Abstract
The interaction of the membrane traversing stator subunits a and b of the rotary ATP synthase was probed by substitution of a single Cys into each subunit with subsequent Cu+2 catalyzed cross-linking. Extensive interaction between the transmembrane (TM) region of one b subunit and TM2 of subunit a was indicated by cross-linking with 6 Cys pairs introduced into these regions. Additional disulfide cross-linking was observed between the N-terminus of subunit b and the periplasmic loop connecting TM4 and TM5 of subunit a. Finally, benzophenone-4-maleimide derivatized Cys in the 2–3 periplasmic loop of subunit a were shown to cross-link with the periplasmic N-terminal region of subunit b. These experiments help to define the juxtaposition of subunits b and a in the ATP synthase.
Keywords: ATP synthase, transmembrane helix, disulfide cross-linking, benzophenone-4-maleimide, subunit a, subunit b
1. Introduction
The ATP synthase is a rotary enzyme embedded in the membranes of mitochondria, chloroplasts and many bacteria. It couples the flow of hydrogen ions across the membrane (or sodium ions in some species) to the synthesis of ATP (for reviews see [1–4]). The enzyme from E. coli, with its eight different subunits α3β3γδεab2c10, [5,6] is considered a prototype of the mammalian enzyme. The 3 nucleotide-binding catalytic sites are formed by the stator subunits α and β, which are connected to the membrane through the peripheral stalk subunits δ and b. The b subunits are each anchored to the membrane through a single transmembrane (TM) helix, and further interact with subunit a, another stator subunit. The third membrane subunit, called c, forms an n-fold symmetrical ring, where n appears to be 10 in E. coli [6]. The c-ring forms a dynamic interface with subunit a, and makes fixed contacts with the other rotor subunits γ and ε. The γ subunit forms a central shaft within the hexamer of α and β subunits, and both γ and ε make multiple contacts with α and β subunits as the rotor goes through 360° of rotation relative to the stator [7].
The complex of a, b, and c subunits is traditionally referred to as Fo, while the remaining subunits form F1. Subunit c is folded as a helical hairpin with a single proton carrier, D61, which is located near the center of the membrane [8]. Proton translocation is facilitated by subunit a, which appears to form 2 offset half-channels that provide pathways for hydrogen ions to access the key aspartate residue of subunit c (D61) from different sides of the membrane [9,10]. The proton pathway is expected to include travel through one half-channel, protonation of c_D61, rotation through nearly 360°, and exit through the other half-channel.
The structures of c-rings are known for enzymes from several different species through x-ray crystallography [11–17], but such information is not available for subunit a. Analysis of monocysteine mutants by chemical labeling studies revealed that subunit a contains 5 TM helices, with the N-terminus in the periplasm and the C-terminus in the cytoplasm [18–20]. The fold of the TM helices has been proposed, based on disulfide bond formation of di-substituted cysteine mutants [21]. Evidence for the existence of the half-channels has been provided by a series of accessibility studies using various sulfhydryl labels [22–27]. Disulfide cross-linking studies have found TM4 and TM5 to be near the c-ring [28,29], and the cytoplasmic loop connecting TM3 and TM4 can be cross-linked to c subunits using a bi-functional cross-linker [30]. Similarly, from a residue in the cytoplasmic loop between TM1 and TM2 of subunit a, K74C, cross-linking to subunit b was observed using a bi-functional cross-linker [31]. Cross-linked a-b products were also found using a bi-functional cross-linker, benzophenone-4-maleimide (BPM), with b_R36C [32], and but no information from disulfide cross-linking between a and b subunits has been reported.
The structure of the first 34 amino acids of subunit b has been solved by NMR in a membrane mimetic organic solvent (chloroform:methanol:water 4:4:1) using a synthetic peptide [33]. Residues 4–22 were found to form an α-helix. Combined with results from disulfide formation of monocysteine mutants, a model of the subunit b dimer was produced. Cysteines at positions 6 and 10 formed disulfides at the highest yield [33]. Results of a previous study had suggested that one face of TM2 in subunit a was conserved and could interact with b subunits [34]. Therefore, a systematic analysis of this possible interaction was carried out using monocysteine mutations in both subunits. In addition, the proximity of the N-terminal residue 2 of subunit b to periplasmic regions of subunit a and c was tested by cross-linking.
2. Materials & Methods
2.1 Materials
Materials were obtained as described in previous work [28,29,35,36].
