). To test how substitutions of Ser-86 to Ala or Asp affect the interaction of phyB Pfr with PIF3, we performed yeast two-hybrid assays under different light conditions. Under nonsaturating light, the β-galactosidase enzyme activity increased in a linear fashion in relation to irradiation time, indicating that neither the amount of phyB(N651) (PHYB N-terminal 1 to 651 amino acids) Pfr nor that of PIF3 was rate limiting. When compared with phyB(N651) and phyB(N651)Ser86Ala, the capacity of phyB(N651)Ser86Asp Pfr to bind PIF3 was greatly reduced (Figure 5A). By contrast, under saturating light conditions, the binding capacities of the various phyB N-terminal fragments to PIF3 were nearly identical, indicating that substitution of Ser-86 with Ala or Asp itself does not physically interfere with or prevent interaction of these proteins (Figure 5A).
Figure 5.
Phosphorylation of Ser-86 Regulates the Dark Reversion of phyB.
(A) Under nonsaturating light conditions, R/FR reversible binding of PIF3 to the phospho-mimic phyBSer86Asp-YFP is reduced when compared with phyB-GFP in vitro. Liquid cultures of yeast cells expressing the AD-PIF3 and PHYB(N651)-BD (black bars) or PHYB(N651)Ser86Ala-BD (gray bars) or PHYB(N651)Ser86Asp-BD (white bars) fusion proteins were grown overnight in darkness in the presence of exogenously supplied chromophore. The overnight cultures were halved and irradiated with the indicated fluence rate of R light for 10 min and then returned again to darkness. The β-galactosidase enzyme activity was measured 4 h after the light pulse. Error bars indicate se of three independent experiments.
(B) Substitution of Ser-86 to Ala or Asp does not alter photoconversion of recombinant phyB photoreceptors in vitro. Liquid yeast cultures supplemented with exogenously added chromophore and expressing the phyB-YFP, phyBSer86Ala-YFP, and phyBSer86Asp-YFP photoreceptors were grown overnight in darkness. Photoconversion of the Pfr conformer of the various fusion proteins to Pr was measured as described by Kunkel et al. (1993). The fluence rate of the reverting FR light (20 µmol m−2 s−1) and the amount of Pfr, as the relative amount of Pfr (%) is shown. Pfr (%) at photoequilibrium = 100%. Error bars indicate se of three independent experiments.
(C) Dark reversion of the Pfr of the phospho-mimic phyBSer86Asp-YFP fusion protein is accelerated compared with phyB-GFP or phyBSer86Ala-YFP in vitro. Liquid yeast cultures expressing the recombinant fusion proteins were grown overnight and reconstituted with phycocyanobilin. Samples of identical density were prepared, irradiated for 5 min with saturating R light, and transferred into darkness. Dark reversion of phyB-YFP, phyBSer86Asp-YFP, and phyBSer86Ala-YFP Pfr was measured as described (Kunkel et al., 1995). The relative amount of Pfr (%) to the total amount of phytochrome (Ptot) is shown. Error bars indicate se of three independent experiments.
(D) The phospho-mimic phyBSer86Asp-YFP Pfr also rapidly dark reverts in planta. Eighty milligrams of 4-d-old etiolated Arabidopsis seedlings were irradiated for 5 min with saturating R light and incubated afterwards in darkness. The total amounts of phyB-GFP (closed triangle), phyBSer86Asp-YFP (closed diamond), and phyBSer86Ala-YFP (closed circle) and dark reversion of phyB-GFP (open triangle), phyBSer86Asp-YFP (open diamond), and phyBSer86Ala-YFP (open circle) Pfr were measured as described (Eichenberg et al., 1999). The relative amount of Pfr (%) to the total amount of phytochrome (Ptot) is shown. Error bars indicate se of three independent experiments.
The Phospho-Mimic phyBSer86Asp-YFP Photoreceptor Displays Accelerated, Whereas the Nonphosphorylatable phyBSer86Ala-YFP Exhibits Decreased, Dark Reversion When Compared with phyB-GFP in Planta
It has been shown that signaling by phyB Pfr is stopped either by FR light–driven conversion of the Pfr into Pr (photoconversion) or by the light-independent relaxation (dark reversion) of the thermodynamically less stable Pfr into the Pr conformer. It is evident that dark reversion can play an important role under conditions when the amount of the Pfr form is limited (i.e., in low-fluence nonsaturating light) (Rockwell et al., 2006). The physiological and molecular analysis of the phyB-YFP mutants demonstrated that phosphorylation of Ser-86 negatively regulates phyB signaling at low fluence rates (Figures 2A and 2B). To test whether Ser86Ala and/or Ser86Asp substitutions affect spectral properties and/or FR-induced photoconversion of the photoreceptor, we compared the absorption spectra and rate of photoconversion of the various phyB fusion proteins in vitro. Our data unambiguously demonstrate that photoconversion (Figure 5B) and difference spectra (see Supplemental Figure 4 online) of these photoreceptor molecules do not significantly differ. To verify the possible involvement of Ser-86 phosphorylation in regulating dark reversion of phyB, we measured the rate of this process in vitro in yeast cells and in planta. For in vivo assays, we introgressed the various P35S:PHYB-YFP transgenes into the phyA-211 phyB-9 double null background to minimize possible misinterpretation of the measured Pfr/Pr values due to high phyA levels in etiolated seedlings. Dark reversion measurements showed that the phospho-mimic mutant phyBSer86Asp-YFP reverts faster and to a greater extent than phyB-GFP in vitro (Figure 5C) and in planta (Figure 5D), whereas substitution of Ser-86 to Ala exerted the opposite effect. In comparison with phyB-GFP, the percentage of dark-reverting phyBSer86Ala-YFP was reduced and the kinetics of the reaction were also slightly slower. We conclude that these results readily explain (1) the decreased rate of nuclear import, (2) the limited formation of phyBSer86Asp-YFP photobodies in vivo, and (3) the reduced amount of PIF3/phyBSer86Asp complexes in vitro, and at least partially define the molecular mechanism by which phosphorylation-modulated dark reversion negatively regulates phyB signaling.
DISCUSSION
Here, we show that Ser-86 of phyB is phosphorylated in vivo and that substitution of this Ser with Ala or Asp dramatically alters the rate of dark reversion of phyB Pfr and thereby R light–induced signaling in transgenic Arabidopsis. Characterization of phyB-401 (Kretsch et al., 2000; Ádám et al., 2011) and phyB-101 (Elich and Chory, 1997) demonstrated that light-independent relaxation of phyB Pfr indeed plays an important role in regulating R light–induced signaling and that substitutions of amino acid residues residing in (Ádám et al., 2011) or deletions (Oka et al., 2004) affecting the so-called light-sensing knot (Kikis et al., 2009) alter the rate of dark reversion. Ser-86 is not part of the light regulatory knot, yet its substitution dramatically alters the dark reversion of phyB in vitro (Figure 5C) and in vivo (Figure 5D). It was reported that interaction of ARABIDOPSIS RESPONSE REGULATOR4 (ARR4) with phyB inhibits the dark reversion of the Pfr conformer (Sweere et al., 2001) and that ARR4 may play a role in integrating phyB and cytokinin signaling (Mira-Rodado et al., 2007). We performed in vivo coimmunoprecipitation assays and found that substitutions of Ser-86 did not affect interaction of phyB with ARR4 (see Supplemental Figure 5 online). This observation suggests that phosphorylation of Ser-86 regulates the dark reversion via a mechanism independent of ARR4. We note that irrespective of the mechanisms by which phosphorylation regulates dark reversion, our data show that the Ser86Ala and Ser86Asp substitutions do not affect the light absorption properties of the photoreceptor as determined by chromophore binding, difference spectra (see Supplemental Figure 4 online), and photoconversion (Figure 5B).