2.2 Plasmids, mutagenesis, and growth
To produce mutations in the genes for both subunits a and c, a new plasmid, pNac, was constructed. The plasmid pFV2-HA [36] was reduced by digesting with PpuMI and XbaI, isolating the 3.7 kb fragment, and ligating with a 15 bp linker (CTCAAGACTGGTGGT and appropriate overhangs) that regenerated the two sites. This plasmid has resistance to ampicillin, and contains the complete genes for subunits a and c, and also codes for the first 53 residues of subunit b, up to the PpuMI site. Individual mutations in the genes for subunits a or b were constructed using Quikchange II mutagenesis (Agilent Technologies), and transferred to pFV2-HA [36] using the 1.3 kb fragment from the digestion at the PflMI site in the gene for subunit a and PpuMI in the gene for subunit b. Double mutants were constructed by using the BspEI site in the HA-tag region at the 5’-end of the gene for subunit a and the MluI site in the gene for gamma. The subunit b mutations were carried on the 3.2 kb fragment, and the subunit a mutations were on the 6.0 kb fragment. The pFV2-HA plasmids were expressed in E. coli strain DK8 [37], which lacks the genes for the ATP synthase. Succinate medium for the analysis of the mutations in pFV2-HA was made from Minimal A, supplemented with 0.2 % succinate, and 0.2 mM L-valine, L-leucine, and L-isoleucine, as described previously [35].
The monocysteine mutants in subunit a from residue 128 to 146 were previously constructed in pLN6HisHA [38]. The double cysteine mutant a_I129C/a_D146C was made by double digestion of the two plasmids using EcoRI and DraIII. The 2.1 kb fragment with the I129C mutation, and the 1.2 kb fragment with the D146C mutation were ligated. These mutations were all expressed in RH305 [39,40], a strain that lacks subunit a. Succinate medium for the analysis of these mutations was the same as above, but lacking the 3 amino acids.
2.3 Membrane preparation, Cross-linking, and Immunoblotting
For disulfide cross-linking of b mutants (residues 4–26) cells were grown to A600 = 1 in LB medium, harvested, and resuspended in TMG buffer (50 mM Tris-HCl, 5 mM MgCl2, 10% glycerol, pH 7.5) according to [28]. Cross-linking reactions were carried out as described [28], except that, when indicated, CuCl2 was used instead of Cu(1,10 phenanthroline)2 SO4. For cross-linking of subunit a to subunit b with benzophenone-4-maleimide (BPM), whole intact cells of the strain RH305 were used. The pellet obtained from cells grown in a 50-ml LB flask was resuspended in 1 ml of TMG buffer and placed into 1.5-ml polypropylene microcentrifuge tubes transparent to UV light. 1 mM of BPM (from a 50 mM stock solution in ethanol) cross-linker was added to cells followed by incubation in darkness on a rotating platform for 1 hr at room temperature. After that the cross-linker was quenched by 15 mM cysteine, and cells were exposed to UV light (3 cm distance) for two hours. For extraction of subunit a, cells were spun down (1 min at 20,000 × g (14,000 RPM) in a microcentrifuge and resuspended in 1 ml of extraction buffer. (50 mM Tris-HCl, pH 8.0, 250 mM sucrose, 100 mM KCl, 30 mM imidazole, 1.5% octylglucoside, 0.5% sodium deoxycholate, 0.5% sodium cholate, 0.5% sodium dodecyl sulfate, 2.5% glycerol.) Extraction was carried out on a rotating platform for 90 min at 4°C. After that samples were centrifuged as above and supernatant was placed into 1 ml tubes containing 150 µl of Ni-NTA resin, equilibrated with the same extraction buffer. After incubation on a rotating platform for 45 min at room temperature the resin was spun down and the supernatant was discarded. 1 ml of fresh extraction buffer was added into the tube and resin was washed for 5 – 10 min on a rotating platform. This step was repeated 3 times. For elution 200 µl of elution buffer (the same extraction buffer with 180 mM imidazole) was added to the resin for 5 min. The supernatant containing isolated subunit a was collected and stored for further use.
Procedures for electrophoresis and immunoblotting of disulfide cross-linked b mutants (4–26) and BPM cross-linked products were previously described [36,38]. The b_NC2 mutants were analyzed according to previously published procedures [28,29].
3. Results
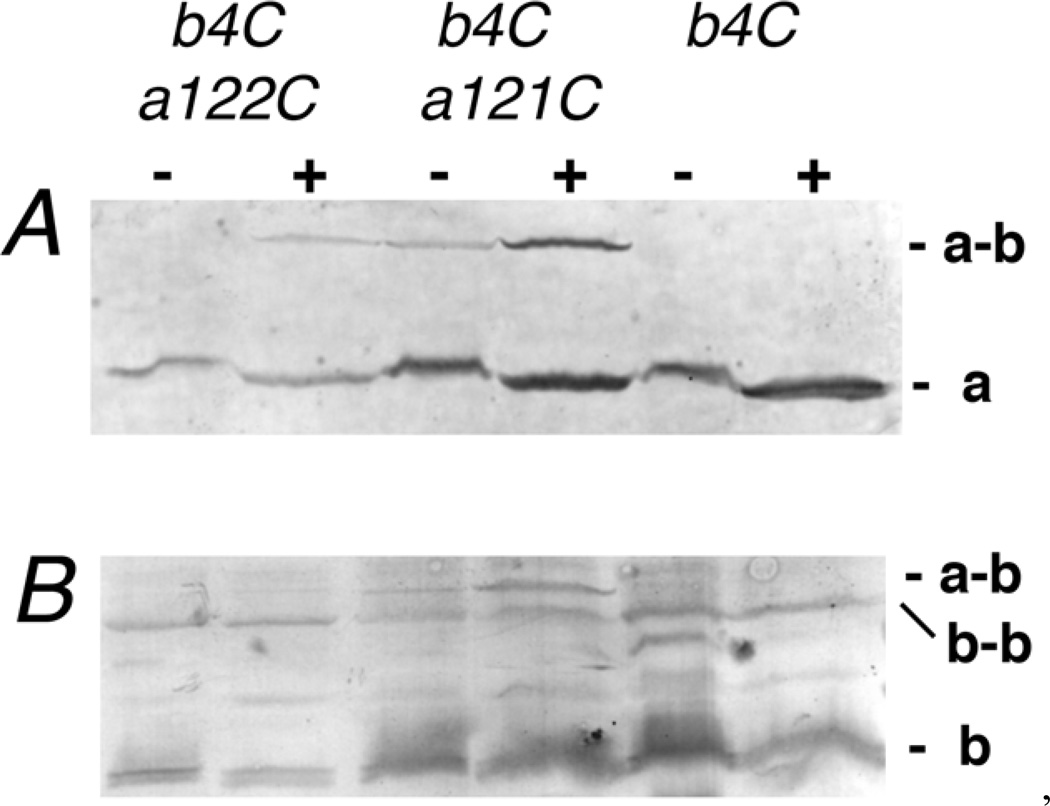
Interactions between the TM region of subunit b and TM2 of subunit a were probed by the introduction of a cysteine substitution into each subunit and subsequent cross-linking trials with the double mutant membranes. A summary of the results for the combinations tested is shown in Table 1. All of the single and double mutants described in this study were found to grow on minimal medium supplemented with succinate as sole carbon source, indicative of a functional ATP synthase. Membrane vesicles were prepared from cell cultures expressing the double mutants, and treated with Cu+2 ions to catalyze disulfide formation. Western blots were probed with antibodies both to subunit b and to the HA peptide, which is carried at the C-terminus of subunit a. Single mutations in the b subunit were also tested, since b-b dimers had previously been shown to form following cysteine substitutions at particular positions in the TM region. Among the 38 double mutants tested, only eight resulted in a cross-linked a-b product. Western blots demonstrating the cross-linked products for b_N4C/a_L121C, b_N4C/a_P122C, b_F14C/a_W111C, b_V15C/a_W111C, b_V18C/a_T107C, and b_M22C/a_L104C, are shown in Figures 1–3. In addition, b_W26C/a_L100C, and b_W26C/a_I101C formed a-b cross-links (results not shown).
Table 1.