The conclusion that phosphorylation of Ser-86 specifically affects only the rate of dark reversion is strongly supported by the pronounced hyposensitivity of P35S:PHYBSer86Asp-YFP and hypersensitivity of P35S:PHYBSer86Ala-YFP transgenic lines at low intensities of light and their nearly identical phenotypes at high intensities. At low fluence rates, where the amount of Pfr is likely to be limited, the accelerated/decreased dark reversion of phyB Pfr profoundly affects the flux of signaling, in contrast with high fluences, where dark reversion is efficiently compensated for by the conversion of Pr to Pfr. The dark reversion–mediated rapid inactivation of the phosphorylated form of phyB Pfr readily explains the impaired light-induced translocation of phyBSer86Asp-YFP into the nucleus and the decreased binding of PIF3 under nonsaturating light conditions, which in turn provides a mechanistic explanation for the observed hyposensitive phenotypes.
The inactivation of phyB signaling is not due to accelerated degradation of the photoreceptor since the Ser86Ala and Ser86Asp substitutions do not alter the stability/degradation of these fusion proteins in planta at low fluences of R light (Figure 4B). By contrast, we show that phosphorylation of Ser-86 dramatically alters subnuclear distribution of phyB as the phospho-mimic phyBSer86Asp-YFP fusion proteins do not associate with photobodies up to 8 µmol m−2 s−1, and even at higher fluence rates only a reduced number of phyBSer86Asp-YFP photobodies can be detected. These observations indicate that phosphorylation of the photoreceptor may regulate formation of phyB photobodies by two mechanisms. It is evident that phosphorylation via accelerated dark reversion limits the amount of phyB Pfr and thus inhibits formation of these nuclear complexes under nonsaturating light conditions. However, our data also demonstrate that phosphorylation negatively affects photobody formation even at saturating light conditions where phyB Pfr levels are high. It is plausible to assume that under this condition, phosphorylation may modify interaction of phyB Pfr with factors required for assembly/stabilization of photobodies. However, at present, neither the composition of photobodies nor the precise function of phyB in regulating formation of these structures is known; thus, validation of the above hypothesis will require further experiments.
Independent of the exact function of phytochrome-associated photobodies in mediating light signaling, our data clearly suggest a new paradigm by which phosphorylation modulates activity of phyB. At present, we are unable to determine whether phosphorylation of Ser-86 is due to phyB autophosphorylation or the activity of an unknown kinase. The major obstacle to defining the mode of Ser-86 phosphorylation is the lack of reliable in vitro systems to generate reproducible data on the autophosphorylation of phyB. Autophosphorylation of phyB was reported a few years ago; however, this observation has not yet been corroborated (Phee et al., 2008). This report concluded that (1) phyB Pr and Pfr autophosphorylate approximately at equal levels in vitro; (2) deletion of the N-terminal region, containing 100 amino acids, prevented phosphorylation; and (3) mapping of phosphorylation sites was not feasible. Our data showing phosphorylation of Ser-86 in vivo provide reasonable support for these conclusions and demonstrate that the extreme N-terminal region of phyB is indeed subject to phosphorylation.
We also note that dark reversion kinetics of wild-type and mutant phyB derivatives are surprisingly similar in plant and yeast cells. Based on this observation, we hypothesize that phosphorylation of the Ser-86 residue might also occur in the heterologous system, but validation of this hypothesis requires additional experiments (Figures 5C and 5D). Moreover, alignment of the identified Ser-86 phosphorylation site across species indicates that this Ser is highly conserved in phyB but not in phyA photoreceptors (see Supplemental Figure 6 online). Our MS analysis also indicates that phyB is phosphorylated on additional sites beside Ser-86, and Zn-Phos-Tag analysis of the transgenic P35S:PHYBSer86Ala-YFP lines in fact corroborated this prediction in planta (see Supplemental Figure 7 online). However, future studies will be needed to get a precise map of phyB phosphorylation and the extent of light regulation of this posttranslational modification. The identification of the kinase(s) and phosphatases regulating the phosphorylation status of phyB is another important challenge.
METHODS
For the purification of phyB-TAP fusion protein, sample preparation for MS analysis, and measurement of differential spectra, see Supplemental Methods 1 online.