Yield of cross-linked a–b product from double mutants
|
a Subunit Monocysteine Mutations |
b Subunit Monocysteine Mutations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N4C | F14C | V15C | L16C | F17C | V18C | L19C | F20C | A21Ca | M22C | W26C | |
| P122C | ++ | ||||||||||
| L121C | + | ||||||||||
| W111C | + | + | − | − | |||||||
| V110C | − | − | − | − | − | ||||||
| F109C | − | − | − | − | − | ||||||
| I108C | − | − | − | − | − | ||||||
| T107C | − | ++ | − | − | − | ||||||
| L106C | − | − | − | ||||||||
| A105C | − | − | − | ||||||||
| L104C | − | − | − | ++ | |||||||
| I101C | + | ||||||||||
| L100C | + | ||||||||||
Residue 21 is natively cysteine, but all cysteines in pFV2-HA have been changed to alanine, so it is a unique cysteine here.
(−) little or no a–b product detected
(+) less than 50% yield (subunit a)
(++) greater than 50% yield (subunit a)
Figure 1.
Disulfide cross-linking of b_N4C to a_L121C and a_P122C. Membrane vesicles were prepared from cells expressing the single b mutation N4C, and double mutants in combination with a_P122C and a_L121C. Each sample was treated with (+) or without (−) CuCl2 to promote disulfide formation. In Panel A the Western blot was probed with the anti-HA antibody to detect subunit a. In panel B the blot was probed with anti-b antibody.
Figure 3.
Disulfide cross-linking of b_V18C to a_T107C, and b_M22C to a_L104C. Membrane vesicles were prepared from cells expressing the single b mutation V18C and the double mutants b_V18C/a_T107C and b_M22C/a_L104C. Each sample was treated with (+) or without (−) CuCl2 to promote disulfide formation. In Panel A the Western blot was probed with the anti-HA antibody to detect subunit a. In panel B the blot was probed with anti-b antibody.
Disulfide cross-links were also demonstrated between b_N2C at the N-terminus of subunit b and substitutions a_G227C and a_L228C in the 4–5 periplasmic loop of subunit a connecting TM helices 4 and 5 (Fig. 4). In addition, the a_G227C and a_L228C sulfhydryls were shown to be close enough to the periphery of the c-ring to form disulfide cross-links with c_V78C (Fig. 5). The formation of the a-b cross-links did not significantly reduce the ATP-driven quinacrine quenching response of treated membranes (as described in ref. [41]), indicating an unperturbed and functional structure, whereas the formation of the b _N2C-c_V78C cross-link does abolish ATPase coupled quinacrine quenching [41], as would be predicted by the rotary model for ATP synthesis [1–4].
Figure 4.
Disulfide cross-linking of b_N2C to a_G227C and to a_L228C. Membrane vesicles were prepared from cells expressing the single b mutation N2C and the double mutants b_N2C/a_G227C and b_N2C/a_L228C. Each sample was treated with (+) or without (−) Cu(1,10 phenanthroline)2 SO4 to promote disulfide bond formation as described [28]. The Western blots in the left and right panels were probed with antibody to subunit b and subunit a respectively.
Figure 5.
Screening for a-c dimer formation between c_V78C and cysteine in the 4–5 loop of subunit a. Membrane vesicles were prepared from cells expressing c_V78C and seven Cys substitutions in the 4–5 loop of subunit a. Each sample was treated with Cu(1,10 phenanthroline)2 SO4 to promote disulfide bond formation as detected by Western blots probed with antibody to subunit a [28]. Dimer formation between subunits a and c was only observed with the c_V78C/a_G227C and c_V78C/a_L228C double mutant pairs.
Using a different approach, the proximity of subunit b to the 2–3 periplasmic loop in subunit a, connecting TM helices 2 and 3, was tested using a photoactivatable-cross-linker benzophenone-4-maleimide (BPM). Mono-cysteine mutations in subunit a had been constructed previously at positions 127–135, 137–138, and 141–146. Since these residues appear to reside in the periplasm or the periplasmic side of TM3 [25], cross-linking was carried with whole cells, which had been partially permeabilized with polymyxin B nonapeptide [20], and subsequent treatment with BPM and then UV radiation. In initial studies, residues 127–134 and residues 142–146 all showed various levels of a–b cross-linking (results not shown). The results for a_I129C, a_V142C, a_S144C, and a_D146C are shown in Figure 6A. Two of the residues that showed efficient cross-linking were selected, 129 and 146, and the 129/146 double mutant was constructed. The results of photoactivated cross-linking of subunit a and b using a_I129C/D146C are shown in Figure 6B. The results indicate that subunit a can be simultaneously cross-linked from this region to both of the b subunits.
Figure 6.
Cross-linking of subunit a to subunit b through benzophenone-4-maleimide. In panel A four different mutants were reacted with BPM, and then incubated in the presence (+) or absence (−) of UV radiation. The Western blots were probed with anti-HA antibody to show the presence of the a-b cross-linked product. Control blots, shown just below, demonstrate a consistent level of subunit a in the membranes initially. In panel B, cross-linking of the single mutant a_D146C and the double mutant a_D146C/a_Y128C are shown using BPM following UV radiation. The blot is probed with anti-b antibody. It shows an a-b product in the single mutant, and both a-b and a-b-b products in the double mutant.
4. Discussion
In this report the structural proximity of the TM helices of subunit b and TM2 of subunit a, and the 2–3 and 4–5 connecting loops, between TM2 and TM3 and TM4 and TM5, have been defined by cross-linking reactions. A view of the TM helices of the subunit b-dimer, as proposed in an NMR based structural model of the TM peptide region [33], is shown in Figure 7A. Residues 6 and 10, which were shown to promote the strongest disulfide bond and homodimer formation after substitution by cysteine, are colored yellow [33]. The residues in the TM helix region of subunit b that form disulfides with matched residues in subunit a, are colored red. This pattern indicates that the cross-linkable residues of TM2 of subunit a are likely to be interacting with only one of the two b subunits, as is discussed further below.
Figure 7.
Model of the b dimer and its placement relative to subunit a. A. One b subunit is colored white, and the other is colored light blue. The structure shown is that predicted by solution NMR of the peptide shown [33]. Residues 6 and 10 when changed to cysteine were shown to form disulfide-linked dimers in the membrane, and are colored yellow. Cys substitutions that form disulfides with Cys introduced into subunit a, as demonstrated here, are colored red. B. A cross-sectional map of TM helices of subunits a, b, and c as viewed from the cytoplasmic side of the membrane. The orientation of TM helices for subunit a is based upon an internal disulfide cross-linking study [21]. No information is available about the relative location of aTM1. The b subunit dimer is placed according to the results presented here. The disulfide cross-links generated in this study are shown by solid black lines. Thin dotted lines between the b subunits represent disulfide cross-links between them [33]. Thick dotted lines between TM helices of subunit a represent periplasmic loops between TM2–3 and TM4–5. The C-terminal helices (C2) of only three subunits from the c10 ring are shown.
On inspection of the cross-sectional view of the b-dimer model (Fig. 7B), it is clear that the cross-linkable face of two b TM helices could not simultaneously interact with the a TM2 cross-linkable region. Given the b-b dimer formation with Cys at positions 6 and 10, this conclusion is true regardless of whether the proposed helix-helix interactions in the NMR model are correct or not. The cross-sectional view also makes it clear that the b_N2C cross-link to the residue 227 and 228 Cys in the 4–5 loop of subunit a likely originates from the b-subunit that does not cross-link with aTM2. The b subunit neighboring aTM5 is also likely to be the one generating the b_N2C/c_V78C cross-link reported previously by Jones et al. [41]. In addition, Jones et al. [41] reported cross-linking of Cys at positions 74, 75 and 78 in subunit c with Cys at positions a227 and a228 in the 4–5 loop, which is consistent with the model shown in Fig 7B.
Cross-linking of b_N4C to a_P122C suggests that only about four residues at the N-terminus of the b subunits are exposed in the periplasm, since a_P122C, but not a_L120C, was shown previously to be accessible to a periplasmic maleimide label [38]. Cysteine substitutions within this four residue span appear to cross-link in several distinct ways. In addition to the cross-link of b_N2C to residues in the TM4-TM5 loop and the C-terminus of subunit c, and the cross-link of b_N4C with the aTM2 peripheral interface as described in the preceding paragraph (Fig. 7A & B), the N-termini of both b subunits appear to be near the TM2-TM3 periplasmic loop of subunit a. This proximity is indicated by the positive BPM catalyzed cross-linking with subunit b from multiple sites within the 2–3 loop of subunit a, and the simultaneous cross-linking from a_I129C and a_D146C to both b subunits. Given the shortness of the exposed N-terminal segment of subunit b, and the relatively short length of the cross-linker (about 6 Å), it seems likely that the periplasmic end of aTM3 is angled toward the N-termini of the b subunits to give the aTM2–5 four-helix bundle a funnel like appearance. A structural answer such as that suggested above seems necessary since the packed diameter of each TM helix shown in Fig. 7B should be in the range of 10 Å, which would make cross-linking between subunit b and residue 146 in aTM3 impossible if all of the alpha-helices were of equal length and packed in parallel.