Purification of phyB from Plants
To facilitate large-scale purification of phyB from plant tissue, the photoreceptor was fused to the improved TAP tag (Rigaut et al., 1999) and expressed under the control of the P35S promoter (transgenic line provided by Andreas Hiltbrunner). The phyB-TAP fusion protein was purified from 3-week-old transgenic plants grown under short-day conditions (8 h light/16 h darkness). Prior to purification, plants were adapted to 24-h dark treatment followed by irradiation with low-intensity R light (0.5 µmol m−2 s−1) for 6 h. For a detailed description of purification of phyB-TAP, see Supplemental Methods 1 online.
Sample Preparation and MS Analysis
After SDS-PAGE, Coomassie Brilliant Blue–stained gel slices were cut into pieces and destained, dried, and then rehydrated with trypsin solution, and one portion of the isolated peptide was enriched for phosphopeptides on TiO2. A more detailed description of sample preparation can be found at http://msf.ucsf.edu/ingel.html. The enriched fraction was analyzed by liquid chromatography–tandem MS on a Waters Q-TOF Premier and an Agilent XCT Plus Iontrap mass spectrometer in information-dependent acquisition mode. Database searches were performed with ProteinProspector (v 5.3.0) against Arabidopsis thaliana sequences in the National Center for Biotechnology Information 2011.03.30 database (62,702 entries). For a more detailed description of the protocols employed, see Supplemental Methods 1 online.
Cloning and Plant Transformation
The full-length PHYB coding region was inserted as an XbaI-StuI fragment between the cauliflower mosaic virus P35S promoter and the YFP-NOS terminator of pPCVB (Bauer et al., 2004). The Ser86Ala and Ser86Asp mutations were created using a site-directed mutagenesis kit (Stratagene) using the following oligonucleotides: 5′-CGTACGTCGTCGTCTTAAGTGCTTGTGAGTAGTCG-3′ and 5′-CGACTACTCACAAGCACTTAAGACGACGACGTACG-3′ to introduce Ser86Ala mutation and 5′-CGACTACTCACAAGATCTTAAGACGACGACGTACG-3′ and 5′-CGTACGTCGTCGTCTTAAGATCTTGTGAGTAGTCG-3′ to introduce Ser86Asp mutation. Every construct was verified by sequencing. phyB-9 mutants (Reed et al., 1993) were transformed by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998), and transgenic plants were selected by their resistance to Basta.
Shade Avoidance and Hypocotyl and Cotyledon Growth Measurements
For shade avoidance, seeds were sown on nylon mesh on half-strength strength Murashige and Skoog plates and stored in the dark for 3 d at 4°C. A germinating, R light treatment (6 h, 50 µmol m−2 s−1) was given at 20°C, and the plates were returned to darkness at 20°C for a further 21 h. Plates were then transferred to constant white light conditions in a Percival Scientific AR-66L growth cabinet (PAR 400 to 700 nm = 130 µmol m−2 s−1, R:FR = 10, 20°C) for 3 d. Plates were then transferred for four more days to constant white light supplemented with FR (low R:FR conditions, R:FR = 0.4) or kept in constant white light as control conditions (high R:FR condition). Plates were kept vertically during the experiment. Hypocotyl length and cotyledon area measurements were performed as described by Ádám et al. (2011). Scanned seedling images were analyzed using MetaMorph (Universal Imaging) or ImageJ (National Institutes of Health) software.