The relative positions of the a, b, γ and ε subunits have been addressed in two studies using FRET (Förster resonance energy transfer) distance measurements [42,43]. The a and b subunits were both located relative to fluorophores at position 56 of ε and position 106 of γ. Subunit a was tagged genetically with green fluorescent protein at its C-terminus [43], while both b subunits were tagged at position 64 by a single fluorophore Cy5bis-C5-maleimide [42]. The results presented here are consistent with these earlier studies, if one considers the location of the C-terminus of subunit a (TM5) relative to the b subunit dimer interface. In a recent study of the I. tartaricus Fo sector using cryoelectron microscopy at 7 Å resolution [17], two helices were located in projection at positions very similar to the that of the two b subunits in Fig. 7B.
In summary, the cross-linking results presented here position one b subunit in proximity to much of the peripheral surface of the membrane spanning region of TM2 of subunit a, whereas the second subunit b is likely to be closer to TM5 and the surface of the c-ring. The N-termini of the b subunits are packed in such a way that contact can be made by one of the two b subunits with periplasmic extensions of aTM2, aTM3 and aTM5.
Figure 2.
Disulfide cross-linking of b_F14C and b_V15C to a_W111C. Membrane vesicles were prepared from cells expressing the single b mutations F14C and V15C, and in combination with a_W111C. Each sample was treated with (+) or without (−) CuCl2 to promote disulfide formation. In Panel A the Western blot was probed with the anti-HA antibody to detect subunit a. In panel B the blot was probed with anti-b antibody.
Highlights.
-
>
Interactions of subunits a and b of the rotary ATP synthase were examined.
-
>
Cys substitutions were used for disulfide formation or with a bi-functional reagent.
-
>
Cross-linked products indicated that one b subunit has contact with TM2 of subunit a.
-
>
Additional cross-links were detected between subunits a and b and between b and c.
-
>
A model is presented in which one b subunit is near aTM2 and the other is near aTM5.
Acknowledgements
This work was supported by grants from NIH (GM40508) and the Welch Foundation (N-1378) to SBV and NIH grant GM23105 to RHF. We thank Oleg Dmitriev (University of Saskatchewan) for providing coordinates for the b-dimer model.
Abbreviations
- TM
transmembrane
- a_W111
residue W111 of subunit a
- BPM
benzophenone-4-maleimide
- HA
Hemagglutinin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vik SB. ATP synthesis by oxidative phosphorylation. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, editors. EcoSal—Escherichia coli and Salmonella: Cellular and molecular biology. Washington DC: ASM Press; 2007. [Google Scholar]
- 2.von Ballmoos C, Cook GM, Dimroth P. Unique rotary ATP synthase and its biological diversity. Annu Rev Biophys. 2008;37:43–64. doi: 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- 3.Nakamoto RK, Baylis Scanlon JA, Al-Shawi MK. The rotary mechanism of the ATP synthase. Arch Biochem Biophys. 2008;476:43–50. doi: 10.1016/j.abb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Ballmoos C, Wiedenmann A, Dimroth P. Essentials for ATP synthesis by F1Fo ATP synthases. Annu Rev Biochem. 2009;78:649–672. doi: 10.1146/annurev.biochem.78.081307.104803. [DOI] [PubMed] [Google Scholar]
- 5.Foster DL, Fillingame RH. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli . J Biol Chem. 1982;257:2009–2015. [PubMed] [Google Scholar]
- 6.Jiang W, Hermolin J, Fillingame RH. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc Natl Acad Sci U S A. 2001;98:4966–4971. doi: 10.1073/pnas.081424898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 8.Girvin ME, Fillingame RH. Hairpin folding of subunit c of F1Fo ATP synthase:1H distance measurements to nitroxide-derivatized aspartyl-61. Biochemistry. 1994;33:665–674. doi: 10.1021/bi00169a006. [DOI] [PubMed] [Google Scholar]