Zn-Phos-Tag PAGE, Phosphatase Treatment, and Immunoblotting
PhyB-GFP–overexpressing plants were grown on half-strength Murashige and Skoog plates (1.2% agar) without Suc for 7 d under the indicated light conditions. Samples were homogenized in liquid N2 and then extracted with 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM DTT supplemented with protease inhibitor cocktail (Sigma-Aldrich). Buffer volumes were adjusted to fresh weight. Aliquots (21.5 μL) of samples were combined with 2.5 μL of 10 mM MnCl2 and either with 1 μL (400 U) λ-protein phosphatase (NEB) or blank enzyme storage buffer. Phosphatase-treated samples were incubated at 30°C for 1 h. Reactions were stopped by adding 25 μL 2× LDS loading buffer with 100 mM DTT (Invitrogen) and denaturation at 70°C for 10 min. Samples were loaded onto 7% 50 μM Phos-tag NuPAGE gels (Kinoshita and Kinoshita-Kikuta, 2011). Gels were blotted to polyvinylidene difluoride membranes using NuPAGE transfer buffer. After blocking with 5% milk in PBST, the upper part of the membrane was probed with α-GFP-HRP (Miltenyi Biotec) for 1 h and the lower part with α-plant actin IgG (Sigma-Aldrich) for 1 h followed by α-mouse IgG-HRP (Promega) for 30 min. After treating the membrane with Immobilon Western Chemiluminescent HRP substrate (Millipore), luminescent signals were detected using a liquid nitrogen–cooled charge-coupled device camera (Micromax; Roper Scientific). Digital images were analyzed and signals were quantified using Metamorph software package (Molecular Devices). Additional immunoblot assays were performed according to Bauer et al. (2004).
Yeast Two-Hybrid Methods
PIF3-pGAD424 and pD153 vectors and yeast propagation techniques are described by Shimizu-Sato et al. (2002). PHYB(N651) and its mutated Ser86Ala and Ser86Asp counterparts were amplified using 5′-AAATCTAGAGAAACAATGGTTTCCGGAGTCGGG-3′ and 5′-GAGCCCGGGTGCACCTAACTCATCAATCCC-3′ oligonucleotides and cloned as XbaI-SmaI fragments to the pD153 vector. Every construct was verified by sequencing.
Photoconversion of the Yeast-Expressed PHYB Versions
A thin layer (2 mL in a 5-cm Petri dish) of yeast suspension was treated by a 30-s-long saturating R light pulse (60 μmol m−2 s−1) and measured directly (0-s FR treatment), or the suspension was further treated by a FR light pulse (6 μmol m−2 s−1) for different time intervals. After the treatment, the Pfr-to-Ptot ratio was measured in 100 μL yeast suspension using a RatioSpec. Every data point was measured at least four times in two independent experiments, and error bars represent sd. The Pfr amount, normalized to photoequilibrium, was plotted against the FR light dose (given as length of the FR light irradiation). As light source, a portable R (Emax – 660 nm) and FR (Emax – 730 nm) LED irradiated box was used.
Epifluorescence Microscopy and Quantification of Nuclear Import
Detailed description of microscopic equipments and filter settings for GFP and YFP fluorescence measurements are described by Bauer et al. (2004). Quantification of the nuclear import was performed according to Pfeiffer et al. (2012). Twelve-bit TIFF images, not containing any saturated pixels, were taken of least 50 nuclei per sample using fluorophore-specific filter sets and applying identical exposure times. Mean gray value intensity of pixels in the examined nuclei was calculated, and background was corrected by subtracting the corresponding vacuolar signal. All calculations were performed using the ImageJ software (National Institutes of Health).
Dark Reversion Assays
In vivo dark reversion measurements were performed with a custom-made dual-wavelength spectrophotometer with measuring beams at 730 and 800 nm. For the measurements, the samples were irradiated with actinic light generated through filters at 650 nm (Balzers KG9) and 756 nm (Schott RG9). Prior to measurements, the samples were irradiated for 40 min with saturating R light (60 µmol m−2 s−1) and incubated in darkness for various times. In vitro dark reversion experiments were performed as described (Kunkel et al., 1995).
Accession Numbers
The ArabidopsisGenome Initiative locus identifier for PHYB is AT2G18790, for PIF3 is AT1G09530, and for ARR4 is AT1G10470.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Domains of PHYB and Identification of Ser-86 Phosphorylation.
Supplemental Figure 2. Cotyledon Expansion Response of phyB-YFP, phyBSer86Ala-YFP, and phyBSer86Asp-YFP Expressing Transgenic Seedlings Is Identical under High-Intensity R Light.
Supplemental Figure 3. Light-Induced Accumulation of phyB-YFP, phyBSer86Ala-YFP, and phyBSer86Asp-YFP Fusion Protein in the Nucleus Is Identical under High-Intensity R Light.