- 9.Vik SB, Antonio BJ. A mechanism of proton translocation by F1Fo ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 10.Pogoryelov D, Krah A, Langer JD, Yildiz Ö, Faraldo-Gómez JD, Meier T. Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases. Nat Chem Biol. 2010;6:891–899. doi: 10.1038/nchembio.457. [DOI] [PubMed] [Google Scholar]
- 11.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus . Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 12.Dimroth P, von Ballmoos C, Meier T. Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series. EMBO Rep. 2006;7:276–282. doi: 10.1038/sj.embor.7400646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pogoryelov D, Yildiz Ö, Faraldo-Gómez JD, Meier T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat Struct Mol Biol. 2009;16:1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- 14.Vollmar M, Schlieper D, Winn M, Büchner C, Groth G. Structure of the c14 Rotor Ring of the Proton Translocating Chloroplast ATP Synthase. J Biol Chem. 2009;284:18228–18235. doi: 10.1074/jbc.M109.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preiss L, Yildiz Ö, Hicks DB, Krulwich TA, Meier T. A New Type of Proton Coordination in an F1Fo-ATP Synthase Rotor Ring. PLoS Biol. 2010;8:e1000443. doi: 10.1371/journal.pbio.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Symersky J, Pagadala V, Osowski D, Krah A, Meier T, Faraldo-Gomez JD, Mueller DM. Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakulinen JK, Klyszejko AL, Hoffmann J, Eckhardt-Strelau L, Brutschy B, Vonck J, Meier T. Structural study on the architecture of the bacterial ATP synthase Fo motor. Proc Natl Acad Sci U S A. 2012;109:E2050–E2056. doi: 10.1073/pnas.1203971109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long JC, Wang S, Vik SB. Membrane topology of subunit a of the F1Fo ATP synthase as determined by labeling of unique cysteine residues. J Biol Chem. 1998;273:16235–16240. doi: 10.1074/jbc.273.26.16235. [DOI] [PubMed] [Google Scholar]
- 19.Valiyaveetil FI, Fillingame RH. Transmembrane topography of subunit a in the Escherichia coli F1Fo ATP synthase. J Biol Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 20.Wada T, Long JC, Zhang D, Vik SB. A novel labeling approach supports the five-transmembrane model of subunit a of the Escherichia coli ATP synthase. J Biol Chem. 1999;274:17353–17357. doi: 10.1074/jbc.274.24.17353. [DOI] [PubMed] [Google Scholar]
- 21.Schwem BE, Fillingame RH. Cross-linking between helices within subunit a of Escherichia coli ATP synthase defines the transmembrane packing of a four-helix bundle. J Biol Chem. 2006;281:37861–37867. doi: 10.1074/jbc.M607453200. [DOI] [PubMed] [Google Scholar]
- 22.Angevine CM, Fillingame RH. Aqueous access channels in subunit a of rotary ATP synthase. J Biol Chem. 2003;278:6066–6074. doi: 10.1074/jbc.M210199200. [DOI] [PubMed] [Google Scholar]
- 23.Angevine CM, Herold KA, Fillingame RH. Aqueous access pathways in subunit a of rotary ATP synthase extend to both sides of the membrane. Proc Natl Acad Sci U S A. 2003;100:13179–13183. doi: 10.1073/pnas.2234364100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angevine CM, Herold KAG, Vincent OD, Fillingame RH. Aqueous access pathways in ATP synthase subunit a: Reactivity of cysteine substituted into transmembrane helices 1, 3, and 5. J Biol Chem. 2007;282:9001–9007. doi: 10.1074/jbc.M610848200. [DOI] [PubMed] [Google Scholar]
- 25.Moore KJ, Angevine CM, Vincent OD, Schwem BE, Fillingame RH. The cytoplasmic loops of subunit a of Escherichia coli ATP synthase may participate in the proton translocating mechanism. J Biol Chem. 2008;283:13044–13052. doi: 10.1074/jbc.M800900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steed PR, Fillingame RH. Subunit a facilitates aqueous access to a membrane-embedded region of subunit c in Escherichia coli F1Fo ATP synthase. J Biol Chem. 2008;283:12365–12372. doi: 10.1074/jbc.M800901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong H, Fillingame RH. Chemical reactivities of cysteine substitutions in subunit a of ATP synthase define residues gating H+-transport from each side of the membrane. J Biol Chem. 2010 doi: 10.1074/jbc.M110.175844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Fillingame RH. Interacting helical faces of subunits a and c in the F1Fo ATP synthase of Escherichia coli defined by disulfide cross-linking. Proc Natl Acad Sci U S A. 1998;95:6607–6612. doi: 10.1073/pnas.95.12.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore KJ, Fillingame RH. Structural interactions between transmembrane helices 4 and 5 of subunit a and the subunit c ring of Escherichia coli ATP synthase. J Biol Chem. 2008;283:31726–31735. doi: 10.1074/jbc.M803848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang D, Vik SB. Close proximity of a cytoplasmic loop of subunit a with c subunits of the ATP synthase from Escherichia coli . J Biol Chem. 2003;278:12319–12324. doi: 10.1074/jbc.M212413200. [DOI] [PubMed] [Google Scholar]
- 31.Long JC, DeLeon-Rangel J, Vik SB. Characterization of the first cytoplasmic loop of subunit a of the Escherichia coli ATP synthase by surface labeling, cross-linking, and mutagenesis. J Biol Chem. 2002;277:27288–27293. doi: 10.1074/jbc.M202118200. [DOI] [PubMed] [Google Scholar]
- 32.McLachlin DT, Coveny AM, Clark SM, Dunn SD. Site-directed cross-linking of b to the α, β, and a subunits of the Escherichia coli ATP synthase. J Biol Chem. 2000;275:17571–17577. doi: 10.1074/jbc.M000375200. [DOI] [PubMed] [Google Scholar]
- 33.Dmitriev O, Jones PC, Jiang W, Fillingame RH. Structure of the membrane domain of subunit b of the Escherichia coli FoF1 ATP synthase. J Biol Chem. 1999;274:15598–15604. doi: 10.1074/jbc.274.22.15598. [DOI] [PubMed] [Google Scholar]
- 34.DeLeon-Rangel J, Zhang D, Vik SB. The role of transmembrane span 2 in the structure and function of subunit a of the ATP synthase from Escherichia coli . Arch Biochem Biophys. 2003;418:55–62. doi: 10.1016/s0003-9861(03)00391-6. [DOI] [PubMed] [Google Scholar]
- 35.Ishmukhametov RR, Pond JB, Al-Huqail A, Galkin MA, Vik SB. ATP synthesis without R210 of subunit a in the Escherichia coli ATP synthase. Biochim Biophys Acta. 2008;1777:32–38. doi: 10.1016/j.bbabio.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae L, Vik SB. A more robust version of the Arginine 210-switched mutant in subunit a of the Escherichia coli ATP synthase. Biochim Biophys Acta. 2009;1787:1129–1134. doi: 10.1016/j.bbabio.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Brusilow WS, Simoni RD. In vivo evidence for the role of the ε subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli . J Bacteriol. 1984;160:1055–1060. doi: 10.1128/jb.160.3.1055-1060.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, Vik SB. Helix packing in subunit a of the Escherichia coli ATP synthase as determined by chemical labeling and proteolysis of the cysteine-substituted protein. Biochemistry. 2003;42:331–337. doi: 10.1021/bi026649t. [DOI] [PubMed] [Google Scholar]
- 39.Humbert R, Brusilow WS, Gunsalus RP, Klionsky DJ, Simoni RD. Escherichia coli mutants defective in the uncH gene. J Bacteriol. 1983;153:416–422. doi: 10.1128/jb.153.1.416-422.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartzog PE, Cain BD. Mutagenic analysis of the a subunit of the F1Fo ATP synthase in Escherichia coli: Gln-252 through Tyr-263. J Bacteriol. 1993;175:1337–1343. doi: 10.1128/jb.175.5.1337-1343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones PC, Hermolin J, Jiang W, Fillingame RH. Insights into the rotary catalytic mechanism of FoF1 ATP synthase from the cross-linking of subunits b and c in the Escherichia coli enzyme. J Biol Chem. 2000;275:31340–31346. doi: 10.1074/jbc.M003687200. [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann B, Diez M, Börsch M, Gräber P. Subunit movements in membrane-integrated EFoF1 during ATP synthesis detected by single-molecule spectroscopy. Biochim Biophys Acta. 2006;1757:311–319. doi: 10.1016/j.bbabio.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 43.Düser MG, Bi Y, Zarrabi N, Dunn SD, Börsch M. The proton-translocating a subunit of FoF1-ATP synthase is allocated asymmetrically to the peripheral stalk. J Biol Chem. 2008;283:33602–33610. doi: 10.1074/jbc.M805170200. [DOI] [PMC free article] [PubMed] [Google Scholar]