Supplemental Figure 4. Difference Spectra of the phyB-GFP, phyBSer86Ala-YFP, and phyBSer86Asp-YFP Fusion Proteins Are Identical.
Supplemental Figure 5. Ser86Ala and Ser86Asp Substitutions Do Not Affect the Interaction between phyB and ARR4.
Supplemental Figure 6. Alignment of the Identified phyB Ser-86 Phosphorylation Site across Species.
Supplemental Figure 7. The phyBSer86Ala-YFP Fusion Protein Is Phosphorylated in Planta.
Acknowledgments
Work in Szeged, Hungary was supported by an Országos Tudományos Kutatási Alapprogramok (81399) grant to F.N. and a Társadalmi Megújulás Operatív Program (4.2.2-0001) grant to K.F.M.; in Freiburg, Germany by the Sonderforschungbereich grant (746) to E.S. and F.N.; in Edinburgh, UK by the Scottish Universities Life Sciences Alliance professorial grant to F.N.; and in Lausanne, Switzerland by grants from the Swiss National Science Foundation (3100A0-112638) and the SystemsX.ch grant “plant growth in a changing environment” to C.F. We thank Klaus Harter for providing the ARR4 polyclonal antibody.
AUTHOR CONTRIBUTIONS
F.N. and E.S. designed research. M.M., J.B., É.Á., A.V., É.K., S.L., P.G., Z.M., C.F., K.F.M., and T.K. performed research. T.K., E.S., and F.N. analyzed data. F.N. wrote the article.
Glossary
- R
red
- FR
far-red
- MALDI-TOF-MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- YFP
yellow fluorescent protein
- GFP
green fluorescent protein
- MS
mass spectrometry
- Col-0
Columbia-0
- cR
constant R
- TAP
tandem affinity purification
References
- Ádám É., Hussong A., Bindics J., Wüst F., Viczián A., Essing M., Medzihradszky M., Kircher S., Schäfer E., Nagy F. (2011). Altered dark- and photoconversion of phytochrome B mediate extreme light sensitivity and loss of photoreversibility of the phyB-401 mutant. PLoS ONE 6: e27250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. (1998). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1: 939–948 [DOI] [PubMed] [Google Scholar]
- Al-Sady B., Ni W.M., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C.S., Adám É., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Shi X., Chen L., Dai M., Zhou Z., Shen Y., Li J., Li G., Wei N., Deng X.W. (2012). Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell 24: 1907–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen M., Schwab R., Chory J. (2003). Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl. Acad. Sci. USA 100: 14493–14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J.R., Hondred D., Walker J.M., Vierstra R.D. (1992). Phytochrome requires the 6-kDa N-terminal domain for full biological activity. Proc. Natl. Acad. Sci. USA 89: 5039–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G., Yi H., Lee J., Kwon Y.K., Soh M.S., Shin B., Luka Z., Hahn T.R., Song P.S. (1999). Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401: 610–613 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., Chen D.L., Yeh K.C., Abel S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124: 1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberg K., Kunkel T., Kretsch T., Speth V., Schäfer E. (1999). In vivo characterization of chimeric phytochromes in yeast. J. Biol. Chem. 274: 354–359 [DOI] [PubMed] [Google Scholar]
- Elich T.D., Chory J. (1997). Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell 9: 2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Yeh K.C., Lagarias J.C., Zhang H., Elich T.D., Chory J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Han Y.J., Kim H.S., Kim Y.M., Shin A.Y., Lee S.S., Bhoo S.H., Song P.S., Kim J.I. (2010). Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiol. 51: 596–609 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Jordan E.T., Marita J.M., Clough R.C., Vierstra R.D. (1997). Characterization of regions within the N-terminal 6-kilodalton domain of phytochrome A that modulate its biological activity. Plant Physiol. 115: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis E.A., Oka Y., Hudson M.E., Nagatani A., Quail P.H. (2009). Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet. 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Shen Y., Han Y.J., Park J.E., Kirchenbauer D., Soh M.S., Nagy F., Schäfer E., Song P.S. (2004). Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E. (2011). Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11: 319–323 [DOI] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Ádám E., Harter K., Schäfer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsch T., Poppe C., Schäfer E. (2000). A new type of mutation in the plant photoreceptor phytochrome B causes loss of photoreversibility and an extremely enhanced light sensitivity. Plant J. 22: 177–186 [DOI] [PubMed] [Google Scholar]
- Kunkel T., Speth V., Büche C., Schäfer E. (1995). In vivo characterization of phytochrome-phycocyanobilin adducts in yeast. J. Biol. Chem. 270: 20193–20200 [DOI] [PubMed] [Google Scholar]
- Kunkel T., Tomizawa K., Kern R., Furuya M., Chua N.H., Schäfer E. (1993). In vitro formation of a photoreversible adduct of phycocyanobilin and tobacco apophytochrome B. Eur. J. Biochem. 215: 587–594 [DOI] [PubMed] [Google Scholar]
- Lapko V.N., Jiang X.Y., Smith D.L., Song P.S. (1997). Posttranslational modification of oat phytochrome A: Phosphorylation of a specific serine in a multiple serine cluster. Biochemistry 36: 10595–10599 [DOI] [PubMed] [Google Scholar]
- Lapko V.N., Jiang X.Y., Smith D.L., Song P.S. (1999). Mass spectrometric characterization of oat phytochrome A: Isoforms and posttranslational modifications. Protein Sci. 8: 1032–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko V.N., Wells T.A., Song P.S. (1996). Protein kinase A-catalyzed phosphorylation and its effect on conformation in phytochrome A. Biochemistry 35: 6585–6594 [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira-Rodado V., Sweere U., Grefen C., Kunkel T., Fejes E., Nagy F., Schäfer E., Harter K. (2007). Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. J. Exp. Bot. 58: 2595–2607 [DOI] [PubMed] [Google Scholar]
- Morelli G., Ruberti I. (2002). Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci. 7: 399–404 [DOI] [PubMed] [Google Scholar]
- Nagy F., Schäfer E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53: 329–355 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Oka Y., Matsushita T., Mochizuki N., Suzuki T., Tokutomi S., Nagatani A. (2004). Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell 16: 2104–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.K., Popp C., Wüst F., Bindics J., Viczián A., Hiltbrunner A., Nagy F., Kunkel T., Schäfer E. (2012). Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phee B.K., Kim J.I., Shin D.H., Yoo J., Park K.J., Han Y.J., Kwon Y.K., Cho M.H., Jeon J.S., Bhoo S.H., Hahn T.R. (2008). A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem. J. 415: 247–255 [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.S., Lagarias J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.S., et al. (2005). Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120: 395–406 [DOI] [PubMed] [Google Scholar]
- Saijo Y., Zhu D., Li J., Rubio V., Zhou Z., Shen Y., Hoecker U., Wang H., Deng X.W. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 31: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Feng S., Li J., Tan-Wilson A., Qu L.J., Wang H., Deng X.W. (2009). Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Stockhaus J., Nagatani A., Halfter U., Kay S., Furuya M., Chua N.H. (1992). Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev. 6: 2364–2372 [DOI] [PubMed] [Google Scholar]
- Sweere U., Eichenberg K., Lohrmann J., Mira-Rodado V., Bäurle I., Kudla J., Nagy F., Schäfer E., Harter K. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294: 1108–1111 [DOI] [PubMed] [Google Scholar]
- Trupkin S.A., Debrieux D., Hiltbrunner A., Fankhauser C., Casal J.J. (2007). The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol. Biol. 63: 669–678 [DOI] [PubMed] [Google Scholar]
- Van Buskirk E.K., Decker P.V., Chen M. (2012). Photobodies in light signaling. Plant Physiol. 158: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.W., Jang I.-C., Henriques R., Chua N.-H. (2009). FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21: 1341–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.C., Lagarias J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95: 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.C., Wu S.H., Murphy J.T., Lagarias J.C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science 277: 1505–1508 [DOI] [PubMed] [Google Scholar]